NOTCH2 mutations in splenic marginal zone lymphoma are associated with poor prognosis.
Abstract
Splenic marginal zone lymphoma (SMZL), the most common primary lymphoma of spleen, is poorly understood at the genetic level. In this study, using whole-genome DNA sequencing (WGS) and confirmation by Sanger sequencing, we observed mutations identified in several genes not previously known to be recurrently altered in SMZL. In particular, we identified recurrent somatic gain-of-function mutations in NOTCH2, a gene encoding a protein required for marginal zone B cell development, in 25 of 99 (∼25%) cases of SMZL and in 1 of 19 (∼5%) cases of nonsplenic MZLs. These mutations clustered near the C-terminal proline/glutamate/serine/threonine (PEST)-rich domain, resulting in protein truncation or, rarely, were nonsynonymous substitutions affecting the extracellular heterodimerization domain (HD). NOTCH2 mutations were not present in other B cell lymphomas and leukemias, such as chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL; n = 15), mantle cell lymphoma (MCL; n = 15), low-grade follicular lymphoma (FL; n = 44), hairy cell leukemia (HCL; n = 15), and reactive lymphoid hyperplasia (n = 14). NOTCH2 mutations were associated with adverse clinical outcomes (relapse, histological transformation, and/or death) among SMZL patients (P = 0.002). These results suggest that NOTCH2 mutations play a role in the pathogenesis and progression of SMZL and are associated with a poor prognosis.
SMZL is an indolent malignancy of splenic B lymphocytes characterized by splenomegaly, peripheral leukocytosis, and cytopenias with a median age of onset of >50 yr (Isaacson et al., 2008). SMZL is the most common primary malignancy of the spleen and represents ∼10% of all lymphomas that involve the spleen (Franco et al., 2003). Although the disease course is usually indolent, with many patients surviving beyond 10 yr, some patients present with more aggressive disease and survival between 1 and 2 yr (Chacón et al., 2002). A “watch and wait” approach to instituting therapy may be considered for patients with favorable clinical prognostic factors (Arcaini et al., 2006); however, as it is difficult to predict subsequent risk of disease aggressiveness or refractoriness, a common first-line therapeutic approach is splenectomy and anti–B lymphocyte biological agents such as the anti-CD20 antibody (rituximab). Refractory cases may then be treated with more toxic chemotherapies, such as alkylating agents or purine analogues. In contrast to many other B cell malignancies, SMZL is not associated with recurrent balanced translocations or genetic mutations. Moreover, little is known about the genetic events underpinning the development of aggressive or refractory disease or the transformation to higher grade disease. Therefore, a detailed understanding of SMZL pathogenesis would provide clinically useful insight into patient prognosis and could inform decision-making regarding early therapeutic intervention versus adoption of a “watch and wait” approach.
The NOTCH family of transmembrane receptor proteins is important for mediating cell fate determination and differentiation in a variety of embryonic and adult tissues. During hematopoietic differentiation, NOTCH1 signaling is known to influence cell fate decisions as lymphocytes differentiate into B or T cells (Pui et al., 1999; Radtke et al., 1999; Robey and Bluestone, 2004). Moreover, NOTCH2 is known to control B lymphocyte specification into cells of marginal zone lineage (Saito et al., 2003; Witt et al., 2003). Whereas defects in NOTCH1 signaling have been implicated in oncogenesis in T-acute lymphoblastic leukemia (Weng et al., 2004; Aster et al., 2011), CLL/SLL (Puente et al., 2011; Del Giudice et al., 2012), and MCL (Kridel et al., 2012), comparatively little is known about the potential role of NOTCH2 signaling defects in the development of malignancies affecting cells of B lymphocyte lineage (Aster et al., 2011).
To better understand the pathogenetic mechanisms involved in SMZL, we performed WGS and targeted Sanger gene sequencing and identified recurrent mutations predominantly clustered in the C-terminal portion of the NOTCH2 gene in SMZL. These mutations are similar to previously defined oncogenic mutations in NOTCH1 and are markers of poor prognosis in SMZL.
RESULTS
Genome sequencing and NOTCH2 mutation confirmation
To gain insight into the pathogenesis of SMZL, we performed WGS on six index cases of SMZL. WGS yielded a mean of 350 ± 10 million mapped reads per sample with an average of 97.6 ± 0.08% genome coverage and 96.4 ± 0.3% fully called exome coverage. The median genomic sequencing depth exceeded 80× in all samples normalized across the entire genome. To enhance our ability to identify somatic alterations that are important in SMZL pathogenesis, we focused on variations that were present in any of the six SMZL genomes and not in the Database of SNPs (dbSNP).
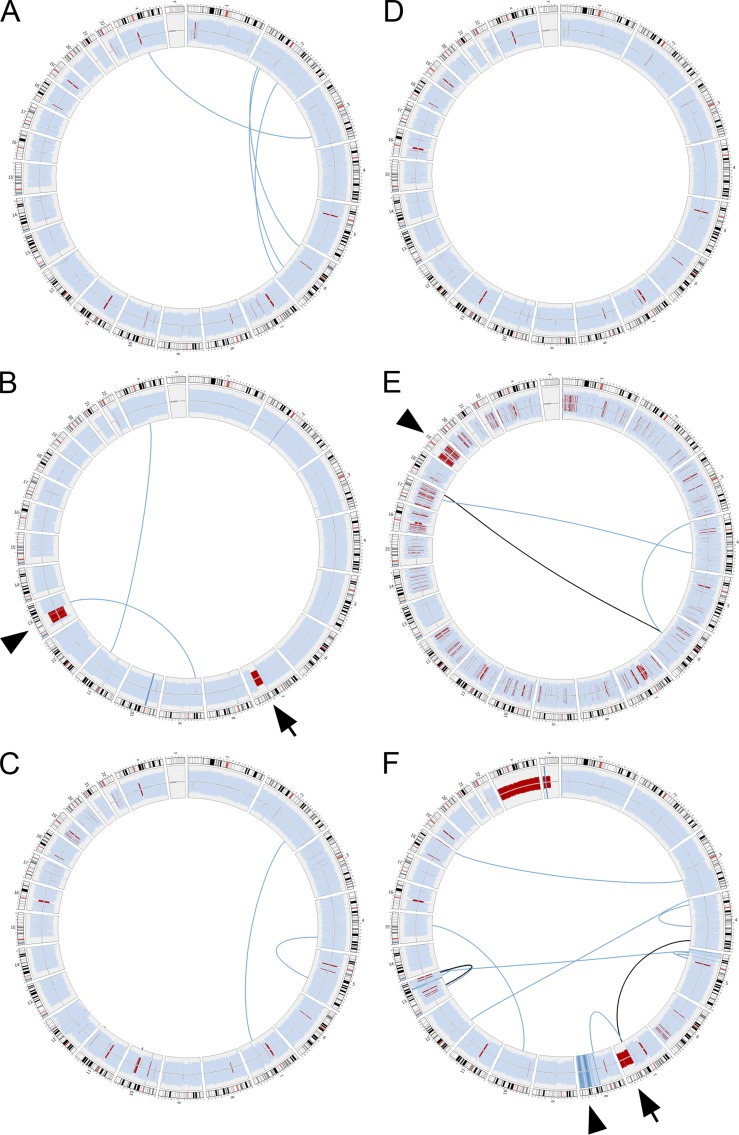
After normalization to publicly available constitutional normal genome sequencing data, relative depth of coverage for distinct chromosomal regions were examined for evidence of recurrent chromosomal gains or losses. Corresponding plots of ploidy for each genome are shown in Fig. 1 (outer data track; light blue indicates regions of euploidy, dark blue indicates region of chromosomal gain, and red indicates region of chromosomal loss). Overall, the SMZL genomes had relatively few large structural alterations affecting chromosomes (Fig. 1 and Table S1). However, in keeping with previous observations (Mateo et al., 1999; Gruszka-Westwood et al., 2003; Salido et al., 2010; Watkins et al., 2010; Rinaldi et al., 2011) recurrent deletions involving the long arm of chromosome 7 (del7q) were seen in two of the six index genomes (Fig. 1, B and F, arrows). Additionally, one of these genomes also showed a partial loss of genetic elements corresponding to the subcentromeric region of chromosome 13 (del13q; Fig. 1 B, arrowhead).
