Abstract
Proliferating cells consume more glucose to cope with the bioenergetics and biosynthetic demands of rapidly dividing cells as well as to counter a shift in cellular redox environment. This study investigates the hypothesis that manganese superoxide dismutase (MnSOD) regulates cellular redox flux and glucose consumption during the cell cycle. A direct correlation was observed between glucose consumption and percentage of S-phase cells in MnSOD wild type fibroblasts, which was absent in MnSOD homozygous knockout fibroblasts. Results from electron paramagnetic resonance spectroscopy and flow cytometry assays showed a significant increase in cellular superoxide levels in S-phase cells, which was associated with an increase in glucose and oxygen consumption, and a decrease in MnSOD activity. Mass spectrometry results showed a complex pattern of MnSOD-methylation at both lysine (68, 89, 122, and 202) and arginine (197 and 216) residues. MnSOD protein carrying a K89A mutation had significantly lower activity compared to wild type MnSOD. Computational-based simulations indicate that lysine and arginine methylation of MnSOD during quiescence would allow greater accessibility to the enzyme active site as well as increase the positive electrostatic potential around and within the active site. Methylation-dependent changes in the MnSOD conformation and subsequent changes in the electrostatic potential around the active site during quiescence vs. proliferation could increase the accessibility of superoxide, a negatively charged substrate. These results support the hypothesis that MnSOD regulates a “metabolic switch” during progression from quiescent through the proliferative cycle. We propose MnSOD as a new molecular player contributing to the “Warburg effect.”
Keywords: MnSOD, methylation, glucose, cell cycle, Warburg effect
Introduction
Normal cell proliferation is an orderly transition from quiescence (G0) to proliferation (G1, S, G2, and M phases) and back to quiescence. Such an orderly transition is needed to maintain the regenerative capacity of cells and to avoid aberrant proliferation. Progression from quiescent to proliferation is regulated by the sequential activation of cyclin dependent kinases (CDKs) (1, 2). Cyclin/CDKs are the positive regulators of cell cycle progression, while CDK inhibitors are the negative regulators of cellular proliferation. Recent evidence suggests that metabolic activities play a significant role linking cellular metabolism to the cell cycle regulatory machinery (3–7). Cellular proliferation is accompanied by an increase in the utilization of glucose and glutamine. Aerobic glycolysis coordinates the bioenergetic and biosynthetic demands of rapidly proliferating cells (4, 5). During aerobic glycolysis, glucose is converted to pyruvate, which is then converted to lactate by lactate dehydrogenase. Pyruvate can enter the tricarboxylic acid cycle and mitochondrial electron transport chain leading to the generation of ATP. The third pathway of glucose utilization involves the pentose phosphate pathway, which generates precursor metabolites needed for cellular macromolecular synthesis. Previous studies have established a regulatory role for glucose-6-phosphate dehydrogenase, the rate limiting step in the pentose phosphate pathway, during proliferation (8, 9). NADPH is one of the products of the pentose phosphate pathway, which is directly coupled to the cellular glutathione and thioredoxin redox systems. It is postulated that increased glycolysis during proliferation could counter a shift in the cellular redox environment facilitating progression through the cell cycle. In fact, we and others have observed a gradual increase in cellular glutathione levels as cells progress through the cell cycle (unpublished observations) (10).
The increase in glutathione during the cell cycle could be in response to a shift in the cellular redox environment towards a more oxidizing environment (11, 12). The redox environment of mitotic cells is 3–4-fold more oxidizing than G1-cells (11). These previously published results support the hypothesis that a redox cycle within the cell cycle regulates progression from quiescent through the proliferative cycle (13). The cellular redox environment is a balance between the production of reactive oxygen species (ROS: superoxide and hydrogen peroxide) and their removal by the antioxidant network. The mitochondrial electron transport chain is the major generator of cellular ROS. Manganese superoxide dismutase (MnSOD) is a redox-enzyme that is encoded by nuclear DNA and localized to the mitochondrial matrix. MnSOD converts mitochondrial generated superoxide to hydrogen peroxide. Recent evidence suggests that MnSOD activity and mitochondrial generated ROS regulate transitions between the quiescent and proliferative cycle (14–16). MnSOD-dependent regulation of quiescent cells entrance into the proliferative cycle and subsequent exit from the proliferative cycle is associated with changes in levels of cyclins (D1 and B1) and CDK inhibitors (p21 and p16) (14, 16). Cyclin D1 integrates mitochondrial function with DNA synthesis (17). Likewise, mitochondrial fusion is linked to the activation of cyclin E at the G1/S border and entry into S-phase (18). Results presented in this study show: (i) a direct correlation between glucose consumption and percent S-phase in MnSOD wild type mouse embryonic fibroblasts; (ii) a periodic change in MnSOD activity during the cell cycle; (iii) an increase in superoxide levels during S-phase; and (iv) a change in MnSOD methylation status in quiescent compared to proliferating cells. These results support the hypothesis that MnSOD regulates a “metabolic switch” during progression from quiescent through the proliferative cycle.
Materials and Methods
Cell culture
Human normal skin fibroblasts (AG01522D, Coriell Cell Repositories), mammary epithelial non-tumorigenic (MCF10A) and adenocarcinoma (MB231) cells (ATCC), wild type and homozygous knockout MnSOD mouse embryonic fibroblasts (MEFs) were cultured following our previously published protocols (16, 19, 20). Adenovirus infections and transgene expression were performed (15, 16, 19, 20), and MnSOD activity was measured using biochemical and gel-electrophoresis based assays (14, 15, 21). Fibroblasts were synchronized by contact inhibition (22). MB231 cells were synchronized at the G1/S border using 2 mM thymidine, washed, and continued in culture for isolation of cells in S- and G2-phases. G1-cells of the daughter generation were obtained by incubating G1/S synchronized MB231 cells with nocodazole (200 ng/mL) and harvesting cells at 4 h post-mitotic shake-off.
Flow cytometry assays
Cell cycle phase distributions were determined by flow cytometry measurements of DNA content (16). Dihydroethidium (DHE), MitoSOX-Red, and MitoTracker-Green fluorescence were used to probe for cellular and mitochondrial ROS levels and mitochondrial mass (15, 16, 22). Mean fluorescence intensity was calculated using Flowjo software (Tree Star, Inc. USA). Auto-fluorescence of cells was used for background correction; fold change calculated relative to control or G1-cells.
Immunoprecipitation assay
Immunoprecipitation was performed following the manufacturer’s supplied protocol (Direct IP from Pierce®, USA). Rabbit polyclonal antibody against methylated-lysine (Abcam Inc.) was coupled to beads (AminoLink Plus Coupling Resin), and incubated with 500 µg of protein extracts. Bound proteins were eluted and MnSOD was identified in the immunoprecipitates via immunoblotting.
Site-directed mutagenesis
QuickChange II Kit (Stratagene) was used to mutate lysine 89 and lysine 202 of MnSOD to alanine. pShooter (Invitrogen) expression vectors carrying human MnSOD cDNAs with wild type and lysine-to-alanine mutations were transfected into MnSOD (−/−) MEFs. MnSOD expression was measured using western blotting and activity assays.
Electron paramagnetic resonance spectroscopy
Cells were incubated with PBS containing 100 mM 5, 5-dimethyl-1-pyrroline N-oxide (DMPO) and EPR spectra recorded using a Bruker EMX-spectrometer with a magnetic field modulation frequency of 100 kHz; modulation amplitude, 1:0 G; scan rate, 60 G/21 s (15, 16). Spectra were results of forty signal-averaged scans collected over about 20 min. EPR peak heights (WinEPR software) were normalized to cell number. The specificity of the superoxide origin of the signal was determined by pre-incubating cells with CuZnSOD (1000 units/mL).
