Abstract
Hepatocellular carcinoma (HCC) represents the most common primary malignancy of the liver with a worldwide increasing incidence. Although the risk factors for HCC are well characterized, the molecular mechanisms responsible for malignant transformation of hepatocytes are not well understood. In this study, a case–control study including 291 HCC patients and 294 healthy controls was conducted to investigate the association between HCC susceptibility and with a 4-bp insertion/deletion polymorphism (rs66465034) in the proximal promoter of CD3G. Logistic regression analysis showed that the heterozygote and the homozygote 4-bp ins/ins confer a significantly increased risk of HCC after controlling for other covariates (adjusted odds ratio [OR]=1.51, 95% confidence interval [C.I.] 1.01–2.27, p=0.040; OR=1.71, 95% C.I. 1.07–2.89, p=0.025, respectively). Carriage of the 4-bp insertion allele was associated with a greatly increased risk of developing the disease (OR=1.30, 95% C.I. 1.02–1.64, p=0.027). Moreover, hepatitis B virus (HBV) stratification analysis showed that the differences between cases and controls were more obvious in HBV-positive than in the HBV-negative population, suggesting a possible role of this polymorphism in the immune regulation during HBV infection. Further, luciferase-based transient transfection assays revealed that rs66465034 can affect promoter activity of CD3G, indicating its possible functional significance. Our data suggested that common genetic polymorphisms in CD3G may influence HCC risk in Chinese population. Considering the relative small sample size, replication in other populations with larger sample size and further functional analysis are required for fully understanding the roles of CD3G polymorphisms in predisposition for HCC.
Not only are viral infections associated with hepatocellular carcinoma, host gene polymorphisms may increase susceptibility. In this study, one of the signaling chains of the T cell receptor, the CD3γ, was shown to be a risk factor in Asians.
Introduction
Hepatocellular carcinoma (hcc) represents the most common primary malignancy of the liver with a worldwide increasing incidence (Rahbari et al., 2011). It ranks third in annual cancer mortality rates and has the highest annual fatality ratio of any tumor. Carcinogenesis of HCC is a multistep and complex process, and it is widely recognized that multiple risk factors, such as hepatitis B virus (HBV) or hepatitis C virus (HCV), alcohol abuse, and exposure to dietary aflatoxin, contribute to hepatocarcinogenesis. Chronic HBV infection is by far the most important risk factor for HCC in humans, accounting for 55% of cases worldwide and 80% or more of those in China and sub-Saharan Africa, the highest incidence regions of HCC (Kew, 2010). Although the risk factors for HCC are well characterized, the molecular mechanisms responsible for malignant transformation of hepatocytes are not well understood. Epidemiological studies have provided evidences that genetic factor is one of important variants for mediating an individual's susceptibility to HCC (Mínguez et al., 2009). Recently, significant progress has been made in our understanding of the genetic predisposition to HCC, thanks to the implementation of genome-wide association studies (GWASs) that are currently routinely being used to identify common polymorphisms that underlie disease susceptibility in the large population (Zhang et al., 2010). However, the genetic basis of susceptibility to HCC is still poorly understood and early detection of HCC is seldom available because of the lack of reliable biological markers.
One recent GWAS has identified genetic variations in the “antigen presentation and processing” pathway as being highly significantly associated with HCC, which suggests that polymorphisms affecting the immune response and differences in T-cell receptor (TCR) processing are involved in HCC susceptibility (Clifford et al., 2010). CD3G gene encodes CD3-gamma polypeptide, which together with CD3-epsilon, -delta, and -zeta, and the TCR alpha/beta and gamma/delta heterodimers, forms the TCR-CD3 complex. This complex plays an important role in coupling antigen recognition to several intracellular signal-transduction pathways. The CD3 gamma chain has long been considered to mediate targeting to lysosomes for degradation (Dietrich et al., 1994; Dietrich and Geisler, 1998). Defects in this gene are associated with T cell immunodeficiency (Guy and Vignali, 2009). Further, it has been proposed as one of the important candidate genes for cancer development due to its pivotal roles in TCR signaling pathway (Landi et al., 2008). The “GGCT” 4-bp insertion/deletion polymorphism (rs66465034) is located in the proximal promoter of CD3G, where is the potential source of polymorphism affecting gene expression. The aim of present study was to investigate whether rs66465034 is associated with the risk of HCC in a Chinese population and to assess the possible functional significance of the polymorphism.
