Abstract
In this study, using the immunofluorescent method, the immunopositive signals to ubiquitin and proteasomes in nucleoli of root meristematic cells of soybean seedlings have been observed. In fact, those signals were present exclusively in nucleolar vacuoles. No signals were observed in the nucleolar territory out of the nucleolar vacuoles or in the nucleoli without vacuoles. The ubiquitin-proteasome system (UPS) may act within the nucleoli of plants with high metabolic activities and may provide an additional level of regulation of intracellular proteolysis via compartment-specific activities of their components. It is suggested that the presence of the UPS solely in vacuolated nucleoli serves as a mechanism that enhances the speed of ribosome subunit production in very actively transcribing nucleoli. On the other hand, nucleolar vacuoles in a cell/nucleus could play additional roles associated with temporary sequestration or storage of some cellular factors, including components of the ubiquitin-proteasome system.
Key words: ubiquitin, proteasome, nucleolar vacuole, soybean root meristematic cell, immunofluorescence.
Introduction
Intracellular protein degradation is important for the regulation of many fundamental cellular processes for both plants and animals. The ubiquitin-proteasome-dependent proteolytic pathway that has been found in all eukaryotes is one of the nonlysosomal protein degradation systems.1,2 The 26S proteasome is a multicatalytic proteinase complex that acts as the ATP-dependent degradation system and digests vast majority of intracellular proteins tagged with a multiubiquitin chain as a degradation signal.3,4 In all eukaryotes the ubiquitin-proteasome system (UPS) degradation determines the turnover rate for a wide range of proteins including long- and short-lived ones as well as regulatory proteins e.g. metabolic key enzymes, transcription factors, cyclins, inhibitors of cyclin dependent kinases as well as regulators of DNA replication, repair and apoptosis.4
Generally in all eukaryotes the UPS is engaged in lots of key processes. Proteasomal proteolysis is engaged in regulation of nuclear structure and functions such as replication, transcription and splicing.5 Cell cycle-dependent changes in the localization of the proteasomes suggest that it may have a regulatory function related to the cell cycle.6,7 Ubiquitin and proteasomes are intimately involved in chromatin structure modifications and gene control that is accompanied with histone ubiquitination.8 The proteasomes play an important role in the processing of antigen presentation.9 Proteasomes can be activated by various unfavourable environmental conditions10 and ageing.11 Degradation, by the proteasomal system, of oxidatively modified proteins plays an important role in the antioxidant defenses.12 This system provides for cellular quality control since it prevents accumulation of misfolded or damaged proteins.13
In plants the UPS has been implicated in the regulation of almost every developmental process from embryogenesis to floral organ production.14 This pathway participates in the regulation of such processes as differentiation of vascular bundles,15 auxin signal transduction pathway,16 apoptosis,17 selective ion uptake through plasma membrane proton pomp,18,19 nitrogen recycling.20 Transcription factors are regulated via the UPS during organogenesis and vascular differentiation.21 The UPS is involved in the replacement of histones with protamine-like proteins during spermatogenesis in algae.22 Ubiquitin-mediated proteolysis regulates hormone biosynthesis, transport, perception and signaling.23 The role of the plant UPS has been established in the defense against pathogen and in disease resistance,24 against abiotic stresses including heat shock and wounding as well as in organ senescence.20,25,26 The ubiquitin-proteasome pathway is also involved in the regulation of programmed cell death (PCD).27
The presence of ubiquitin and proteasomes in all parts of plants has been documented. However, the highest amount was observed in the root and shoot apical meristems, in leaf primordia and vascular tissue of young sunflower21 and rice plants28 suggesting that proteolysis mediated by the 26S proteasomes may be involved in organ formation. Hence, an enhanced level of ubiquitin and proteasomes can be observed at the onset of organogenesis.21
Generally in animal cells the proteasomes are more abundant in cytosol than in nucleoplasm, although the UPS is an active component of the cell nucleus29 and this proportion varies with the cell type and growth conditions.30 Localization of the proteasome is often cell cycle-dependent.6 Nuclear distribution of proteasomes is species-specific and differs in higher versus lower eukaryotes.29
The UPS has been implicated in various aspects of nucleolar function. Recently involvement of the UPS in ribosome biogenesis has been suggested in mammalian cells. An important function of the nuclear UPS is to control the supply of ribosomal proteins for the assembly of new ribosomal subunits in the nucleolus. Ubiquitin and ubiquitin-like proteins play a role in ribosome biosynthesis and control the flow of materials to and from a nucleolus.31 It was shown that complexes associated with pre-rRNA processing factors were ubiquitinated and immunofluorescence revealed that ubiquitin was present within nucleoli. Inhibition of proteasome activity resulted in nucleolar morphology disruption and accumulation of 90S preribosome, it also affected the distribution of rRNA-processing proteins in nuclei, resulting in impaired rRNA processing.32 Moreover, fibrillarin and nucleophosmin, main nucleolar proteins participating in early and late rRNA processing, respectively, undergo ubiquitination.33,34 That is why the aim of the present work was to check, by immunofluorescence, the presence of ubiquitin and proteasomes within plant root meristematic cell nucleoli. To date, localization of the UPS elements has been reported neither in animal nor in plant normal nucleoli, except in the above mentioned examples.