Glucose and oxygen consumption assays
Cultures were incubated for 4–6 h with Dulbecco's Modified Eagle Medium and glucose concentration was measured using a Bayer Glucometer Elite ® with Bayer Ascensia Elite® Blood glucose test strips (23). Glucose consumption rate (GCR) was calculated per cell, and fold-change was determined relative to the GCR of G1-cells.
Oxygen consumption rate (OCR) of cells was measured polarographically (Yellow Springs Instrument Co). Measurements were performed at 37°C on 3 mL samples, air-saturated culture media without serum with 5–8 × 106 cells. OCR was expressed as attomoles of oxygen consumed cell−1 s−1, using an initial oxygen concentration of 192 µM (24).
Mass spectrometry
Total cellular protein extracts prepared from quiescent and proliferating cultures of normal human fibroblasts were resolved using gel-electrophoresis. Coomassie Blue stained gel slices were excised and subjected to tandem mass spectrometry (MS2) analysis.
Molecular Modeling
Molecular modeling simulations were performed using Sybyl-X software (Version 1.2; Tripos, Inc., St. Louis, MO). The crystallographic coordinates of the 2.3 Å human MnSOD structure (PDB: 2GDS), were obtained from the RCSB Protein Data Bank (25), and the Asp54 residues in the tetramer were replaced with His residues. The MMFF94s force-field and MMFF94 charges were applied to the un-methylated MnSOD tetramer and the structure was minimized using the Powell method (1000 iterations; termination gradient of 0.001 kcal/mol) (26). The proliferative and quiescent forms of MnSOD were further minimized using the Powell method to examine structural changes induced by lysine and arginine mono- and di-methylation.
Statistical analysis
Statistical analysis was done using the one and two-way analysis of variance with Tukey’s honestly significant difference test (SigmaPlot computer software version 11.0).
Results
MnSOD activity regulates glucose consumption during cellular proliferation
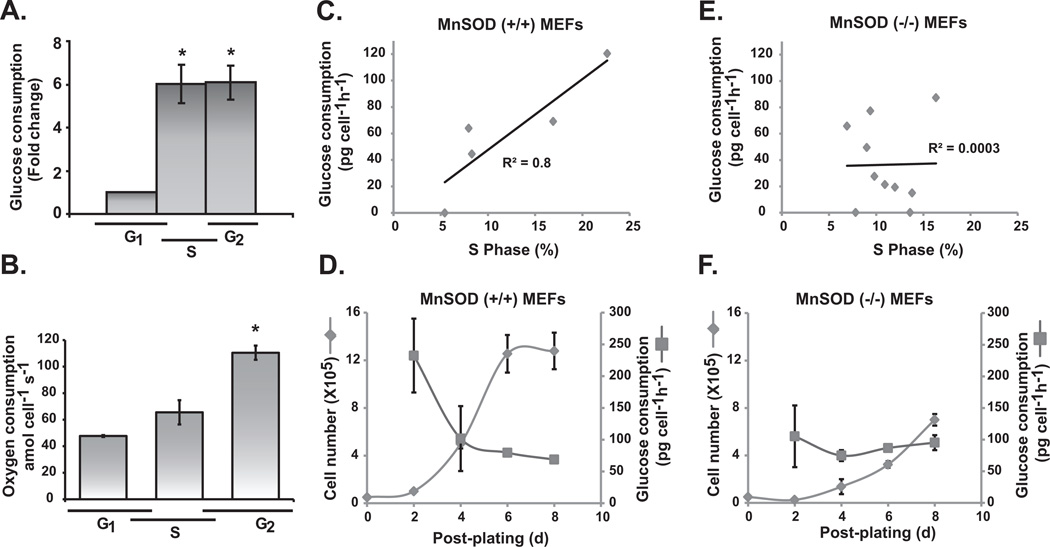
Cellular proliferation is accompanied by an increase in glucose consumption to support the high demand of bioenergetics and biosynthetic processes. MnSOD activity and mitochondrial ROS have been shown to regulate transitions between the quiescent and proliferative states. To determine if MnSOD activity coordinates glucose consumption rate (GCR) during transition from the quiescent to the proliferative state, we initially determined if GCR varies during the cell cycle. GCR increased approximately 6-fold in S- and G2-cells compared to G1-cells (Figure 1A). Oxygen consumption rate (OCR) of G1-cells was determined to be approximately 50 amol cell−1 s−1, and G2-cells exhibited more than 100 amol cell−1 s−1, a 2-fold increase in OCR (Figure 1B). GCR in MnSOD wild type MEFs exhibited a direct correlation with the percentage of S-phase cells (R2 = 0.8; Figure 1C). GCR was approximately 40 pg cell−1 h−1 in cultures with 10% S-phase, and 120 pg cell−1 h−1 in cultures with 25% S-phase cells (Figure 1C). GCR decreased as cells exit the proliferative cycle and entered quiescence (Figure 1D). It is interesting to note that such a correlation between the percentage of S-phase cells and GCR was absent in homozygous knockout MnSOD MEFs (R2 = 0.0003; Figure 1E). As reported previously, MnSOD (−/−) MEFs did not exit the proliferative cycle (16), and the inability to exit the proliferative cycle was associated with a relatively constant GCR (Figure 1F).
Figure 1. A direct correlation between glucose consumption and percentage of S-phase cells in MnSOD wild type mouse embryonic fibroblasts.
Quiescent MEFs were replated at a lower cell density and harvested at different times for flow cytometry measurements of cell cycle phases. Cells from replicate dishes were used for glucose (A) and oxygen (B) consumption assays. Asterisk indicates statistical significance relative to G1-cells; n = 3, P <0.05. MEFs with wild type (+/+) (C and D) and knockout (−/−) (E and F) MnSOD were cultured to obtain cells with varying percentage in S-phase. Cells from replicate dishes were used to measure glucose consumption rate. Regression plots show the correlation between the rate of glucose consumption and percentage of S-phase. In a separate set of experiments, cell numbers and glucose consumption rates were determined at different days post-plating (D and F).