Materials and Methods
Study populations
The case–control study was performed on genomic DNA extracted from peripheral blood of newly diagnosed incident HCC cases together with controls matched for sex and age after obtaining informed consent. All subjects recruited were unrelated ethnic Han Chinese. The case series comprised of 291 HCC patients diagnosed, hospitalized, and treated in the affiliated hospitals of Soochow University from 2005 to 2008. A total of 294 controls were cancer-free individuals selected from a community nutritional survey that was conducted in the same regions during the same period as recruitment of cancer patients. The diagnosis of the cases, the inclusion and exclusion criteria for the cases and controls, and the definition of smokers and drinkers were described in detail previously (Gao et al., 2009; Chen et al., 2010; He et al., 2010). Briefly, the diagnosis of these patients was confirmed by a pathological examination combined with positive imaging (magnetic resonance imaging and/or computerized tomography). Patients were excluded who were suffering from (1) primary or secondary biliary cirrhosis or Budd-Chiari syndrome, (2) autoimmune hepatitis or toxic hepatitis, (3) recurrence of HCC, (4) other tumors except HCC, and (5) liver disease due to parasitosis, diabetes, fatty liver, metabolism disorders, and severe cardiovascular diseases. Tumor stages were determined according to a modified American Joint Committee on Cancer and international union against cancer standard. The subjects who smoked more than two cigarettes per day for more than 1 year were classified as smokers. Others were defined as non-smokers. Subjects were considered as alcohol drinkers, if they drank at least once per week. All participants were negative for antibodies to HCV, hepatitis D virus, or HIV. The design of the study was approved by the Ethical Committee of Soochow University.
DNA extraction and genotyping
A Chelex method was used for extracting genomic DNA of blood samples (Walsh et al., 1991). DNA fragments containing the polymorphism were amplified with the forward primer 5′-TGGGTTCTTGCCTTCTCTCAA-3′ and reverse primer 5′-CATGTCAGTCTCTGTCCTCCG-3′. Polymerase chain reaction (PCR) was performed in a total volume of 20 μL, including 2.0 μL 10×PCR buffer, 1.5 mM MgCl2, 0.25 mM dNTPs, 0.5 mM each primer, 50 ng of genomic DNA, and 1.0 U of Taq DNA polymerase. The PCR conditions were 94°C for 5 min, followed by 35 cycles of 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C, with a final elongation at 72°C for 5 min. The PCR products were analyzed by 7% non-denaturing polyacrylamide gel electrophoresis (PAGE) and visualized by silver staining (Allen et al., 1989). The allelic discrimination of rs66465034 was determined by the numbers and the positions of the band on the gels. The 4-bp deletion allele yielded a 132-bp band and the insertion allele yielded a 136-bp band. To validate the genotyping method, we analyzed 20 randomly selected DNA samples by both direct sequencing and PCR method; the concurrence rate of these two methods was 100%, suggesting that the PCR method is reliable. Genotyping was performed without knowledge of the case or control status. A 10% random sample was tested in duplicate by different researchers, and the reproducibility was 100%.
