The design of molecules that bind tightly and selectively to a specific site on a protein constitutes a fundamental challenge in molecular recognition. Finding high-affinity ligands for protein surfaces that bind to other proteins has proven to be particularly difficult.[1] Foldamers, oligomers with discrete folding propensities,[2] represent an unconventional source of ligands for protein-recognition surfaces,[3] but realizing this potential requires that we learn how to design sequences that contain unnatural building blocks and effectively mimic one of the surfaces involved in a given protein-protein interaction. Here we show that systematic backbone modification throughout a natural protein-binding domain, a process we refer to as sequence-based design, can expeditiously generate foldamers that bind tightly and selectively to target protein surfaces.
Structure-based design has been employed in previous efforts to address the problem of engineering biological function into foldamers.[2,3] Such efforts have begun with the identification of non-natural oligomers that have a strong propensity to adopt a specific secondary structure. This secondary structure then serves as a conformational scaffold for the design of foldamers that are intended to mimic a target protein surface. We have previously applied the structure-based design approach to develop foldamer ligands for the BH3-recognition cleft of Bcl-xL.[3c,4,5] Bcl-xL is a member of the Bcl-2 family, which controls programmed cell death pathways and includes both anti-apoptotic members (e.g., Bcl-2, Bcl-xL, Mcl-1) and pro-apoptotic members (e.g., Bak, Bad, Puma).[6] Bcl-2 and its anti-apoptotic homologues display a long cleft that accommodates an α-helical BH3 domain from a pro-apoptotic family member.[7]
Structural information available for Bcl-xL and its complexes with BH3 domains[7] suggested that the α-helical BH3 domain is a plausible target for mimicry by a foldamer helix. However, we had to prepare and evaluate more than 200 β-peptides[8] and α/β-peptides[9] designed to mimic the α-helical Bak BH3 domain before identifying an oligomer with significant affinity for Bcl-xL.[3c,4] Ultimately, success in this endeavor was achieved when we moved away from pure foldamer backbones to chimeric (α/β+α)-peptides with a 1:1 α:β residue alternation in the N-terminal segment and exclusively α residues at the final six positions. The best (α/β+α)-peptide showed impressive affinity for Bcl-xL (Ki = 2 nM), but this oligomer failed to bind to Mcl-1, an anti-apoptotic Bcl-2 family member targeted by the Bak BH3 peptide that served as the prototype for foldamer design.[4a,10] This limitation is significant as the utility of non-natural ligands designed to mimic BH3 domains is greatly enhanced if those ligands bind to multiple anti-apoptotic Bcl-2 family proteins.[11] Furthermore, the C-terminal α-peptide segment in the chimeric (α/β+α)-peptide was highly susceptible to proteolysis (the α/β segment was impervious to proteases).[4b]
Here we describe sequence-based design of α/β-peptide ligands for BH3-recognition clefts, a strategy that differs fundamentally from the structure-based design approaches to foldamer ligands previously pursued by us and others.[3] This approach involves replacing subsets of regularly spaced α residues with β3 residues bearing the original side chains; each α→β replacement introduces an extra methylene into the backbone. The sequence-based approach does not directly aim to recapitulate the folded structure of an α-peptide prototype, although conformational mimicry is apparently achieved as a by-product of the replacement strategy we have employed. We have recently demonstrated that sequence-based design can be used to generate helix-bundle foldamer quaternary structure from an α-peptide prototype.[12] Here we ask whether this method can be used to mimic the protein-binding behavior of an α-helical BH3 domain. Our results show that sequence-based design is more efficient than structure-based design for generating foldamers that bind tightly to anti-apoptotic Bcl-2 family proteins, and that sequence-based design can deliver α/β-peptides that display significant resistance to proteolytic degradation.