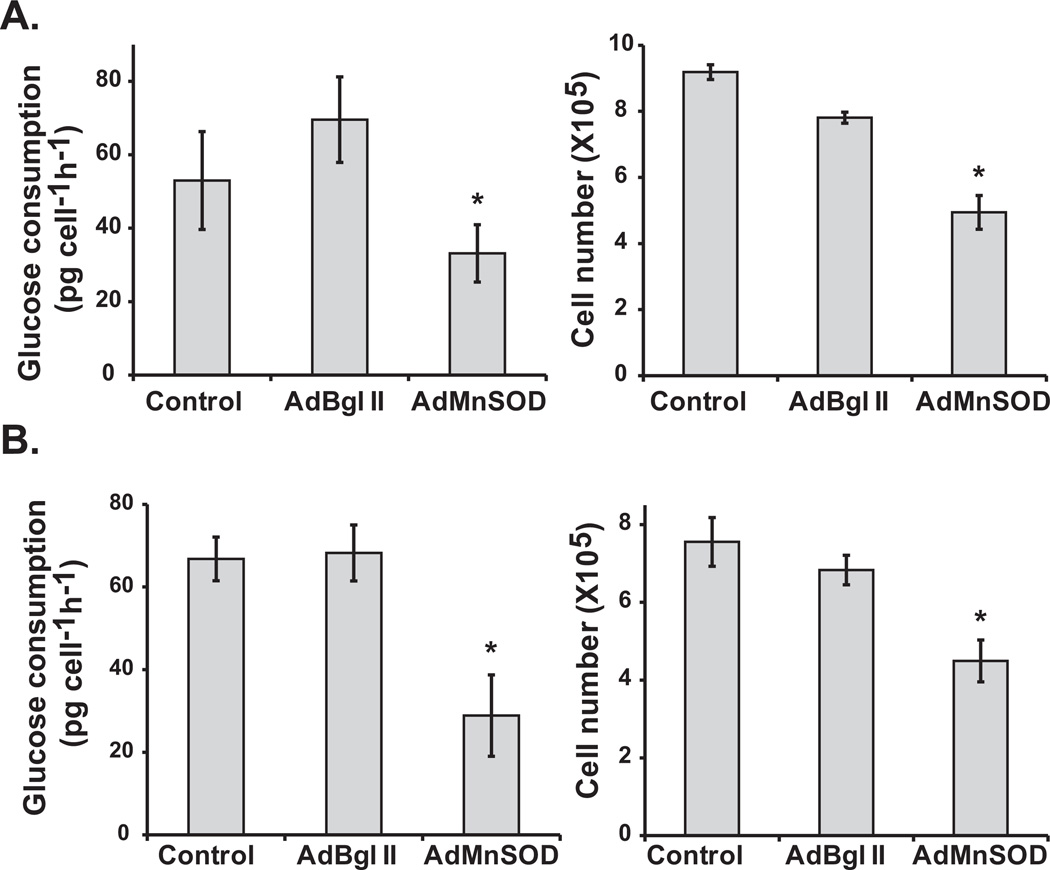
To further determine if MnSOD activity regulates GCR, MnSOD (−/−) MEFs were infected with 100 MOI (multiplicity of infection) of AdMnSOD (an adenoviral construct containing the cDNA for MnSOD). AdMnSOD infection increased MnSOD protein levels and activity (Figure 4E, Supplemental Figure 1). The GCR in control and AdBgl II infected cells at 96 h post-infection was approximately 50 pg cell−1 h−1, and a 2-fold decrease in GCR was observed in AdMnSOD infected cells (Figure 2A, left panel). An increase in MnSOD activity and subsequently a decrease in GCR were accompanied with a decrease in cell number (Figure 2A, right panel). The generality of this phenomenon was further evaluated by repeating the experiment using MB231 cells. GCR in control and AdBgl II infected MB231 cells was approximately 65 pg cell−1 h−1 and overexpression of MnSOD suppressed GCR by approximately 2-fold (Figure 2B, left panel). As previously shown (Figure 2A, right panel), overexpression of MnSOD and subsequent decrease in GCR inhibited cell growth (Figure 2B, right panel). Likewise, a decrease in MnSOD activity in normal human fibroblasts overexpressing a dominant-negative form of MnSOD increased GCR and percentage of S-phase cells (Supplemental Figure 2). These results support the hypothesis that MnSOD activity regulates a “metabolic switch” facilitating a redox environment that is conducive for cell cycle progression.
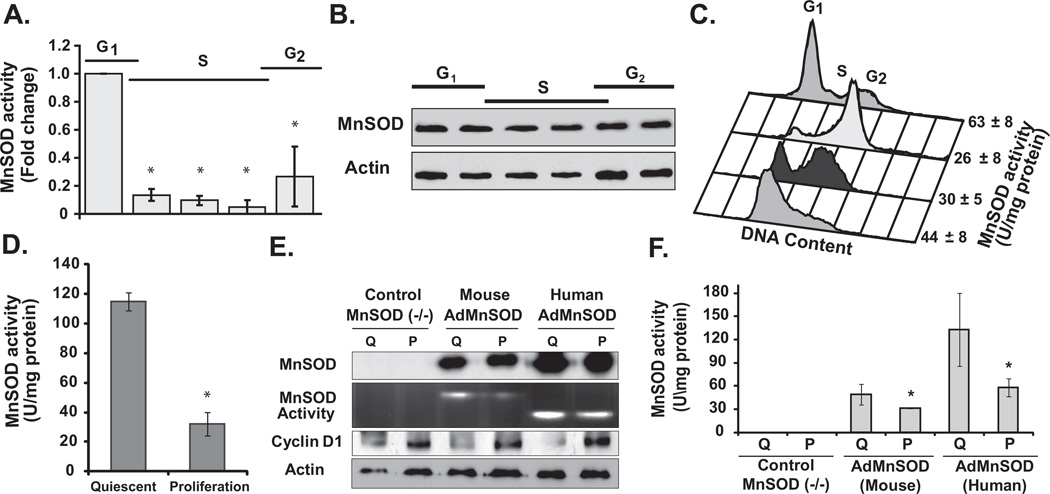
Figure 4. Periodic change in MnSOD activity during the cell cycle.
Synchronized MEFs were harvested at different times post-replating, and analyzed for MnSOD (A) activity and (B) protein levels. Cells in replicate dishes were used for flow cytometry measurements of cell cycle phase distributions. (C) Representative histograms of DNA content exhibiting cell cycle phase distributions when cells in replicate dishes were harvested for biochemical measurements of MnSOD activity: MB231 cells at the time of release from the thymidine block, and later in S- and G2-phase. Cells in G1-phase of the daughter generation were collected at 4 h post-mitotic shake-off. (D) MnSOD activity in quiescent and proliferating MCF10A cells. Asterisk indicates statistical significance in MnSOD activity relative to quiescent cells; n = 3, P <0.05. (E) Quiescent (Q) and proliferating (P) cultures of MnSOD (−/−) MEFs were infected with 50 MOI of adenoviruses carrying CMV-promoter driven mouse or human MnSOD cDNA; immunoblotting assay to determine protein levels of MnSOD, cyclin D1, and actin. A gel-electrophoresis-based method was used to measure MnSOD activity. (F) Cells from replicate dishes were harvested for a spectroscopy-based assay to measure MnSOD activity. Asterisk indicates statistical significance in MnSOD activity relative to quiescent (Q) cultures; n = 3, P <0.05.
Figure 2. MnSOD activity regulates a “metabolic switch” influencing cellular proliferation.
One hundred MOI of adenoviruses carrying CMV-promoter driven mouse or human MnSOD cDNA were used to infect asynchronous cultures of (A) MnSOD (−/−) MEFs and (B) MB231 cells. AdBgl II represents non-replicative adenoviral vector without MnSOD cDNA. It was included as a control for adenoviral infections (14). Glucose consumption and cell numbers were measured at 96 h post-infection for MEFs (2A), and 72 h post-infection for MB231 (2B). Asterisk indicates significant differences in glucose consumption rate and cell numbers in MnSOD overexpressing vs. control and AdBgl II infected cells; n = 3, P <0.05.