Construction of reporter plasmids and luciferase assays
The promoter region (from −467 to +32 bp relative to the translation start site) containing either insertion or deletion allele of rs66465034 was amplified with forward primer 5′-ACAAGATCTCCTGAATGAAGGCCTGGACTGAGGTGG-3′ and reverse primer 5′-CTAAAGCTTGTCAGTCTCTGTCCTCCGGCAAAAGCG-3′ from two homozygous human genomic DNA samples. The PCR products were separated in agarose gel and extracted, purified, and cloned with TA cloning kit (Promega). Finally, the above-prepared fragments were subcloned into pGL3-basic vector using restriction enzymes BglII and HindIII. The resulting constructs were verified by sequencing.
HepG2 and sk-Hep-1 cells were maintained in Dulbecco's modified Eagle's medium with high glucose (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Gibco) and 50 μg/mL streptomycin (Gibco) at 37°C in an incubator supplemented with 5% CO2. Cells were seeded at 1×105 cells per well in 24-well plates (BD Biosciences). Sixteen hours after the plating, cells were transfected by lipofectamin 2000 according to manufacturer's suggestion. In each well, 500 ng of constructed pGL3-basic vector and 50 ng of pRL-TK vector (Promega) were co-transfected into cells. The un-constructed pGL3-basic vector was added as negative control. After transfection for 24 h, cells were harvested by the addition of 100 μL passive lysis buffer. Firefly luciferase activities in cell lysate were measured with the Dual Luciferase assay system (Promega) in TD-20/20 luminometer (Turner Biosystems) and were normalized with the Renilla luciferase activities. Six replicates for each group and the experiments were repeated at least three times.
Statistical analysis
Hardy–Weinberg equilibrium was assessed using a goodness-of-fit χ2 test for biallelic markers. The adjusted odds ratios (ORs) with their 95% confidence intervals (C.I.s) of the association between polymorphism and HCC risk were estimated by multiple logistic regression models after controlling for sex, age, smoking status, drinking status, tumor stage, and HBV infection. In all cases, homozygosis for the most common allele (i.e., del/del) was used as the reference category. As HBV infection was one of the major risk factors for HCC, a stratified analysis by HBV infection status was performed using binary logistic regression model. Student's t test was used to examine the differences in luciferase reporter gene expression. The statistical analyses were implemented in Statistic Analysis System software (version 8.0; SAS Institute). p<0.05 was used as the criterion of statistical significance. The statistical power was calculated by PS software (http://biostat.mc.vanderbilt.edu/twiki/bin/view/Main/PowerSampleSize) and Epi Info software (http://openepi.com).
Results
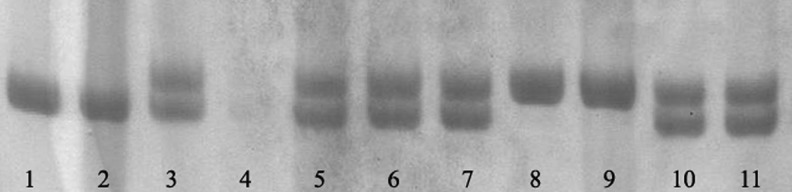
The statistical analysis of demographic characteristics of the 291 HCC patients and 294 controls was summarized in Table 1. There were no statistically significant differences between cases and controls in terms of the frequency distribution of sex, age, smoking, and drinking status. As expected, HBV infection was a significant risk factor for HCC. About 74.2% of the cases were HBsAg positive, which was significantly higher than that of the controls (11.6%, p<0.0001). PAGE analysis of the polymorphism was shown in Figure 1. Genotype distributions had no deviation from Hardy–Weinberg equilibrium in both case and control groups (p=0.088 and p=0.591, respectively). We use a C.I. at 95% to obtain a power of 0.65 by Epi Info software and an OR of 1.6 with α set at 0.05 to obtain a power of 0.66 by PS software.
Table 1.