Figure 1.
Structural alterations in index SMZL cases. Circos diagrams of genomic complexity identified in six index SMZL cases. Outer data track represents the relative sequencing coverage of chromosomal regions normalized to publicly available genome sequencing data of 49 healthy individuals corresponding to ploidy at these regions. Light blue indicates region of euploidy, dark blue indicates region of chromosomal gain and red indicates region of chromosomal loss. Arrows indicate deletion portions of the long arm of chromosome 7 (del7q) in two of the six index genomes. Arrowheads indicate other regions of chromosomal loss or gain. Inner data tracks represent large structural alterations between spatial distinct genomic regions affecting the coding regions of one or more genes. Light blue lines indicate a single gene involved in structural alteration. Black lines indicate two genes involved in structural alteration. The three index genomes with mutations in NOTCH2 are shown in A–C.
Individual sequencing reads that mapped to two spatially separated regions of the reference genome were used to identify putative gene fusion or gene disruption events. To reduce the number of candidate structural alterations likely to be pathogenetic, we filtered these data to exclude structural alterations that did not affect coding elements of the involved genes (Fig. 1, inner tracks; light blue lines indicate a single gene involved in structural alteration; black lines indicate that two genes from nonadjacent genomic regions were involved in structural alteration). This analysis revealed no evidence of recurrent chromosomal translocation or chimeric fusions in the six index cases. A complete list of all structural alterations identified through genomic sequencing involving one or more genes is presented in Table S1.
In total, 2,995 candidate genes were identified with at least one previously undocumented single-nucleotide polymorphism (SNP) or small insertion/deletion event (indel) in at least one of the six SMZL genomes (comparison to dbSNP; not shown). Of these, 232 genes showed novel alterations in at least two of the six SMZL index genomes (Table S2). These included mutations in epigenetic modifiers, including MLL2 and MLL3, which have been previously reported to occur in follicular and diffuse large B cell lymphomas, but not in marginal zone lymphomas (Morin et al., 2011; Pasqualucci et al., 2011). Among the recurrently altered genes, we prioritized NOTCH2 as a candidate gene likely to be important to SMZL pathogenesis based on its known role in murine marginal zone B lymphocyte development (Saito et al., 2003; Witt et al., 2003). In three of six index SMZL cases, variant call analysis identified NOTCH2 mutations predicted to lead to protein truncation in the distal C-terminal region in the transactivation (TAD) and proline/glutamate/serine/threonine-rich (PEST) domains (Table 1). Two of these cases harbored the same p.R2400X nonsense amino acid substitution mutation, and one case harbored a length-affecting mutation leading to a frameshift at residue p.I2304 (Fig. 2 and Table 2).
Table 1.
NOTCH2 mutations identified in SMZL and MALT-L samples
| First mutation | Second variation | Confirmed | ||||||
| Cohort | Disease | Identifier | Gene | Protein | Gene | Protein | Consequence | Somatic |
| Discovery | SMZL | D-1 | c.6909dupC | p.I2304fsX9 | FS | Yes | ||
| Discovery | SMZL | D-2 | c.7198C>T | p.R2400X | NS | Yes | ||
| Discovery | SMZL | D-3 | c.7198C>T | p.R2400X | NS | Yes | ||
| Validation | SMZL | V-1 | c.4999G>A | p.V1667I | MS | Yes | ||
| Validation | SMZL | V-2 | c.6304A>T | p.K2102X | NS | Yes | ||
| Validation | SMZL | V-3 | c.6824C>A | p.A2275D | MS | Yes | ||
| Validation | SMZL | V-4 | c.6834delinsGCACG | p.T2280fsX12 | FS | Yes | ||
| Validation | SMZL | V-5 | c.6853C>T | p.Q2285X | NS | Yes | ||
| Validation | SMZL | V-6 | c.6853C>T | p.Q2285X | NS | Yes | ||
| Validation | SMZL | V-7 | c.6868G>A | p.E2290X | NS | N/A | ||
| Validation | SMZL | V-8 | c.6873delG | p.K2292fsX3 | FS | Yes | ||
| Validation | SMZL | V-9 | c.6909delC | p.I2304fsX2 | FS | N/A | ||
| Validation | SMZL | V-10 | c.6909delC | p.I2304fsX2 | FS | N/A | ||
| Validation | SMZL | V-11 | c.6909delC | p.I2304fsX2 | c.7072A>G | p.M2358V | FS/MS | N/A |
| Validation | SMZL | V-12 | c.6909dupC | p.I2304fsX9 | FS | Yes | ||
| Validation | SMZL | V-13 | c.6910delinsCCC | p.I2304fsX3 | FS | Yes | ||
| Validation | SMZL | V-14 | c.6973C>T | p.Q2325X | NS | N/A | ||
| Validation | SMZL | V-15 | c.7198C>T | p.R2400X | NS | Yes | ||
| Validation | SMZL | V-16 | c.7198C>T | p.R2400X | NS | Yes | ||
| Validation | SMZL | V-17 | c.7198C>T | p.R2400X | NS | Yes | ||
| Validation | SMZL | V-18 | c.7198C>T | p.R2400X | NS | Yes | ||
| Validation | SMZL | V-19 | c.7198C>T | p.R2400X | NS | Yes | ||
| Validation | SMZL | V-20 | c.7198C>T | p.R2400X | NS | Yes | ||
| Validation | SMZL | V-21 | c.7198C>T | p.R2400X | NS | N/A | ||
| Validation | SMZL | V-22 | c.7231G>T | p.E2411X | NS | Yes | ||
| Specificity | MALT-L | S-1 | c.7198C>T | p.R2400X | NS | N/A | ||
All NOTCH2 mutations identified through either whole genomic sequencing (discovery cohort) or targeted Sanger sequencing (validation and specificity cohorts) of the exonic regions of the NOTCH2 gene C terminus are shown. Where constitutional tissue was available for sequencing, somatic acquisition of each mutation was confirmed. One sample from the validation cohort of SMZL samples had two separate mutations. All other mutations were heterozygous. NS, non-sense; MS, missense; FS, frameshift. N/A indicates that constitutional tissue was not available for a given sample.
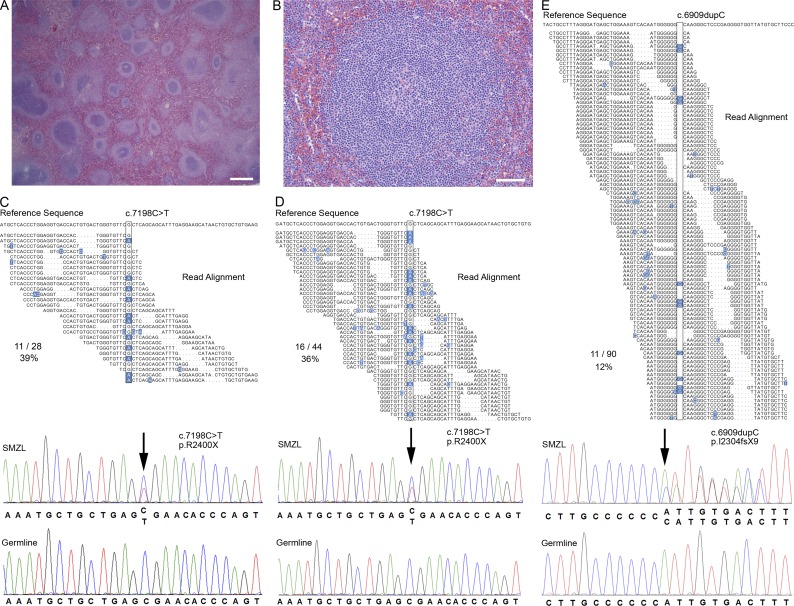
Figure 2.