Cell cycle phase specific accumulation of ROS levels
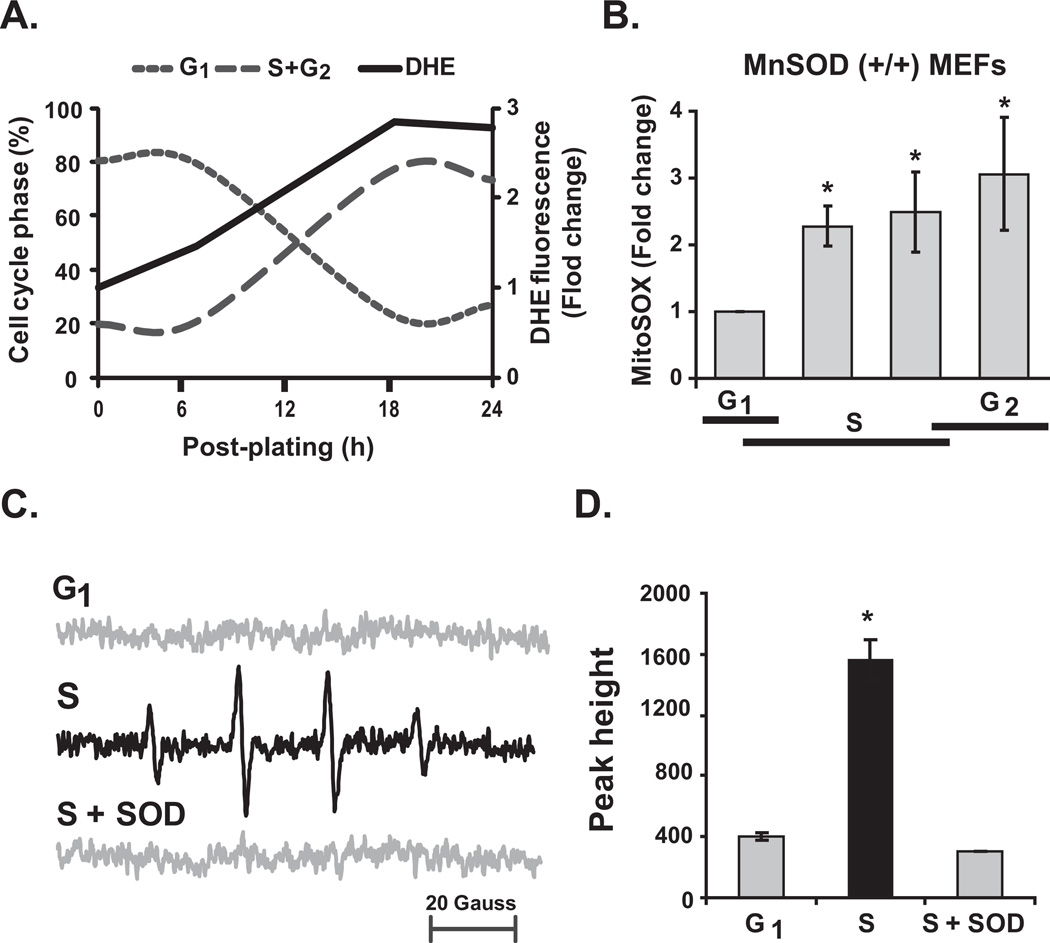
MnSOD is a redox enzyme that is known to regulate the cellular redox environment by converting superoxide to hydrogen peroxide. To determine if changes in GCR and MnSOD activity (see below, Figure 4) during the cell cycle were associated with changes in the cellular redox environment, flow cytometry and EPR measurements of cellular ROS levels were performed in synchronized MEFs. The percentage of G0/G1 cells at the time of post-replating was approximately 80% and decreased to 20% at 20 h post-replating when 60% of the cells progressed to S- and G2-phases (Figure 3A). Cellular ROS levels, measured by DHE-oxidation, increased approximately 3-fold at 20–24 h post-replating compared to the cells at the time of re-plating. A 2- to 3-fold increase in MitoSOX-oxidation, an indicator of an increase in mitochondrial ROS levels (15, 16, 22), was observed in S- and G2-cells compared to G1 cells (Figure 3B). It is interesting to note that MitoSOX fluorescence did not show any significant change during the cell cycle of MnSOD (−/−) MEFs (Supplemental Figure 3 and Table I). These results suggest that MnSOD activity shifts the cellular (presumably mitochondrial) redox environment towards a more oxidizing environment as cells progress through the cell cycle.
Figure 3. Increase superoxide levels in S-phase cells.
Quiescent MEFs were replated at a lower cell density and harvested at different times for (A) flow cytometry measurements of cell cycle phases and DHE-oxidation; (B) cells in replicate dishes were used for measurements of MitoSOX-oxidation. Asterisk indicates statistical significance relative to G1-cells; n = 3, P <0.05. (C) Representative EPR spectra of DMPO-OH in G1- and S-phase MEFs, and S-phase MEFs incubated with CuZnSOD. (D) EPR peak heights were normalized to cell number; peak heights in G1 and S+SOD represent the noise level of the spectra. Asterisk indicates statistical significance relative to G1-cells; n = 3, P <0.05.
Flow cytometry measurements of DHE-oxidation were repeated to distinguish superoxide from other oxidants (27). These results indicate that the increase in DHE-oxidation during S- and G2-phase was primarily due to an increase in cellular superoxide levels (Supplemental Figure 4). These results are also consistent with results obtained from EPR spectroscopy (Figure 3C and 3D). A 1:2:2:1 spectrum of DMPO-OH was observed in synchronized S-phase cells. EPR peak height measurements showed an approximately 4-fold increase in DMPO-OH in S-phase compared to G1-phase cells. A significant suppression of EPR peak height in S-phase cells pre-treated with SOD suggests that the observed DMPO-OH species is derived from superoxide.
Periodic changes in MnSOD activity during the cell cycle
Mitochondria are the major source of cellular ROS and MnSOD activity is known to regulate mitochondrial ROS levels. To determine if MnSOD activity itself varies during the cell cycle, synchronized MEFs representative of G1-, S-, and G2-phases were assayed for MnSOD activity (21). MnSOD activity was higher in G1-phase and decreased significantly as cells progress through S- and G2-phases (Figure 4A). The periodic changes in MnSOD activity were not due to a corresponding change in MnSOD protein levels (Figure 4B). To determine the generality of this observation, MnSOD activity was measured in synchronized cell populations of MB231 (Figure 4C). MnSOD activity in the G1/S synchronized cells was approximately 44 U mg−1 protein, which decreased to 30 and 26 U mg−1 protein in S- and G2-phases, respectively. Surprisingly, following cell division, MnSOD activity in the G1-phase of the daughter generation increased to 63 U mg−1 protein. Furthermore, MnSOD activity in quiescent MCF10A cells was approximately 120 U mg−1 protein, which significantly decreased to 30 U mg−1 protein at 24 h post-replating when approximately 20% cells progressed to S-phase (Figure 4D). These results showed a periodic change in MnSOD activity during the parental generation that is faithfully preserved in the daughter generation.
Post-translational modifications of MnSOD during quiescence and proliferation
Since the periodic changes in MnSOD activity through the cell cycle were not due to change in its protein levels (Figure 4B), we investigated whether these changes were related to a post-translational modification of the protein. First, quiescent and proliferating cultures of MnSOD (−/−) MEFs were infected with adenoviruses containing CMV-promoter driven mouse or human MnSOD cDNA. Cyclin D1 protein levels were assessed to evaluate cellular quiescence and proliferation. MnSOD protein levels were comparable in AdMnSOD infected quiescent and proliferating cells (Figure 4E). However, MnSOD activity showed a significant increase in quiescent compared to proliferating cells (Figure 4E and 4F). These results indicate that a post-translational modification of MnSOD regulates its activity during transitions between quiescence and proliferation.