Root meristematic cells of soybean seedlings seem to be an attractive object to study plant nucleoli. There is a single, quite big nucleolus in the nucleus of this cell. Nucleoli of soybean, a plant from subtropical climate, clearly responded to chilling showing differences in ultrastructure and considerably reduced transcriptional activity.35,36 Moreover, in the nucleoli of plants subjected to recovery after chilling stress transcriptional activity and transport of ribosomal subunits to the cytoplasm were even more intensive than in the control and then characteristic, single, huge, and centrally located nucleolar vacuoles appeared in them.37 It seems that the UPS contributes indirectly to changes of nucleolar activity and structure in soybean cells quickly responding to environmental conditions. Current studies showed that immunosignals against 20S proteasomes and ubiquitin were present in nucleolar vacuole territories.
Materials and Methods
Material
Seeds of soybean (Glycine max (L.) Merr.) cv. Aldana (obtained from Plant Breeding and Acclimatization Institute in Radzików, Poland) were germinated for three days at 25°C in darkness in Petri dishes on filter paper moistened with distilled water (control).
To obtain nucleoli with vacuoles the control seedlings were treated with chilling (10°C) for four days, and then the plants were transferred to 25°C for one day.
Immunofluorescence
Root apical meristems were cut off from the seedlings and fixed in 4% paraformaldehyde in 0.01 M phosphate buffered saline (PBS), pH 7.4 for 45 min at room temperature (RT) and then washed in PBS. Next the material was treated with a digestion mixture (1% pectinase, 1% cellulase, 1% pectolyase in PBS) for 30 min at RT. After washing in PBS squashed preparations were made. Slides were air-dried and then incubated in 5% BSA with 0.5% Triton X-100 in PBS for 60 min at RT, washed in PBS with 0.5% triton X-100. The preparations were covered with the mouse monoclonal antibody to 20S proteasomes (10 µg/mL diluted in PBS with 0.1% Triton) or with the rabbit polyclonal antibody to ubiquitin (1:300 diluted in DAKO solution; DAKO, Chełmniec, Poland) and both left for 12 h at 4°C then washed five times in PBS with Triton. The secondary antibodies conjugated with FITC - anti-mouse Ig (1:30; Sigma Aldrich, Poznan, Poland) and anti-rabbit Ig (1:80; Sigma) to proteasomes and ubiquitin, respectively, were incubated for 2 h in darkness at RT. After this, the slides were washed in PBS with 0.5% Triton and once only in PBS. Dried preparations were covered with PBS:glycerol mixture (9:1) with 2.3% DABKO (Sigma). Observations were performed using Optiphot-2 epifluorescence microscope equipped with B-2A filter (blue light; λ=470 nm) for FITC. Images were recorded using DXM 1200 CCD Nicon camera.
Slides incubated only in the secondary antibody served as a negative control. Moreover, the control tests with other antibodies, i.e. polyclonal rabbit tri-methyl histone H3 (lys4) antibody (H3K4met3) and rabbit polyclonal acetyl histone H3 (lys9) antibody (H3K9acetyl) (Cell Signaling Technology, Inc., Danvers, MA, USA) were carried out. Immunofluorescencent procedures were similar to those for proteasome and ubiquitin.
Semi-thin section preparation
Root tips of each variant were fixed in 2% glutaraldehyde in 1% cacodylate buffer (pH 7.2–7.4) for 3 h at 4°C. Then they were postfixed in 1% OsO4 in the same buffer. After dehydration in the ethanol series, the material was embedded in a medium containing the mixture of Epon 812 and Spurr's resin. Semi-thin sections were placed on microscopic slides and stained with toluidine blue. Nucleolar vacuoles were observed by means of light microscope Jenamed-2 (Carl Zeiss, Jena, Germany) and CCD CAMERA MTV - 1801 CB connected to a microscope and computer-aided IMAL-512 system.
Statistic
Meristematic cells with nucleoli showing high activity, i.e. those from the control and post-chilling recovered plants were taken into account to check both the number of vacuolated nucleoli and for immunofluorescent studies. Five root tips for each experimental variant and 150 cells from each root tip were examined. The number of cells with vacuolated nucleoli was counted on semi-thin sections, while the number of cells with nucleoli with immunosignals was estimated on squashed preparations. Significance of difference between vacuolated and immunolabeled vacuolated nucleoli as well as between nucleoli immunolabeled with the antibodies against proteasome and ubiquitin was estimated for the control and recovered seedlings. Standard deviations for each value (±SD) were evaluated. Statistical significance of differences were done using the Student's t-test (P<0.05).