WGS identifies NOTCH2 mutations in SMZL. (A and B) A representative case of SMZL with typical histopathological features of SMZL at low and high power (Bars: (A) 400 µm; (B) 50 µm) including expansion of pale staining marginal zones surrounding splenic follicles in a biphasic pattern. (C–E) Reverse complement sequence reads (Read Alignment) mapped to the reference genome (Reference Sequence) from three index samples with mutations in NOTCH2 (boxed) with deviations from reference genome highlighted in blue. Bottom panels show Sanger sequencing electropherograms confirming mutations in the index cases (SMZL) and the absence of the mutations in matched normal constitutional tissue (Germline). One frameshift p.I2304fsX9 mutation and two nonsense p.R2400X mutations were identified in three patients among the six index cases (arrows). The total number of reads containing the indicated mutation compared with the total number of reads mapping to this region is shown (C, 11/28; D, 16/44; E, 11/90). Genome sequencing was performed once for each index case. Sanger sequencing confirmation of somatic acquisition was performed in at least two independent replicates.
Table 2.
Patient and disease characteristics of SMZL patients according to NOTCH2 mutational status
| Total | Positive | Negative | Student’s t test | |||||||
| Average | St Dev | n | Average | St Dev | n | Average | St Dev | n | P-value | |
| Percent male | 35% | 71 | 22% | 18 | 40% | 53 | 0.19 | |||
| Age at diagnosis | 62 | 12 | 71 | 63 | 9 | 18 | 61 | 13 | 53 | 0.63 |
| Age at splenectomy | 63 | 12 | 71 | 65 | 10 | 18 | 63 | 12 | 50 | 0.62 |
| Stage at diagnosis | 3.7 | 0.8 | 56 | 3.5 | 1.1 | 13 | 3.8 | 0.7 | 43 | 0.39 |
| International Prognostic | 2.4 | 0.9 | 43 | 2.3 | 1.0 | 9 | 2.5 | 0.9 | 34 | 0.68 |
| Index/Follicular Lymphoma | ||||||||||
| International Prognostic | ||||||||||
| Index | ||||||||||
| Hemoglobin, g/deciliter | 11.8 | 2.0 | 51 | 11.7 | 1.7 | 11 | 11.9 | 2.1 | 40 | 0.77 |
| Lactate dehydrogenase (U/liter) | 328 | 154 | 42 | 321 | 122 | 8 | 330 | 162 | 34 | 0.89 |
| Albumin (g/deciliter) | 4.2 | 0.5 | 19 | 4.4 | 0.4 | 4 | 4.2 | 0.5 | 15 | 0.63 |
| White blood cell (K/mm3) | 18.7 | 23.9 | 21 | 11.2 | 6.8 | 5 | 21.0 | 26.9 | 16 | 0.44 |
| Platelet (K/mm3) | 201 | 109 | 19 | 160 | 50 | 4 | 213 | 119 | 15 | 0.41 |
| β2-microglobulin (mg/liter) | 3.8 | 1.5 | 19 | 3.5 | 1.9 | 4 | 3.9 | 1.4 | 15 | 0.68 |
Summary of available clinical, histopathological, and laboratory parameters for SMZL patients is shown according to NOTCH2 mutational status. No statisticlly significant differences were noted. St Dev, standard deviation.
These mutations in NOTCH2 result in deletion of known or predicted degradation motifs that regulate protein stability (Fryer et al., 2004; Chiang et al., 2006; Kopan and Ilagan, 2009). Moreover, NOTCH2 is known to drive development toward the marginal zone B cell lineage (Saito et al., 2003). Therefore, we focused our effort to further characterize NOTCH2 mutations in SMZL as they are likely to be important to the pathogenesis of this disease. Using Sanger sequencing, we confirmed the presence of these mutations in the index tumor samples (Fig. 2, SMZL) and their somatic acquisition by testing matched constitutional tissues (Germline).
Prevalence of NOTCH2 mutations in SMZL
To establish the prevalence of NOTCH2 mutations among a larger SMZL cohort, targeted Sanger sequencing of the region comprising all domains known to be important for intracellular NOTCH family signaling (exons 25 through 34; Fig. 3) was performed. We focused on this region based on the location of mutations in NOTCH2 identified in our initial screen and the analogous location of gain-of-function mutations known to contribute to other mature B cell lymphoproliferative disorders such as CLL/SLL (Del Giudice et al., 2012) and MCL (Kridel et al., 2012), as well as T-acute lymphocytic leukemia (T-ALL; Weng et al., 2004). These exons comprise three Lin-12-NOTCH repeat (LNR) domains (prevent ligand-independent activation), the HD (regulates ligand-independent activation), a single-pass trans-membrane region, RBP-Jκ–associated module domain (binds the CBF1/RBP-Jκ/suppressor of hairless/LAG-1 [CSL] transcription factor), six ankyrin repeats (bind CSL and Mastermind), the TAD, and the PEST domain important for regulating degradation of the NOTCH2 intracellular domain (NICD2; Fig. 4 A).
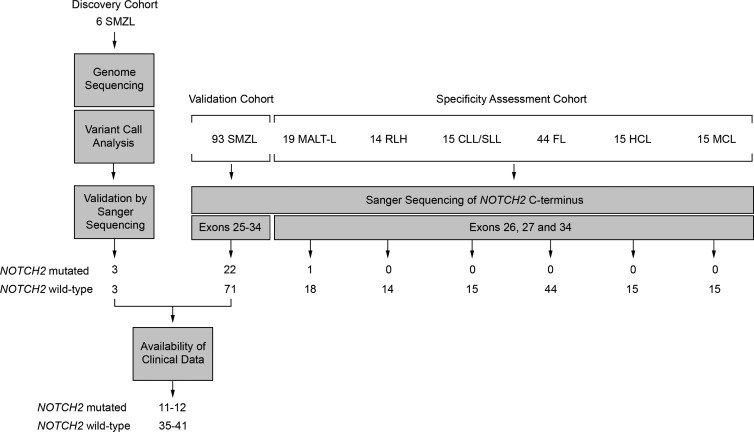
Figure 3.
Discovery, validation, and specificity assessment of NOTCH2 mutations in SMZL and other B cell lymphomas. A summary of the experimental design and results illustrates initial NOTCH2 mutation discovery in three of six index SMZL cases through WGS, all of which were confirmed as somatic mutations by traditional Sanger sequencing. Sanger sequencing of the C-terminal of NOTCH2 comprising exons 25–34 was performed in 93 additional SMZL cases comprising the validation cohort to establish the rate of recurrence of NOTCH2 mutations in SMZL. In total, 22 additional SMZL cases harbored mutations in the HD and PEST domains of NOTCH2. To assess the specificity of NOTCH2 mutations among other abnormal B cell proliferations, Sanger sequencing of NOTCH2 regions with recurrent mutations identified in discovery and validation samples of SMZL (exons 26, 27 and 34) was performed for 19 cases of nonsplenic marginal zone lymphoma (MALT-L), 14 cases of RLH, 15 chronic lymphocytic lymphoma (CLL/SLL), 44 low-grade follicular cell lymphoma (grade 1–2; FL), 15 HCL, and 15 MCL. To assess the consequences of NOTCH2 mutation on disease characteristics, clinical data were collected on a total of 46–53 SMZL cases including 11–12 cases with NOTCH2 mutation.
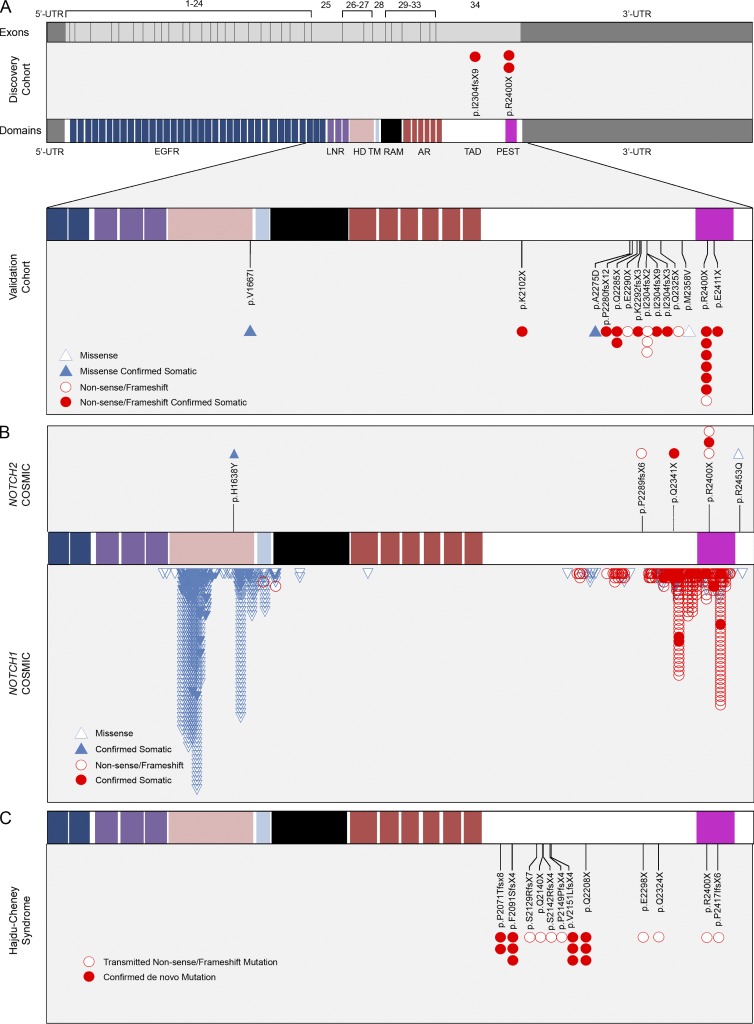
Figure 4.