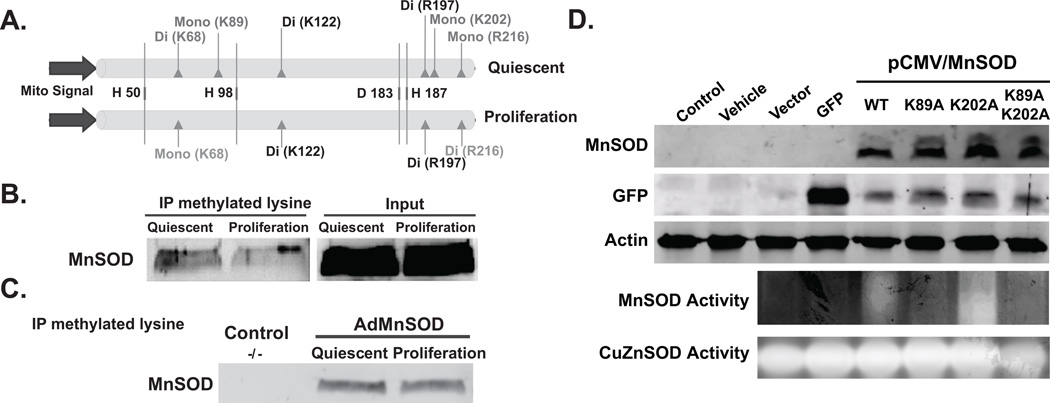
To characterize specific post-translational modifications of MnSOD total cellular proteins isolated from quiescent and proliferative cultures were resolved using one-dimensional gel electrophoresis and coomassie-stained bands corresponding to MnSOD were excised for tandem mass spectrometry (MS2) analysis. The MS2 spectra were evaluated for common post-translational modifications such as phosphorylation, acetylation, methylation, sumoylation, and ubiquitination. Methylation was identified as the only post-translational modification of MnSOD during quiescence and proliferation (Supplemental Figure 5A). MnSOD is methylated at both lysine (68, 89, 122, and 202) and arginine (197 and 216) residues (Figure 5A). In quiescent fibroblasts MnSOD is dimethylated at K68 and K122, and mono-methylated at K89 and K202. Arginine is dimethylated at R197 and mono-methylated in R216. In proliferating cells, MnSOD is mono-methylated at K68, while K89 and K202 were un-methylated. MnSOD K122- and R197-di-methylation did not change between quiescent and proliferative growth-state. R216, which was mono-methylated in quiescent cells, became di-methylated in proliferating cells. It is interesting to note the conservation of MnSOD lysine (68, 89, 122, and 202) and arginine (R197 and 216) among different species (Supplemental Figure 5B).
Figure 5. Lysine and arginine methylation of MnSOD during quiescence and proliferation.
(A) An illustration of the MnSOD methylation pattern identified by mass spectrometry in quiescent and proliferating cultures. MnSOD is methylated at both lysine (68, 89, 122, and 202) and arginine (197 and 216). K68 is di-methylated during quiescence and mono-methylated during proliferation. K89 and K202 are mono-methylated specifically during quiescence. The histidines representing MnSOD active sites were included in this illustration for reference. (B) Total cellular protein extracts prepared from quiescent and proliferating cultures of normal human fibroblasts were used to immunoprecipitate methylated proteins using antibodies to methylated lysine; immunoblotting was used to identify MnSOD in the immunoprecipitates. (C) The assay in (B) was repeated in protein extracts prepared from control, 50 MOI AdmouseMnSOD infected quiescent and proliferating cultures of MnSOD (−/−) MEFs. (D) MnSOD protein levels and activity in MnSOD (−/−) MEFs transfected with pShooter plasmid DNAs containing wild-type or K-to-A mutant carrying human MnSOD cDNAs. Un-transfected control, cells transfected with the transfecting agent alone (vehicle), cells transfected with plasmid DNA without an insert (vector), and cells transfected with plasmid DNA containing GFP were included for comparison. For the co-transfection experiments, one-fourth of the GFP-plasmid DNA was used. Actin protein levels and CuZnSOD activity were used for comparison.
The MS2 results were validated using an immunoprecipitation-immunoblotting assay (Figure 5B and 5C). Total cellular protein extracts prepared from quiescent and proliferating fibroblasts were incubated with antibodies against methylated-lysine (Abcam, USA). An immunoblotting assay was performed to identify MnSOD in the immunoprecipitates. Consistent with the MS2 results, results from the immunoprecipitation-immunoblotting assay identified MnSOD as being methylated in both quiescent and proliferating fibroblasts (Figure 5B). The amount of the methylated form of MnSOD appears to decrease in proliferating vs. quiescent fibroblasts. Comparable results were also observed in MnSOD overexpressing quiescent and proliferating MnSOD (−/−) MEFs (Figure 5C).
The role of methylation regulating MnSOD activity was further evident from the results shown in Figure 5D. Site-directed mutagenesis was used to mutate lysine (89 and 202, individually as well as double mutation; Supplemental Figure 6) to alanine. MnSOD protein levels and activity were measured in MnSOD(−/−) MEFs transfected with pShooter plasmid DNAs containing wild type and K-to-A mutant carrying human MnSOD cDNAs. It is interesting to note that while MnSOD protein was present in cells transfected with wild-type and K-to-A mutant carrying human MnSOD cDNAs, K89A mutation significantly decreased MnSOD activity (Figure 5D). The specificity of K89-methylation regulating MnSOD activity was also evident in cells transfected with the K89A-K202A double mutation. Furthermore, the rate of glucose consumption increased in cells expressing K89A-MnSOD (122 pg cell−1 h−1) compared to wt-MnSOD (87 pg cell−1 h−1). These results demonstrate that (a) methylation status of K89 has a significant role in regulating MnSOD activity; and (b) MnSOD activity influences glucose consumption.