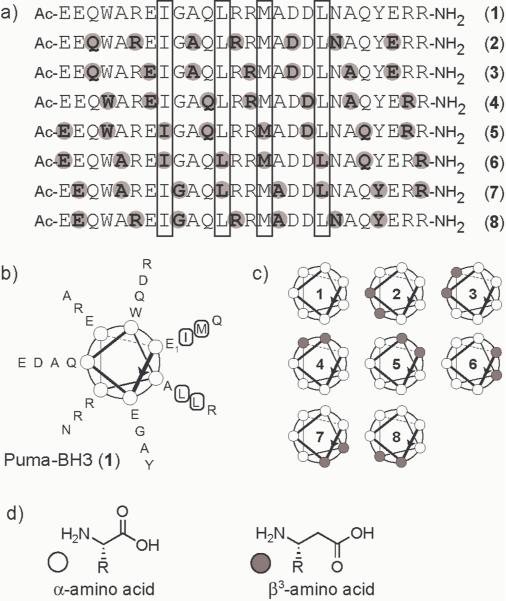
Puma is a Bcl-2 homologue that binds promiscuously to anti-apoptotic family members.[13] We prepared a 26-residue α-peptide corresponding to the Puma BH3 domain (1) and seven α/β-peptide analogues (2–8) with the same primary sequence of side chains displayed on different α/β-peptide backbones (Figure 1). Each α/β-peptide contains an ααβαααβ backbone repeat, which is derived from the heptad pattern common among α-peptide sequences that form α-helices with a well-developed `stripe' of hydrophobic side chains running along one side (Figure 1B). Recent crystal structures demonstrate that the ααβαααβ backbone allows formation of an α-helix-like conformation.[12] α/β-Peptides 2–8 represent all possible isomers of the Puma BH3 sequence with the ααβαααβ backbone pattern; these oligomers can be viewed as a series of analogues of Puma in which a band of β residues moves around the helical periphery (Figure 1C).
Figure 1.
a) Primary sequence of Puma BH3 peptide (1) and α/β-peptide analogues 2–8 (grey circles and bold letters indicate β3 residues). b) Helical wheel diagram of 1; boxed residues in (a) and (b) indicate hydrophobic positions most important for binding based on sequence homology.[7,13] c) Schematic representations of 1–8, drawn in the same orientation as (b), with white and grey circles indicating heptad positions occupied by α and β3 residues, respectively. d) Structure of an α-amino acid and a β3-amino acid.
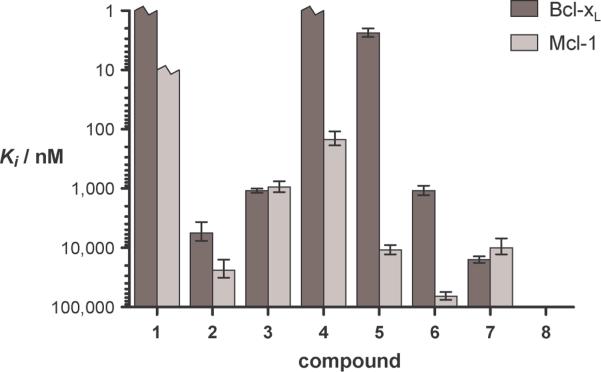
We tested 1–8 for the ability to bind to two distinct Bcl-2 family targets, Bcl-xL and Mcl-1. Inhibition constants (Ki) for each compound were determined by competition fluorescence polarization (FP) assays (Figures 2) with a fluorescently-labeled Bak-BH3 peptide as the tracer.[3c] The Puma-BH3 α-peptide (1) shows affinities for Bcl-xL and Mcl-1 that are tighter than can be measured with these FP assays, which is consistent with previous work.[13] Ki values for α/β-peptides 2–8 vary from <1 nM to >100 mM. Variation in the position of β residue incorporation causes considerable changes in affinity for each protein, >100,000-fold for Bcl-xL and >700-fold for Mcl-1.
Figure 2.
Inhibition constants for displacement of a fluorescently labeled Bak BH3 peptide bound to Bcl-xL or Mcl-1 by compounds 1–8. Broken bars indicate compounds binding tighter than discernable in the assay. The values for 8 were weaker than 100 μM for both proteins.