Results and Discussion
Genes encoding subunits of the ubiquitin-proteasome system (UPS) are present in organisms ranging from some prokaryotes to all studied eukaryotes, and appear to have been highly conserved throughout evolution,38 hence in the current work the antibodies to core subunits of the human 20S proteasomes and to the human ubiquitin were used to identify proteasomes and ubiquitin in the plant material. In this study the immunocytofluorescent microscopy showed nuclear and cytoplasmic labeling with the antibodies against 20S proteasomes and ubiquitin in all studied cells of soybean root meristems suggesting that both elements of the UPS were present in the nucleus as well as in the cytoplasm. In other eukaryotes both components of the UPS are also found in the cytoplasm and within the nucleus. Proteasomes alone are found in the cytoplasm, both free and attached to the cytoplasmic reticulum, and in Golgi apparatus,30,39 associated with cytoskeletal elements and in the nuclei of eukaryotic cells.40 In the cell nucleus proteasomes are present throughout nucleoplasm, concentrating in subnuclear structures such as splicing speckles,33 promyelocytic leukemia (PML) nuclear bodies41 and clastosomes.42 In all mammalian cells analysed so far endogenous proteasomes were distributed throughout the cytoplasm and nucleoplasm, however they were detectable neither in the nuclear envelop nor in nucleoli.5
It is interesting that in these studies immunopositive signals were also present in the nucleoli of soybean cells. However, immunostaining was not observed in every nucleolus and those which were stained were labeled only partially. Immunosignals for proteasomes and ubiquitin were present solely in these nucleoli which most probably contained nucleolar vacuoles. The nucleoli without vacuoles were excluded from immunostaining (Figure 1A,A'). Observations showed that the distribution and pattern of labeling for both proteasomes and ubiquitin in the soybean root meristematic cells were almost identical (Figure 1 A,A',B,B',C,C',D,D'). Dimensions of the labeled areas in particular nucleoli were various depending, as we suppose, on sizes of nucleolar vacuoles. The labeling was most frequently located in the central part of a nucleolus and occupied relatively big area (Figure 1 B,B'). There were also immunosignals which occupied single, small areas (Figure 1 C,C') and a few small foci in one nucleolus (Figure 1 D,D'). The sizes of immunosignal areas and their location were in agreement with the sizes and location of nucleolar vacuoles (Figure 2 A,B,C,D). In order to show that occurrence of cells with nucleolar vaculoles as well as labeling of these vacuoles are not rare or accidental the rows of neighbouring cells, as they occur in root meristems, were demonstrated in Figure 3 A,B,C. Earlier and present studies showed that in the cells of control plants there were 18–26% vacuolated nucleoli while in the cells of plants recovered after 4-day chilling there were many more nucleoli with vacuoles, 40–54%. In most cases the nucleolar vacuoles were huge, single and centrally located in those nucleoli43 (Table 1). Current studies showed that higher number of nucleoli with immunosignals was observed in the cells of plants subjected to recovery in comparison to the control plants. These experiments, on the basis of sizes and localization of immunosignals, indeed, would confirm the fact that immunopositive reactions were localised to nucleolar vacuole territories. In this paper only nucleoli of the recovered plants were demonstrated in Figures, although nucleoli of the control plants also contained nucleolar vacuoles with immunosignals, but these were less spectacular than those of the recovered plants.
Figure 1.
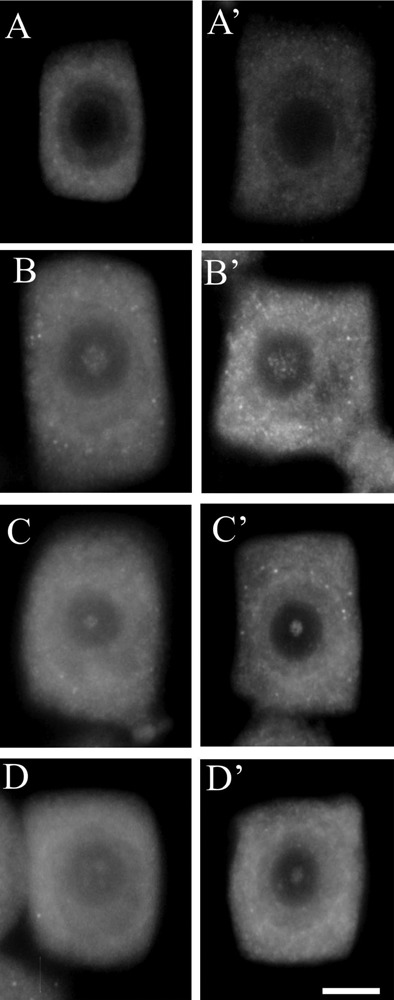
Immunofluorescent detection of proteasomes (A, B, C, D) and ubiquitin (A', B', C', D') in root meristematic cells of soybean; A,A'), nucleoli free of immunosignals; B,B'), nucleoli with one big, centrally located labeled area; C,C'), nucleoli with one small labelled area; D,D'), nucleoli with a few small labelled areas. Scale bar: 15 µm.
Figure 2.
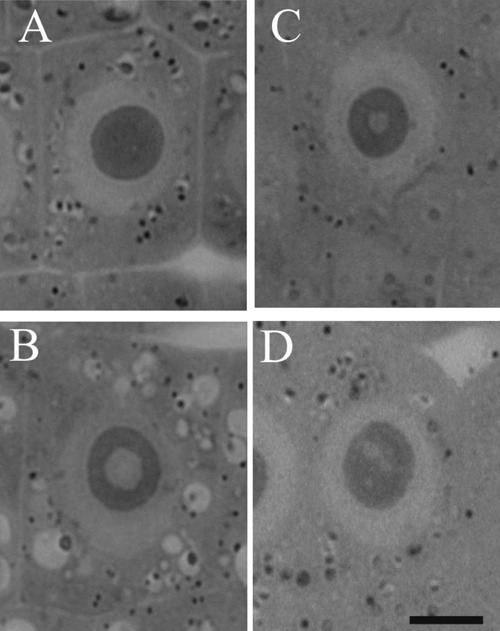
Nucleoli of root meristematic cells of soybean on semi-thin sections. A), nucleoli without vacuoles; B), nucleoli with one, big, centrally located vacuole, characteristic of plants recovered after chilling stress; C), nucleolus with a small vacuole; D), nucleolus with a few small vacuoles. Scale bar: 10 µm.
