Abstract
RANTES (CCL5) is a chemokine implicated in many human diseases. We previously showed that the transcription factor Kruppel-like factor 13 (KLF13) controls the late (3-5 days after activation) expression of RANTES in T lymphocytes and that KLF13 itself is translationally regulated through the 5′-untranslated region of its mRNA. Here, we show that KLF13 levels are further regulated by ubiquitination and degradation. KLF13 protein is undetectable in resting human T lymphocytes, but treatment with either proteosomal or lysosomal inhibitors increases KLF13 protein levels. Glycogen synthase kinase 3β (GSK3β)–mediated phosphorylation of KLF13 triggers the ubiquitination of KLF13 by the E3 ligase Fbw7γ, resulting in KLF13 protein degradation. Knockdown of either Fbw7γ or GSK3β by small interfering RNA increases KLF13 expression in resting human T lymphocytes. In contrast, in murine T lymphocytes, KLF13 protein is abundant because of the absence of Fbw7γ. Treatment of unactivated human lymphocytes with lysosomal inhibitors stabilizes KLF13 protein, resulting in an increase of RANTES mRNA and protein. Taken together, these studies found that tightly regulated control of both synthesis and degradation allows rapid changes in the level of KLF13 in human T lymphocytes.
Introduction
RANTES (CCL5) was first identified as a gene expressed “late” after T-cell activation,1 but soon it was recognized as a CC chemokine, attracting T cells, dendritic cells, eosinophils, natural killer (NK) cells, mast cells, and basophils to sites of inflammation and infection. RANTES is highly expressed in patients with HIV who do not progress to full-blown AIDS, and its receptor, CCR5, is a coreceptor for HIV entry into T lymphocytes.2 Although RANTES was originally thought to be a T cell–specific protein, it was soon realized that RANTES is expressed rapidly after activation in a variety of cell types, including platelets, macrophages, eosinophils, fibroblasts, and endothelial, epithelial, and endometrial cells.3–7 Although expression in non-T cells occurs in minutes to hours and only requires rel proteins p50 and p65 for induction, regulation in T lymphocytes is complex and expression occurs 3-5 days after activation.8
The late expression of RANTES in T lymphocytes is transcriptionally regulated by Kruppel-like factor 13 (KLF13). We identified KLF13 as RANTES factor of late activated T lymphocytes 1 (RFLAT-1) by expression cloning in a search for the transactivator regulating the “late” expression of RANTES in T lymphocytes.9 Our studies further found that KLF13 serves as a lynchpin, recruiting a large trans-acting complex (enhancesome), including the MAPK Nemo-like kinase, p300/CREB binding protein (CBP), p300/CBP-associated factor, and the ATPase Brahma-related gene 1 to the RANTES promoter in T lymphocytes.10 Klf13−/− mice show decreased RANTES expression in vivo as well as enlarged thymi and spleens, resulting from decreased apoptosis of lymphocytes because of increased expression of Bcl-XL.11 Thus, KLF13 is a positive regulator of RANTES expression in T cells and a negative regulator of Bcl-XL, suggesting a dual role for KLF13 in inflammation in both inducing leukocyte recruitment and dampening the immune response via apoptosis.
Although KLF13 protein is not expressed in T lymphocytes until 3-5 days after activation, mimicking RANTES expression, KLF13 mRNA is present at all time points, including in resting T lymphocytes.12 We previously showed that KLF13 is translationally regulated through its 5′-untranslated region (UTR) in a cell type–specific manner in T lymphocytes.12 Overexpression of the eukaryotic translation initiation factor 4E (eIF4E) increased KLF13 protein expression, whereas inhibition of Mnk1, which phosphorylates eIF4E, reduced KLF13 production, indicating cap-dependent translational regulation. KLF13 translation is further regulated by miR-125a, through its 3′-UTR in T lymphocytes.13 Additional post-translational modifications that control KLF13 protein expression have not been reported.
A number of transcription factors involved in control of cell growth are unstable proteins targeted for degradation by the ubiquitin-proteasome system. The SKP1/CUL1/F-box (SCF) complex is among the best-studied multiple subunit Really Interesting New Gene (RING)–finger ligases that regulate cell cycle control proteins in many types of cells. The ubiquitin ligase F-box and WD repeat domain containing 7 (Fbw7) is a subunit of the SCF complex that regulates the cell cycle in T lymphocytes by degrading cell cycle regulators such as cyclin E and Notch.14 We show here that KLF13 is ubiquitinated by Fbw7 and degraded in resting human T lymphocytes. In addition to translational regulation in activated T lymphocytes, a ubiquitin-dependent degradation pathway provides T lymphocytes an additional mechanism for precise and rapid control of KLF13 expression in response to environmental stimuli. Because KLF13 has dual roles as an activator and a repressor in immune responses, steady state levels of KLF13 are closely regulated by several mechanisms, including (1) cap-dependent translation, (2) microRNA, and (3) ubiquitination-mediated degradation.
Methods
T-cell isolation and culture
Human and mouse tissues and blood were obtained with protocols approved by the National Institutes of Health/National Cancer Institute Institutional Review Board and Animal Care and Use Committee. Human PBMCs were prepared as described.12 PBMCs were cultured in RPMI 1640 supplemented with 10% FBS (Hyclone), 100 U/mL penicillin/streptomycin, 2mM l-glutamine, and 10mM HEPES. T cells were enriched from PBMCs or freshly isolated mouse splenocytes by negative selection with the use of a T-cell enrichment kit (StemCell Technologies) according to the manufacturer's directions. HEK293T cells were cultured in DMEM supplemented with 10% FBS and 100 U/mL penicillin/streptomycin. ED1 cells were kindly provided by Dr Ethan Dmitrovsky (Geisel School of Medicine at Dartmouth University)15 and cultured in RPMI 1640 supplemented with 10% FBS (Hyclone), 100 U/mL penicillin/streptomycin, 2mM l-glutamine, and 10mM HEPES. For inhibitor treatment, cells were incubated for 4 hours or overnight in media containing MG-132 (EMD Millipore), lactacystin (EMD Millipore), concanamycin A (CMA; Sigma-Aldrich), or glycogen synthase kinase 3 (GSK3) inhibitors (XV, VIII; EMD Millipore). T cells were activated with anti-CD3 plus anti-CD28 Ab for up to 7 days. The cells were lysed in cell lysis buffer (100mM Tris-HCl, pH 8.0, 100mM NaCl, 5mM EDTA, 5% glycerol, 0.1% NP-40, 1mM DTT, 50mM NaF, and 1mM Na3VO4) and subjected to Western blot analysis or immunoprecipitation.
Plasmids and transfection
cDNAs encoding KLF13 and T7-tagged and V5-tagged human KLF13 were subcloned into pcDNA3.1, and cDNAs encoding FLAG-tagged human KLF13 variants [wild-type (WT), S107G, S111G, S119G, S123G, S289G, S293G, S297G], FLAG-tagged Fbw7 variants (WT, ΔF, ΔF/R505A) were subcloned into pCMVtag2b. cDNA encoding V5-tagged mouse KLF13 was subcloned into pcDNA3.1. A plasmid encoding HA-ubiquitin was kindly provided by Dr Stanley Lipkowitz (National Cancer Institute, National Institutes of Health). Plasmids were transfected to HEK293T cells or ED1 cells with the use of lipofectamin 2000 (Invitrogen) according to the manufacturer's directions.
Antibodies
Anti-KLF13 antibodies for immunoprecipitation were purified from rabbit sera.9 Anti-KLF13 antibodies for immunoblotting were purchased from Santa Cruz Biotechnology. Monoclonal antibodies against T7 and HA tags and their HRP-conjugated antibodies were purchased from Roche Applied Science. Monoclonal antibody against FLAG (M2) was purchased from Sigma-Aldrich. Anti-V5 Ab and anti-GAPDH Ab were purchased from Invitrogen and Millipore, respectively. Fluorescence-labeled anti-CD3 and anti-CD56 Abs were purchased from BD Bioscience. FITC-conjugated RANTES Ab and its IgG2B for isotype control were purchased from Caltag Laboratories and used for intracellular staining. For immunoblotting, cells were lysed in lysis buffer. After centrifugation, supernatant fluids were subject to SDS-PAGE, and transferred to polyvinylidene difluoride membranes. Protein bands were detected by ECL (GE Healthcare).