For both protein targets, 4 is the tightest binding foldamer, with Ki < 1 nM for Bcl-xL and Ki = 150 nM for Mcl-1. It is noteworthy that α/β-peptide 5, which contains β modification at critical hydrophobic residues in the Puma BH3 sequence, shows nM affinity for Bcl-xL. Our data demonstrate that the location of β residue incorporation strongly influences Bcl-xL vs. Mcl-1 selectivity among the Puma-derived α/β-peptide isomers, in addition to affinity for these protein targets. For example, 3 shows equal affinity for the two proteins, but 5 displays >4,000-fold selectivity for Bcl-xL over Mcl-1. We tested the validity of our conclusions regarding affinity and selectivity derived from FP competition assays for α-peptide 1 and α/β-peptides 4 and 5 by performing direct-binding FP measurements with analogues in which the N-terminal acetyl group is replaced with a BODIPY-TMR fluorophore. The Kd values determined by direct binding are consistent with the Ki values obtained from competition data (Table 1); differences in absolute values of Kd vs. Ki may reflect modest contributions of the appended fluorophore to affinity as measured in the direct binding mode.
Table 1.
Binding affinity and protease stability data for α-peptide 1 and α/β-peptides 4, 5.
| Ki (nM)a | Kd (nM)b | t1/2 (min.)c | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Bcl-xL | Mcl-1 | Bcl-xL | Mcl-1 | Prot. K | Pronase | |
| 1 | < 1 | < 10 | < 1 | < 2 | 0.7 | 1 |
| 4 | < 1 | 150 | 2.2 | 110 | > 3,000 | 100 |
| 5 | 2.4 | 11,000 | 1 | 1100 | 170 | 3.5 |
Inhibition constants determined by competition FP.
Dissociation constants of BODIPY-labeled analogues determined by direct binding FP.
Measured half life of a 50 μM solution of peptide or α/β-peptide in the presence of 10 μg/mL proteinase K or 5 μg/mL pronase.
Having established that certain α/β analogues of the Puma BH3 domain can bind with high affinity to the natural protein partners, we were curious to determine whether the α/β-peptides would be recognized and processed by proteolytic enzymes. We tested α-peptide 1 and α/β-peptides 4 and 5 for their susceptibility to two proteases with broad substrate profiles: proteinase K, a non-specific serine protease that tends to cleave C-terminal to hydrophobic residues, and pronase, an aggressive mixture of endopeptidases and exopeptidases that digests proteins into individual amino acids.[14] The results (Table 1) show that the ααβαααβ backbone can confer substantial resistance to proteolytic degradation. α/β-Peptide 4, which binds tightly to both Bcl-xL and Mcl-1, showed a >4,000-fold improvement in stability to proteinase K and a 100-fold improvement in stability to pronase relative to α-peptide 1. Analysis of the cleavage products by mass spectrometry indicated that the β residues tend to protect nearby amide groups from proteolysis,[15] which is consistent with previous reports for isolated α→β3 insertions.[16,17] α/β-Peptide 5 is more susceptible than is isomer 4 to proteolytic degradation, but 5 nevertheless shows significant improvement relative to α-peptide 1.
Previous work has suggested that the α-helical propensity of BH3-derived α-peptides may be an important determinant of affinity for anti-apoptotic Bcl-2 family proteins.[6,18] We employed circular dichroism (CD) spectroscopy to probe for conformational differences among two of the tight binding α/β-peptides (4 and 5) and one of the weakest binding analogues (7). Qualitative comparison of CD spectra for 4, 5, and 7 indicates that the large differences in binding affinity among these three isomers cannot be explained by differences in helical propensity.[15] Each of these three α/β-peptides shows a CD minimum at ~202 nm with per-residue ellipticity between −13,000 and −15,000 deg cm2 dmol−1 res−1 in aqueous solution. We have previously shown that helix formation in the ααβαααβ backbone is reflected by a strong CD minimum at 206 nm with a maximum magnitude of ~ −40,000 deg cm2 dmol−1 res−1.[12] Thus, the CD data for 4, 5 and 7 alone in aqueous solution suggest relatively low population of the helical state. Similarly, the CD signature for Puma α-peptide 1 in aqueous solution ([θ]222 = −10,000 deg cm2 dmol−1 res−1) suggests little α-helical content. Based on the established precedent for induction of α-helix formation upon binding of BH3 domain α-peptides to Bcl-xL and Mcl-1,[7] we hypothesize that α/β-peptides such as 4 and 5 are induced to adopt helical conformations upon binding to protein partners.
