Abstract
Diffuse large B-cell lymphoma (DLBCL), the most common type of non-Hodgkin lymphoma, remains a partially curable disease. Genetic alterations affecting components of NF-κB signaling pathways occur frequently in DLBCL. Almost all activated B cell–like (ABC) DLBCL, which is the least curable group among the 3 major subtypes of this malignancy, and a substantial fraction of germinal center B cell–like (GCB) DLBCL exhibit constitutive NF-κB pathway activity. It has been demonstrated that ABC-DLBCL cells require such activity for proliferation and survival. Therefore, inhibition of NF-κB activation in DLBCL may provide an efficient and targeted therapy. In screening for small-molecule compounds that may inhibit NF-κB activation in DLBCL cells, we identified a compound, NSC697923, which inhibits the activity of the ubiquitin-conjugating (E2) enzyme Ubc13-Uev1A. NSC697923 impedes the formation of the Ubc13 and ubiquitin thioester conjugate and suppresses constitutive NF-κB activity in ABC-DLBCL cells. Importantly, NSC697923 inhibits the proliferation and survival of ABC-DLBCL cells and GCB-DLBCL cells, suggesting the Ubc13-Uev1A may be crucial for DLBCL growth. Consistently, knockdown of Ubc13 expression also inhibited DLBCL cell survival. The results of the present study indicate that Ubc13-Uev1A may represent a potential therapeutic target in DLBCL. In addition, compound NSC697923 may be exploited for the development of DLBCL therapeutic agents.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is an aggressive and heterogeneous disease comprising at least 3 major subtypes with distinct molecular, biologic, and clinical properties: activated B cell–like DLBCL (ABC-DLBCL), germinal center B cell–like DLBCL (GCB-DLBCL), and primary mediastinal B-cell lymphoma.1,2 Although the overall cure rate for DLBCL reaches more than 50% with the current therapies such as R-CHOP (rituximab plus cyclophosphamide, hydroxydaunorubicin, oncovin, and prednisone/prednisolone), less than 40% of ABC-DLBCL patients are cured.2–4 Therefore, new therapy approaches efficient for this and other DLBCL subtypes are highly desirable.
The transcription factor NF-κB controls the expression of a wide range of genes involved in cell proliferation, survival, stress response, angiogenesis, and inflammation.5,6 NF-κB activity is tightly regulated by multiple signaling pathways, and abnormal NF-κB activation has been linked to cancer development and progression.7–9 Constitutive NF-κB activation has been observed in high frequency in all of the main DLBCL subtypes, especially in ABC-DLBCL, with more than 90% of the tumors showing nuclear NF-κB, the hallmark of its activation.9–14 A recent genomic study revealed that more than 60% of ABC-DLBCLs and approximately 30% of GCB-DLBCLs harbor somatic mutations in multiple components of NF-κB signaling pathways, such as the BCR, CD40, and TLR pathways.15 Significantly, it has been demonstrated that constitutive NF-κB signaling is required for the proliferation and survival of ABC-DLBCL cell lines.11,13,16,17 All of these observations suggest a primary role for constitutive NF-κB signaling in the pathogenesis of DLBCL and, therefore, the NF-κB signaling pathway may represent a rational therapeutic target in DLBCL.9,11,18
Ubiquitination, the covalent attachment of the ubiquitin (Ub) molecule to target proteins, regulates diverse cellular processes.19 Ubiquitination proceeds through a stepwise enzymatic cascade involving 3 classes of enzymes: a Ub-activating enzyme (E1), a Ub-conjugating enzyme (E2), and a Ub ligase (E3). The E1 enzyme activates Ub in an ATP-dependent manner and transfers the activated Ub to an E2 enzyme through the formation of a thioester bond between the carboxy terminus of Ub and the active site cysteine of the E2, generating an E2 and Ub thioester conjugate (referred to as E2∼Ub). The E2 then cooperates with an E3 to attach the Ub to a lysine residue of a substrate. Ub itself can serve as a substrate and the process can undergo multiple rounds, resulting in the formation of polyubiquitin chains.19,20 Because Ub has 7 lysine residues and any one of them can be conjugated to another Ub, polyubiquitin chains of different linkages with distinct functional properties are formed in cells. For example, lysine 48 (K48)–linked polyubiquitin chains typically target substrates for proteasomal degradation, whereas K63-linked polyubiquitin chains function as scaffolds to assemble protein complexes in NF-κB signaling and DNA repair.21–24