NOTCH2 mutations in SMZL. (A, top) The 34 exons of NOTCH2 are shown as gray boxes flanked by the 5′- and 3′-untranslated (UTR) regions of exons 1 and 34, respectively, above the protein domain structure of NOTCH2 including 36 epidermal growth factor-like repeats (EGFR, blue; mediates ligand binding), three LNR domains (purple; prevents ligand-independent activation), the HD (pink; prevents ligand-independent activation), a single-pass transmembrane region (TM, light blue), RBP-Jκ–associated module (RAM, black; required for NOTCH signaling), six ankyrin repeats (AR, red; bind the CSL transcription factor), the transactivation domain (TAD, white), and the proline-, glutamate-, serine- and threonine-rich domain (PEST, magenta). (A, Middle) Three mutations in the TAD and the PEST domain downstream of the AR region were identified in the SMZL discovery cohort. (A, Lower) Targeted Sanger sequencing of the SMZL validation cohort uncovered the same as well as additional missense (blue triangles), nonsense and frameshift (red circles) mutations in the HD, TAD and PEST domains. Constitutional tissue from a total of 19 patients confirmed somatic acquisition of these mutations in all samples (solid symbols). Sanger sequencing confirmation was performed in at least two independent replicates. (B) The locations of mutations in hematolymphoid malignancies in the NOTCH2 (7 total; top) and NOTCH1 (>800 total; bottom) genes in the COSMIC database are displayed adjacent to NOTCH2 and NOTCH1 amino acid sequence alignment. Mutations within both genes cluster in the HD and TAD/PEST domains. Specific amino acid residues and the predicted consequence of all NOTCH2 mutations in COSMIC are also displayed. (C) Mutations in HCS are confined to the C-terminal of the NOTCH2 protein and cluster within the TAD and PEST domains. The p.R2400X mutation seen in 9 cases of SMZL is also present in at least one case of HCS as an inherited mutation (open symbol). De novo mutations are displayed as solid symbols.
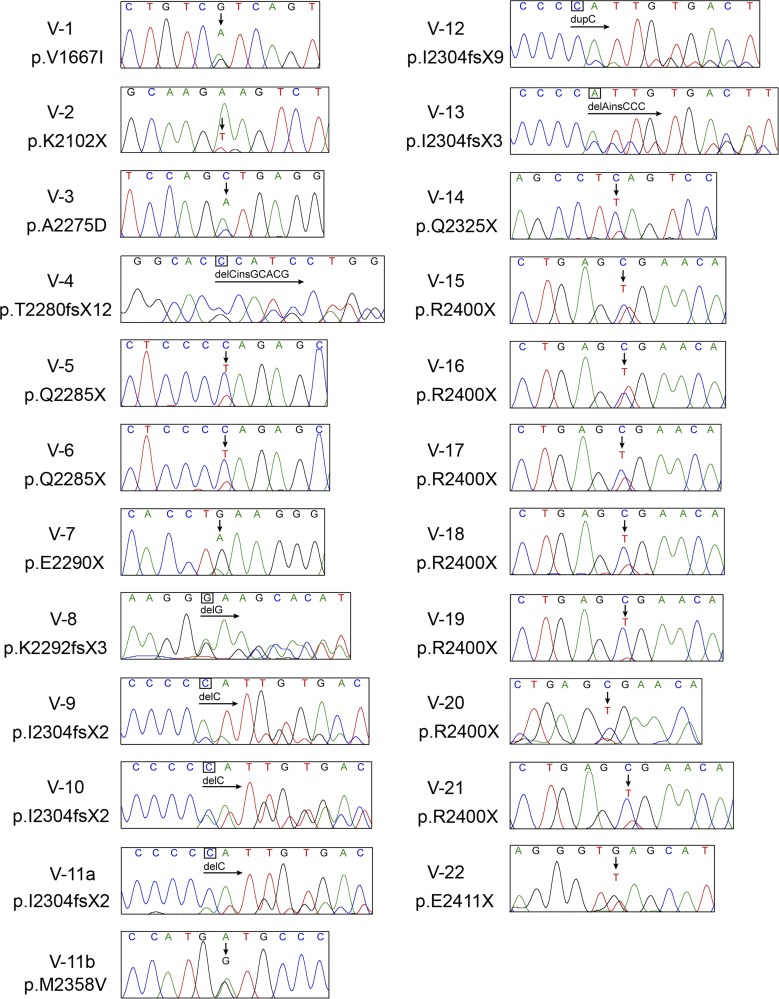
In total, 93 additional SMZL cases were screened by Sanger sequencing for mutations in NOTCH2. A total of 11 novel mutations, 7 additional p.R2400X, and 5 additional frameshift mutations affecting the p.I2304 residue were discovered in these SMZL cases (Fig. 4 A and Fig. 5 and Table 1). These mutations were largely truncating mutations (either frameshift or nonsense mutations) confined to the distal TAD and PEST domains and are predicted to eliminate degradation signals in the PEST domain, thereby increasing the stability of the NICD2. A single missense mutation (p.V1667I) located in the HD is analogous to the p.V1722I NOTCH1 mutation in T-ALL associated with ligand-independent NOTCH1 activation (Malecki et al., 2006; Gordon et al., 2007). Overall, 25 of 99 SMZL cases (25.3%) harbored NOTCH2 mutations. Whereas most of these mutations were single heterozygous mutations, 1 of 25 SMZL patients had two distinct NOTCH2 mutations, including both a truncating mutation (p.I2304fsX2) and a missense variant (p.M2358V; although constitutional tissue was not available to assess somatic acquisition). Of the 25 cases with NOTCH2 mutations, 19 patients had corresponding matched normal tissue. None of the constitutional tissues harbored sequence variants, indicating that the detected mutations in NOTCH2 are somatically acquired.
Figure 5.
Sanger sequencing identification of NOTCH2 mutations in SMZL validation cohort. Sanger sequencing results for each NOTCH2-mutated sample are shown. Arrows indicate site of nucleotide change. Amino acid change predicted for each mutation is indicated below sample label. Traces shown are representative of at least two independent amplification and sequencing reactions.
Having established a high frequency of NOTCH2 mutations in our validation cohort, we queried our initial genomic sequencing screening data for the existence of alterations affecting other genes in the NOTCH signaling pathway. This investigation identified predicted protein coding alterations affecting MAML2, an essential cofactor of the NOTCH2 transcriptional complex, in the three genomes that did not have NOTCH2 mutations. These alterations included previously reported p.Q237R and p.V836I variants, as well as a novel p.G25W mutation. Sanger sequencing confirmed the variants in the corresponding tumor samples. However, the previously reported variants were present in corresponding constitutional tissue, and thus were not somatically acquired. The novel p.G25W mutation was confirmed to be somatically acquired by direct Sanger sequencing (Table S2). The mutation affects an amino acid with the N-terminal region of the MAML2 protein known to mediate protein–protein interactions with NOTCH family members. We therefore sought to assess the prevalence of additional MAML2 mutations in our validation cohort. This identified a single additional somatic mutation in MAML2 (p.A11S) in a genome without an identified NOTCH2 mutation. Overall, the prevalence of putative impactful somatic mutations in MAML2 was therefore 2 out of 99 cases (2.0%). The identification of mutations affecting MAML2 is intriguing and warrants further investigation. No mutations were found in FBW7 or other NOTCH pathway–related genes in the discovery cohort.