Figure 3.
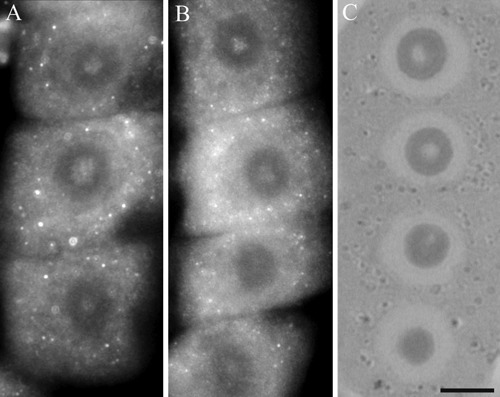
Rows of neighbouring cells in soybean root meristems with: immunodetection of proteasomes (A) and ubiquitin (B) as well as a semithin section of cells with nucleolar vacuoles (C). Scale bar: 7.5 µm.
Table 1. Frequency of vacuolated and of immunolabeled vacuolated nucleoli in control and recovered soybean seedlings.
| Growth conditions | Frequency (%) of | ||
|---|---|---|---|
| Vacuolated nucleoli | Vacuolated nucleoli immunolabeled with antibody against | ||
| Proteasome | Ubiquitin | ||
| Control (3d/25°C) | 26.3±3.1 | 23.8±3.6 | 25.1±3.9 |
| Recovery(3d/25°C + 4d/10°C + 1d/25°C) | 53.7±5.9 | 49.3±6.2 | 51.5±5.8 |
Mean values ± SD; there is no significance of difference between vacuolated and immunolabeled vacuolated nucleoli as well as between nucleoli immunolabeled with antibody to proteasome and ubiquitin for both control and recovered experimental variants, P<0.05.
Although the number of vacuolated nucleoli was somewhat higher than of those with immunosignals in both experimental variants, the statistical analysis showed that there was no significance of difference between them (Table 1). Similar situation occurred in the case of the number of nucleoli labeled with the antibodies to proteasome and ubiquitin (Table 1). On the basis of the obtained results a conclusion could be drawn that all vacuolated nucleoli contained immunosignals for proteasomes or ubiquitin.
Some experimental procedures, especially fixation, may induce artefact formation.29 In order to check that immunosignals present in nucleolar vacuoles were not artifacts some control tests were carried out. The negative controls without primary antibodies did not show any specific immunosignals at the whole cell areas (Figure 4A). Tests with the use of antibodies to nuclear proteins, H3K4met3 and H3K9acetyl, and the same procedure as in the case of proteasome and ubiquitin, did not show any immunosignals at the nucleolar territory. (Figure 4 B,C).
Figure 4.
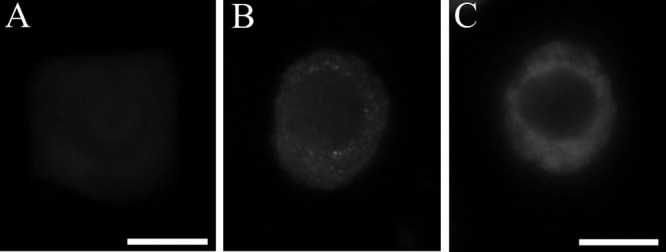
Control tests: A), without primary antibody, scale bar: 15 µm; B), with nuclear antibodies H3K4met2 and C), with H4K12acetyl, scale bar: 10 µm.
