Abstract
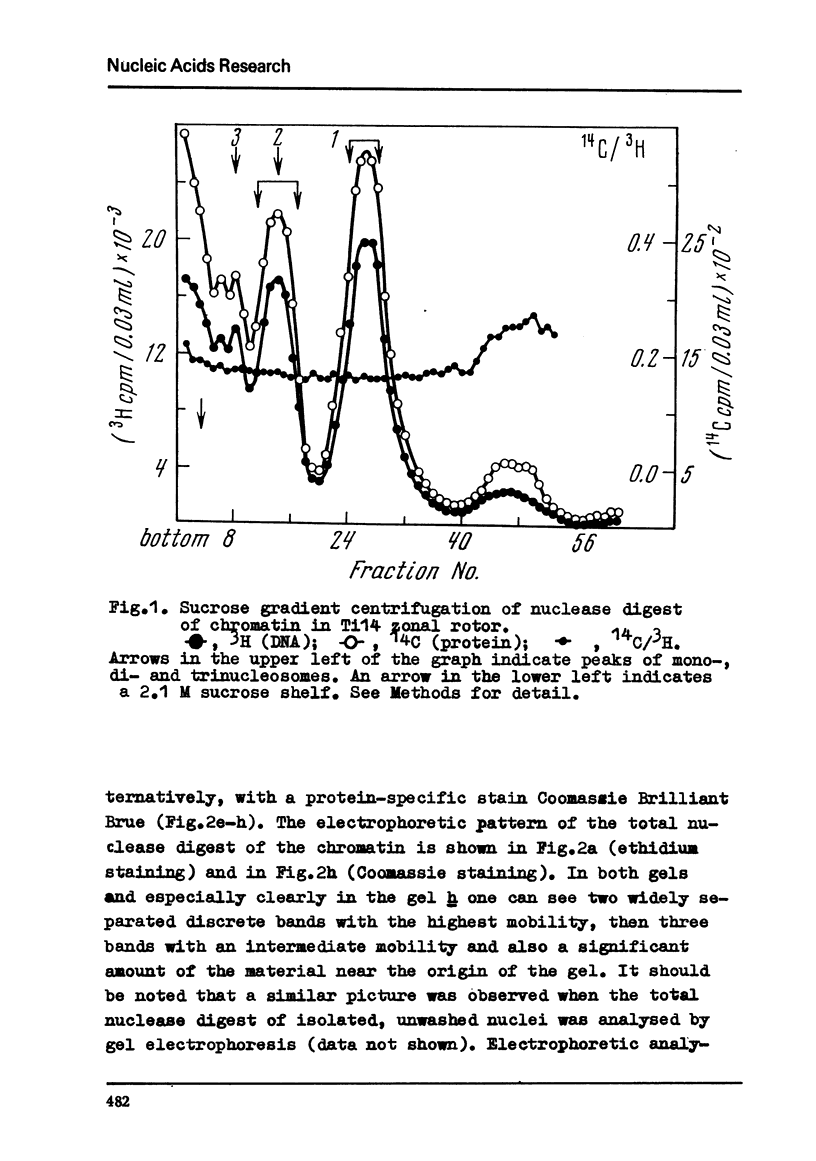
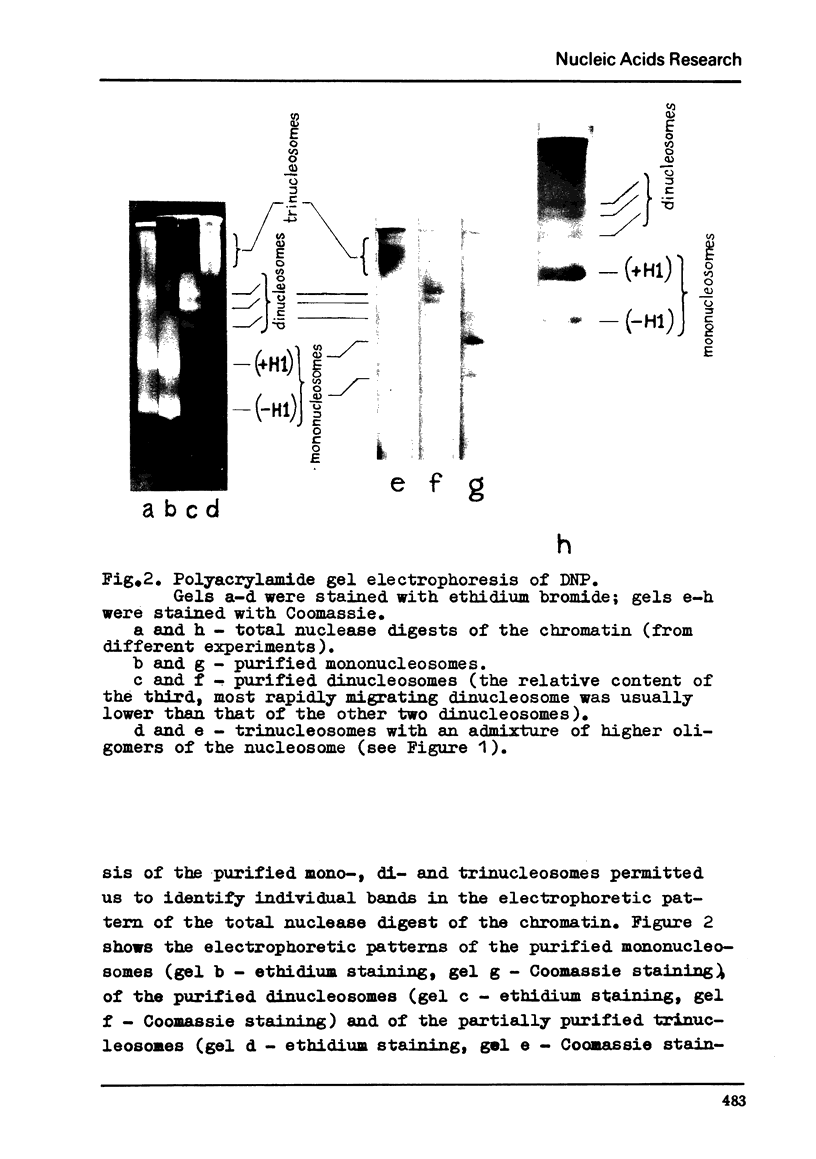
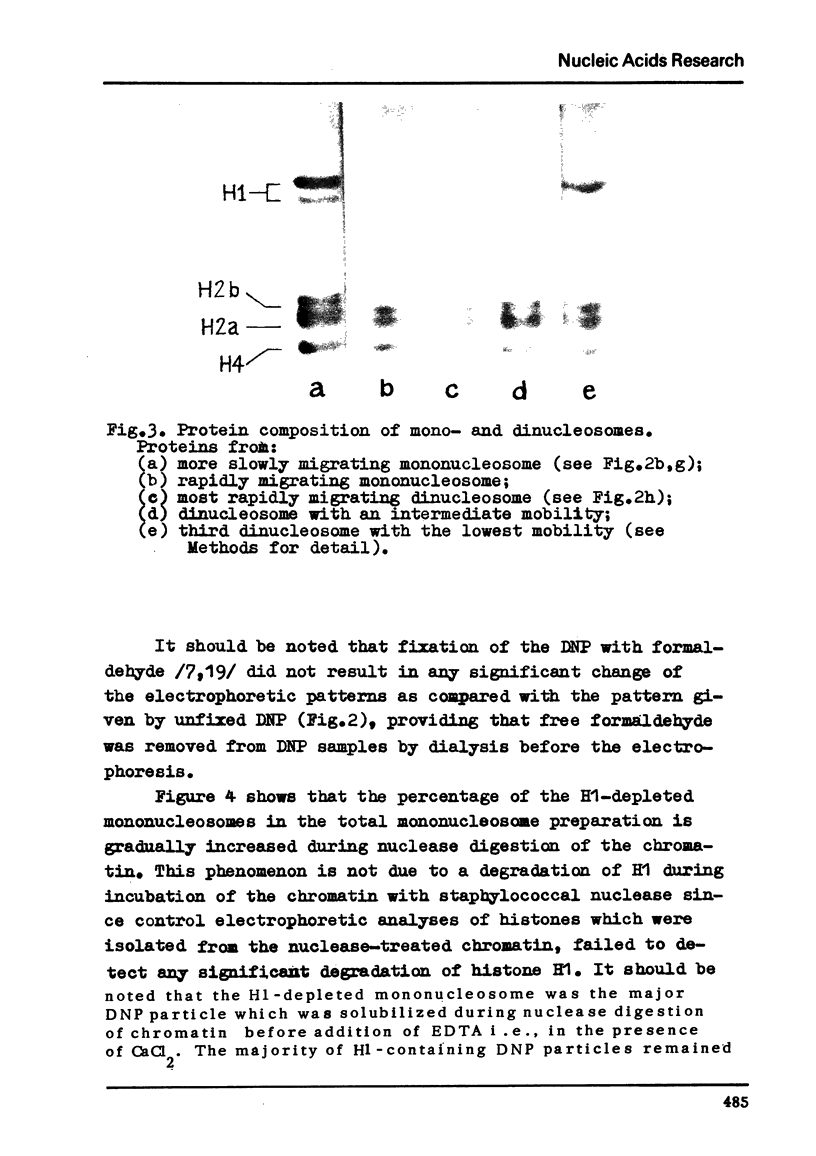
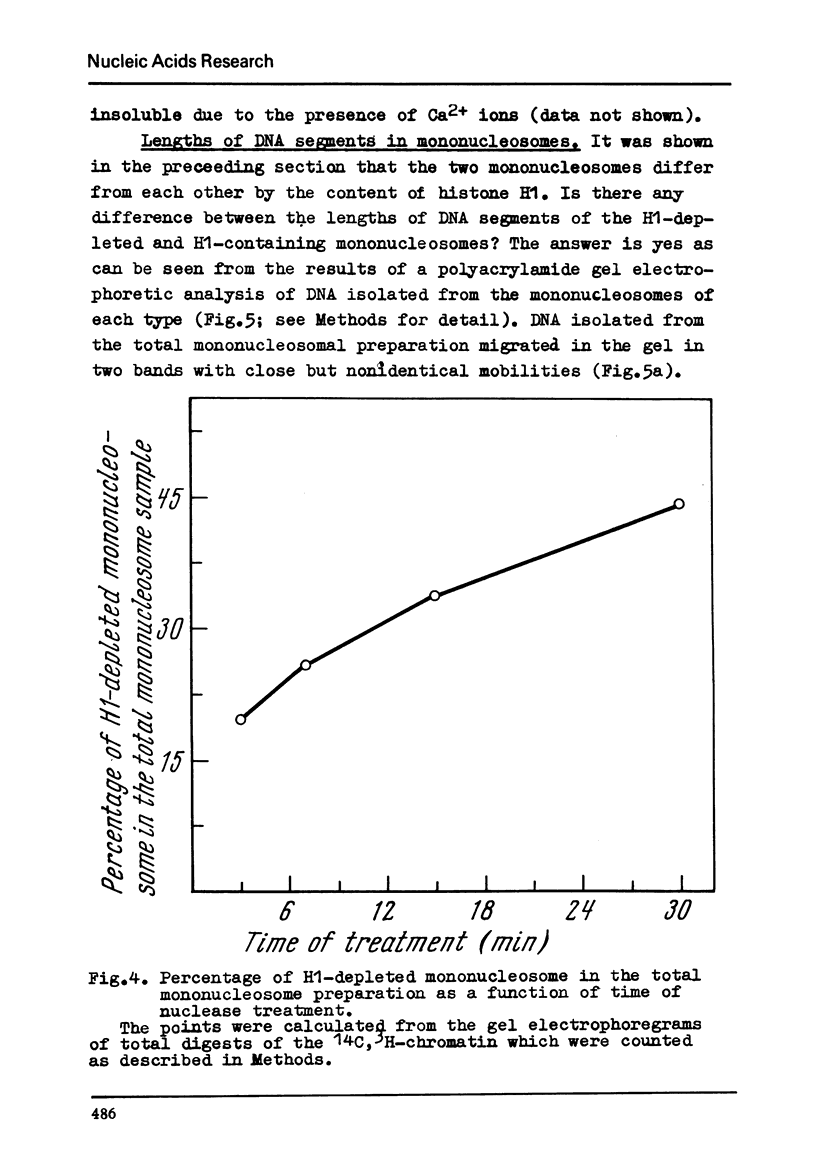
Chromatin subunits ("nucleosomes") which were purified by sucrose gradient centrifugation of a staphylococcal nuclease digest of chromatin have been studied. We found that such a preparation contains nucleosomes of two discrete types which can be separated from each other by polyacrylamide gel electrophoresis. Nucleosome of the first type contains all five histones and a DNA segment of approximately 200 base pairs long, whereas nucleosome of the second type lacks histone H1 and its DNA segment is approximately 170 base pairs long, i.e., about 30 base pairs shorter than the DNA segment of the nucleosome of the first type. Purified dimer of the nucleosome also can be fractionated by gel electrophoresis into three discrete bands which correspond to dinucleosomes containing two molecules of histone H1, one and no H1. These and related findings strongly suggest that the H1 molecule is bound to a short (approximately 30 base pairs) terminal stretch of the nucleosomal DNA segment which can be removed by nuclease (possibly in the form of H1-DNA complex) without any significant disturbance of main structural features of the nucleosome.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R., Melchior W., Jr, Sollner-Webb B., Felsenfeld G. Specific sites of interaction between histones and DNA in chromatin. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4101–4105. doi: 10.1073/pnas.71.10.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakayev V. V., Melnickov A. A., Osicka V. D., Varshausky A. J. Studies on chromatin. II. Isolation and characterization of chromatin subunits. Nucleic Acids Res. 1975 Aug;2(8):1401–1419. doi: 10.1093/nar/2.8.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin J. P., Boseley P. G., Bradbury E. M., Ibel K. The subunit structure of the eukaryotic chromosome. Nature. 1975 Jan 24;253(5489):245–249. doi: 10.1038/253245a0. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Pollard H. B. The presence of F3-F2a1 dimers and F1 oligomers in chromatin. Biochem Biophys Res Commun. 1975 May 5;64(1):282–288. doi: 10.1016/0006-291x(75)90250-8. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Noll M., Kornberg R. D. Electron microscopy of defined lengths of chromatin. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3320–3322. doi: 10.1073/pnas.72.9.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Langmore J. P., Wooley J. C. Chromatin architecture: investigation of a subunit of chromatin by dark field electron microscopy. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2691–2695. doi: 10.1073/pnas.72.7.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Polisky B., McCarthy B. Location of histones on simian virus 40 DNA. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2895–2899. doi: 10.1073/pnas.72.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rill R. L., Oosterhof D. K., Hozier J. C., Nelson D. A. Heterogeneity of chromatin fragments produced by micrococcal nuclease action. Nucleic Acids Res. 1975 Sep;2(9):1525–1538. doi: 10.1093/nar/2.9.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasrabuddhe C. G., Van Holde K. E. The effect of trypsin on nuclease-resistant chromatin fragments. J Biol Chem. 1974 Jan 10;249(1):152–156. [PubMed] [Google Scholar]
- Schuerch A. R., Mitchell W. R., Joklik W. K. Isolation of intact individual species of single- and double-stranded RNA after fractionation by polyacrylamide gel electrophoresis. Anal Biochem. 1975 May 12;65(1-2):331–345. doi: 10.1016/0003-2697(75)90517-5. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. Cleavable cross-links in the analysis of histone-histone associations. FEBS Lett. 1975 Oct 15;58(1):353–358. doi: 10.1016/0014-5793(75)80296-1. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Ilyin Y. V., Georgiev G. P. Very long stretches of free DNA in chromatin. Nature. 1974 Aug 16;250(467):602–606. doi: 10.1038/250602a0. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Release of discrete subunits after nuclease and trypsin digestion of chromatin. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1212–1216. doi: 10.1073/pnas.72.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]