Ubc13 (also known as UBE2N) is the active subunit of an E2 enzyme that catalyzes the synthesis of K63-linked polyubiquitin chains. It functions together with one of its 2 cofactors, Uev1A (UBE2V1) and Mms2 (UBEV2), E2 variants that lack the active-site cysteine residues.20,23 In response to the engagement of membrane receptors such as TLR and TCR, Ubc13-Uev1A, in conjunction with the E3 enzyme TRAF6, catalyzes the formation of K63-linked polyubiquitin chains that interact with both TAK1 and IKK complexes and thereby bring these 2 kinases into proximity. Consequently, the activated TAK1 phosphorylates and activates the IKK complex, which in turn phosphorylates IκB proteins, leading to IκB protein degradation and subsequent NF-κB activation.21,22 In complex with the Mms2 cofactor, Ubc13 promotes the K63-linked ubiquitination at sites of DNA double-strand breaks, leading to the recruitment of repair proteins to the DNA lesions.23,24 In addition to participating in NF-κB activation and DNA double-strand break repair, Ubc13 regulates other cellular processes, including nuclear localization of the tumor suppressor p53 protein and MAPK activation.25,26 Given the in vivo function of Ubc13, the potential of targeting this E2 enzyme has been proposed, and several compounds that inhibited the complex formation between Ubc13 and Uev1A have been described previously.27,28 The utility of these compounds for the development of therapeutics, however, has been questioned because of their chemical properties.29
Chronic active BCR signaling,11 together with MYD88-medated signaling,13 is largely responsible for the constitutive NF-κB activity in ABC-DLBCL cells and controls the proliferation and survival of these cells. Therefore, inhibition of this pathway may provide new therapeutic strategies. Indeed, small-molecule inhibitors of kinases, such as Syk and PKC-β, which mediate NF-κB activation in the BCR signaling pathway,30,31 have been tested in clinical trials with some efficacy.32 Because both Syk and PKCβ function upstream of the CARMA1 (CARD11)–BCL10-MALT1 (CBM) complex in the pathway,30,31 inhibition of these kinases may have little effect on the constitutive NF-κB activation in more than 10% of DLBCLs that harbor mutations in the CBM complex.10,12,15,33 We have devised a cell-based assay to screen compound libraries for small molecules that inhibit PKCβ signaling. In the present study, we report that one of the identified compounds, NSC697923, specifically inhibits the activity of Ubc13-Uev1A. We show that this compound impedes NF-κB activation in ABC-DLBCL cells and demonstrate that the compound inhibits the proliferation and survival of both ABC-DLBCL and GCB-DLBCL cell lines and survival of primary DLBCL cells. In addition, consistent with the observations from our studies on the compound, we show herein that knockdown of Ubc13 expression induces cell death of DLBCL cells. These findings suggest that Ubc13-Uev1A may be an attractive therapeutic target for DLBCL therapy. Compound NSC697923 may be a valuable pharmacologic agent for studying the functions of Ubc13 and may be explored for the development of therapeutic agents targeting Ubc13-Uev1A.
Methods
Cells, chemical compounds, and antibodies
HEK293T cells were grown in DMEM supplemented with 10% FBS. The DLBCL cell lines SUDHL-6, OCI-Ly3, OCI-Ly7, OCL-Ly10, and HBL-1 were cultured as described previously.16,34 RAW264.7 cells were cultured in α-MEM medium supplemented with 10% FBS. Mouse embryo fibroblasts were prepared from day 14 embryos and those with less than 4 passages were used for the experiments described herein. Primary DLBCL cells were prepared from fresh patient tumor specimens. Tumors were washed in sterile PBS, mechanically disrupted, and filtered through a 75-μm nylon cell strainer to make a single-cell suspension. The primary DLBCL cells that remained alive after being cultured for 7 days were used for the experiments described herein. All procedures with primary DLBCL cells were carried out with a protocol approved by the University of Rochester Research Subjects Review Board.