Presence of the UPS components in a nucleolus was rarely documented. The nucleoli themselves are generally thought to be devoid of proteasomes under normal growth condition40 as detailed proteomic analysis of human nucleoli showed44 and the role of nucleolus in the proteasomal activity in degradation of the nucleolar proteins has not been documented to date,5,40 but ubiquitin is present in normal nucleoli.32 However, SUMO proteases have been identified in the nucleoli of HeLa cells.45 Moreover, sumoylation acting in coordination with the UPS regulates the maintenance of nucleolar integrity. A nucleolus is also the key organelle in which SUMO-1 conjugates accumulate in response to proteasome inhibition.46 In some cases, under particular conditions, components of the UPS can be found within a nucleolus. For example, proteasomes and their potential substrates accumulated in nucleoli during inhibition of proteasomal protein degradation by MG 132 in human cells,47 during increased Myc expression48 and when altered localization of the nuclear protein PML occurred,47 suggesting that nucleoli are the site of protein degradation. Proteasome inhibition exerts specific effects on certain components of nucleolar architecture and on early and late processing factors of pre-ribosome biosynthesis, as well as it disrupts multiple processes in ribosome biogenesis and substantially influences Pol I transcription.32 Even in the soybean seedlings treated with epoxomycin, an inhibitor of the proteolytic activity of proteasomes, alterations in nucleolar ultrastructure can be seen (Stępiński, unpublished data). Cic1p/Nsa3p yeast protein is associated with 26S proteasome and localised to nucleolus. This protein is required for early pre-rRNA processing and for release of pre-60S from the nucleolus into the nucleoplasm.49 Additionally the functions of ubiquitin and nucleoli seem to be connected. A possible role for ubiquitin in nucleolar disassembly was suggested.50 Moreover, several proteins associated with ribosome biosynthesis are ubiquitinated. Also ubiquitin ligase is known to regulate the processing and nuclear export of tRNA, mRNA as well as rRNA in yeast.51 Ubiquitin has been localized within both the nucleolus and the pre-ribosomal complexes of mammalian cells.32 Since biosynthesis of ribosomal precursors is regulated at multiple levels including the transcription of ribosomal genes, phosphorylation, methylation and acetylation of nucleolar factors,52 the levels of these factors could be controlled by the UPS, hence, it is suggested that the UPS is required for multiple steps of ribosome biogenesis.32
More speculatively, one could believe that presence of both components of the UPS, ubiquitin and proteasomes in a nucleolus establishes a functional degradation pathway of nucleolar and ribosomal proteins in this organelle so their transport to distinct subnuclear domains or to the cytoplasm would not be necessary. However, the nucleolar protein, fibrillarin, was described to be degraded in nucleoplasmic foci under conditions that specifically inhibit nucleolar transcription. It means that nucleolar proteins are degraded out of nucleoli.33 In mammalian cells upon inhibition of proteasome-dependent proteolysis, nucleolar proteins, fibrillarin and UBF, did not accumulate so it is suggested that they do not represent proteasomal substrates and they generally do not co-localize with protein degradation foci in interphase nuclei.5 However, more recent data show that USP36, a deubiquitinat ing enzyme, plays an essential role in regulating nucleolar structure and function and specifically localizes to nucleoli together with its substrates, namely fibrillarin and nucleophosmin, nucleolar proteins which undergo deubiquitination. Moreover, USP36 in cooperation with B23 regulates the level of nucleolar protein ubiquitination.53 Nucleoli of all eukaryotic organisms, including plants, play the same general function, i.e. ribosome subunit production, and contain almost identical set of factors/proteins to act the function, hence, it is possible that components of UPS might be present also in nucleoli of plant cells.
It is interesting and intriguing that ubiquitin and proteasomes have been localised exclusively in nucleoli possessing vacuoles. Vacuolated nucleoli are less common in animal cells than in plant ones. In addition nucleolar vacuoles appear most of all in very active nucleoli. This might be the reason why ubiquitin and proteasomes have not been identified in other objects so far. In the case of soybean, vacuolated nucleoli appeared in plants grown at optimal temperature of 25°C as well as in those subjected to 4-day chilling stress (when rRNA transcription considerably slowed down) and subsequently recovered at optimal temperature. In the latter case the nucleoli with big vacuoles appeared and rRNA transcription proceeded even more intensively than in the control.37 So, presence of the UPS components solely in the vacuolated nucleoli proves the fact that the ubiquitin-proteasome pathway is present in nucleoli with intensified activity. It cannot be excluded that nucleolar and ribosomal proteins predestined to be degraded in poorly active, non-vacuolated nucleoli, are transported to the extranucleolar nucleoplasm or to the cytoplasm and their proteolysis takes place there. On the other hand in very active, vacuolated nucleoli, where there is no time for transport of proteins predestined for degradation out of nucleoli the whole machinery of the UPS is brought to nucleoli and proteolysis occurs inside nucleolar vacuoles. Under normal conditions synthesis and import of ribosomal proteins is in excess in relation to needs.54 The more so as the excess of such proteins may occur in the most active cells, i.e. in plants subjected to recovery. Ribosomal proteins that have failed to assemble into ribosomal subunits in nucleoli and excessive ribosomal proteins are also degraded by the proteasome.54
Moreover, in mammalian nucleoli the structures called nucleolar aggresomes, which contain proteins and other compounds are formed. Such aggregation of proteins often results from insufficient protein degradation by the UPS. Aggresomes are also formed under various forms of stress as well as when cells are treated with poteasome inhibitors. Most proteins accumulating in nucleolar aggrosomes are degraded in proteasomes but a question whether this process takes place in nucleoli remains opened.55 Increased nuclear load of proteins which are destined to be degraded enhances aggresome formation. The more so as such a situation could take place in soybean plants under chilling stress during which damage of proteins could appear.
Moreover, communication between ribosome assembly factors and proteasome activity could be an important mechanism to synchronize ribosome synthesis with cell growth and division. Especially it might refer to the cells with active, vacuolated nucleoli where metabolic processes proceed with high speed and the activity of the factors controlling biochemical processes has to be quickly regulated also by the UPS. It is accepted that nucleolar vacuoles appear when transport of ribosomal subunits exceeds their synthesis.56,57 However, the UPS may help form or enlarge nucleolar vacuoles by digestion of nucleolar matrix proteins at the site of nucleolar vacuole arising.