Immunoprecipitation
For coimmunoprecipitation, HEK293T cells were transfected with plasmids that expressed T7-tagged KLF13 or KLF13 and FLAG-tagged Fbw7 (WT, ΔF, or ΔF/R505A). Twenty-four hours after transfection, cells were lysed with lysis buffer; immunoprecipitated with anti-T7, anti-KLF13, or anti-FLAG antibodies; and subjected to immunoblotting.
Ubiquitination assays
KLF13 ubiquitination in vivo was detected by transfecting HEK293T cells with plasmids that expressed KLF13 and/or HA-ubiquitin with or without plasmids that expressed FLAG-Fbw7. Cells were lysed and immunoprecipitated with anti-KLF13 antibodies. Ubiquitination of KLF13 was analyzed by Western blot analysis with the use of anti-HA antibodies. The in vitro ubiquitnation assay was performed with 35S-labeled in vitro–translated KLF13 (from wheat germ extract) as substrate. The reaction was performed in a volume of 10 μL that contained ubiquitylation reaction buffer (50mM Tris, pH 7.6, 5mM MgCl2, 1.0mM DTT), 2mM ATP, 1.5 ng/μL E1 (Boston Biochem), 10 ng/μL Ubc3, 10 ng/μL Ubc5, 2.5 μg/μL ubiquitin (Sigma-Aldrich), 1μM ubiquitin aldehyde, 0.15μM Okadaic Acid (Sigma-Aldrich), 100 ng of recombinant GSK3β, and SCF (Fbw7γ) purified from HEK293T cells. For ligase purification, FLAG-tagged Fbw7γ (or an empty vector) was cotransfected into HEK293T cells along with Myc-tagged Skp1, Cul1, and Rbx1. Forty-eight hours after transfection, cells were lysed, and FLAG-Fbw7γ was immunoprecipitated with the use of anti-FLAG M2 agarose beads. The beads were washed 4 times with lysis buffer, twice in ubiquitylation reaction buffer, and added to the ubiquitylation reactions. Reactions were incubated at 30°C, stopped by adding SDS loading buffer at the indicated times, resolved by SDS-PAGE, and analyzed by autoradiography.
siRNA
Fbw7 isoform-specific small interfering RNAs (siRNAs), which were previously described,14 and a universal negative control for siRNA were obtained from Integrated DNA Technologies. GSK3β-depleting SMARTpool siRNA oligonucleotides and nontargeting siRNA oligonucleotides were purchased from Dharmacon. Freshly isolated human T cells were transfected with siRNAs with the use of a 96-well shuttle Nucleofector (Lonza) according to the manufacturer's directions.
Real-time PCR
Total RNA was isolated from resting or anti-CD3/anti-CD28 antibody-activated human T cells after 1, 2, 3, and 5 days with the RNeasy kit with genomic DNA elimination step by DNase I treatment, according to the manufacturer's protocol (QIAGEN). Total RNA (1 μg) was used for reverse transcription to cDNAs with the use of reverse transcriptase from Promega. Real-time quantitative PCR was performed with SYBR Green Supermix Kit (Applied Biosystems). Relative expression of KLF13 or Fbw7 isoforms was normalized to β-glucuronidase or hypoxanthine-guanine phospho ribosyltransferase (HPRT) expression. Primers were designed as described10; hKLF13 (forward, 5′-GTTTACGGGAAATCTTCGCA-3′; reverse, 5′-GCGAACTTCTTGTTGCAGTC-3′), hFbw7 (forward, 5′-TGGGATATCAAAACAGGACAGTGT-3′; reverse, 5′-TAAACAGGTCACAGCACTCTGATG-3′), hFbw7α (forward, 5′-CGAACTCCAGTAGTATTGTGGACCT-3′; reverse, 5′-TTCTTTTCATTTTTGTTGTTTTTGTATAGA-3′), hFbw7β (forward, 5′-CAGCCGGACACACGGG-3′; reverse, 5′-GGTCCAACTTTCTTTTCATTTTGTAAA-3′), hFbw7γ (forward, 5′-AAAAGTGCAAAAGAGCCTCTACCA-3′; reverse, 5′-TGGGCAATGATGCTAATGCTAA-3′), β-glucuronidase (forward, 5′-CTCATTTGGAATTTTGCCGATT-3′; reverse, 5′-CCGAGTGAAGATCCCCTTTTTA-3′), hHPRT (forward, 5′-GACTTTGCTTTCCTTGGTCAGG-3′; reverse, 5′-AGTCTGGCTTATATCCAACACTTCG-3′), mFbw7α (forward, 5′-CTCACCAGCTCTCCTCTCCATT-3′; reverse, 5′-GCTGAACATGGTACAAGGCCA-3′), mFbw7β (forward, 5′-TTGTCAGAGACTGCCAAGCAG-3′; reverse, 5′-GACTTTGCATGGTTTCTTTCCC-3′), mFbw7γ (forward, 5′-AACCATGGCTTGGTTCCTGTTG-3′; reverse, 5′-CAGAACCATGGTCCAACTTTC-3′), mHPRT (forward, 5′-GCCTAAGATGAGCGCAAGTTG-3′; reverse, 5′-TACTAGGCAGATGGCCACAGG-3′), and hRANTES (forward, 5′-AGTCGTCTTTGTCACCCGAAA-3′; reverse, 5′-AGCTCATCTCCAAAGAGTTGATG-TAC-3′).
Immunohistochemistry
KLF13 immunohistochemistry was performed on 5-μm thick paraffin-embedded sections with the use of goat anti-KLF13 antibody (Santa Cruz Biotechnology). Antigen retrieval for deparaffinized tissue sections was performed in citrate buffer for 20 minutes with a steamer. After blocking of endogenous peroxidase, paraffin-embedded sections were submitted to immunohistochemistry with the use of the Vectastain Elite ABC system (Vector Laboratories).
In vitro kinase assay
GSK3β was purchased from New England Biolabs. The in vitro kinase reaction was performed according to the manufacturer's instructions. Briefly, 10 μg of GST-KLF13 or GST control protein was incubated with GSK3β in the presence of 5 μCi [γ-32P]ATP and 200μM cold ATP in the kinase reaction buffer for 30 minutes. The reaction was stopped by the addition of NuPAGE LDS sample buffer. The proteins were resolved by SDS-PAGE, and phosphorylation was detected by autoradiography.
Cell staining and flow cytometry
PBMCs from 4 donors were treated for 16 hours with or without CMA (1 μg/mL) and stained for surface expression of FITC-CD3 and PE-CD56 (BD Bioscience). Cells were then permeabilized with BD Cytofix/Cytoperm and stained with APC-VL1 (RANTES; eBioscience). Mean fluorescence was analyzed with FlowJo Version 7.5.3 software (TreeStar).