Demographic Characteristics Among Hepatocellular Carcinoma Cases and Controls
| |
Cases |
Controls |
|
||
|---|---|---|---|---|---|
| Characteristics | N=291 | Frequencies (%) | N=294 | Frequencies (%) | p-Value |
| Age (mean±SD) | 50.9±8.7 | 51.1±9.2 | 0.89a | ||
| Gender | |||||
| Male | 190 | 65.3 | 197 | 67.0 | 0.66b |
| Female | 101 | 34.7 | 97 | 33.0 | |
| Smoking status | |||||
| Smokers | 92 | 31.6 | 91 | 31.0 | 0.86b |
| Nonsmokers | 199 | 68.4 | 203 | 69.0 | |
| Drinking status | |||||
| Drinker | 123 | 42.3 | 120 | 40.8 | 0.72b |
| Nondrinker | 168 | 57.7 | 174 | 59.2 | |
| Tumor stages | |||||
| Ia+Ib | 202 | 69.4 | |||
| IIa+IIb | 56 | 19.2 | |||
| IIIa+IIIb | 33 | 11.3 | |||
| HBsAg, N (%) | |||||
| Positive | 216 | 74.2 | 34 | 11.6 | <0.0001b |
| Negative | 75 | 25.8 | 260 | 88.4 | |
Cases indicate patients with HCC and controls are non-cancerous patients.
Two-sided two-sample t-test between cases and controls.
χ2 test for differences between cases and controls.
SD, standard deviation; HCC, hepatocellular carcinoma.
FIG. 1.
Polyacrylamide gel electrophoresis analysis of 4-bp insertion/deletion CD3G polymorphism. The deletion allele corresponds to the 132-bp fragment, and the insertion allele corresponds to the 136-bp fragment. Lanes 1, 2: homozygous (deletion/deletion); lanes 3, 5–7, 10, 11: heterozygous (insertion/deletion); lanes 8, 9: homozygous (insertion/insertion); lane 4, negative control.
Our results showed that rs66465034 was significantly associated with HCC susceptibility, at both the allele and genotype levels (Table 2). In the co-dominant model, compared with the reference (del/del), we found that the heterozygote and homozygote ins/ins of rs66465034 were associated with a significantly increased risk of HCC after controlling for other covariates, such as age, sex, drinking status, smoking status, and HBV infection (adjusted OR=1.51, 95% C.I. 1.01–2.27, p=0.040; OR=1.71, 95% C.I. 1.07–2.89, p=0.025, respectively). In the dominant model (ins/ins+ins/del vs. del/del), significant associations were also observed between cases and controls (adjusted OR=1.56, 95% C.I. 1.07–2.27, p=0.018). Meanwhile, frequencies of 4-bp deletion or insertion allele were significantly different between HCC and control groups. In the additive model, each additional copy of the 4-bp insertion allele was associated with a 30% increased risk of developing the disease (OR=1.30, 95% C.I. 1.02–1.64, p=0.027). Further, based on HBV stratification analysis, the overall trend is that the differences between cases and controls were more obvious in HBV-positive than in the HBV-negative population (Table 3).
Table 2.
Genotypic and Allelic Frequencies of rs66465034 in Cases and Controls, and Risk of Hepatocellular Carcinoma
| Genotype/allele | Cases (n) | % | Controls (n) | % | ORa(95% CI) | p-Value |
|---|---|---|---|---|---|---|
| del/del | 66 | 22.7 | 92 | 31.0 | 1.00 (Reference) | — |
| ins/del | 160 | 55.0 | 149 | 50.7 | 1.51 (1.01–2.27) | 0.040 |
| ins/ins | 65 | 22.3 | 53 | 18.0 | 1.71 (1.07–2.89) | 0.025 |
| ptrend | 0.022 | |||||
| del | 292 | 50.2 | 333 | 56.6 | 1.00 (Reference) | — |
| ins | 290 | 49.8 | 255 | 43.4 | 1.30 (1.02–1.64) | 0.027 |
Adjusted for sex, age, smoking status, drinking status, HBV infection, and tumor stage.
OR, odds ratio; CI, confidence interval; HBV, hepatitis B virus.
Table 3.