Specificity of NOTCH2 mutations
Having established the frequency of NOTCH2 mutations in SMZL, we next sought to assess the specificity of these mutations for SMZL. We performed targeted Sanger sequencing on CLL/SLL, FL, HCL, MCL, and reactive lymphoid hyperplasia (RLH) samples. No evidence of NOTCH2 mutations was identified in any of 103 cases of CLL/SLL, FL, HCL, MCL or RLH (95% confidence interval 0–2.9%; Fig. 3 and Fig. 6 A). Given the relatively small number of non-SMZL cases examined here for each disease category, low prevalence of NOTCH2 mutations in these other disease entities cannot be ruled out.
Figure 6.
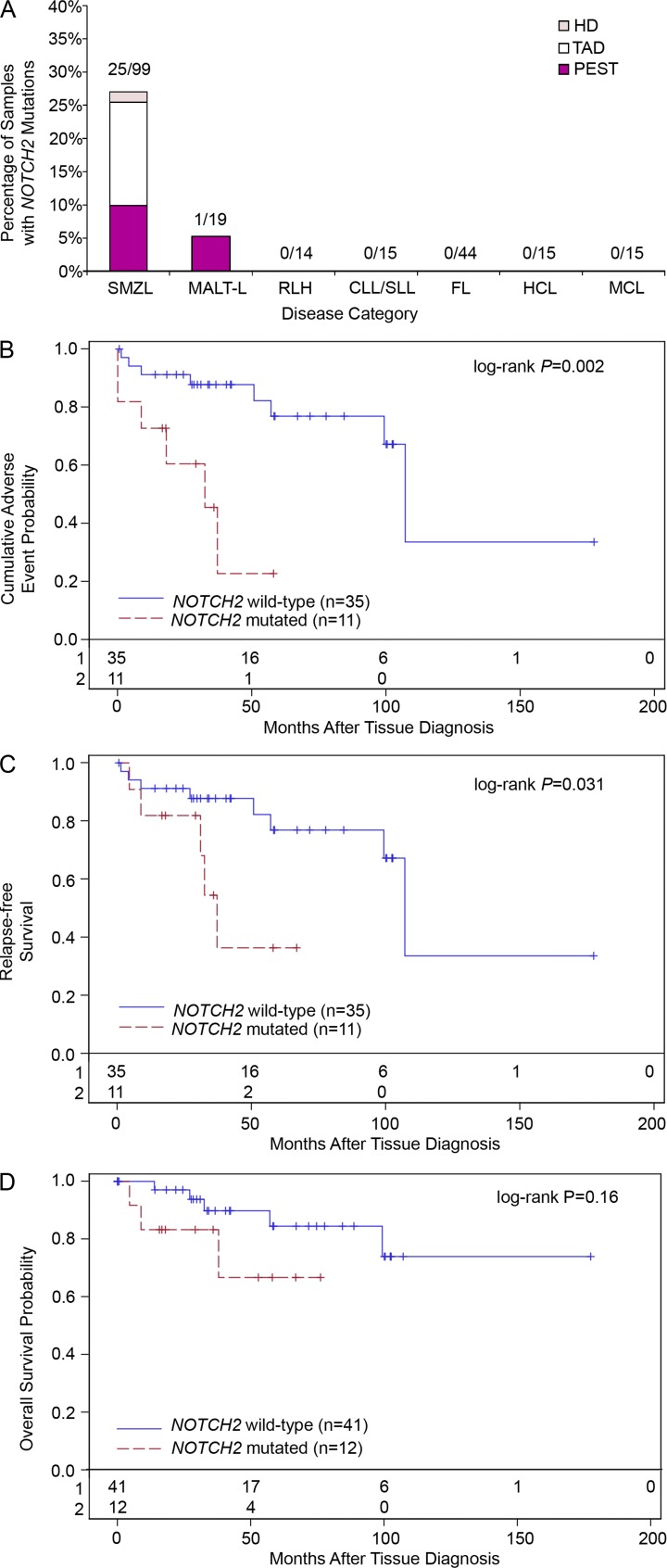
Impact of NOTCH2 mutations on clinical outcome in SMZL. (A) Frequency of NOTCH2 mutations in SMZL, MALT-L, and other B cell proliferative disorders divided among the different domains of the NOTCH2 protein (color corresponds to domain in which mutations were located). For each separate disease, the number of samples with NOTCH2 mutations compared with total number of samples analyzed is displayed above each bar. (B) Cumulative probability of relapse, transformation or death from time of tissue diagnosis for patients with NOTCH2-mutated (red lines) and NOTCH2-wild-type (blue lines) SMZL. (C) Relapse-free survival from tissue diagnosis. (D) Overall survival from tissue diagnosis. The total number of patients in each analysis is displayed along the x-axes of each graph.
In addition to assessing 99 SMZL cases, we also assessed 19 nodal and extranodal marginal zone lymphomas for the presence of NOTCH2 mutations and identified one sample (an extranodal marginal zone B cell lymphoma of the breast mucosa-associated lymphoid tissue lymphoma [MALT-L]) that also harbored a heterozygous p.R2400X mutation (Fig. 6 A). These data indicate a high frequency of NOTCH2 mutations in SMZLs and a lower (5.3%) frequency in nonsplenic MZL. Collectively, these data indicate a high predilection of activating mutations in NOTCH2 among MZLs.
Impact of NOTCH2 mutations on clinical outcome
Having demonstrated the presence of NOTCH2 mutations in a subset of SMZL cases, we next sought to determine whether the presence of these mutations influenced clinical outcomes. Time to adverse outcome, defined from tissue diagnosis to relapse, transformation, or death was compared between patients harboring NOTCH2 mutations and those with wild-type NOTCH2. Survival data were available for 46 patients from this study, including 11 patients with NOTCH2 mutations and 35 patients with wild-type NOTCH2 with a median follow up of 40 mo (range: 0.7–177 mo). Patients with NOTCH2 mutations had significantly shorter time to adverse outcome compared with patients with wild-type NOTCH2 (the median time to adverse outcome was 32.6 mo in NOTCH2-mutated patients versus 107.2 mo in patients without NOTCH2 mutations (P = 0.002; Fig. 6 B). After controlling for patient gender, performance status, age and stage at diagnosis, harboring a NOTCH2 mutation is associated with shorter time to adverse outcome (hazard ratio = 5.57; P = 0.057). Furthermore, patients with NOTCH2 mutations also had significantly shorter relapse-free survival, defined from tissue diagnosis to relapse or death (P = 0.031; Fig. 6 C). Altogether, these results demonstrate that the presence of NOTCH2 mutation at diagnosis indicates worse patient outcome.
DISCUSSION
We performed WGS in six cases of SMZL and identified NOTCH2 mutations in half of these cases. Sanger sequencing of 93 additional SMZLs and 103 other types of B cell lymphoma or leukemia or reactive lymphoid hyperplasia showed NOTCH2 mutations in 22 additional SMZL patients, yielding an overall frequency of 25.3%. No mutations were identified in other non-MZL B cell lymphomas and leukemias analyzed. Moreover, in 19 patients with NOTCH2-mutated SMZL, constitutional DNA was available for assessment and was confirmed to be wild-type, thus indicating somatic acquisition of NOTCH2 mutation in SMZL.
In total, we identified 26 NOTCH2 mutations in 25 SMZL patients. These mutations represented six unique types of nonsense mutations, five unique types of frameshift mutations, and three unique types of missense mutations. 25 of these mutations affected the TAD or PEST domains, with 23 predicted to yield protein truncation at or upstream of the PEST domain. The remaining case harbored a somatic p.V1667I mutation in the HD. All of these mutations were identified in the same protein domains as have been reported for NOTCH1 in T-ALL, CLL/SLL, and MCL. However, NOTCH1 mutations in T-ALL are more prevalent in the HD than the TAD and PEST domain (Fig. S3). Disruption of the C-terminal PEST domain renders NOTCH less susceptible to regulation by ubiquitin-mediated proteolysis, and thus results in increased activation of the NOTCH pathway (Gupta-Rossi et al., 2001; Oberg et al., 2001; Wu et al., 2001). Using reporter assays for assessment of NOTCH activation, we confirmed that representative mutations affecting either the PEST or HD indeed resulted in NOTCH2 transcriptional hyperactivation (not depicted).