Results
KLF13 expression is differentially regulated in mouse and human T lymphocytes
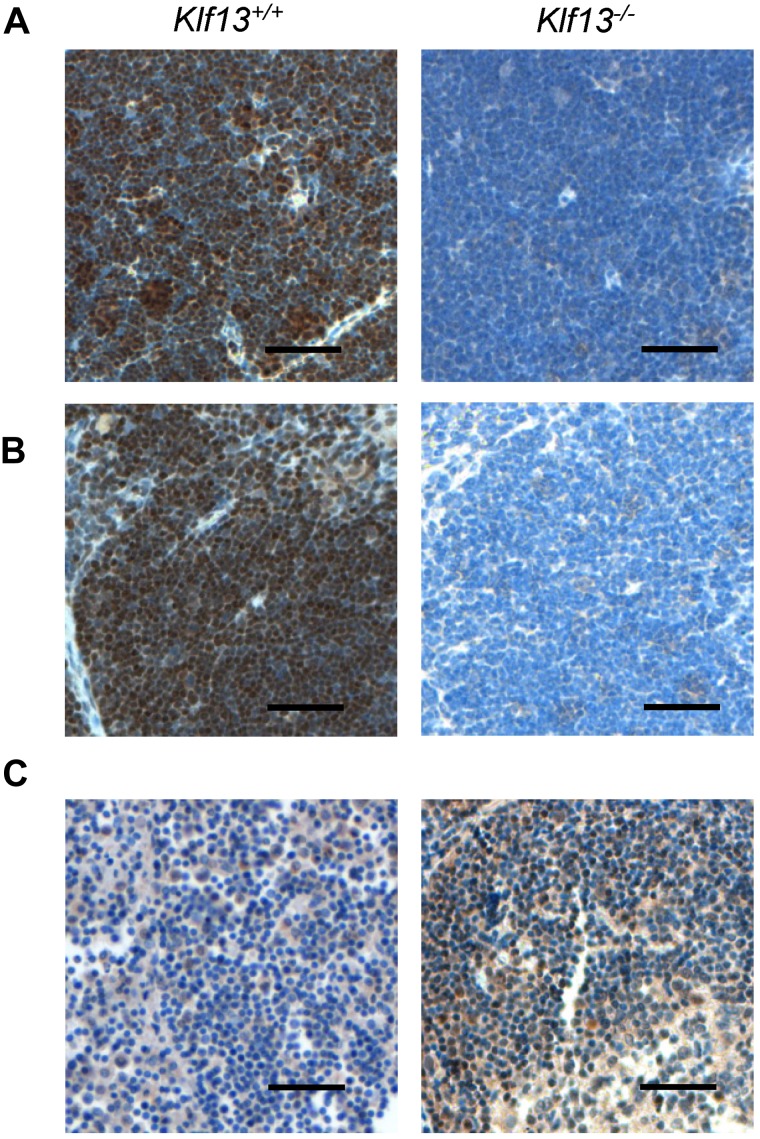
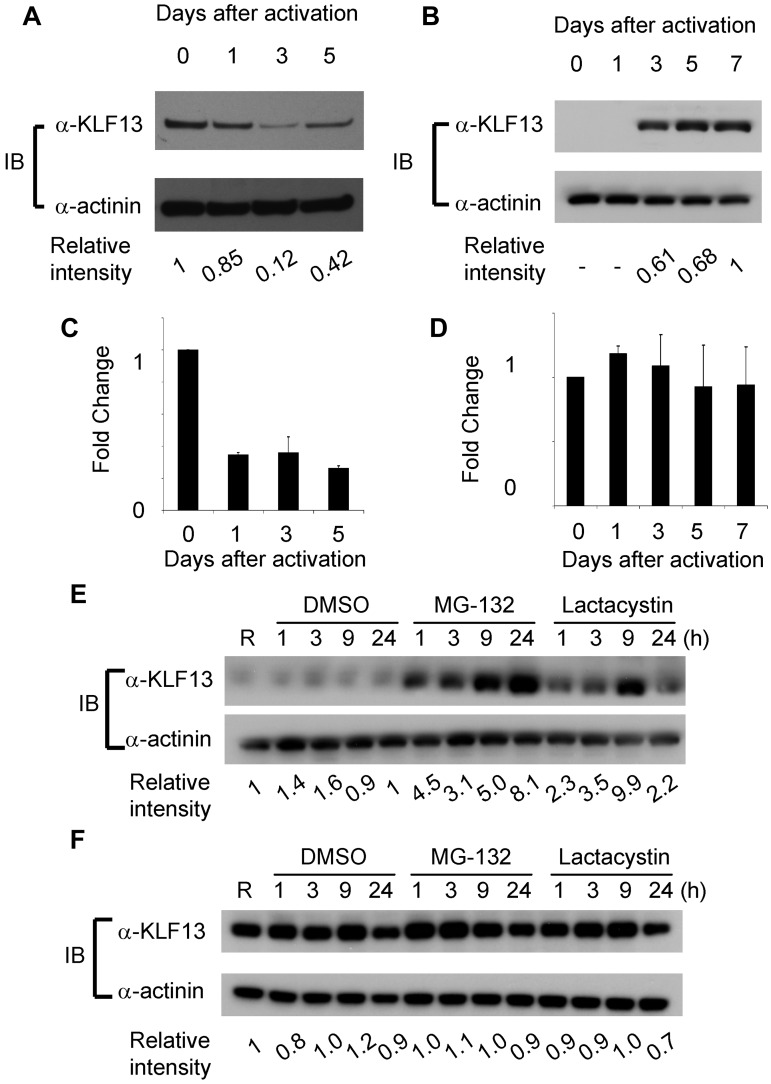
To compare KLF13 expression in the human and mouse, immunohistochemistry was performed with mouse and human tissues as well as specimens from Klf13−/− mice.11 KLF13 is highly expressed in normal mouse thymus and spleen (Figure 1A-B), whereas KLF13 is minimally detected in human lymph node (Figure 1C, left panel) but is highly expressed in tonsil (Figure 1C, right panel). Western blot analysis confirmed that KLF13 protein is highly expressed in both resting and activated mouse T lymphocytes (Figure 2A) but only in activated human T lymphocytes (Figure 2B). KLF13 mRNA levels decreased to some extent after activation of mouse T cells (Figure 2C) but remained constant in human T cells (Figure 2D). We previously showed that this distinct kinetic of human KLF13 expression is partially because of the eIF4E-dependent translational regulation.12 To examine whether proteasome-mediated protein degradation also is involved in regulating levels of KLF13, mouse and human T cells were treated with MG-132 or lactacystin. These inhibitors increased KLF13 protein in human T cells (Figure 2E) but did not affect KLF13 levels in murine T cells (Figure 2F).
Figure 1.
Immunohistochemistry of KLF13 in mouse and human tissues. (A) Murine thymus. (B) Murine spleen. (C) Human lymph node (left panel) and tonsil (right panel). Scale bar is 50 μm.
Figure 2.
Differential regulation of KLF13 protein in human and mouse T lymphocytes. (A-B) Western blot analysis of KLF13 expression in mouse (A) or human (B) T cells activated with anti-CD3 and anti-CD28 Abs. (C-D) RT-PCR of KLF13 mRNA levels in mouse (C) and human (D) T cells activated with anti-CD3 and anti-CD28 Abs. (E-F) Western blot analysis of KLF13 expression in human (E) or mouse (F) T cells treated with DMSO (vehicle control), MG-132 (5 μg/mL) or lactacystin (10μM). Relative intensity was determined by ImageJ Version 1.46 software (National Institutes of Health). Data are representative of 3-7 similar experiments. IB indicates immunoblot. Data are mean ± SD.