Logistic Regression Analyses for the Association Between rs66465034 and Risk of Hepatocellular Carcinoma in Hepatitis B Virus Positive and Negative Groups
| |
HBV positive |
HBV negative |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Cases | % | Controls | % | ORa(95% C.I.) | Cases | % | Controls | % | ORa(95% CI) |
| del/del | 40 | 18.5 | 14 | 41.2 | 1.00 (Reference) | 17 | 22.7 | 81 | 31.2 | 1.00 (Reference) |
| ins/del | 111 | 51.4 | 13 | 38.2 | 2.98 (1.21–7.46) | 41 | 54.7 | 132 | 50.8 | 1.49 (0.78–2.95) |
| ins/ins | 65 | 30.1 | 7 | 20.6 | 3.25 (1.11–9.84) | 17 | 22.7 | 47 | 18.1 | 1.74 (0.79–3.97) |
| ptrend | p=0.014 | p=0.148 | ||||||||
Adjusted for sex, age, smoking status, drinking status, and tumor stage.
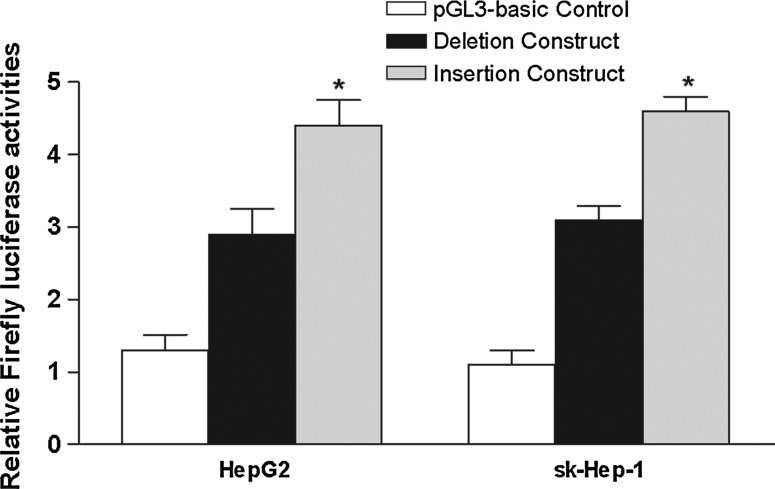
To further investigate whether rs66465034 would likely affect the level of transcriptional activity of CD3G, two luciferase reporter gene constructs were generated by PCR, and they were used to transiently transfect HCC cell lines. As shown in Figure 2, we found that CD3G promoter containing insertion allele drove a 1.48–1.52-fold increased reporter expression compared with the deletion allele containing counterpart in HepG2 and sk-Hep-1 cells.
FIG. 2.
Luciferase expression of two constructs in HepG2 and sk-Hep-1 cells co-transfected with pRL-TK to standardize transfection efficiency. *p<0.01 compared with deletion construct.
Discussion
To the best of our knowledge, this is the first molecular epidemiological study to investigate the associations of CD3G polymorphism with risk of HCC in Chinese population. The genotyping of 291 HCC patients and 294 healthy control individuals showed significant associations with the 4-bp insertion/deletion variant within proximal promoter of CD3G with HCC susceptibility. Moreover, using in vitro assays, we found that the insertion variant allele significantly increased the transcription activity of the CD3G gene compared with the deletion allele.
Promoter sequences are potential sources of polymorphism affecting gene expression. In the current study, the luciferase-based transient transfection system is used to examine the effect of this polymorphism on the promoter activity. Significant difference is observed between two constructs concerning firefly luciferase activities. Therefore, it is possible that increased spacing by a 4-bp insertion may create flexibility in the DNA chain, facilitating the bindings of transcription factors, thus increasing the CD3G promoter activity.