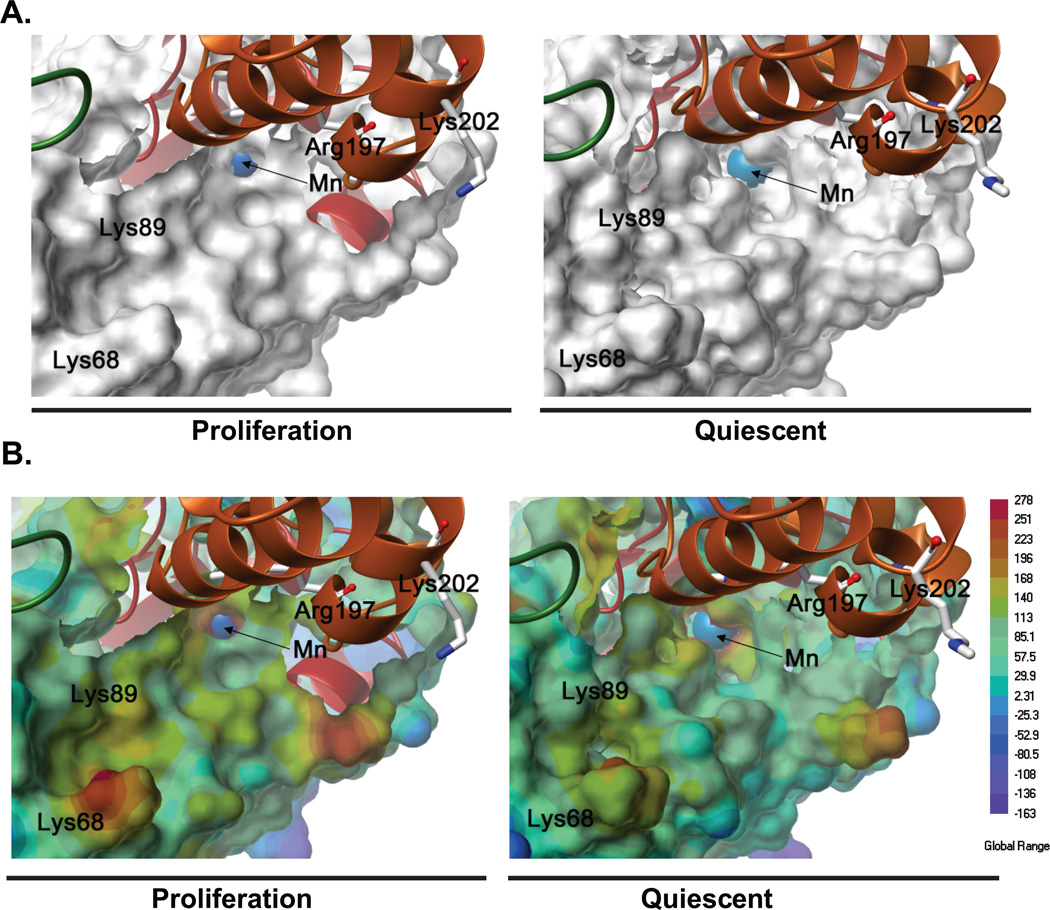
Results from the molecular biology approach were comparable to computer modeling of MnSOD-methylation influencing its activity. Models of methylated tetrameric MnSOD were constructed using computational-based molecular modeling, based on our mass spectrometry data of the methylated state of MnSOD during proliferation and quiescence (Supplemental Figure 7A and B). The methylated residues (Lys68, 89, 122, and 202; Arg197 and 216) are well distributed in each monomeric unit of the MnSOD tetramer. Although no large global changes in conformation were observed between the proliferative and quiescent MnSOD models, energy minimization of these models revealed conformational differences in and around the active site, which may modulate the accessibility of substrates and enzyme activity. In the quiescent MnSOD-methylation model, the Mn ion in the active site was more accessible than in the proliferative MnSOD-methylation model (Figure 6A). In addition, changes in electrostatic potential were also observed around the active site of quiescent MnSOD-methylation model (Figure 6B). The Lys68 and 89 residues within the monomer and the residues Arg197 and Lys202 from the adjacent monomer were in close proximity to the active site, but are not close enough to sterically affect the accessibility of the substrate. However, the differences in the methylation of residues in proliferative and quiescent MnSOD seem to induce distinct conformational changes in the accessible surface and the electrostatic potential around the active site. The overall contraction of the catalytic residues H50, H98, D183, and H187 around the Mn ion in the active site of the quiescent MnSOD model compared with the proliferative model (Supplemental Figure 7C) appears to enlarge the entrance to active site cavity, which in addition to increasing the accessibility to the Mn ion also increases the surface area of positive electrostatic potential around the Mn ion. Furthermore, differences in the orientation of amino acid side chains, particularly His50 and 54, in the MnSOD models may also contribute to the enlargement of the active site cavity and changes in electrostatic potential. Therefore, these methylation-dependent changes in MnSOD conformation and electrostatic potential during quiescence may increase the accessibility of superoxide, a negatively charged substrate, to the enzyme active site.
Figure 6. Structural and electrostatic changes in MnSOD during quiescence and proliferation.
(A) Connolly molecular surface representations of a monomeric unit of the methylated-MnSOD tetramer in proliferation and quiescence; the manganese (Mn) ion (blue) is shown as a space fill representation. (B) Electrostatic potential energy surfaces of the methylated-MnSOD tetramer in proliferation and quiescence; surfaces colored blue are negative and the Mn ion (blue) is shown as a space fill representation.
Discussion
Recent evidence suggests that a redox cycle within the mammalian cell cycle could coordinate cellular metabolism to the cell cycle regulatory machinery (13, 28). We have shown previously that MnSOD activity and mitochondrial generated ROS regulate transitions between quiescent and proliferative states favoring superoxide signaling that facilitates proliferation and hydrogen peroxide signaling that supports quiescence (15, 16). Results from the present study showed a significant increase in cellular superoxide levels during S-phase, which correlated with a decrease in MnSOD activity and increase in GCR and OCR. The periodic change in MnSOD activity during the cell cycle was associated with a complex pattern of MnSOD lysine and arginine methylation. Most importantly, a direct correlation was observed between GCR and percentage of S-phase cells in MnSOD (+/+) MEFs, which was absent in MnSOD (−/−) MEFs.
GCR increased approximately 6-fold in S- and G2-cells compared to G1-cells, indicating an increase in aerobic glycolysis as cells progress through the cell cycle (Figure 1). Aerobic glycolysis was originally reported by Warburg as a key pathway of energy generation in cancer vs. normal cells (6). However, several recent studies report that aerobic glycolysis is central to rapidly dividing non-cancerous as well as cancer cells (4, 5, 29–31). It is hypothesized that the increase in glucose consumption is necessary to cope with the high bioenergetic demand and biosynthesis of rapidly dividing cells. Previous studies have shown a role for glycolytic enzymes in the regulation of cellular proliferation, e.g. M2 isoform of pyruvate kinase, lactate dehydrogenase, 6-phosphofructo-2-kinase/fructose-2 biphosphophosphatase isoform, and ubiquitin ligase anaphase promoting complex cyclosome-Cdh1 (4, 29–31). Consistent with these observations, we have shown previously that cyclin D1 and cyclin B1 protein levels during the cell cycle correlate with MnSOD activity, changes in cellular redox environment, and mitochondrial function (16). These previous reports, along with the results presented in Figures 1 and 2, suggest a link between cellular metabolism and cell cycle regulatory machinery.
Progression through the cell cycle was also associated with a gradual increase in OCR (Figure 1B). An increase in OCR and GCR was associated with a significant increase in cellular (presumably of mitochondrial origin) ROS levels (Figure 3A and 3B). Synchronized fibroblasts in S-phase appear to have a 4-fold higher steady-state level of superoxide compared to G1-cells (Figure 3C, 3D, and Supplemental Figure 4). It appears that rapidly proliferating cells simultaneously exhibit both glycolysis and respiration during S- and G2-phases, yet the fold-change shifts more towards glycolysis (6-fold increase) than respiration (2-fold increase). While the downstream pathway of superoxide-signaling needs to be investigated further, we speculate that a shift towards a more oxidizing environment during S- and G2-phase could facilitate one- and two-electron reduction reactions in preparation for a successful cell division. Metal co-factors in cell cycle regulatory kinases and phosphatases could be the site for one-electron reductions, while cysteine residues in proteins could be the site for two-electron reductions (13, 16, 28, 32, 33). The two electron reduction reactions of CDC25C has been shown to regulate its phosphatase activity (34). In fact, a previous study used bioinformatics to predict that the thiol-disulfide exchange reactions of cell cycle regulatory proteins could be maximal in G2-cells compared to G1-cells (10). Thus, periodic changes in ROS generated from metabolic processes could coordinate with the redox-sensitive cell cycle pathways facilitating progression through the cell cycle.