Phorbol 12-myristate 13-acetate (PMA) and lipopolysaccharide (LPS) were from Sigma-Aldrich. TNFα and RANKL were from PeproTech. Compound NSC697923 was obtained from the National Cancer Institute (NCI) Developmental Therapeutic Program. Methyl 5-nitro-2-furoate, nitrofuroxazide, nitrofurantoin, furazolidone, nitrofurazone, and nitrofuroxime were purchased from Fisher Scientific. The compounds were dissolved in DMSO, and controls contained the same amounts of this solvent used for the highest tested concentrations of NSC697923. Antibodies specific for phospho-IκBα (2859), phospho-IKKα/β (2694), and cleaved caspase 3 (9661) were from Cell Signaling Technology. Antibodies specific for IκBα (sc-371), IKKα (sc-7605), Mcl-1 (sc-819), actin (sc-1616), Ub (sc-8017), Uev1A (sc-47 556), PARP (sc-7150), and caspase 3 (sc-748) were from Santa Cruz Biotechnology. p100/p52 antibody (05-361) was from Upstate Biotechnology. Anti-Ubc13 (37-1100) and anti-UbcH5 (A-615) were from Invitrogen and Boston Biochem, respectively. Anti-GST was kindly provided by Dr Ed Harlow (Harvard Medical School, Boston, MA).
Generation of the NF-κB–luciferase reporter cell line
HEK293T cells were transfected with the pNF-κB luciferase reporter construct, which contains 4 copies of the NF-κB–binding site upstream of the coding sequence for firefly luciferase, together with the pBabe-puro for selection. Individual clones, selected with 1.5 μg/mL of puromycin, were assessed for NF-κB–luciferase reporter activation by TNFα and PMA. A responsive clone was then infected with a lentivirus expressing green fluorescent protein (GFP), and the resulting 293T cells, referred to as 293T–NF-luc cells, carrying the stably integrated NF-κB–luciferase reporter and constitutively expressing GFP were used for compound screening.
Compound library and compound screening
The NCI Mechanistic Set, which consists of 879 selected small-molecule compounds, was obtained from the NCI Developmental Therapeutic Program. For compound screening, 293T–NF-luc cells were seeded in 96-well plates at 30 000 cells per well in 100 μL of DMEM supplemented with 10% FBS and incubated overnight. Compounds in 100 μL of culture medium were then added into each well at a final concentration of 2μM 1.5 hours before the addition of PMA (100 ng/mL) or TNFα (10 ng/mL). Six hours after stimulation by PMA/TNFα, the cells were lysed in a solution containing 50mM HEPES, pH 7.4, 250mM NaCl, 1% NP40, 10% glycerol, and 1mM DTT. After lysis, the green fluorescence of the lysates was measured using a PerkinElmer Victor 2 Multilabel counter. Firefly luciferase activity of the lysates was assayed using the Promega luciferase assay reagents following the manufacturer's instructions, and normalized to the green fluorescence readings, which were directly proportional to the cell numbers in the wells under our experimental conditions.
Preparation of GST-Ubc13 and GST-TRAF6 proteins
A plasmid expressing GST-Ubc13 was constructed by cloning the full-length human Ubc13 coding sequence generated by PCR using pFlagCMV2-UbcH13 (Addgene) as the template into a pGEX vector (Amersham). The construct was verified by DNA sequencing. The plasmid pGEX-TRAF6, which expresses the GST-TRAF6 fusion protein,35 was kindly provided by Dr Bryant Darnay (M D Anderson Cancer Center, Houston, TX). GST-Ubc13 and GST-TRAF6 fusion proteins were expressed in Escherichia coli BL21 and purified with glutathione-agarose beads (Sigma-Aldrich), as described previously.36
In vitro ubiquitination assay
The purified recombinant proteins used in the in vitro ubiquitination reactions were all purchased from Boston Biochem, except GST-TRAF6, which was expressed and purified as described earlier in this study. The reactions were carried out at 37°C for 40 minutes in a buffer containing 50mM Tris-HCl, pH 7.5, 5mM MgCl2, 200μM ATP, 120μM Ub (U-100H), and 0.1μM E1 (E-304). For Ubc13-mediated Ub chain synthesis, the reaction mixture included 0.2μM Ubc13 (E2-600) and 0.2mM Uev1A (E2-662) with or without GST-TRAF6. For UbcH5c-catalyzed ubiquitination, UbcH5c (E2-627) instead of Ubc13 and Uev1A was used in the reaction. Compound NSC697923 was added into the reaction mixtures at the indicated concentrations. The reactions were terminated with an equal volume of SDS-PAGE sample buffer and the products were analyzed by Western blotting with a Ub-specific antibody.