Fbw7 catalyzes KLF13 ubiquitination
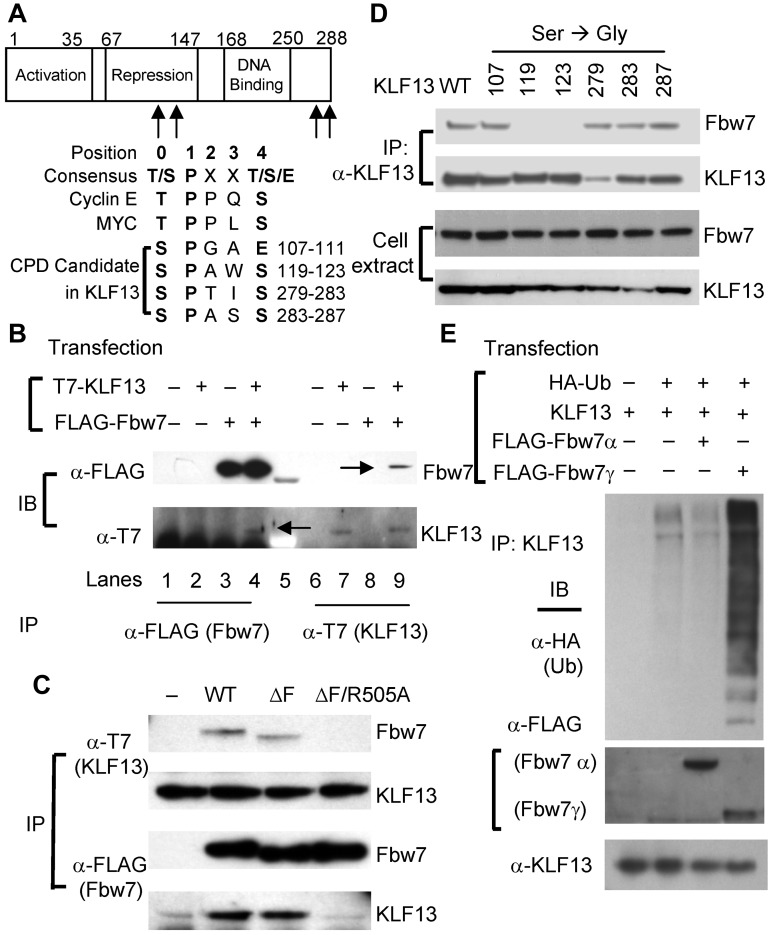
Sequence analysis showed that KLF13 contains 4 potential CDC4-phosphodegron (CPD) candidates shared by several substrates of the ubiquitin ligase Fbw7 (Figure 3A) but none of the known motifs for the ubiquitin ligases Itch, Nedd4, Cbl-b, or β-TrCP. To examine the possible interaction between KLF13 and Fbw7, coimmunoprecipitation experiments were performed in HEK293T cells. After transfecting HEK293T cells with plasmids encoding T7-tagged KLF13 and FLAG-tagged Fbw7, anti-FLAG Ab for Fbw7 or anti-T7 Ab for KLF13 was used for immunoprecipitation. KLF13 is coimmunoprecipitated with anti-Flag Ab (Fbw7), and Fbw7 coimmunoprecipitates with anti-T7 Ab (KLF13; Figure 3B), showing that KLF13 is associated with Fbw7 in transfected HEK293T cells. To further delineate this association, Fbw7 mutants were generated for coimmunoprecipitation experiments. Mutant Fbw7ΔF prevents substrate degradation, increasing the interaction between enzyme and substrate, whereas mutant Fbw7ΔF/R505A prevents the enzyme-substrate interaction.16 As expected, KLF13 is coimmunoprecipitated with Fbw7ΔF, but not with Fbw7ΔF/R505A (Figure 3C), indicating that KLF13 interacts with Fbw7 through its known substrate binding domain. Phosphorylation of serine or threonine residues in CPD domains is critical to the interaction between Fbw7 and its substrates.17 There are 6 possible phosphorylation sites in the 4 candidate CPD sites in KLF13 (Figure 3A). To identify the responsible CPD(s) in KLF13, we individually mutated these 6 serine residues to glycine in KLF13 (S107G, S119G, S123G, S279G, S283G, and S287G) and performed a coimmunoprecipitation assay in HEK293T cells. S119G and S123G mutants fail to interact with Fbw7, whereas WT and the remaining KLF13 mutants interact with Fbw7 (Figure 3D). This finding indicates that the second CPD (residues 119-123, SPAWS) is responsible for KLF13 interaction with Fbw7. There are 3 isoforms of Fbw7 (α, β, and γ) that result from alternative splicing.18 Fbw7α and Fbw7γ are mainly localized in the nucleus, whereas Fbw7β is localized in the cytoplasm.19 To examine whether Fbw7 facilitates ubiquitination of KLF13, an in vivo ubiquitination assay was performed with HEK293T cells transfected with plasmids that express KLF13, FLAG-tagged Fbw7α or Fbw7γ, and HA-tagged ubiquitin. Poly-ubiquitination was observed in cells transfected with plasmids that express KLF13 and ubiquitin. Moreover, ubiquitination of KLF13 was enhanced when the cells were cotransfected with Fbw7γ but not with Fbw7α (Figure 3E), indicating that KLF13 is a substrate of Fbw7 and its ubiquitination depends on Fbw7γ expression rather than on Fbw7α expression.
Figure 3.
Fbw7 interacts with KLF13 and catalyzes its ubiquitination. (A) CPD candidate sequences in KLF13 are compared with CPD consensus sequences and CPD sequences in cyclin E and Myc. (B) HEK293T cells were transiently transfected with plasmids that express T7-tagged KLF13 and/or FLAG-tagged Fbw7. Cell lysates were immunoprecipitated (IP) with anti-T7 Ab or anti-FLAG Ab and analyzed by Western blot analysis. Co-IP bands are marked by arrows in lanes 4 and 9. (C) HEK293T cells were transiently transfected with plasmids that express T7-tagged KLF13 and/or WT and mutated forms (ΔF and ΔF/R505A) of FLAG-tagged Fbw7. Whole-cell extracts were IP and analyzed by immunoblot (IB). (D) HEK293T cells were transiently transfected with plasmids expressing KLF13 (WT or S/G-mutated forms at indicated amino acid residue) and FLAG-tagged Fbw7. Whole-cell extracts were IP by anti-KLF13 Ab and analyzed by IB. (E) HEK293T cells were transiently transfected with plasmids that express KLF13, FLAG-tagged Fbw7 (α or γ) and HA-tagged ubiquitin (Ub). Cell lysates were IP with anti-KLF13 Ab for Western blot analysis with anti-HA Ab. Whole-cell extracts were analyzed by Western blot analysis with anti-FLAG Ab for Fbw7 or anti-KLF13 Ab.
Fbw7γ regulates KLF13 ubiquitination in human T lymphocytes
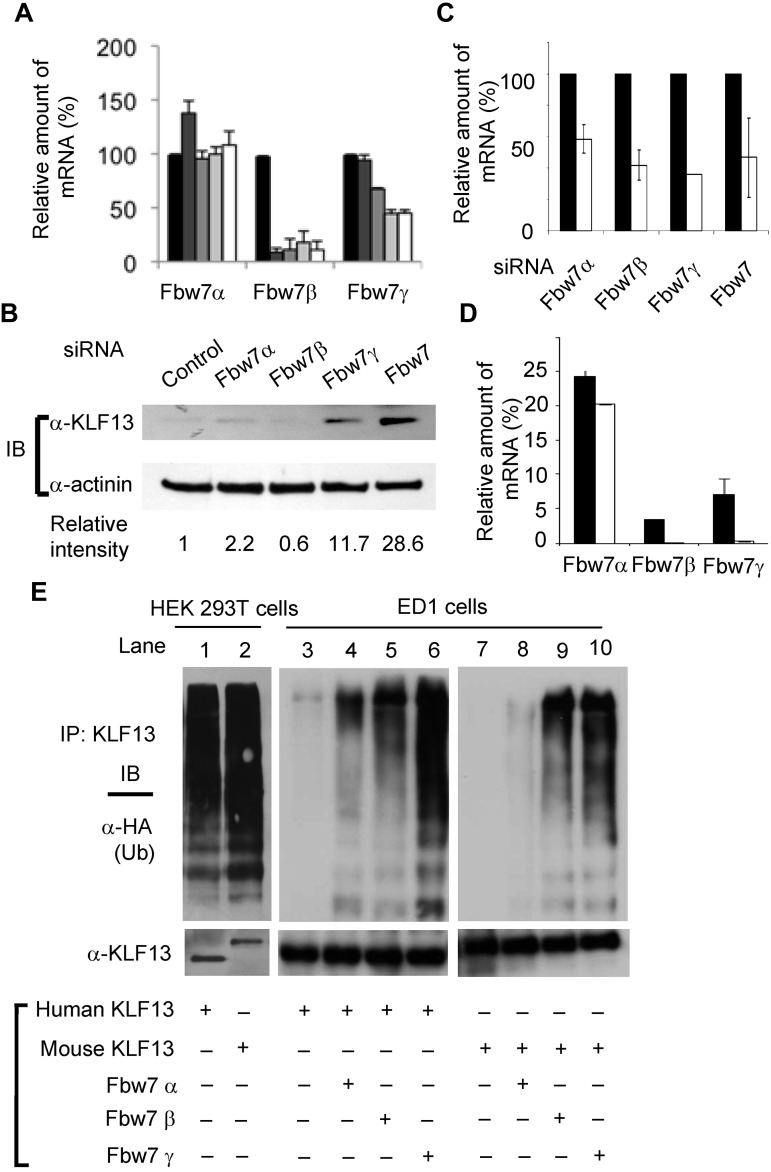
The expression kinetics of the 3 isoforms of Fbw7 was examined by RT-PCR in activated T cells. mRNA levels of Fbw7β and Fbw7γ were reduced at days 3 and 5 after T-cell activation (Figure 4A), consistent with higher KLF13 protein expression after T-cell activation. To examine the direct effect of Fbw7 on KLF13 protein, freshly isolated human T cells were transfected with siRNAs specific for each of the 3 Fbw7 isoforms, and the whole-cell lysates were then analyzed by immunoblotting. KLF13 was detected in resting T cells transfected with siRNA specific for Fbw7α (although very little), Fbw7γ, or Fbw7 but not Fbw7β (Figure 4B). The levels of knockdown achieved with different Fbw7 siRNA oligonucleotides are shown in Figure 4C. The mRNA levels of Fbw7 isoforms in both human and mouse T cells were determined by RT-PCR. Fbw7α mRNA levels are similar in human and mouse T cells when normalized with HPRT expression. In contrast, Fbw7β and Fbw7γ transcripts are hardly detectable in mouse T lymphocytes but are present in human T cells (Figure 4D). The relative expression level of Fbw7γ mRNA was < 0.3% of that of HPRT in mouse T cells but > 7% of that of HPRT in human T cells. These results indicate that KLF13 protein expression is regulated by different mechanisms in human and mouse T lymphocytes.