The immune system plays a pivotal role in the development of HCC as chronic inflammation is one of the most important causes of HCC (Moradpour and Blum, 2005). Chronic hepatitis is characterized by an inefficient T cell response unable to completely clear HBV or HCV from the liver, which consequently sustains continuous cycles of low-level cell destruction (Guidotti and Chisari, 2006.). A three-step process of “hepatitis (liver inflammation)–liver fibrosis/cirrhosis–HCC” is believed to be involved in hepatocarcinogenesis although how fibrosis promotes HCC remains unexplained (Elsharkawy and Mann, 2007). It has been suggested that host genetics factors are associated with response to hepatitis B vaccine (Davila et al., 2010). Of particular interest, the association between rs66465034 and HCC susceptibility was preferentially observed for HBV-positive patients in the present study. During HBV infection, T cells require an antigen receptor-driven signal to become activated, proliferate, and exert their effector function. These events are mediated by the mature TCR complex in which the CD3 complex plays a key role in transmitting signals after TCR engagement (Wange and Samelson, 1996). It has been demonstrated that T cell responses were reduced or abrogated in T cells derived from CD3-gamma null-mutant mice, probably because of decreased expression levels of the mature TCR complex lacking CD3-gamma (Haks et al., 2001). Moreover, CD3 proteins are dispensable for NK cell development (Wang et al., 1999), which has been suggested to be involved in the early, non-specific immune response to clear HBV virus (Li et al., 2011). Therefore, it is possible that polymorphisms within CD3G may influence its expression levels which in turn disturb CD3 complex assembling.
On the other hand, host immunity can either protect or promote tumor growth by the predominance and activation of certain subsets of immune cells (Fatourou and Koskinas, 2009). It has been suggested that antigens, such as alpha-fetoprotein (AFP) and glypican 3, which are highly expressed in HCC, are potential targets for T-cell responses (Capurro et al., 2003; Butterfield et al., 2007). Several studies have come to the conclusion that cytotoxic T-cell infiltration of the tumors is indicative of a better survival, whereas the predominance of suppressor cells is associated with a worse outcome and lower survival rates (Mizukoshi et al., 2006; Gao et al., 2007). In line of this, aberrant CD3-gamma expression potentially due to polymorphisms residing in the promoter region may contribute to host immunity against HCC.
Taken together, our data suggest that common genetic polymorphisms in CD3G may influence HCC risk in Chinese population. However, the presence of this HCC-associated polymorphism does not immediately imply that this polymorphism is causative since there are such possibilities that another genetic variation linked to rs66465034 is the true causal mutation driving the association. Finally, we have to acknowledge that the statistical power for the current study is not completely adequate because of relative small sample size, especially for stratification analysis, which would prevent us to effectively detect the association between HCC incidence and CD3G indel polymorphism. Replications in other populations with larger sample size and further functional analysis are required for fully understanding the roles of CD3G polymorphisms in predisposition for HCC.
Acknowledgments
This study is supported by grants from National Natural Science Foundation of China (No. 81171893 and No. 30800621), China Postdoctoral Science Foundation (No. 20080431121 and No. 200902530), and Undergraduates Innovating Experimentation Project of Soochow University (No. 11cxxj072). The authors would like to thank Dr. Haiyan Liu for her helpful discussions.
Disclosure Statement
No competing financial interests exist.