Mitochondria are the major source of cellular ROS generation. Results presented in Figures 1 and 2 suggest that MnSOD activity could regulate GCR during proliferation. A direct correlation was observed between GCR and the percentage of S-phase cells in MnSOD (+/+) MEFs (Figure 1C). Such a correlation was absent in MnSOD (−/−) MEFs (Figure 1E). Overexpression of MnSOD in MnSOD (−/−) MEFs resulted in a decrease in GCR and cell proliferation (Figure 2). Results presented in Figure 1D and 1F as well as our earlier published results (16) showed that MnSOD (−/−) MEFs failed to exit the proliferative cycle while MnSOD (+/+) cells exited from the proliferative cycle and entered into quiescence. The rate of GCR decreased as MnSOD (+/+) MEFs entered quiescence. It is interesting to note that the inability of MnSOD (−/−) MEFs to exit the proliferative cycle was associated with a relatively constant GCR. These results further support our hypothesis that MnSOD regulates a metabolic switch during the cell cycle.
Cancer cells, in general, exhibit a significant decrease in MnSOD activity, and overexpression of MnSOD delays cancer cell proliferation (35–39). It is worth noting that overexpression of MnSOD in rapidly proliferating MB231 cells also showed a decrease in GCR and an inhibition in cellular proliferation (Figure 2B). MnSOD activity in MB231 cells is 17 U/mg and cell population doubling of 23 h (40). MCF10A is a non-malignant human mammary epithelial cell line with high MnSOD activity (36 U/mg) and a cell population doubling time of 31 h (40). The difference in cell population doubling time correlated with 24% S-phase in MCF10A and 43% S-phase in MB231 cells (41). It is interesting to note that the GCR in MCF10A cells was 33 pg cell−1 h−1 (23) compared to 65 pg cell−1 h−1 in MB231 cells (Figure 2B). Taken together, these results clearly showed an inverse correlation of MnSOD activity with GCR, and with the percentage of S-phase cells. There is a direct correlation between MnSOD activity and cell doubling time, higher activity was associated with an increase in cell doubling time, while lower MnSOD activity decreases cell doubling time. These previously published results combined with the results presented here further support the hypothesis that MnSOD regulates a “metabolic switch” coordinating GCR with cell cycle progression.
MnSOD activity exhibited a significant change during the cell cycle (Figure 4). MnSOD activity in S-phase fibroblasts and MB231 cancer cells was significantly lower than cells in G1-phase (Figure 4A and 4C). Following cell division, MnSOD activity increased again in G1-phase of the daughter generation. Overexpression of MnSOD in quiescent and proliferating fibroblasts also showed growth-state related variations in MnSOD activity (Figure 4E and 4F), indicating a post-translational modification of MnSOD regulating its activity. Results from mass spectrometry analysis revealed a complex pattern of lysine and arginine methylation of MnSOD in quiescent and proliferating fibroblasts (Figure 5A and Supplemental Figure 5). Consistent with the mass spectrometry analysis, methylation of MnSOD is also evident from the results obtained from the immunoprecipitation-immunoblotting assay (Figure 5B and 5C). Results from site-directed mutagenesis demonstrate that K89A mutation significantly decreased MnSOD activity (Figure 5D), suggesting that K89-methylation status influences MnSOD activity. Computational-based molecular modeling simulations based on the mass spectrometry data indicate that lysine and arginine methylation of MnSOD during quiescence would increase the accessibility of the active site and the surface area of positive electrostatic potential around and within the active site (Figure 6; Supplemental Figure 7). These changes could increase the accessibility of superoxide, a negatively charged substrate, to the enzyme active site more during quiescence compared to the proliferative state.
In summary, our results support the hypothesis that MnSOD regulates a “metabolic switch” during the cell cycle. Loss of this MnSOD-dependent metabolic control could lead to aberrant proliferation. We propose MnSOD as a new molecular player that contributes to the “Warburg effect.”
Supplementary Material
Acknowledgments
Acknowledgements and Grant support
We thank Jaimee Eckers, Adam Nicholson, Monali Goswami, and Wusheng Xiao for technical assistance, Dr. Sarita G. Menon for cell synchronization protocol, Dr. Ming Zhou for assistance with mass spectrometry, and the staff at the Flow Cytometry and Radiation and Free Radical Research Core Facilities. Funding from NIH CA111365 and NIEHS P42 ES 013661 supported this work. The mass spectrometry efforts of this project have been funded in whole or in part with NCI Contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States Government.
Footnotes
Key finding: Studies of cells deficient in MnSOD, a mitochondrial enzyme that controls cellular redox flux show that MnSOD regulates glucose consumption during transit through the cell cycle, implying a role in the Warburg effect.
References
- 1.Grana X, Reddy EP. Cell cycle control in mammalian cells: role of cyclins, cyclin dependent kinases (CDKs), growth suppressor genes and cyclin-dependent kinase inhibitors (CKIs) Oncogene. 1995;11:211–219. [PubMed] [Google Scholar]
- 2.Sherr CJ. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 3.Kondoh H, Lleonart ME, Nakashima Y, Yokode M, Tanaka M, Bernard D, et al. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 2007;9:293–299. doi: 10.1089/ars.2006.1467. [DOI] [PubMed] [Google Scholar]
- 4.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vazquez A, Liu J, Zhou Y, Oltvai ZN. Catabolic efficiency of aerobic glycolysis: the Warburg effect revisited. BMC Syst Biol. 2010;4:58. doi: 10.1186/1752-0509-4-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warburg O, Posener K, Negelein E. Ueber den Stoffwechsel der Tumoren. Biochemische Zeitschrift. 1924;152:319–344. [Google Scholar]
- 7.Zu XL, Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem Biophys Res Commun. 2004;313:459–465. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 8.Ho HY, Cheng ML, Lu FJ, Chou YH, Stern A, Liang CM, et al. Enhanced oxidative stress and accelerated cellular senescence in glucose-6-phosphate dehydrogenase (G6PD)-deficient human fibroblasts. Free Radic Biol Med. 2000;29:156–169. doi: 10.1016/s0891-5849(00)00331-2. [DOI] [PubMed] [Google Scholar]
- 9.Tian WN, Braunstein LD, Apse K, Pang J, Rose M, Tian X, et al. Importance of glucose-6-phosphate dehydrogenase activity in cell death. Am J Physiol. 1999;276:C1121–C1131. doi: 10.1152/ajpcell.1999.276.5.C1121. [DOI] [PubMed] [Google Scholar]
- 10.Conour JE, Graham WV, Gaskins HR. A combined in vitro/bioinformatic investigation of redox regulatory mechanisms governing cell cycle progression. Physiol Genomics. 2004;18:196–205. doi: 10.1152/physiolgenomics.00058.2004. [DOI] [PubMed] [Google Scholar]
- 11.Goswami PC, Sheren J, Albee LD, Parsian A, Sim JE, Ridnour LA, et al. Cell cycle-coupled variation in topoisomerase IIalpha mRNA is regulated by the 3'-untranslated region. Possible role of redox-sensitive protein binding in mRNA accumulation. J Biol Chem. 2000;275:38384–38392. doi: 10.1074/jbc.M005298200. [DOI] [PubMed] [Google Scholar]