The work reported here demonstrates that a straightforward principle of sequence-based design can be used to convert a helical α-peptide ligand into an α/β-peptide with comparable binding affinity for protein targets and substantially improved proteolytic stability. The strategy we describe represents a fundamental departure from previous work on the development of foldamer-based inhibitors of protein-protein interactions,[2–4] and, at least in this case, the sequence-based approach has proven to be more efficient than the structure-based approach for generating foldamer mimics of α-helices. Evaluation of a series of just seven α/β-peptides designed based purely on primary sequence information led to a compound that rivals the best of our previously described chimeric (α/β+α) ligands[3c,4] in binding affinity for Bcl-xL. Moreover, the best α/β-peptide discussed here binds moderately well to Mcl-1, a biomedically important Bcl-2 family protein that is not targeted by oligomers identified via structure-based design. Our results also raise interesting questions, such as the origin of the large differences in binding behavior among Puma BH3 isomers with the same ααβαααβ backbone pattern.
It has long been known that insertion of a β-residue into an α-peptide hinders enzymatic cleavage of the amide bonds near the insertion site, and one or two α→β backbone modifications have been incorporated into biologically active α-peptides to generate analogues with increased proteolytic stability.[16,17,19] Previous work has shown also that one or two β-residues can be accommodated into helical peptides without disrupting secondary structure.[20] However, our implementation of multiple and systematic α→β3 replacements throughout a peptide sequence (7 of 26 positions substituted in the Puma BH3 domain) constitutes a significant advance beyond these precedents in the design of bioactive, proteolytically stable oligomers. Our finding that one version of this substitution pattern is well tolerated in terms of binding to anti-apoptotic proteins is surprising and noteworthy.
The sequence-based design illustrated here can be implemented with commercially available α and β-amino acid monomers and standard automated peptide synthesis methods; therefore, it will be easy for others to undertake analogous efforts. The α/β-peptides we describe do not represent an endpoint in ligand design, but rather a starting point for optimization via further backbone and/or sequence modification. Such optimization could improve affinity, alter selectivity, and enhance proteolytic stability. The results we have achieved so far suggest that systematic exploration of backbone composition could prove to be broadly effective for generating foldamers with useful properties from biologically active α-peptides.
Supplementary Material
Footnotes
This work was supported by the NIH (GM56414 to S.H.G. and CA119875 to W.S.H.). We thank Peptech for providing protected β-amino acids at a discounted price and Prof. York Tomita for providing plasmids used for expression of Bcl-xL and Mcl-1.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- [1].Arkin MR, Wells JA. Nat. Rev. Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- [2].(a) Gellman SH. Acc. Chem. Res. 1998;31:173–180. [Google Scholar]; (b) Goodman CM, Choi S, Shandler S, DeGrado WF. Nat. Chem. Biol. 2007;3:252–262. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].For selected examples, see: Werder M, Hauser H, Abele S, Seebach D. Helv. Chim. Acta. 1999;82:1774–1783.; Kritzer JA, Lear JD, Hodsdon ME, Schepartz A. J. Am. Chem. Soc. 2004;126:9468–9469. doi: 10.1021/ja031625a.; Sadowsky JD, Schmitt MA, Lee HS, Umezawa N, Wang SM, Tomita Y, Gellman SH. J. Am. Chem. Soc. 2005;127:11966–11968. doi: 10.1021/ja053678t.; English EP, Chumanov RS, Gellman SH, Compton T. J. Biol. Chem. 2006;281:2661–2667. doi: 10.1074/jbc.M508485200..
- [4].a) Sadowsky JD, Fairlie WD, Hadley EB, Lee HS, Umezawa N, Nikolovska-Coleska Z, Wang SM, Huang DCS, Tomita T, Gellman SH. J. Am. Chem. Soc. 2007;129:139–154. doi: 10.1021/ja0662523. [DOI] [PubMed] [Google Scholar]; b) Sadowsky JD, Murray JK, Tomita Y, Gellman SH. ChemBioChem. 2007;8:903–916. doi: 10.1002/cbic.200600546. [DOI] [PubMed] [Google Scholar]
- [5].For other examples of chemically modified peptides binding to Bcl-2 family proteins, see: Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191.; Wang DY, Liao W, Arora PS. Angew. Chem. 2005;117:6683–6687.; Angew. Chem. Int. Ed. 2005;44:6525–6529. doi: 10.1002/anie.200501603..