References
- Allen R.C. Graves G. Budowle B. Polymerase chain reaction amplification products separated on rehydratable polyacrylamide gels and stained with silver. Biotechniques. 1989;7:736–744. [PubMed] [Google Scholar]
- Butterfield L.H. Ribas A. Potter D.M. Economou J.S. Spontaneous and vaccine induced AFP-specific T cell phenotypes in subjects with AFP-positive hepatocellular cancer. Cancer Immunol Immunother. 2007;56:1931–1943. doi: 10.1007/s00262-007-0337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurro M. Wanless I.R. Sherman M. Deboer G. Shi W. Miyoshi E. Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125:89–97. doi: 10.1016/s0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- Chen S. He Y. Ding J. Jiang Y. Jia S. Xia W. Zhao J. Lu M. Gu Z. Gao Y. An insertion/deletion polymorphism in the 3′ untranslated region of beta-transducin repeat-containing protein (betaTrCP) is associated with susceptibility for hepatocellular carcinoma in Chinese. Biochem Biophys Res Commun. 2010;391:552–556. doi: 10.1016/j.bbrc.2009.11.096. [DOI] [PubMed] [Google Scholar]
- Clifford R.J. Zhang J. Meerzaman D.M. Lyu M.S. Hu Y. Cultraro C.M. Finney R.P. Kelley J.M. Efroni S. Greenblum S.I. Nguyen C.V. Rowe W.L. Sharma S. Wu G. Yan C. Zhang H. Chung Y.H. Kim J.A. Park N.H. Song I.H. Buetow K.H. Genetic variations at loci involved in the immune response are risk factors for hepatocellular carcinoma. Hepatology. 2010;52:2034–2043. doi: 10.1002/hep.23943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila S. Froeling F.E. Tan A. Bonnard C. Boland G.J. Snippe H. Hibberd M.L. Seielstad M. New genetic associations detected in a host response study to hepatitis B vaccine. Genes Immun. 2010;11:232–238. doi: 10.1038/gene.2010.1. [DOI] [PubMed] [Google Scholar]
- Dietrich J. Geisler C. T cell receptor zeta allows stable expression of receptors containing the CD3gamma leucine-based receptor-sorting motif. J Biol Chem. 1998;273:26281–26284. doi: 10.1074/jbc.273.41.26281. [DOI] [PubMed] [Google Scholar]
- Dietrich J. Hou X. Wegener A.M. Geisler C. CD3 gamma contains a phosphoserine-dependent di-leucine motif involved in down-regulation of the T cell receptor. EMBO J. 1994;13:2156–2166. doi: 10.1002/j.1460-2075.1994.tb06492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsharkawy A.M. Mann D.A. Nuclear factor-kappaB and the hepatic inflammation-fibrosis-cancer axis. Hepatology. 2007;46:590–597. doi: 10.1002/hep.21802. [DOI] [PubMed] [Google Scholar]
- Fatourou E.M. Koskinas J.S. Adaptive immunity in hepatocellular carcinoma: prognostic and therapeutic implications. Expert Rev Anticancer Ther. 2009;9:1499–1510. doi: 10.1586/era.09.103. [DOI] [PubMed] [Google Scholar]
- Gao Q. Qiu S.J. Fan J. Zhou J. Wang X.Y. Xiao Y.S. Xu Y. Li Y.W. Tang Z.Y. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol. 2007;25:2586–2593. doi: 10.1200/JCO.2006.09.4565. [DOI] [PubMed] [Google Scholar]
- Gao Y. He Y. Ding J. Wu K. Hu B. Liu Y. Wu Y. Guo B. Shen Y. Landi D. Landi S. Zhou Y. Liu H. An insertion/deletion polymorphism at miRNA-122-binding site in the interleukin-1alpha 3′ untranslated region confers risk for hepatocellular carcinoma. Carcinogenesis. 2009;30:2064–2069. doi: 10.1093/carcin/bgp283. [DOI] [PubMed] [Google Scholar]
- Guidotti L.G. Chisari F.V. Immunobiology and pathogenesis of viral hepatitis. Annu Rev Pathol. 2006;1:23–61. doi: 10.1146/annurev.pathol.1.110304.100230. [DOI] [PubMed] [Google Scholar]