On the one hand, these observations can suggest that the UPS may act within the nucleolus and may provide additional level of regulation of intracellular proteolysis via compartment-specific activities of its components. Possibly, the localization of proteasomes and ubiquitin is associated with the need for enhanced proteolysis in individual cell compartments, in this case in a very active nucleolus, to regulate nucleolar processes as well as to clean up unwanted proteins produced as the consequence of stress. Stress conditions may create situations in which massive amounts of ribosomal proteins become unemployed and thus become potential substrates for the nuclear UPS.31 On the other hand, it is worth noticing that immunopositive signals for ubiquitin and proteasomes present both in nucleolus and in the other cellular compartments refer to free ubiquitin and its various conjugates. Proteins modified with ubiquitin are not necessarily predestined for degradation but their activity and localization can be also regulated through ubiquitylation.58 That is why the presence of ubiquitin in a nucleolus does not necessarily mean that degradation of ubiquitinated proteins takes place in a nucleolus, however it may indicate that nucleoli, precisely the nucleolar vacuoles, may not merely be useless empty space formed in active nucleoli as a consequence of intensive transport of pre-ribosomal particles out of the nucleoli, but they could be the site of temporary sequestration or storage of the UPS components or ubiquitin conjugates as well as other biochemical cellular components in plant cells,59 similarly as different discrete, focal regions in animal nucleoli are used as storage compartments for certain enzymatic and regulatory proteins, which can be released in a regulated manner to exert their function elsewhere.60,61
Acknowledgements:
the author is grateful to Prof. Maria Kwiatkowska (Department of Cytophysiology, University of Łódź, Poland) for her valuable advice and comments during preparation of the manuscript. Special thanks to Dr. Agnieszka Wojtczak from the same Department who supplied the antibodies used. This work was partially supported by grant from University of Łódź, No. 505/379.
References
- 1.Von Kampen J, Wettern M, Schulz M. The ubiquitin system in plants. Physiol Plant. 1996;97:618–24. [Google Scholar]
- 2.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–60. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glickman MH. Getting in and out of the proteasome. Cell Dev Bio. 2000;11:149–58. doi: 10.1006/scdb.2000.0161. [DOI] [PubMed] [Google Scholar]
- 4.Abramova EB, Sharova NP, Karpov VL. The proteasome: destroy to live. Mol Biol. 2002;36:613–24. [PubMed] [Google Scholar]
- 5.Rockel TD, Stuhlmann D, von Mikecz A. Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J Cell Sci. 2005;118:5231–42. doi: 10.1242/jcs.02642. [DOI] [PubMed] [Google Scholar]
- 6.Palmer A, Mason GG, Paramio JM, Knecht E, Rivett AJ. Changes in proteasome localization during the cell cycle. Eur J Cell Biol. 1994;64:163–75. [PubMed] [Google Scholar]
- 7.Yanagawa Y, Kimura S. Cell cycle regulation through ubiquitin/proteasome-mediated roteolysis in plants. Japan Agric Res Quarterly. 2005;39:1–4. [Google Scholar]
- 8.Kinyamu HK, Chen J, Archer TK. Linking the ubiquitin-proteasome pathway to chromatin remodeling/modification by nuclear receptors. J Mol Endocrinol. 2005;34:281–97. doi: 10.1677/jme.1.01680. [DOI] [PubMed] [Google Scholar]
- 9.Lehner PJ, Cresswell P. Processing and delivery of peptides presented by MHC class I molecules. Curr Opin Immunol. 1996;8:59–67. doi: 10.1016/s0952-7915(96)80106-3. [DOI] [PubMed] [Google Scholar]
- 10.Mathew A, Mathur SK, Morimoto RI. Heat shock response and protein degradation: regulation of HSF2 by the ubiuquitin-proteasome pathway. Mol Cell Biol. 1998;18:5091–8. doi: 10.1128/mcb.18.9.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bregegere F, Milner Y, Friguet B. The ubiquitin-proteasome system at the crossroads of stress-response and ageing pathways: a handle for skin care? Ageing Res Rev. 2006;5:60–90. doi: 10.1016/j.arr.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Ullrich O, Reinheckel T, Sitte N, Hass R, Grune T. poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. PNAS USA. 1999;96:6223–8. doi: 10.1073/pnas.96.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–9. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 14.Moon J, Parry G, Estelle M. The ubiquitin-proteasome pathway and plant development. Plant Cell. 2004;16:3181–3195. doi: 10.1105/tpc.104.161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachmair A, Becker F, Masterson RV, Schell J. Perturbation of the ubiquitin system causes leaf curling, vascular tissue alteration and necrotic lessions in a higher plant. EMBO J. 1990;9:4543–9. doi: 10.1002/j.1460-2075.1990.tb07906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray WM, Estelle M. Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem Sci. 2000;25:133–8. doi: 10.1016/s0968-0004(00)01544-9. [DOI] [PubMed] [Google Scholar]