- [6].Adams JM, Cory S. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].For selected structures, see: Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983.; Czabotar PE, Lee EF, van Delft MF, Day CL, Smith BJ, Huang DCS, Fairlie WD, Hinds MG, Colman PM. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6217–6222. doi: 10.1073/pnas.0701297104..
- [8].Cheng RP, Gellman SH, DeGrado WF. Chem. Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- [9].a) Hayen A, Schmitt MA, Ngassa FN, Thomasson KA, Gellman SH. Angew. Chem. Int. Ed. 2004;43:505–510. doi: 10.1002/anie.200352125. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2004;116:511–516. [Google Scholar]; b) De Pol S, Zorn C, Klein CD, Zerbe O, Reiser O. Angew. Chem. Int. Ed. 2004;43:511–514. doi: 10.1002/anie.200352267. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2004;116:517–520. [Google Scholar]; Sharma GVM, Nagendar P, Jayaprakash P, Krishna PR, Ramakrishna KVS, Kunwar AC. Angew. Chem. Int. Ed. 2005;44:5878–5882. doi: 10.1002/anie.200501247. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2005;117:6028–6032. [Google Scholar]
- [10].Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, Adams JM, Huang DCS. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Certo M, Moore VD, Nishino M, Wei G, Korsmeyer S, Armstrong SA, Letai A. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- [12].Horne WS, Price JL, Keck JL, Gellman SH. J. Am. Chem. Soc. 2007;129:4178–4180. doi: 10.1021/ja070396f. [DOI] [PubMed] [Google Scholar]
- [13].Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DCS. Mol. Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- [14].a) Ebeling W, Hennrich N, Klockow M, Metz H, Orth HD, Lang H. Eur. J. Biochem. 1974;47:91–97. doi: 10.1111/j.1432-1033.1974.tb03671.x. [DOI] [PubMed] [Google Scholar]; b) Narahashi Y, Shibuya K, Yanagita M. J. Biochem. 1968;64:427–437. doi: 10.1093/oxfordjournals.jbchem.a128914. [DOI] [PubMed] [Google Scholar]
- [15].See Supporting Information.
- [16].For a review, see: Steer DL, Lew RA, Perlmutter P, Smith AI, Aguilar MI. Curr. Med Chem. 2002;9:811–822. doi: 10.2174/0929867024606759..
- [17].a) Guichard G, Zerbib A, Le Gal FA, Hoebeke J, Connan F, Choppin J, Briand JP, Guillet JG. J. Med. Chem. 2000;43:3803–3808. doi: 10.1021/jm000909s. [DOI] [PubMed] [Google Scholar]; b) Reinelt S, Marti M, Dedier S, Reitinger T, Folkers G, de Castro JAL, Rognan D. J. Biol. Chem. 2001;276:24525–24530. doi: 10.1074/jbc.M102772200. [DOI] [PubMed] [Google Scholar]; c) Webb AI, Dunstone MA, Williamson NA, Price JD, de Kauwe A, Chen WS, Oakley A, Perlmutter P, McCluskey J, Aguilar MI, Rossjohn J, Purcell AW. J. Immunol. 2005;175:3810–3818. doi: 10.4049/jimmunol.175.6.3810. [DOI] [PubMed] [Google Scholar]
- [18].Other work has suggested that binding affinity among α-peptides is not a simple function of helical propensity. See ref. [13].
- [19].Koglin N, Zorn C, Beumer R, Cabrele C, Bubert C, Sewald N, Reiser O, Beck-Sickinger AG. Angew. Chem. Int. Ed. 2003;42:202–205. doi: 10.1002/anie.200390078. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2003;115:212–215. [Google Scholar]
- [20].Roy RS, Karle IL, Raghothama S, Balaram P. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16478–16482. doi: 10.1073/pnas.0407557101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.