Figure 4.
Fbw7γ regulates KLF13 in human T cells. (A) Total RNA was prepared from human T cells activated with anti-CD3 plus anti-CD28 Abs for 0 (black bar), 1 (dark gray bar), 2 (medium gray bar), 3 (light gray bar), or 5 (open bar) days. The mRNA level of each Fbw7 isoform was determined by RT-PCR and normalized to the level in day 0 resting T cells. (B-C) Human T cells were transfected with control siRNA or siRNA for each isoform of Fbw7. After 24 hours, the cells were analyzed by immunoblot assay with the use of anti-KLF13 Ab (B) or by RT-PCR (C). (D) Relative expression levels of human and mouse Fbw7 were determined by RT-PCR and normalized to HPRT expression. (E) HEK293T cells (lanes 1-2) were transiently transfected with human KLF13 (lane 1) or mouse V5-tagged KLF13 (lane 2) and HA-tagged ubiquitin (Ub). The cell lysates were immunoprecipitated (IP) with anti-KLF13 Ab for Western blot analysis with anti-HA Ab or anti-KLF13 Ab. ED1 cells (lanes 3-10) were transiently transfected with plasmids expressing human (lanes 3-6) or mouse V5-tagged KLF13 (lanes 7-10), Flag-tagged Fbw7α (lanes 4,8), Fbw7β (lanes 5,9), Fbw7γ (lanes 6,10) and HA-tagged Ub (all lanes). The cell lysates were IP with anti-V5 Ab for Western blot analysis with anti-HA Ab or V5 Ab. Data are mean ± SD.
The differences in KLF13 levels in resting mouse and human T lymphocytes could be a function of sequence or species differences. To establish which is responsible, we performed in vivo ubiquitination assays with the use of human HEK293T cells and murine ED1 cells. Expression of Fbw7 isoforms in ED1 cells as measured by RT-PCR is similar to that in murine T cells; Fbw7α is highly expressed (relative level of mRNA = 203), whereas Fbw7β is expressed at a much lower level (relative level of mRNA = 12) and Fbw7γ is undetectable (relative level of mRNA = 1). Human HEK293T cells transfected with mouse or human KLF13 and immunoprecipitated KLF13 was then probed for ubiquitin by Western blot analysis (Figure 4E lanes 1,2). Both mouse and human KLF13 were ubiquitinated in human cells. In contrast, neither mouse nor human KLF13 was ubiquitinated after transfection into murine ED1 cells (Figure 4E lanes 3,7). Ubiquitination of both human and mouse KLF13 was greatly increased in ED1 cells cotransfected with Fbw7γ (Figure 4E lanes 6,10), somewhat with Fbw7β (Figure 4E lanes 5,9), and minimally with Fbw7α (Figure 4E lanes 4,8). This is consistent with the observation that Fbw7γ can facilitate KLF13 ubiquitination in vivo and in vitro (Figure 3) and that knockdown of Fbw7γ affects KLF13 levels in resting human T lymphocytes more than that of Fbw7α (Figure 4B). Thus, KLF13 protein expression is regulated by Fbw7γ-mediated ubiquitination and degradation in human, but not murine, T lymphocytes.
Inhibition of KLF13 degradation induces RANTES expression in resting T lymphocytes
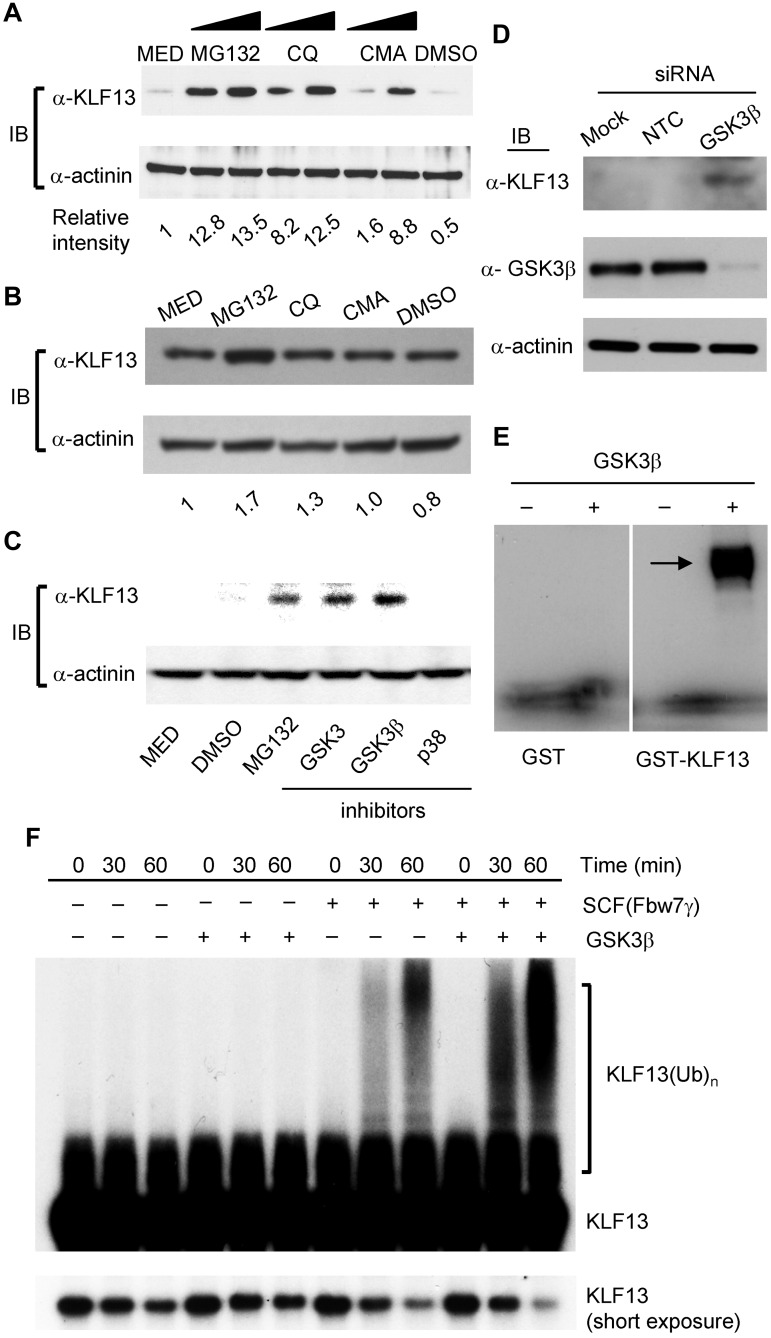
Ubiquitination targets intracellular proteins for degradation in proteasomes and lysosomes.20 To determine which of these compartments degrades KLF13, resting T cells were treated for 4 hours with increasing concentrations of the proteasome inhibitor MG-132 or with the lysosomal inhibitors chloroquine or CMA. Increasing levels of KLF13 were detected with increased concentration of either proteasome or lysosomal inhibitors (Figure 5A). However, proteasomal or lysosomal inhibitors had only a minimal effect on KLF protein levels in activated T cells (Figure 5B). GSK3β is often the priming kinase for targets of Fbw7. Therefore, we asked whether GSK3 kinase inhibitors can also stabilize KLF13 protein in human T lymphocytes. KLF13 protein was stabilized to a similar extent by MG132 or by GSK-3 or GSK-3β inhibitors. However, no stabilization was observed with a p38 inhibitor (Figure 5C). These findings suggest that GSK3β is involved in Fbw7-mediated regulation of KLF13 expression. To further investigate this, GSK3β was knocked down in resting human T cells with the use of GSK3β-specific siRNA or a nontargeting control siRNA (Figure 5D). Western blot analysis confirmed the loss of GSK3β protein and a corresponding increase in KLF13 protein. That GSK3β does in fact phosphorylate KLF13 was shown in an in vitro phosphorylation assay (Figure 5E). Recombinant GSK3β was able to phosphorylate recombinant GST-KLF13 but not recombinant GST. Finally, the role of Fbw7γ and GSK3β in ubiquitination of KLF13 was directly confirmed by an in vitro ubiquitination assay. Ubiquitination of KLF13 protein is directly catalyzed by Fbw7γ and further enhanced by GSK3β-mediated phosphorylation (Figure 5F).