- 17.Wójcik C. Proteasomes in apoptosis: villains or guardians? Cell Mol Life Sci. 1999;56:908–17. doi: 10.1007/s000180050483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samuels AL, Fernando M, Glass ADM. Immunofluorescent localization of plasma membraneH+-ATPase in barley roots and effects on K nutrition. Plant Physiol. 1992;99:1509–14. doi: 10.1104/pp.99.4.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De La Fuente N, Maldonado AM, Portillo F. Glucose activation of the yeast plasma membrane H+-ATPase requires the ubiquitin-proteasome proteolytic pathway. FEBS Lett. 1997;411:308–12. doi: 10.1016/s0014-5793(97)00721-7. [DOI] [PubMed] [Google Scholar]
- 20.Belknap WR, Garbarino JE. The role of ubiquitin in plant senescence and stress responses. Trends Plant Sci. 1996;10:331–5. [Google Scholar]
- 21.Ingvardsen C, Veierskov B, Joshi PA. Immunohistochemical localisation of ubiquitin and the proteasome in sunflower (Helianthus annuus cv. Giganteus) Planta. 2001;213:333–41. doi: 10.1007/s004250000511. [DOI] [PubMed] [Google Scholar]
- 22.Wojtczak A, Kwiatkowska M. Immunocytochemical and ultrastructural analyses of the function of the ubiquitin-proteasome system during spermiogenesis with the use of the inhibitors of proteasome proteolytic activity in the alga, Chara vulgaris. Biol Reprod. 2008;78:577–85. doi: 10.1095/biolreprod.107.062901. [DOI] [PubMed] [Google Scholar]
- 23.Dreher K, Callis J. Ubiquitin, hormones and biotic stress in plants. Ann Bot. 2007;99:787–822. doi: 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devato A, Muskett PR, Shirasu K. Role of ubiquitination in the regulation of plant defence against pathogens. Curr Opin Plant Biol. 2003;6:307–311. doi: 10.1016/s1369-5266(03)00060-8. [DOI] [PubMed] [Google Scholar]
- 25.Bahrami AR, Gray JE. Expression of a proteasome α-type subunit gene during tobacco development and senescence. Plant Mol Biol. 1999;39:325–33. doi: 10.1023/a:1006102110889. [DOI] [PubMed] [Google Scholar]
- 26.Ortiz C, Cardemil L. Heat-shock responses in two leguminous plants: a comparative study. J Exp Bot. 2001;52:1711–9. [PubMed] [Google Scholar]
- 27.Kim M, Ahn JW, Jin UH, Choi D, Paek KH, Pai HS. Activation of the programmed cell death pathway by inhibition of proteasome function in plants. J Biol Chem. 2003;278:19406–5. doi: 10.1074/jbc.M210539200. [DOI] [PubMed] [Google Scholar]
- 28.Yanagawa Y, Kimura S, Takase T, Sakaguchi K, Umeda M, Komamine A, et al. Spatial distribution of the 26S proteasome in meristematic tissues and primordia of rice (Oryza sativa L.) Planta. 2002;214:703–707. doi: 10.1007/s00425-001-0676-2. [DOI] [PubMed] [Google Scholar]
- 29.Scharf A, Rockel TD, von Mikecz A. Localization of proteasomes and proteasomal proteolysis in the mammalian intherphase cell nucleus by systemic application of immunocytochemistry. Histochem Cell Biol. 2007;127:591–601. doi: 10.1007/s00418-006-0266-2. [DOI] [PubMed] [Google Scholar]
- 30.Palmer A, Rivett AJ, Thomson S, Hendil KB, Butcher GW, Fuertes G, et al. Subpopulations of proteasomes in rat liver nuclei microsomes and cytosol. Biochem J. 1996;316:401–7. doi: 10.1042/bj3160401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shcherbik N, Pestov DG. Ubiquitin and ubiquitin-like proteins in the nucleolus: multitasking tools for a ribosome factory. Genes Cancer. 2010;1:681–9. doi: 10.1177/1947601910381382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stavreva DA, Kawasaki M, Dundr M, Koberna K, Mueller WG, Tsujimura-Takahashi T, et al. Potential roles for ubiquitin and the proteasome during ribosome biogenesis. Mol Cell Biol. 2006;26:5131–45. doi: 10.1128/MCB.02227-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen M, Rockel T, Steinweger G, Hemmerich P, Risch J, von Mikecz A. Subcellular recruitment of fibrillarin to nucleoplasmic proteasomes: implications for processing of a nucleolar autoantigen. Mol Biol Cell. 2002;13:3576–87. doi: 10.1091/mbc.02-05-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, et al. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12:1151–64. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 35.Stępiński D. Effect of short- and long-lasting chilling on pre-rRNA synthesis and transport in root meristematic cells of three soybean cultivars. Acta Soc Bot Pol. 2003;72:319–24. [Google Scholar]
- 36.Stępiński D, Kwiatkowska M. Autoradiographic and ultrastructural studies of the effect of chilling on soybean root meristem nucleoli. Acta Biol Cracov Ser Bot. 2003;45:35–42. [Google Scholar]
- 37.Stępiński D. Ultrastructural and autoradiographic studies of the role of nucleolar vacuoles in soybean root meristem. Folia Histochem Cytobiol. 2004;42:57–61. [PubMed] [Google Scholar]