For detecting the E2-Ub (Ubc13∼Ub or UbcH5c∼Ub) thioester conjugates, the reactions were carried out as described in “In vitro ubiquitination assay” without GST-TRAF6. The reactions were terminated by the addition of the SDS-PAGE sample buffer without a reducing agent unless specified. The products were analyzed by Western blotting with an anti-Ubc13 or anti-UbcH5c antibody.
Assay for in vitro interaction between Ubc13 and Uev1A
GST-Ubc13 bound on glutathione-agarose beads was incubated with cell lysates prepared from OCI-Ly10 cells with or without the presence of NSC697923 at 4°C for 1 hour. The beads were washed 3 times with the lysis buffer (50mM Tris-HCl, pH 8.0, 250mM NaCl, 5mM EDTA, 1mM DTT, 0.2% NP-40, 5% glycerol, 0.4mM AEBSF, 0.5mM benzamidine-HCl, 5 μg/mL of leupeptin, 5 μg/mL of aprotinin, 5 μg/mL of pepstatin, 10mM NaF, 0.1mM Na3VO4, and 50mM β-glycerophosphate). The amounts of GST-Ubc13 and Uev1A proteins bound to the beads were then analyzed by Western blotting.
Analyses of cell viability, apoptosis, and cell-cycle progression
Cell viability was measured by trypan blue exclusion assay (Invitrogen) following the manufacturer's instructions. Apoptosis was assayed using the 7-amino-actinomycin D (7AAD)–annexin V apoptosis kit (BD Biosciences) as described previously.34 Analysis of the cell-cycle distribution of the treated cells was carried out with propidium iodide and bromodeoxyuridine double staining as described previously.37
Suppression of Ubc13 expression by RNA interference
The lentiviral pGIPZ constructs expressing an shRNAmir for human Ubc13 (shUbc13, RHS4430-98818339) or a nonsilencing shRNAmir (shControl, RHS4346) were purchased from Open Biosystems. Production of pseudotyped lentiviruses for shRNA expression was carried out in 293TN cells (System Biosciences) by cotransfection of pGIPZ with the viral-packing plasmids (Thermo Fisher). Cell culture supernatants were collected at 48 and 72 hours after transfection and filtered (0.45-μm filter). The viruses were then concentrated by centrifugation at 66 188g (Beckman SW28 rotor) for 2 hours and the virus pellets were resuspended in a small volume of the desired culture medium. OCI-Ly10 or OCI-Ly7 cells were infected with the lentiviruses expressing shControl and shUbc13, respectively. Sixty-eight hours after transfection, the infected cells expressing GFP from the pGIPZ vector were sorted and grown in their respective culture medium. The expression of Ubc13 was determined by Western blotting and the cell viability at the indicated times was analyzed as described in “Analyses of cell viability, apoptosis, and cell-cycle progression.”
Results
Screening for selective small-molecule compound inhibitors of NF-κB activation
To screen for compounds that inhibit NF-κB activation, we established a 293T-derived cell line that carries a stably integrated NF-κB signaling responsive luciferase reporter. Because the phorbol ester PMA, a potent PKC activator, induces NF-κB activation through a PKC-mediated pathway that relies on the CARMA1-BCL10-MALT1 complex,38,39 compounds that inhibit PMA-induced NF-κB activation may impede NF-κB activation induced by chronic active BCR signaling. We therefore set out to identify such compounds through compound library screening using inhibition of PMA-induced luciferase activity in the 293T–NF-luc cells as the readout. Our goal was to identify compounds with some selectivity, rather than a general inhibitor of NF-κB activation such as an IKK inhibitor, because a nonselective inhibition of NF-κB activity might generate severe side effects. To eliminate such compounds, we devised a counter screening in which compounds initially identified from the primary screening were tested for their effect on TNFα-induced NF-κB activation. Compounds that inhibit PMA-induced, but not TNFα-induced, NF-κB activation could be selective NF-κB pathway inhibitors.