- Guy C.S. Vignali D.A. Organization of proximal signal initiation at the TCR:CD3 complex. Immunol Rev. 2009;232:7–21. doi: 10.1111/j.1600-065X.2009.00843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haks M.C. Cordaro T.A. van den Brakel J.H. Haanen J.B. de Vries E.F. Borst J. Krimpenfort P. Kruisbeek A.M. A redundant role of the CD3 gamma-immunoreceptor tyrosine-based activation motif in mature T cell function. J Immunol. 2001;166:2576–2588. doi: 10.4049/jimmunol.166.4.2576. [DOI] [PubMed] [Google Scholar]
- He Y. Ni J. Chen S. Jiang Y. Jia S. Gao Y. The vascular endothelial growth factor-2549 insertion/deletion polymorphism is not associated with susceptibility to hepatocellular carcinoma in Chinese. DNA Cell Biol. 2010;29:393–396. doi: 10.1089/dna.2009.1015. [DOI] [PubMed] [Google Scholar]
- Kew M.C. Epidemiology of chronic hepatitis B virus infection, hepatocellular carcinoma, and hepatitis B virus-induced hepatocellular carcinoma. Pathol Biol (Paris) 2010;58:273–277. doi: 10.1016/j.patbio.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Landi D. Gemignani F. Barale R. Landi S. A catalog of polymorphisms falling in microRNA-binding regions of cancer genes. DNA Cell Biol. 2008;27:35–43. doi: 10.1089/dna.2007.0650. [DOI] [PubMed] [Google Scholar]
- Li J. Han Y. Jin K. Wan Y. Wang S. Liu B. Liu Y. Lu S. Huang Z. Dynamic changes of cytotoxic T lymphocytes (CTLs), natural killer (NK) cells, and natural killer T (NKT) cells in patients with acute hepatitis B infection. Virol J. 2011;8:199. doi: 10.1186/1743-422X-8-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mínguez B. Tovar V. Chiang D. Villanueva A. Llovet J.M. Pathogenesis of hepatocellular carcinoma and molecular therapies. Curr Opin Gastroenterol. 2009;25:186–194. doi: 10.1097/MOG.0b013e32832962a1. [DOI] [PubMed] [Google Scholar]
- Mizukoshi E. Nakamoto Y. Marukawa Y. Arai K. Yamashita T. Tsuji H. Kuzushima K. Takiguchi M. Kaneko S. Cytotoxic T cell responses to human telomerase reverse transcriptase in patients with hepatocellular carcinoma. Hepatology. 2006;43:1284–1294. doi: 10.1002/hep.21203. [DOI] [PubMed] [Google Scholar]
- Moradpour D. Blum H.E. Pathogenesis of hepatocellular carcinoma. Eur J Gastroenterol Hepatol. 2005;17:477–483. doi: 10.1097/00042737-200505000-00002. [DOI] [PubMed] [Google Scholar]
- Rahbari N.N. Mehrabi A. Mollberg N.M. Müller S.A. Koch M. Büchler M.W. Weitz J. Hepatocellular carcinoma: current management and perspectives for the future. Ann Surg. 2011;253:453–469. doi: 10.1097/SLA.0b013e31820d944f. [DOI] [PubMed] [Google Scholar]
- Walsh P.S. Metzger D.A. Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- Wang B. Wang N. Whitehurst C.E. She J. Chen J. Terhorst C. T lymphocyte development in the absence of CD3 epsilon or CD3 gamma delta epsilon zeta. J Immunol. 1999;162:88–94. [PubMed] [Google Scholar]
- Wange R.L. Samelson L.E. Complex complexes: signaling at the TCR. Immunity. 1996;5:197–205. doi: 10.1016/s1074-7613(00)80315-5. [DOI] [PubMed] [Google Scholar]
- Zhang H. Zhai Y. Hu Z. Wu C. Qian J. Jia W. Ma F. Huang W. Yu L. Yue W. Wang Z. Li P. Zhang Y. Liang R. Wei Z. Cui Y. Xie W. Cai M. Yu X. Yuan Y. Xia X. Zhang X. Yang H. Qiu W. Yang J. Gong F. Chen M. Shen H. Lin D. Zeng Y.X. He F. Zhou G. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet. 2010;42:755–758. doi: 10.1038/ng.638. [DOI] [PubMed] [Google Scholar]