Figure 5.
Inhibitors of lysosomal degradation and GSK3β increase KLF13 expression in resting human T cells. (A) Resting human T lymphocytes were treated with MG-132 (2.5 or 5 μg/mL), chloroquine (CQ; 50nM or 100nM), or CMA (0.5 or 1 μg/mL) for 4 hours, and the cell lysates were subjected to Western blot analysis. (B) Human T lymphocytes activated by anti-CD3 and CD28 Ab for 5 days were treated with MG-132 (5 μg/mL), CQ (100nM), or CMA (1 μg/mL) for 4 hours, and the cell lysates were subjected to Western blot analysis. (C) Resting T lymphocytes were treated with MG-132 (1 μg/mL), GSK3 inhibitors (1nM for GSK3 and 0.2mM for GSK3β), or p38 inhibitor (1 μg/mL) for 4 hours, and the cell lysates were subjected to Western blot analysis. (D) Resting human T cells were transfected with nontargeting control (NTC) or GSK3β-depleting siRNA. Cells were harvested after 24 hours, and cell lysates were assayed for GSK3β and KLF13 by Western blot analysis. (E) Recombinant GSK3β was incubated with recombinant GST or GST-KLF13 in the presence of [γ-32P]ATP. The protein kinase reaction products were resolved by SDS-PAGE, and phosphorylation was detected by autoradiography. (F) 35S-in vitro–translated KLF13 was incubated at 30°C in a ubiquitination reaction mix containing SCF(Fbw7γ) purified from HEK293T and/or recombinant GSK3β as indicated. Reactions were stopped by adding SDS loading buffer at the indicated times, resolved by SDS-PAGE, and analyzed by autoradiography.
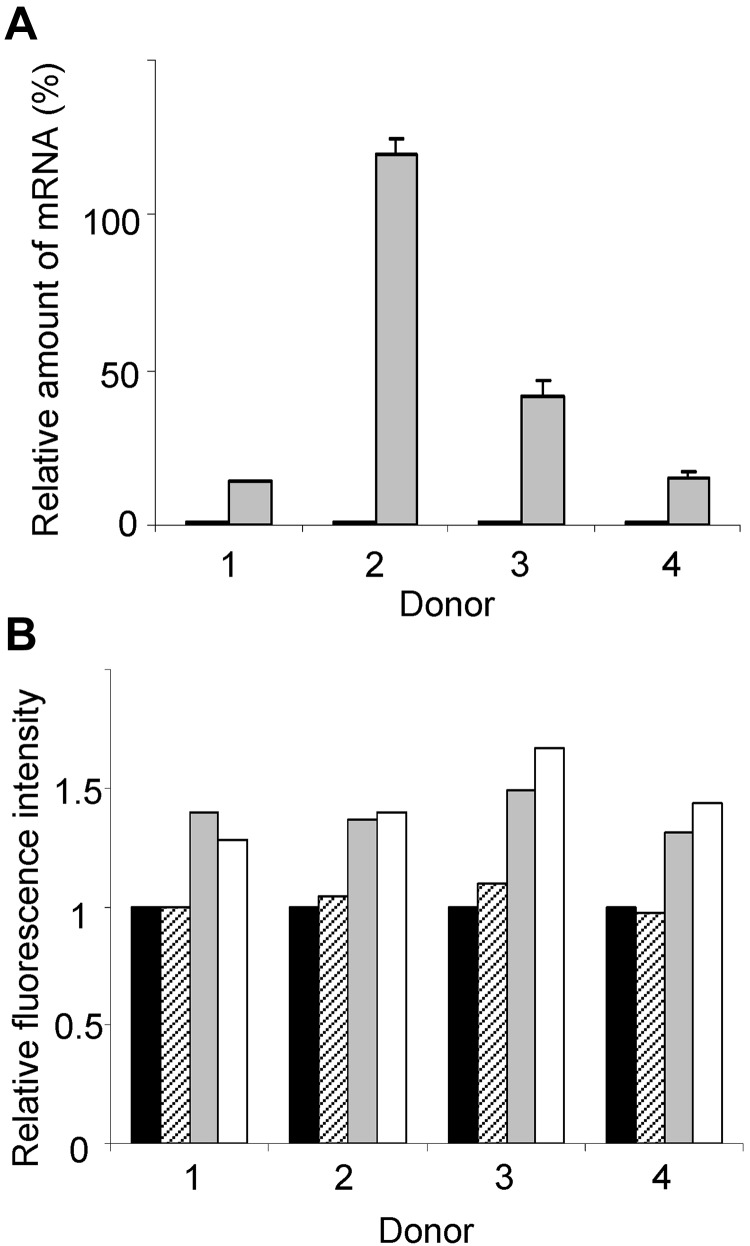
To assess the functional consequence of increased KLF13, PBMCs from 4 different donors were incubated for 16 hours with CMA, and then RANTES mRNA and protein levels were measured. RANTES mRNA levels increased significantly in PBMCs treated with CMA (Figure 6A). RANTES protein, measured by intracellular staining and FACS, increased in T cells and NK T cells treated with CMA (Figure 6B). In contrast, treatment with CMA did not alter RANTES levels in NK cells, suggesting that these cells use a different mechanism to regulate RANTES expression. We were unable to directly assess the role of the proteasome in RANTES expression because PBMCs cultured overnight with MG-132 showed markedly reduced viability.
Figure 6.
Elevated KLF13 expression correlates with increased RANTES expression in resting T lymphocytes. (A) PBMCs from 4 donors were treated for 16 hours with (gray) or without (black) CMA (1 μg/mL), and RANTES mRNA levels were determined by real-time quantitative PCR. (B) PBMCs from 4 donors were treated with CMA for 16 hours, and RANTES expression was determined by intracellular staining and flow cytometry. Mean fluorescence is expressed relative to expression in cells cultured in medium alone (black bar). RANTES expression in CD56+ NK cells (hatched bars), CD56+CD3+ NK T cells (gray bars), and CD3+ T cells (open bars) is shown. Data are mean ± SD.
Discussion
We first described KLF13 as RFLAT-1.9 Although rel proteins regulate RANTES expression in most cell types, the “late” expression in T lymphocytes is regulated by a large transactivating complex composed of MAPK Nemo-like kinase, p300/CBP, p300/CBP-associated factor, and the ATPase Brahma-related gene 1,10 with KLF13 serving as its lynchpin.21 KLF13 protein, like RANTES, is not expressed in T cells until 3-5 days after activation. Intriguingly, however, mRNA for KLF13 is expressed at the same level in both resting and activated T cells. We previously showed that this discrepancy between mRNA and protein expression is in part because of translational regulation in a T cell–specific manner.12 In this study we show that human KLF13 protein expression kinetic is different from that of mouse and that degradation of KLF13 via Fbw7-mediated ubiquitination plays a main role in human T lymphocytes.