- 38.Hilt W, Wolf DH. Proteasomes: destruction as a programme. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- 39.Rivett AJ. Intracellular distribution of proteasomes. Curr Opin Immunol. 1998;10:110–4. doi: 10.1016/s0952-7915(98)80040-x. [DOI] [PubMed] [Google Scholar]
- 40.Wójcik C, DeMartino GN. Intracellular localization of proteasomes. Int J Biochem Cell mBiol. 2003;35:579–89. doi: 10.1016/s1357-2725(02)00380-1. [DOI] [PubMed] [Google Scholar]
- 41.Condemine W, Takahashi Y, Le Bras M, de The H. A nucleolar targeting signal in PML-1 address PML to nucleolar caps in stressed or senescent cells. J Cell Sci. 2007;120:3219–27. doi: 10.1242/jcs.007492. [DOI] [PubMed] [Google Scholar]
- 42.Lafarga M, Berciano MT, Pena E, Mayo L, Castano JG, Bohmann D, et al. Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and substrates of proteasome. Mol Biol Cell. 2002;13:2771–82. doi: 10.1091/mbc.E02-03-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stępiński D. Nucleolar vacuolation in soybean root meristematic cells during recovery after chilling. Biol Plant. 2008;52:507–12. [Google Scholar]
- 44.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, et al. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 45.Gong L, Yeh ETH. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem. 2006;281:15869–77. doi: 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- 46.Matafora V, D'Amato A, Mori S, Blasi F, Bachi A. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol Cell Proteomics. 2009;8:2243–55. doi: 10.1074/mcp.M900079-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mattsson K, Pokrovskaja K, Kiss C, Klein G, Szekely L. Proteins associated with the promyelocytic leukemia gene product (PML)-containing nuclear body move to the nucleolus upon inhibition of proteasome-dependent protein degradation. PNAS USA. 2001;98:1012–7. doi: 10.1073/pnas.031566998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arabi A, Rustum C, Hallberg E, Wright APH. Accumulation of c-Myc and proteasomes at the nucleoli of cells containing elevated c-Myc protein levels. J Cell Sci. 2003;116:1707–7. doi: 10.1242/jcs.00370. [DOI] [PubMed] [Google Scholar]
- 49.Fatica A, Oeffinger M, Tollervey D, Bozzoni I. Cic1p/Nsa3p is required for synthesis and nuclear export of 60S ribosomal subunits. RNA. 2003;9:1431–6. doi: 10.1261/rna.5130503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sudha T, Tsuji H, Sameshima M, Matsuda Y, Kaneda Y, Nagai Y, et al. Abnormal integrity of the nucleolus associated with cell cycle arrest owing to the temperature-sensitive ubiquitin-activating enzyme E1. Chromosome Res. 1995;3:115–23. doi: 10.1007/BF00710672. [DOI] [PubMed] [Google Scholar]
- 51.Neumann S, Petfalski E, Brugger B, Grosshans H, Wieland F, Tollervey D, et al. Formation and nuclear export of tRNA, rRNA and mRNA is regulated by the ubiquitin ligase Rsp5p. EMBO Rep. 2003;4:1156–62. doi: 10.1038/sj.embor.7400026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leary DJ, Huang S. Regulation of ribosome biogenesis within the nucleolus. FEBS Lett. 2001;509:145–50. doi: 10.1016/s0014-5793(01)03143-x. [DOI] [PubMed] [Google Scholar]
- 53.Endo A, Kitamura N, Komada M. Nucleophosmin/B23 regulates ubiquitin dynamics in nucleoli by recruiting deubiquitylating enzyme USP36. J Biol Chem. 2009;284:27918–23. doi: 10.1074/jbc.M109.037218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar proteins dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17:749–60. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Latonen L. Nucleolar aggresomes as counterparts of cytoplasmic aggresomes in proteotoxic stress. Bioessays. 2011;33:386–95. doi: 10.1002/bies.201100008. [DOI] [PubMed] [Google Scholar]
- 56.Deltour R, de Barsy T. Nucleolar activation and vacuolation in embryo radicle cells during early germination. J Cell Sci. 1985;76:67–83. doi: 10.1242/jcs.76.1.67. [DOI] [PubMed] [Google Scholar]
- 57.Jennane A, Thiry M, Diouri M, Goessens G. Fate of the nucleolar vacuole during resumption of cell cycle in pea cotyledonary buds. Protoplasma. 2000;210:172–8. [Google Scholar]
- 58.Gill G. SUMO and ubiquitin in the nuckleus: different functions, similar mechanisms? Genes Dev. 2008;18:2046–59. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 59.Kim SH. Plant nucleolar dynamics. J Plant Biol. 2009;52:193–201. [Google Scholar]
- 60.Visintin R, Amon A. The nucleolus: the magician's hat for cell cycle tricks. Curr Opin Cell Biol. 2000;12:372–7. doi: 10.1016/s0955-0674(00)00102-2. [DOI] [PubMed] [Google Scholar]
- 61.Costanzo M, Cisterna B, Zharskaya OO, Zatsepina OV, Biggiogera M. Discrete foci containing RNase A are found in nucleoli of HeLa cells aged in culture. Eur J Histochem. 2011;55:e15–e15. doi: 10.4081/ejh.2011.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]