Interestingly, in contrast to the activating effect in SMZL and other lymphoid malignancies, recent studies have suggested that the NOTCH pathway can also function as a tumor suppressor. Loss-of-function mutations in the NOTCH pathway (NCSTN, MAML1, APH1A, and NOTCH2) were recently identified in chronic myelomonocytic leukemia (Klinakis et al., 2011). Other studies have identified oncogenic mutations within the epidermal growth factor repeat region of NOTCH1 in head and neck cancer (Agrawal et al., 2011; Stransky et al., 2011). Loss-of-function mutations affecting NOTCH family and pathway genes have also been implicated in skin and lung cancers (Wang et al., 2011). Finally animal and in vitro studies suggest a tumor suppressor role in hepatocellular carcinoma (Viatour et al., 2011), pancreatic carcinoma (Mazur et al., 2010), and neuroblastoma (Zage et al., 2012). In contrast, mutations identified in lymphoid malignancies (T-ALL, B-CLL/SLL, mantle cell, and diffuse large B cell lymphoma) have all been gain-of-function mutations confined to the C-terminal region extending from exon 25 to exon 34, as was the case in our initial study.
Pathogenic germline mutations in the TAD/PEST domain of NOTCH2 have been reported in Hajdu-Cheney syndrome (HCS), a rare autosomal dominant skeletal disorder characterized by facial anomalies, acroosteolysis, and osteoporosis (Isidor et al., 2011; Simpson et al., 2011). The NOTCH2 mutations in HCS include one report of a transmitted p.R2400X mutation (Simpson et al., 2011; Fig. 4 C). No predilection for lymphoma or B lymphocyte dysfunction in HCS patients has been reported to date. Interpretation of the significance of this is confounded by the extreme rarity of this disease. Nonetheless, it is reasonable to speculate that additional genetic alterations may be required for SMZL development.
With regard to neoplasia, isolated NOTCH2 mutations have been reported in a single case of SMZL and a single case of extranodal MZL in a previous study (Trøen et al., 2008), as well as in a small proportion of cases of diffuse large B cell lymphoma (Lee et al., 2009) or Richter’s transformation (Fabbri et al., 2011), but no evidence for prognostic implications for NOTCH2 mutations was presented in any of these studies. NOTCH2 shares significant homology with NOTCH1, and transforming capacity has been demonstrated for truncated alleles of both proteins (Ellisen et al., 1991; Rohn et al., 1996; Capobianco et al., 1997). Intriguingly, loss-of-function mutations affecting NOTCH family and pathway genes have recently been implicated in the pathogenesis of myeloid (Klinakis et al., 2011) and epithelial malignancies (Mazur et al., 2010; Agrawal et al., 2011; Stransky et al., 2011; Viatour et al., 2011; Wang et al., 2011), as well as in neuroblastoma (Zage et al., 2012). These studies highlight the context-dependent roles of NOTCH and its signaling partners, which upon mutation, may contribute to the pathogenesis of neoplasia via different mechanisms in diverse cell types. Altogether, these findings suggest that the 26 NOTCH2 mutations we identified are likely to be pathogenic events contributing to aberrant NOTCH2 signaling in malignant SMZL cells.
Examination of NOTCH2 mutational status in nonsplenic MZLs revealed mutation in ∼5% of cases analyzed. The NOTCH2 mutation identified in a single case of extranodal MZL of the breast was a p.R2400X nonsense mutation. This mutation was also identified in 9 of 99 (9.1%) SMZL cases. The selectivity of NOTCH2 mutations for malignancies of marginal zone B cells is in keeping with the known role of NOTCH2 in marginal zone cell fate determination (Saito et al., 2003; Witt et al., 2003). It is noteworthy that NOTCH1 dictates T cell fate and that supraphysiological NOTCH1 signaling induces T-ALL (Weng et al., 2004). We speculate that because NOTCH2 specifies marginal zone B cell fate, supraphysiological NOTCH2 signaling may analogously play a role in the pathogenesis of MZL.
Somatic mutations affecting specific genes that impact SMZL prognosis are largely unknown. Although previous studies have implicated a role for mutations targeting genes in the NF-κB pathway in a subset of SMZL (Rossi et al., 2011), only TP53 alterations affecting a small minority of cases have been demonstrated to impact SMZL prognosis (Salido et al., 2010; Rinaldi et al., 2011). TP53 mutations were not identified in our initial WGS screen of six index cases of SMZL. We have found that the presence of NOTCH2 mutations in SMZLs at time of diagnosis predicted an adverse disease course characterized either by refractoriness to therapy, histological transformation to higher grade disease, or an otherwise aggressive clinical course. Assessment of NOTCH2 mutation status in cases of SMZL may thus be useful to predict the risk of aggressive disease. This finding may also inform clinical decision-making at diagnosis, with the presence of NOTCH2 mutation being an indication for more aggressive therapy. By analogy with pathogenetic mechanisms of NOTCH1 mutation in T-ALL, it is tempting to speculate that similar downstream targets promoting proliferation, survival and deregulated metabolic pathways are also deregulated in SMZL. In addition to predicting a more aggressive disease course with an increased tendency to relapse, there is a trend toward reduced overall survival (i.e., time to death) among patients with NOTCH2-mutated SMZL (Fig. 6 D). However, this trend in overall survival did not reach the level of statistical significance, presumably because of the small sample size in this study (P = 0.16).
In summary, we used WGS to reveal high-frequency recurrent somatic mutations involving NOTCH2 in SMZL. NOTCH2 mutations appear to be specific for marginal zone lymphomas as compared with other B cell leukemias and lymphomas. Additionally, our initial studies indicate an adverse outcome for patients with NOTCH2-mutated SMZL. Therefore, we have identified a biomarker specific for a subset of SMZL patients that may have value in both diagnosis and prognosis of patients with SMZL. Our findings therefore expand the spectrum of recurrent genetic alterations affecting genes in the NOTCH pathway in human malignancy and suggest potential therapies targeting NOTCH2 in the treatment of SMZL. Our studies further underscore the ability of unbiased large-scale screening approaches to uncover novel molecular mechanisms in neoplasia.
MATERIALS AND METHODS
Patients and samples.
Six SMZL samples from the University of Michigan were selected as index cases for WGS. To assess the prevalence of NOTCH2 mutations in SMZL, an additional 93 SMZL cases were obtained from the University of Texas MD Anderson Cancer Center (31 cases), the University of Utah Health Sciences Center (25 cases), the Southern California Permanente Medical Group (20 cases), the University of Michigan (15 cases), and the University of Wisconsin (2 cases). Approval from the University of Michigan Hospital institutional review board (HUM00023256) was obtained for these studies. To assess the specificity of NOTCH2 mutations in SMZL, genomic DNA was extracted from additional tissues representing non-SMZL diseases, including 15 cases of CLL/SLL, 15 cases of MCL, 44 cases of grade 1–2 FL, 15 cases of HCL, and 14 cases of RLH. In addition, 19 cases of nonsplenic MZL (i.e., nodal and extranodal/mucosa-associated lymphoid tissue lymphoma) were analyzed.
Pathology review.
All specimens were reviewed independently and confirmed by consensus among three hematopathologists (MSL, NGB, and KEJ) according to World Health Organization classification criteria (Isaacson et al., 2008) without knowledge of NOTCH2 mutational status. Only cases containing adequate neoplastic tissue were included in subsequent analyses.
WGS and targeted NOTCH2 DNA sequencing.