- 12.Menon SG, Sarsour EH, Spitz DR, Higashikubo R, Sturm M, Zhang H, et al. Redox regulation of the G1 to S phase transition in the mouse embryo fibroblast cell cycle. Cancer Res. 2003;63:2109–2117. [PubMed] [Google Scholar]
- 13.Menon SG, Goswami PC. A redox cycle within the cell cycle: ring in the old with the new. Oncogene. 2007;26:1101–1109. doi: 10.1038/sj.onc.1209895. [DOI] [PubMed] [Google Scholar]
- 14.Sarsour EH, Agarwal M, Pandita TK, Oberley LW, Goswami PC. Manganese superoxide dismutase protects the proliferative capacity of confluent normal human fibroblasts. J Biol Chem. 2005;280:18033–18041. doi: 10.1074/jbc.M501939200. [DOI] [PubMed] [Google Scholar]
- 15.Sarsour EH, Goswami M, Kalen AL, Goswami PC. MnSOD activity protects mitochondrial morphology of quiescent fibroblasts from age associated abnormalities. Mitochondrion. 2010;10:342–349. doi: 10.1016/j.mito.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarsour EH, Venkataraman S, Kalen AL, Oberley LW, Goswami PC. Manganese superoxide dismutase activity regulates transitions between quiescent and proliferative growth. Aging Cell. 2008;7:405–417. doi: 10.1111/j.1474-9726.2008.00384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakamaki T, Casimiro MC, Ju X, Quong AA, Katiyar S, Liu M, et al. Cyclin D1 determines mitochondrial function in vivo. Mol Cell Biol. 2006;26:5449–5469. doi: 10.1128/MCB.02074-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci U S A. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhuri L, Sarsour EH, Kalen AL, Aykin-Burns N, Spitz DR, Goswami PC. Polychlorinated biphenyl induced ROS signaling delays the entry of quiescent human breast epithelial cells into the proliferative cycle. Free Radic Biol Med. 2010;49:40–49. doi: 10.1016/j.freeradbiomed.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalen AL, Sarsour EH, Venkataraman S, Goswami PC. Mn-superoxide dismutase overexpression enhances G2 accumulation and radioresistance in human oral squamous carcinoma cells. Antioxid Redox Signal. 2006;8:1273–1281. doi: 10.1089/ars.2006.8.1273. [DOI] [PubMed] [Google Scholar]
- 21.Spitz DR, Oberley LW. An assay for superoxide dismutase activity in mammalian tissue homogenates. Anal Biochem. 1989;179:8–18. doi: 10.1016/0003-2697(89)90192-9. [DOI] [PubMed] [Google Scholar]
- 22.Sarsour EH, Kumar MG, Kalen AL, Goswami M, Buettner GR, Goswami PC. MnSOD activity regulates hydroxytyrosol-induced extension of chronological lifespan. Age (Dordr) 2011 doi: 10.1007/s11357-011-9223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkatesha VA, Kalen AL, Sarsour EH, Goswami PC. PCB-153 exposure coordinates cell cycle progression and cellular metabolism in human mammary epithelial cells. Toxicol Lett. 2010;196:110–116. doi: 10.1016/j.toxlet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner BA, Venkataraman S, Buettner GR. The rate of oxygen utilization by cells. Free Radical Biology and Medicine. doi: 10.1016/j.freeradbiomed.2011.05.024. In Press, Corrected Proof. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramilo CA, Leveque V, Guan Y, Lepock JR, Tainer JA, Nick HS, et al. Interrupting the hydrogen bond network at the active site of human manganese superoxide dismutase. J Biol Chem. 1999;274:27711–27716. doi: 10.1074/jbc.274.39.27711. [DOI] [PubMed] [Google Scholar]
- 26.Powell MJD, et al. Restart procedures for the conjugate gradient method. Mathematical Programming. 1977;12:241–254. [Google Scholar]
- 27.Zhao H, Joseph J, Fales HM, Sokoloski EA, Levine RL, Vasquez-Vivar J, et al. Detection and characterization of the product of hydroethidine and intracellular superoxide by HPLC and limitations of fluorescence. Proc Natl Acad Sci U S A. 2005;102:5727–5732. doi: 10.1073/pnas.0501719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarsour EH, Kumar MG, Chaudhuri L, Kalen AL, Goswami PC. Redox control of the cell cycle in health and disease. Antioxid Redox Signal. 2009;11:2985–3011. doi: 10.1089/ars.2009.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almeida A, Bolanos JP, Moncada S. E3 ubiquitin ligase APC/C-Cdh1 accounts for the Warburg effect by linking glycolysis to cell proliferation. Proc Natl Acad Sci U S A. 2010;107:738–741. doi: 10.1073/pnas.0913668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colombo SL, Palacios-Callender M, Frakich N, De Leon J, Schmitt CA, Boorn L, et al. Anaphase-promoting complex/cyclosome-Cdh1 coordinates glycolysis and glutaminolysis with transition to S phase in human T lymphocytes. Proc Natl Acad Sci U S A. 2010;107:18868–18873. doi: 10.1073/pnas.1012362107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 32.Buettner GR. Superoxide dismutase in redox biology: the roles of superoxide and hydrogen peroxide. Anticancer Agents Med Chem. 2011;11:341–346. doi: 10.2174/187152011795677544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buettner GR, Ng CF, Wang M, Rodgers VG, Schafer FQ. A new paradigm: manganese superoxide dismutase influences the production of H2O2 in cells and thereby their biological state. Free Radic Biol Med. 2006;41:1338–1350. doi: 10.1016/j.freeradbiomed.2006.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savitsky PA, Finkel T. Redox regulation of Cdc25C. J Biol Chem. 2002;277:20535–20540. doi: 10.1074/jbc.M201589200. [DOI] [PubMed] [Google Scholar]
- 35.Li N, Oberley TD, Oberley LW, Zhong W. Inhibition of cell growth in NIH/3T3 fibroblasts by overexpression of manganese superoxide dismutase: mechanistic studies. J Cell Physiol. 1998;175:359–369. doi: 10.1002/(SICI)1097-4652(199806)175:3<359::AID-JCP14>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 36.Ough M, Lewis A, Zhang Y, Hinkhouse MM, Ritchie JM, Oberley LW, et al. Inhibition of cell growth by overexpression of manganese superoxide dismutase (MnSOD) in human pancreatic carcinoma. Free Radic Res. 2004;38:1223–1233. doi: 10.1080/10715760400017376. [DOI] [PubMed] [Google Scholar]
- 37.Venkataraman S, Jiang X, Weydert C, Zhang Y, Zhang HJ, Goswami PC, et al. Manganese superoxide dismutase overexpression inhibits the growth of androgen-independent prostate cancer cells. Oncogene. 2005;24:77–89. doi: 10.1038/sj.onc.1208145. [DOI] [PubMed] [Google Scholar]
- 38.Wang M, Kirk JS, Venkataraman S, Domann FE, Zhang HJ, Schafer FQ, et al. Manganese superoxide dismutase suppresses hypoxic induction of hypoxia-inducible factor-1alpha and vascular endothelial growth factor. Oncogene. 2005;24:8154–8166. doi: 10.1038/sj.onc.1208986. [DOI] [PubMed] [Google Scholar]
- 39.Weydert CJ, Zhang Y, Sun W, Waugh TA, Teoh ML, Andringa KK, et al. Increased oxidative stress created by adenoviral MnSOD or CuZnSOD plus BCNU (1,3-bis(2-chloroethyl)-1-nitrosourea) inhibits breast cancer cell growth. Free Radic Biol Med. 2008;44:856–867. doi: 10.1016/j.freeradbiomed.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weydert CJ, Waugh TA, Ritchie JM, Iyer KS, Smith JL, Li L, et al. Overexpression of manganese or copper-zinc superoxide dismutase inhibits breast cancer growth. Free Radic Biol Med. 2006;41:226–237. doi: 10.1016/j.freeradbiomed.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 41.Menon SG, Coleman MC, Walsh SA, Spitz DR, Goswami PC. Differential susceptibility of nonmalignant human breast epithelial cells and breast cancer cells to thiol antioxidant-induced G(1)-delay. Antioxid Redox Signal. 2005;7:711–718. doi: 10.1089/ars.2005.7.711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.