Other members of the KLF family, including KLF1, KLF2, KLF4, KLF5, KLF6, and KLF10, are known to be ubiquitinated and degraded by a proteasome-dependent pathway,22–27 indicating that ubiquitination is a general mechanism for controlling expression of KLF proteins. Of special interest, KLF2 is ubiquitinated and degraded on T-cell activation, the inverse expression profile compared with KLF13. We have stressed the yin-yang relation of KLF13 and KLF2 expression and function in T lymphocytes.28
Conkright et al first suggested that WW domain containing E3 ubiquitin protein ligase 1 (WWP1), another ubiquitin ligase, inhibited the transcriptional activity of KLF2 and that WWP1 targets KLF2 for ubiquitin-mediated proteasomal degradation.29 Recently, KLF5 was shown to be a substrate of WWP1 and Fbw7 and that both ubiquitin ligases promote KLF5 ubiquitination and degradation.23,30 KLF10 is also ubiquitinated by Itch in T cells and regulates Foxp3 expression by a nonproteolytic pathway, implying complex regulation of KLF family proteins by ubiquitination.26 In contrast, the ubiquitin E3 ligases responsible for KLF1, KLF4, and KLF6 ubiquitination have as yet not been identified. As shown here, KLF13 is the sixth member of this family to be regulated by E3 ligase and ubiquitin-mediated degradation. Of note, other transcription factor families, including Smad, p53, RUNX and Jun, undergo similar degradation.31–34
Fbw7 is the substrate recognition component of the SCF complex that serves as an evolutionarily conserved ubiquitin ligase known to be important in T-cell development.35 All Fbw7 substrates contain a phosphodegron motif that is well conserved from yeast to human.35 There are 4 potential phosphodegron motifs for Fbw7 in KLF13. KLF13 binds to Fbw7 through its substrate binding domain with the second CPD (residues 119-123, SPAWS) and the ubiquitination of KLF13 is facilitated by Fbw7 both in vivo and in vitro (Figure 3). Fbw7 is a known tumor suppressor that degrades substrates such as cyclin, c-Myc, c-Jun, and Notch.14 Like these other Fbw7 substrates, KLF13 may have a role in tumor development because KLF13 negatively regulates Bcl-XL expression.11
There are 3 isoforms of Fbw7, each with its own promoter and with various alternative splicing of the first exon.36 The resulting differences in the amino terminal protein sequence affect tissue expression and intracellular localization of the isoforms.18,36 Thus, Fbw7α and Fbw7γ are localized to the nucleus, whereas the β isoform is confined to the cytoplasm.19 KLF13 is localized to the nucleus, implicating Fbw7α or Fbw7γ in its ubiquitination. However, the γ isoform much more efficiently catalyzes KLF13 ubiquitination than the α isoform in 293T cells. Although no Fbw7 isoform-specific antibodies are available, mRNA for the γ isoform decreases 3-5 days after T-cell activation, coinciding with the increased expression of both KLF13 and RANTES. With the use of Fbw7 siRNA, we observed that Fbw7γ siRNA caused a dramatic increase in KLF13 expression in resting T cells, whereas a minor increase in KLF13 was detected with Fbw7α siRNA. Furthermore, Fbw7γ mRNA is barely detectable in mouse T cells compared with in human T cells. Collectively, these results suggest that Fbw7γ-mediated ubiquitination negatively regulates KLF13 levels in resting human T cells.
Most Fbw7 substrates are phosphorylated by GSK3β. We show here that both GSK3β inhibitors and GSK3β-depleting siRNA stabilize KLF13 protein in resting T cells. Furthermore, GSK3β can directly phosphoylate KLF13 and enhance the ubiquitination of KLF13 in vitro. Because GSK3β activity is inhibited by the PI3 Kinase-AKT pathway on T-cell activation,37 we suggest that priming of KLF13 protein by GSK3β is an additional mechanism to decrease KLF13 expression in resting T lymphocytes.
Mouse KLF13 is > 89% homologous with human KLF13 and contains the critical phosphodegron motif [residues 125-129 (SPAWS) corresponding to residues 119-123 of human KLF13]. As expected, both human and mouse KLF13 were ubiquitinated in human HEK293 T cells. The requirement for Fbw7γ in KLF13 ubiquitination is further confirmed by the in vivo ubiquitination assay in the ED1 cell line. When human and mouse KLF13 were transfected into mouse ED1 cells that lack Fbw7γ, neither was ubiquitinated. Ubiquitination was restored in ED1 cells when either mouse or human KLF13 was cotransfected with Fbw7γ, clearly showing that KLF13 protein ubiquitination and degradation depend on Fbw7γ. We also observed that Fbw7α and Fbw7β promoted some ubiquitination of KLF13 in these transfected cells, but we believe these results are from overexpression becauseFbw7β is expressed in the cytosol and should not have access to KLF13, which is found exclusively in the nucleus. Fbw7γ has previously been reported to affect 2 other substrates: Cyclin E is first modified by Fbw7α and then sequentially ubiquitinated by Fbw7γ,38 and c-Myc expression is regulated by Fbw7γ in nucleoli.39 Although it is possible that, like cyclin E, KLF13 ubiquitination requires both the α and γ isoforms of Fbw7, the extremely high levels of Fbw7α make the γ isoform the limiting factor in the system. The expression level of Fbw7γ is much lower than Fbw7α in human T lymphocytes and is not detectable in murine T cells. Fbw7α mRNA is expressed at much higher levels than either Fbw7β or Fbw7γ in most if not all human cell lines and primary cells.36 It has been shown that Fbw7α mRNA is present in all tissues, whereas Fbw7β mRNA is detected only in brain and testis, and Fbw7γ mRNA is found only in heart and skeletal muscles in the mouse.40 KLF13 was stabilized by proteasome or lysosomal inhibitors in resting T cells but not in activated T cells, which suggests that KLF13 expression is more strongly regulated by protein degradation in resting T cells. Stabilization of KLF13 by lysosomal inhibitors in resting human T cells increases both mRNA and protein level of RANTES, which underscores the critical role of KLF13 in regulating RANTES expression.
Two recent reports highlight the in vivo significance of regulation of KLF13. Zhao et al reported that miR-125a is reduced in patients with systemic lupus erythematosus (SLE).13 KLF13 is a predicted target for miR-125a, and KLF13 expression is increased in patients with SLE, consistent with the decrease in miR-125a. The investigators overexpressed miR-125a and found a significant decrease in both KLF13 and RANTES expression. Remarkably, introduction of miR-125a into T cells from patients with SLE alleviated the elevated RANTES levels. These findings suggest that KLF13 regulates RANTES expression in vivo in a human disease. RANTES is an important chemokine involved in chemoattraction of T cells, eosinophils, and basophils and is HIV-suppressive because CCR5, one of its receptors, is a coreceptor for HIV entry into T cells.2–6 It is an important pharmaceutical target with several inhibitory drugs under investigation.41 Most recently, Ooi et al showed that KLF13 also controls hematopoietic stem cell survival and can promote lymphoid fate decisions regulated by miRNA-125b.42
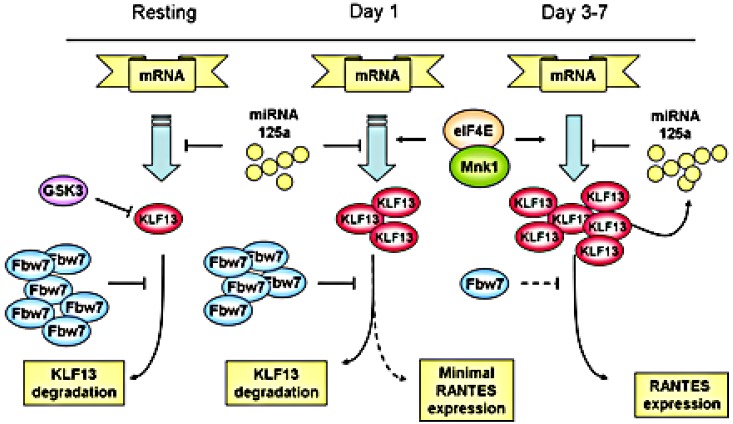
We propose a schematic view of the complicated regulation of KLF13 and RANTES expression in T lymphocytes (Figure 7). RANTES expression in T lymphocytes is controlled by a complex mechanism in which KLF13 is a key. Translational regulation occurs at the 5′-UTR of KLF13 mRNA and is controlled by the translation initiation complex and, in particular, both the amount and phosphorylation of eIF4E. Of note, eIF4E levels are also increased after T-cell activation.12 The 3′-UTR of KLF13 mRNA is also translationally regulated by miRNA 125a.13 Our work here shows that protein degradation because of ubiquitination also contributes to the expression patterns of KLF13 in T cells. In the resting state, KLF13 expression is suppressed by GSK3β and Fbw7-mediated ubiquitination-dependent protein degradation. After T-cell activation both Fbw7γ expression is reduced and GSK3β activity is inhibited, resulting in an increase in KLF13 expression and in turn RANTES levels. Our findings provide another piece in the biologic puzzle of the “late” expression of RANTES in T lymphocytes and may prove to be of clinical relevance if this pathway can be specifically targeted.
Figure 7.
Multiple mechanisms regulate KLF13 expression during human T-cell activation.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and the Center for Cancer Research.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: D.S.K., W.Z., B.J.H., S.J.K., S.E.M., M.P., and C.C. conducted experiments, analyzed data, and prepared figures; and D.S.K., C.C., and A.M.K. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alan M. Krensky, Laboratory of Cellular and Molecular Biology, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892; e-mail: krenskya@mail.nih.gov.