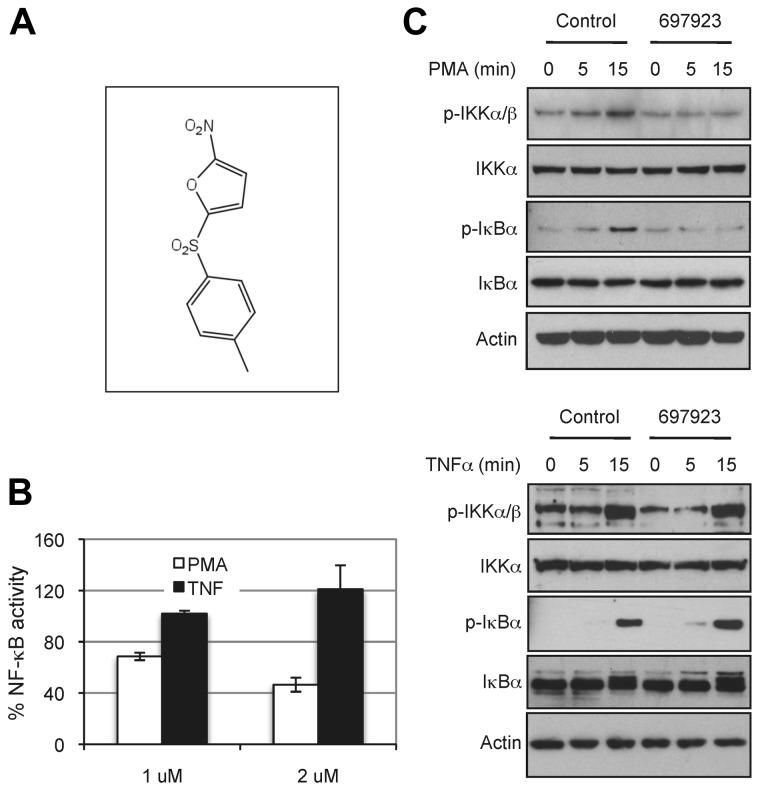
We screened the NCI Mechanistic Set at a final concentration of 2μM. In the primary screening, 109 compounds inhibited PMA-induced NF-κB activation by more than 50%. These compounds were then tested for their inhibition on NF-κB activation induced by TNFα in parallel with their effect on PMA-induced NF-κB activation. Of these, 5 compounds inhibited PMA-induced, but not TNFα-induced, NF-κB activation in the reporter assay. One of the compounds, NSC697923, reproducibly exhibited selective inhibitory effect on NF-κB reporter activation and on endogenous NF-κB activation (Figure 1). Whereas NSC697923 inhibited PMA-induced NF-κB activation, it had no inhibitory effect on PMA-stimulated ERK activation (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), indicating that this compound is a selective inhibitor for NF-κB activation rather than a general inhibitor of PMA-mediated signaling.
Figure 1.
Compound NSC697923 inhibits NF-κB activation induced by PMA, but not by TNFα. (A) The chemical structure of NSC697923. (B) 293T–NF-luc cells were pretreated with the indicated concentrations of NSC697923 or DMSO (final concentration 0.2%; control) for 1 hour. The cells were then stimulated with PMA (100 ng/mL) or TNFα (10 ng/mL) for 6 hours, and the green fluorescence (from GFP for normalization) and luciferase activity in the cell lysate were measured. Data are expressed as the percentage activity of the control (mean ± SD; n = 3). (C) 293T–NF-Luc cells were pretreated with 1μM NSC697923 for 1 hour before being stimulated with PMA or TNFα as indicated. The cells were harvested at the indicated times after stimulation, and phosphorylation of IKK and IκBα were analyzed by Western blotting. Analysis of actin was used as the loading control.