From each of six index SMZL cases, 10 µg of high-molecular-weight genomic DNA was extracted from fresh frozen tumor tissue using the QIAamp DNA extraction kit (QIAGEN) and subjected to WGS by Complete Genomics, Inc. (CGI; Mountain View, CA). CGI performs massively parallel short-read sequencing using a combinatorial probe-anchor ligation (cPAL) chemistry coupled with a patterned nanoarray-based platform of self-assembling DNA nanoballs (Drmanac et al., 2010). Library generation, read-mapping to the NCBI reference genome (Build 37, RefSeq Accession nos. CM000663-CM00686), local de novo assembly and variant-calling protocols were performed as previously described (Drmanac et al., 2010; Roach et al., 2010). Initial read mapping and variant calling were performed using CGAtools v1.3.0 (http://cgatools.sourceforge.net/docs/1.3.0/). Additional downstream bioinformatic analyses were performed using custom designed PERL processing routines. Targeted sequencing of the NOTCH2 C-terminal coding exons 25 to 34 was performed using Sanger sequencing for the SMZL samples in the validation cohort. For all other samples, sequencing was confined to exons 26, 27, and 34, where all confirmed mutations in SMZL samples occurred. Somatic acquisition of each mutation was also assessed when matched constitutional tissue was available for analysis. Genomic DNA from index cases and genomic DNA corresponding to matched constitutional tissue were subjected to Sanger sequencing of regions of the NOTCH2 where mutations were observed through WGS. For targeted sequencing of exons 25–34 in the NOTCH2 C-terminal region in the validation and specificity cohort samples, genomic DNA was extracted using both the QIAGEN BioRobot EZ1 and QIAamp FFPE DNA extraction kits (QIAGEN). For all Sanger sequencing reactions, PCR amplification was performed using Phusion DNA polymerase (New England Biolabs) followed by conventional Sanger sequencing technology using BigDye version 3.1 chemistry run on an Applied Biosystems 3730xl DNA Sequencer at the University of Michigan DNA sequencing Core. All sequencing reactions were performed using nested sequencing primers. Sequencing trace analysis was performed using Mutation Surveyor software. All mutations were verified in at least two independent PCR amplification and sequencing reactions. cDNA nucleotide numbering of coding sequence is based on GenBank accession NG_008163.1. Protein amino acid numbering is based on GenBank accession NP_077719.2. Detailed primer sequences for targeted exon sequencing can be found in Table S3-S4.
Statistical analysis of clinical outcomes.
Clinical outcomes data (time to transformation, relapse or death) were analyzed using standard survival analysis. Survival plots were generated using Kaplan-Meier method and Log-rank tests were used to compare survival times between patients with NOTCH2 mutations and patients with wild-type NOTCH2. Cox-proportional hazards regression analysis was conducted to compare the two groups of patients after adjusting for age, gender, performance status, and stage at diagnosis. Statistical analyses were performed with SAS version 9.3.
Online supplemental material.
Table S1 details genes involved in large scale structural alterations identified by whole genomic sequencing of six index SMZL cases. Table S2 details genes with novel alterations identified in two or more of the index SMZL genomes by genomic sequencing. Tables S3 and S4 detail the primer sequences used for Sanger sequencing confirmation of NOTCH2 mutations. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120910/DC1.
Supplementary Material
Acknowledgments
We thank Drs. Eric Fearon and David Lombard for critically reading the manuscript and helpful suggestions. We also thank Farah Keyoumarsi, Constance Eaves, Robert Lyons and James Cavalcoli for expert technical assistance.
This work was supported in part by National Institutes of Health grants R01 DE119249 and R01 CA136905 (K. Elenitoba-Johnson), R01 CA140806 (M. Lim), and the Department of Pathology at the University of Michigan.
The authors have no competing financial interests.
Footnotes
Abbreviations used:
- CLL/SLL
- chronic lymphocytic leukemia/small lymphocytic lymphoma
- CSL
- CBF1/RBP-Jκ/suppressor of hairless/LAG-1
- FL
- follicular lymphoma
- HCL
- hairy cell leukemia
- HCS
- Hajdu-Cheney syndrome
- HD
- heterodimerization domain
- LNR
- Lin-12-NOTCH repeat
- MALT
- mucosa-associated lymphoid tissue
- MALT-L
- MALT lymphoma
- MCL
- mantle cell lymphoma
- MZL
- marginal zone lymphoma
- NS
- nonsense
- PEST
- proline/glutamate/serine/threonine
- RLH
- reactive lymphoid hyperplasia
- SMZL
- splenic marginal zone lymphoma
- SNP
- single nucleotide polymorphism
- T-ALL
- T-acute lymphocytic leukemia
- WBC
- white blood cell
- WGS
- whole-genome sequencing
References
- Agrawal N., Frederick M.J., Pickering C.R., Bettegowda C., Chang K., Li R.J., Fakhry C., Xie T.X., Zhang J., Wang J., et al. 2011. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 333:1154–1157 10.1126/science.1206923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcaini L., Lazzarino M., Colombo N., Burcheri S., Boveri E., Paulli M., Morra E., Gambacorta M., Cortelazzo S., Tucci A., et al. ; Integruppo Italiano Linfomi 2006. Splenic marginal zone lymphoma: a prognostic model for clinical use. Blood. 107:4643–4649 10.1182/blood-2005-11-4659 [DOI] [PubMed] [Google Scholar]
- Aster J.C., Blacklow S.C., Pear W.S. 2011. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J. Pathol. 223:262–273 10.1002/path.2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capobianco A.J., Zagouras P., Blaumueller C.M., Artavanis-Tsakonas S., Bishop J.M. 1997. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol. Cell. Biol. 17:6265–6273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón J.I., Mollejo M., Muñoz E., Algara P., Mateo M., Lopez L., Andrade J., Carbonero I.G., Martínez B., Piris M.A., Cruz M.A. 2002. Splenic marginal zone lymphoma: clinical characteristics and prognostic factors in a series of 60 patients. Blood. 100:1648–1654 [PubMed] [Google Scholar]
- Chiang M.Y., Xu M.L., Histen G., Shestova O., Roy M., Nam Y., Blacklow S.C., Sacks D.B., Pear W.S., Aster J.C. 2006. Identification of a conserved negative regulatory sequence that influences the leukemogenic activity of NOTCH1. Mol. Cell. Biol. 26:6261–6271 10.1128/MCB.02478-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Giudice I., Rossi D., Chiaretti S., Marinelli M., Tavolaro S., Gabrielli S., Laurenti L., Marasca R., Rasi S., Fangazio M., et al. 2012. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica. 97:437–441 10.3324/haematol.2011.060129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G., et al. 2010. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 327:78–81 10.1126/science.1181498 [DOI] [PubMed] [Google Scholar]
- Ellisen L.W., Bird J., West D.C., Soreng A.L., Reynolds T.C., Smith S.D., Sklar J. 1991. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 66:649–661 10.1016/0092-8674(91)90111-B [DOI] [PubMed] [Google Scholar]
- Fabbri G., Rasi S., Rossi D., Trifonov V., Khiabanian H., Ma J., Grunn A., Fangazio M., Capello D., Monti S., et al. 2011. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J. Exp. Med. 208:1389–1401 10.1084/jem.20110921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco V., Florena A.M., Iannitto E. 2003. Splenic marginal zone lymphoma. Blood. 101:2464–2472 10.1182/blood-2002-07-2216 [DOI] [PubMed] [Google Scholar]
- Fryer C.J., White J.B., Jones K.A. 2004. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell. 16:509–520 10.1016/j.molcel.2004.10.014 [DOI] [PubMed] [Google Scholar]
- Gordon W.R., Vardar-Ulu D., Histen G., Sanchez-Irizarry C., Aster J.C., Blacklow S.C. 2007. Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 14:295–300 10.1038/nsmb1227 [DOI] [PubMed] [Google Scholar]
- Gruszka-Westwood A.M., Hamoudi R., Osborne L., Matutes E., Catovsky D. 2003. Deletion mapping on the long arm of chromosome 7 in splenic lymphoma with villous lymphocytes. Genes Chromosomes Cancer. 36:57–69 10.1002/gcc.10142 [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N., Le Bail O., Gonen H., Brou C., Logeat F., Six E., Ciechanover A., Israël A. 2001. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 276:34371–34378 10.1074/jbc.M101343200 [DOI] [PubMed] [Google Scholar]