References
- 1.Schall TJ, Jongstra J, Dyer BJ, et al. A human T cell-specific molecule is a member of a new gene family. J Immunol. 1988;141(3):1018–1025. [PubMed] [Google Scholar]
- 2.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270(5243):1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 3.Dahinden CA, Geiser T, Brunner T, et al. Monocyte chemotactic protein 3 is a most effective basophil- and eosinophil-activating chemokine. J Exp Med. 1994;179(2):751–756. doi: 10.1084/jem.179.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kameyoshi Y, Dorschner A, Mallet AI, Christophers E, Schroder JM. Cytokine RANTES released by thrombin-stimulated platelets is a potent attractant for human eosinophils. J Exp Med. 1992;176(2):587–592. doi: 10.1084/jem.176.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rot A, Krieger M, Brunner T, Bischoff SC, Schall TJ, Dahinden CA. RANTES and macrophage inflammatory protein 1 alpha induce the migration and activation of normal human eosinophil granulocytes. J Exp Med. 1992;176(6):1489–1495. doi: 10.1084/jem.176.6.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schall TJ, Bacon K, Toy KJ, Goeddel DV. Selective attraction of monocytes and T lymphocytes of the memory phenotype by cytokine RANTES. Nature. 1990;347(6294):669–671. doi: 10.1038/347669a0. [DOI] [PubMed] [Google Scholar]
- 7.Taub DD, Sayers TJ, Carter CR, Ortaldo JR. Alpha and beta chemokines induce NK cell migration and enhance NK-mediated cytolysis. J Immunol. 1995;155(8):3877–3888. [PubMed] [Google Scholar]
- 8.Ortiz BD, Krensky AM, Nelson PJ. Kinetics of transcription factors regulating the RANTES chemokine gene reveal a developmental switch in nuclear events during T-lymphocyte maturation. Mol Cell Biol. 1996;16(1):202–210. doi: 10.1128/mcb.16.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song A, Chen YF, Thamatrakoln K, Storm TA, Krensky AM. RFLAT-1: a new zinc finger transcription factor that activates RANTES gene expression in T lymphocytes. Immunity. 1999;10(1):93–103. doi: 10.1016/s1074-7613(00)80010-2. [DOI] [PubMed] [Google Scholar]
- 10.Ahn YT, Huang B, McPherson L, Clayberger C, Krensky AM. Dynamic interplay of transcriptional machinery and chromatin regulates “late” expression of the chemokine RANTES in T lymphocytes. Mol Cell Biol. 2007;27(1):253–256. doi: 10.1128/MCB.01071-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou M, McPherson L, Feng D, et al. Kruppel-like transcription factor 13 regulates T lymphocyte survival in vivo. J Immunol. 2007;178(9):5496–5504. doi: 10.4049/jimmunol.178.9.5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikolcheva T, Pyronnet S, Chou SY, et al. A translational rheostat for RFLAT-1 regulates RANTES expression in T lymphocytes. J Clin Invest. 2002;110(1):119–126. doi: 10.1172/JCI15336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao X, Tang Y, Qu B, et al. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 2010;62(11):3425–3435. doi: 10.1002/art.27632. [DOI] [PubMed] [Google Scholar]
- 14.Onoyama I, Nakayama KI. Fbxw7 in cell cycle exit and stem cell maintenance: insight from gene-targeted mice. Cell Cycle. 2008;7(21):3307–3313. doi: 10.4161/cc.7.21.6931. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Sempere LF, Galimberti F, et al. Uncovering growth-suppressive MicroRNAs in lung cancer. Clin Cancer Res. 2009;15(4):1177–1183. doi: 10.1158/1078-0432.CCR-08-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413(6853):316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 17.Koepp DM, Schaefer LK, Ye X, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294(5540):173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 18.Grim JE, Gustafson MP, Hirata RK, et al. Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase. J Cell Biol. 2008;181(6):913–920. doi: 10.1083/jcb.200802076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye X, Nalepa G, Welcker M, et al. Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J Biol Chem. 2004;279(48):50110–50119. doi: 10.1074/jbc.M409226200. [DOI] [PubMed] [Google Scholar]
- 20.Herrmann J, Lerman LO, Lerman A. Ubiquitin and ubiquitin-like proteins in protein regulation. Circ Res. 2007;100(9):1276–1291. doi: 10.1161/01.RES.0000264500.11888.f0. [DOI] [PubMed] [Google Scholar]
- 21.Krensky AM, Ahn YT. Mechanisms of disease: regulation of RANTES (CCL5) in renal disease. Nat Clin Pract Nephrol. 2007;3(3):164–170. doi: 10.1038/ncpneph0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banck MS, Beaven SW, Narla G, Walsh MJ, Friedman SL, Beutler AS. KLF6 degradation after apoptotic DNA damage. FEBS Lett. 2006;580(30):6981–6986. doi: 10.1016/j.febslet.2006.10.077. [DOI] [PubMed] [Google Scholar]
- 23.Chen C, Sun X, Guo P, et al. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280(50):41553–41561. doi: 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- 24.Chen ZY, Wang X, Zhou Y, Offner G, Tseng CC. Destabilization of Kruppel-like factor 4 protein in response to serum stimulation involves the ubiquitin-proteasome pathway. Cancer Res. 2005;65(22):10394–10400. doi: 10.1158/0008-5472.CAN-05-2059. [DOI] [PubMed] [Google Scholar]
- 25.Quadrini KJ, Bieker JJ. EKLF/KLF1 is ubiquitinated in vivo and its stability is regulated by activation domain sequences through the 26S proteasome. FEBS Lett. 2006;580(9):2285–2293. doi: 10.1016/j.febslet.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 26.Venuprasad K, Huang H, Harada Y, et al. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol. 2008;9(3):245–253. doi: 10.1038/niXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Srinivasan SV, Lingrel JB. WWP1-dependent ubiquitination and degradation of the lung Kruppel-like factor, KLF2. Biochem Biophys Res Commun. 2004;316(1):139–148. doi: 10.1016/j.bbrc.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 28.Lai DZJ, Wang T, Hu-Li J, et al. KLF13 sustains thymic memory-like CD8+ T cells in BALB/c mice by regulating IL-4-generating invariant natural killer T cells. J Exp Med. 2011;208(5):1093–1103. doi: 10.1084/jem.20101527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conkright MD, Wani MA, Lingrel JB. Lung Kruppel-like factor contains an autoinhibitory domain that regulates its transcriptional activation by binding WWP1, an E3 ubiquitin ligase. J Biol Chem. 2001;276(31):29299–29306. doi: 10.1074/jbc.M103670200. [DOI] [PubMed] [Google Scholar]
- 30.Zhao D, Zheng HQ, Zhou Z, Chen C. The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 2010;70(11):4728–4738. doi: 10.1158/0008-5472.CAN-10-0040. [DOI] [PubMed] [Google Scholar]
- 31.Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001;20(4):723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78(5):787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2001;98(3):974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3(11):973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 35.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8(2):83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 36.Spruck CH, Strohmaier H, Sangfelt O, et al. hCDC4 gene mutations in endometrial cancer. Cancer Res. 2002;62(16):4535–4539. [PubMed] [Google Scholar]
- 37.Diehn M, Alizadeh AA, Rando OJ, et al. Genomic expression programs and the integration of the CD28 costimulatory signal in T cell activation. Proc Natl Acad Sci U S A. 2002;99(18):11796–11801. doi: 10.1073/pnas.092284399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sangfelt O, Cepeda D, Malyukova A, van Drogen F, Reed SI. Both SCF(Cdc4alpha) and SCF(Cdc4gamma) are required for cyclin E turnover in cell lines that do not overexpress cyclin E. Cell Cycle. 2008;7(8):1075–1082. doi: 10.4161/cc.7.8.5648. [DOI] [PubMed] [Google Scholar]
- 39.Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol. 2004;14(20):1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 40.Matsumoto A, Onoyama I, Nakayama KI. Expression of mouse Fbxw7 isoforms is regulated in a cell cycle- or p53-dependent manner. Biochem Biophys Res Commun. 2006;350(1):114–119. doi: 10.1016/j.bbrc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 41.Proudfoot AE, Power CA, Schwarz MK. Anti-chemokine small molecule drugs: a promising future? Expert Opin Investig Drugs. 19(3):345–355. doi: 10.1517/13543780903535867. [DOI] [PubMed] [Google Scholar]
- 42.Ooi AG, Sahoo D, Adorno M, Wang Y, Weissman IL, Park CY. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci U S A. 2010;107(50):21505–21510. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]