Structure-activity relationship analysis
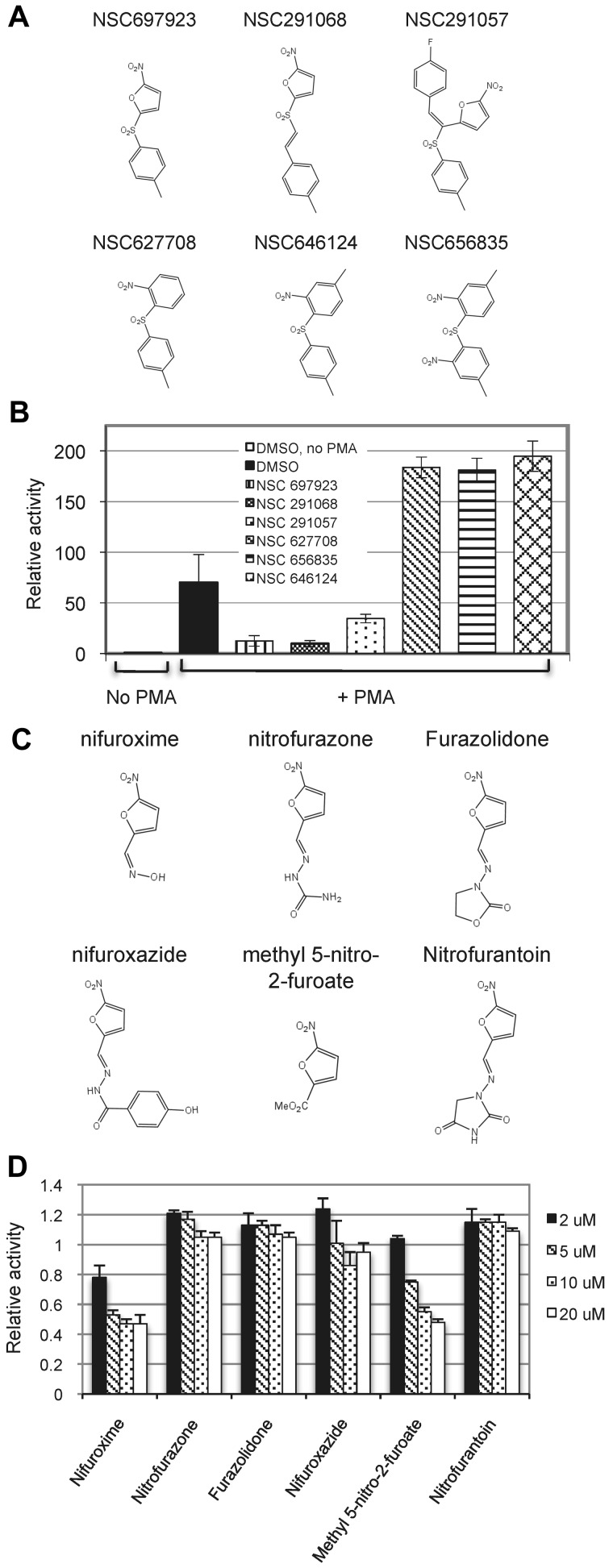
To probe the structural basis for the action of NSC697923, we examined whether several related analogs of this compound, which are available from the NCI, had a similar inhibitory property. Compounds that contain a nitrofuran moiety inhibited PMA-induced NF-κB activation, whereas the compounds without this group had no such inhibitory activity (Figure 2A-B), suggesting that the nitrofuran moiety is important for NF-κB inhibition by NSC697923. To further understand the relevance of nitrofuran group in the inhibition of NF-κB activation, we evaluated a panel of commercially available nitrofuran-containing compounds. Only some of these compounds showed an inhibitory effect on PMA-induced NF-κB activation (Figure 2C-D), indicating that an additional structural domain in NSC697923, apart from the nitrofuran moiety, is critical for its NF-κB–inhibitory activity.
Figure 2.
Effect of NSC697923 related compounds on NF-κB activation induced by PMA. (A) Structures of the tested NSC697923 analogs. (B) 293T–NF-luc cells were pretreated with the indicated compounds (2μM) or DMSO (0.2%) for 1 hour. The cells were then stimulated with PMA for 20 hours or left untreated as indicated. Shown are the relative luciferase activities, which were normalized to GFP reading (mean ± SD; n = 4). The luciferase activity of the unstimulated samples was set at 1. (C) Structures of the tested nitrofuran-containing compounds. (D) 293T–NF-luc cells were pretreated with DMSO (0.02%) or various nitrofuran-containing compounds at the indicated concentrations for 1 hour and then stimulated with PMA for 6 hours. The means of the luciferase activities relative to the DMSO pretreated (control) samples from the triplicates are shown.