- Isaacson P.G., Piris M.A., Berger F., Muller-Hermelink H.K., Catovsky D., Swerdlow S., Nathwani B., Montserrat E., Harris N.L. 2008. Splenic marginal zone lymphoma. WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues. Swerdlow S.H., Campo E., Harris N.L., Jaffe E.S., Pileri S.A., Stein H., Thiele J., Vardiman J.W., IARC, Lyon: 185–187 [Google Scholar]
- Isidor B., Lindenbaum P., Pichon O., Bézieau S., Dina C., Jacquemont S., Martin-Coignard D., Thauvin-Robinet C., Le Merrer M., Mandel J.L., et al. 2011. Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat. Genet. 43:306–308 10.1038/ng.778 [DOI] [PubMed] [Google Scholar]
- Klinakis A., Lobry C., Abdel-Wahab O., Oh P., Haeno H., Buonamici S., van De Walle I., Cathelin S., Trimarchi T., Araldi E., et al. 2011. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 473:230–233 10.1038/nature09999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Ilagan M.X. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 137:216–233 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kridel R., Meissner B., Rogic S., Boyle M., Telenius A., Woolcock B., Gunawardana J., Jenkins C., Cochrane C., Ben-Neriah S., et al. 2012. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 119:1963–1971 10.1182/blood-2011-11-391474 [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Kumano K., Nakazaki K., Sanada M., Matsumoto A., Yamamoto G., Nannya Y., Suzuki R., Ota S., Ota Y., et al. 2009. Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer Sci. 100:920–926 10.1111/j.1349-7006.2009.01130.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki M.J., Sanchez-Irizarry C., Mitchell J.L., Histen G., Xu M.L., Aster J.C., Blacklow S.C. 2006. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol. Cell. Biol. 26:4642–4651 10.1128/MCB.01655-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo M., Mollejo M., Villuendas R., Algara P., Sanchez-Beato M., Martínez P., Piris M.A. 1999. 7q31-32 allelic loss is a frequent finding in splenic marginal zone lymphoma. Am. J. Pathol. 154:1583–1589 10.1016/S0002-9440(10)65411-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur P.K., Einwächter H., Lee M., Sipos B., Nakhai H., Rad R., Zimber-Strobl U., Strobl L.J., Radtke F., Klöppel G., et al. 2010. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc. Natl. Acad. Sci. USA. 107:13438–13443 10.1073/pnas.1002423107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R.D., Mendez-Lago M., Mungall A.J., Goya R., Mungall K.L., Corbett R.D., Johnson N.A., Severson T.M., Chiu R., Field M., et al. 2011. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 476:298–303 10.1038/nature10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg C., Li J., Pauley A., Wolf E., Gurney M., Lendahl U. 2001. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 276:35847–35853 10.1074/jbc.M103992200 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Trifonov V., Fabbri G., Ma J., Rossi D., Chiarenza A., Wells V.A., Grunn A., Messina M., Elliot O., et al. 2011. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 43:830–837 10.1038/ng.892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente X.S., Pinyol M., Quesada V., Conde L., Ordóñez G.R., Villamor N., Escaramis G., Jares P., Beà S., González-Díaz M., et al. 2011. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 475:101–105 10.1038/nature10113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui J.C., Allman D., Xu L., DeRocco S., Karnell F.G., Bakkour S., Lee J.Y., Kadesch T., Hardy R.R., Aster J.C., Pear W.S. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 11:299–308 10.1016/S1074-7613(00)80105-3 [DOI] [PubMed] [Google Scholar]
- Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H.R., Aguet M. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558 10.1016/S1074-7613(00)80054-0 [DOI] [PubMed] [Google Scholar]
- Rinaldi A., Mian M., Chigrinova E., Arcaini L., Bhagat G., Novak U., Rancoita P.M., De Campos C.P., Forconi F., Gascoyne R.D., et al. 2011. Genome-wide DNA profiling of marginal zone lymphomas identifies subtype-specific lesions with an impact on the clinical outcome. Blood. 117:1595–1604 10.1182/blood-2010-01-264275 [DOI] [PubMed] [Google Scholar]
- Roach J.C., Glusman G., Smit A.F., Huff C.D., Hubley R., Shannon P.T., Rowen L., Pant K.P., Goodman N., Bamshad M., et al. 2010. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 328:636–639 10.1126/science.1186802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey E.A., Bluestone J.A. 2004. Notch signaling in lymphocyte development and function. Curr. Opin. Immunol. 16:360–366 10.1016/j.coi.2004.03.009 [DOI] [PubMed] [Google Scholar]
- Rohn J.L., Lauring A.S., Linenberger M.L., Overbaugh J. 1996. Transduction of Notch2 in feline leukemia virus-induced thymic lymphoma. J. Virol. 70:8071–8080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D., Deaglio S., Dominguez-Sola D., Rasi S., Vaisitti T., Agostinelli C., Spina V., Bruscaggin A., Monti S., Cerri M., et al. 2011. Alteration of BIRC3 and multiple other NF-κB pathway genes in splenic marginal zone lymphoma. Blood. 118:4930–4934 10.1182/blood-2011-06-359166 [DOI] [PubMed] [Google Scholar]
- Saito T., Chiba S., Ichikawa M., Kunisato A., Asai T., Shimizu K., Yamaguchi T., Yamamoto G., Seo S., Kumano K., et al. 2003. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 18:675–685 10.1016/S1074-7613(03)00111-0 [DOI] [PubMed] [Google Scholar]
- Salido M., Baró C., Oscier D., Stamatopoulos K., Dierlamm J., Matutes E., Traverse-Glehen A., Berger F., Felman P., Thieblemont C., et al. 2010. Cytogenetic aberrations and their prognostic value in a series of 330 splenic marginal zone B-cell lymphomas: a multicenter study of the Splenic B-Cell Lymphoma Group. Blood. 116:1479–1488 10.1182/blood-2010-02-267476 [DOI] [PubMed] [Google Scholar]
- Simpson M.A., Irving M.D., Asilmaz E., Gray M.J., Dafou D., Elmslie F.V., Mansour S., Holder S.E., Brain C.E., Burton B.K., et al. 2011. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat. Genet. 43:303–305 10.1038/ng.779 [DOI] [PubMed] [Google Scholar]
- Stransky N., Egloff A.M., Tward A.D., Kostic A.D., Cibulskis K., Sivachenko A., Kryukov G.V., Lawrence M.S., Sougnez C., McKenna A., et al. 2011. The mutational landscape of head and neck squamous cell carcinoma. Science. 333:1157–1160 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trøen G., Wlodarska I., Warsame A., Hernández Llodrà S., De Wolf-Peeters C., Delabie J. 2008. NOTCH2 mutations in marginal zone lymphoma. Haematologica. 93:1107–1109 10.3324/haematol.11635 [DOI] [PubMed] [Google Scholar]
- Viatour P., Ehmer U., Saddic L.A., Dorrell C., Andersen J.B., Lin C., Zmoos A.F., Mazur P.K., Schaffer B.E., Ostermeier A., et al. 2011. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. J. Exp. Med. 208:1963–1976 10.1084/jem.20110198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N.J., Sanborn Z., Arnett K.L., Bayston L.J., Liao W., Proby C.M., Leigh I.M., Collisson E.A., Gordon P.B., Jakkula L., et al. 2011. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc. Natl. Acad. Sci. USA. 108:17761–17766 10.1073/pnas.1114669108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins A.J., Huang Y., Ye H., Chanudet E., Johnson N., Hamoudi R., Liu H., Dong G., Attygalle A., McPhail E.D., et al. 2010. Splenic marginal zone lymphoma: characterization of 7q deletion and its value in diagnosis. J. Pathol. 220:461–474 [DOI] [PubMed] [Google Scholar]
- Weng A.P., Ferrando A.A., Lee W., Morris J.P., IV, Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271 10.1126/science.1102160 [DOI] [PubMed] [Google Scholar]
- Witt C.M., Won W.J., Hurez V., Klug C.A. 2003. Notch2 haploinsufficiency results in diminished B1 B cells and a severe reduction in marginal zone B cells. J. Immunol. 171:2783–2788 [DOI] [PubMed] [Google Scholar]
- Wu G., Lyapina S., Das I., Li J., Gurney M., Pauley A., Chui I., Deshaies R.J., Kitajewski J. 2001. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 21:7403–7415 10.1128/MCB.21.21.7403-7415.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zage P.E., Nolo R., Fang W., Stewart J., Garcia-Manero G., Zweidler-McKay P.A. 2012. Notch pathway activation induces neuroblastoma tumor cell growth arrest. Pediatr. Blood Cancer. 58:682–689 10.1002/pbc.23202 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.