Compound NSC697923 inhibits NF-κB activation by multiple stimuli
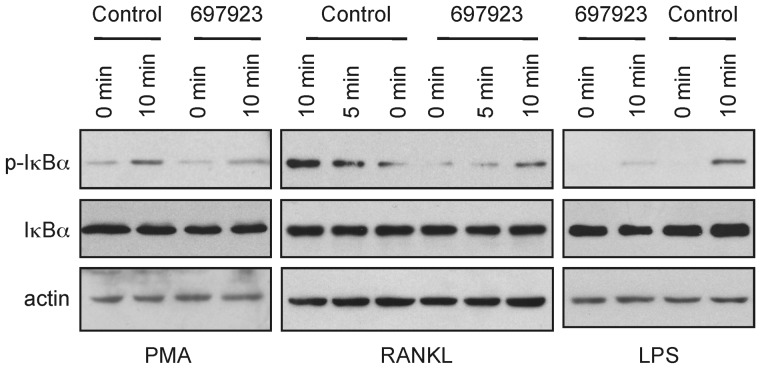
To determine the selectivity of NSC697923 further, we examined the effect of the compound on NF-κB activation induced by other stimuli. In addition to inhibiting PMA-induced NF-κB activation, NSC697923 inhibited IκBα phosphorylation by both RANKL and LPS (Figure 3). Therefore, the inhibitory activity of this compound is not limited to PMA-induced NF-κB activation; rather, the compound inhibits NF-κB activation by multiple, albeit not all, stimuli.
Figure 3.
NSC697923 appears to inhibit NF-κB activation by multiple stimuli. SUDHL-6, RAW264.7, and MEF cells were treated with NSC697923 or DMSO (control) for 30 minutes before stimulated with the PMA (100 ng/mL), RANKL (15 ng/mL), and LPS (1.5 μg/mL) for the indicated times. The levels of IκBα phosphorylation and total IκBα in the indicated samples were assessed by Western blotting.
NSC697923 is an inhibitor of the Ub-conjugating enzyme (E2) complex Ubc13-Uev1A
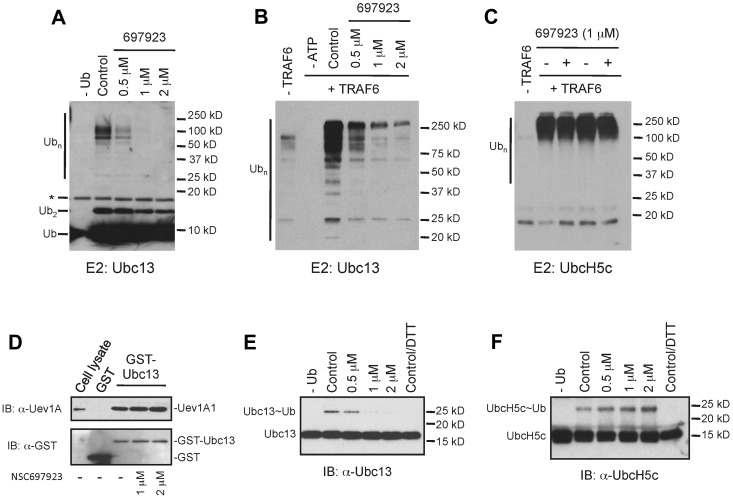
Given that the Ub-conjugating (E2) enzyme Ubc13-Uev1A complex is involved in NF-κB activation by multiple stimuli, including PMA, RANKL, and LPS, but not in the TNFα-induced NF-κB activation in some cells,21,22 we reasoned that compound NSC697923 might inhibit the function of this E2 enzyme. Ubc13-Uev1A, together with a Ub-activating enzyme (E1), can catalyze the synthesis of K63-linked polyubiquitin chains without an E3 in vitro.28 Therefore, we investigated whether NSC697923 has any effect on Ubc13-Uev1A–mediated polyubiquitin chain synthesis in an in vitro assay. As shown in Figure 4A, the compound inhibited polyubiquitin chain formation catalyzed by the Ubc13-Uev1A complex in a dose-dependent manner. Although Ubc13-Uev1A can catalyze Ub chain formation without an E3 enzyme, the efficiency of polyubiquitin chain synthesis is greatly enhanced in the presence of an E3 such as TRAF6.40 Therefore, we also examined the effect of NSC697923 on Ub chain formation catalyzed by Ubc13-Uev1A together with TRAF6. The ubiquitination reaction in the presence of TRAF6 was similarly inhibited by NSC697923 in a dose-dependent manner (Figure 4B).
Figure 4.
NSC697923 specifically inhibits Ubc13-mediated polyubiquitin chain synthesis in vitro. (A) NSC697923 inhibits the synthesis of polyubiquitin chains catalyzed by Ubc13-Uev1A in vitro. The in vitro reaction, which contains purified E1, Ub, Ubc13, and Uev1A, was carried out with or without the indicated concentrations of NSC697923. The reaction products were analyzed by an anti-Ub Ab on Western blots. The asterisk (*) denotes the signal from the cross-reaction of the Ab with the recombinant Ubc13 protein. (B) Same as in panel A, except that purified GST-TRAF6 was included as the E3 in the complete reaction as indicated. (C) NSC697923 exhibits no inhibitory effect on UbcH5c-mediated polyubiquitin chain synthesis. The assay was carried out as in panel B, except that UbcH5c instead of Ubc13-Uev1A was used as the E2 in the ubiquitination reaction. (D) NSC697923 has no inhibitory effect on the formation of Ubc13-Uev1A complex in vitro. Purified GST or GST-Ubc13 protein was incubated with the cell extracts prepared from OCI-Ly10 cells with or without the presence of NSC697923 as indicated. The amounts of Uev1A protein associated with GST or GST-Ubc13 were analyzed by Western blotting. The first lane was loaded with 8% of the input cell lysate. (E) NSC697923 inhibits the formation of the Ubc13∼Ub thioester conjugate. The assay was carried out as described in panel A, except that the loading buffer for SDS-PAGE gel contained no reducing agents unless otherwise specified and the reaction was analyzed by Western blotting with a Ubc13-specific Ab. (F) Formation of the UbcH5c∼Ub conjugate is not inhibited by NSC697923. The assay was carried out as in panel E, except that UbcH5c instead of Ubc13 and Uev1A was used in the reaction and a UbcH5c-specific Ab was used for Western blot analysis.
To investigate the specificity of NSC697923, we examined the effect of this compound on polyubiquitin chain synthesis mediated by UbcH5c, which has high sequence homology to Ubc13 and can also catalyze the lysine 63-linked polyubiquitin chain formation in conjunction with TRAF6.41 In contrast to the inhibition of Ubc13-Uev1A–catalyzed ubiquitination, the UbcH5c-mediated polyubiquitin chain formation was not inhibited by NSC697923 (Figure 4C). Therefore, NSC697923 is a selective inhibitor of Ubc13-Uev1A, rather than a general inhibitor of ubiquitination.
NSC697923 inhibits the formation of the Ubc13-Ub thioester conjugate
Having shown that NSC697923 is an inhibitor of the Ubc13-Uev1A E2 enzyme, we next explored the mechanism of inhibition by this compound. Because both Ubc13 and UevA1 subunits are essential for the K63-linked polyubiquitin chain synthesis catalyzed by this E2 enzyme,20,23 NSC697923 could inhibit Ubc13-Uev1A function by blocking the dimer formation of these 2 subunits. To test this possibility, we examined the effect of NSC697923 on the interaction between Ubc13 and Uev1A in a GST pull-down assay. The compound showed no inhibitory effect on the complex formation between Ubc13 and Uev1A (Figure 4D).
A critical step in ubiquitination is the formation of a thioester bond between Ub and the active site cysteine of an E2. This conjugate, E2∼Ub, is the active form of an E2 enzyme and an essential intermediate for the transfer of Ub to substrates.20,23 We therefore investigated whether NSC697923 has any effect on Ubc13∼Ub formation. Whereas the Ubc13∼Ub conjugate was readily detectable in an in vitro reaction consisting of an E1, Ubc13-Uev1A, Ub, and ATP, formation of this thioester conjugate was inhibited by NSC697923 (Figure 4E). Consistent with the observation that UbcH5c-mediated ubiquitination is not blocked by NSC697923, formation of the UbcH5c∼Ub conjugate was also not inhibited by this compound (Figure 4F). Therefore, NSC697923 specifically inhibits the formation of Ubc13∼Ub conjugate and thereby selectively suppresses Ubc13-mediated ubiquitination.
NSC697923 inhibits constitutive NF-κB signaling in ABC-DLBCL cells
Because Ubc13 is involved in NF-κB activation by multiple signaling pathways, and because NSC697923 inhibits Ubc13 activity, we investigated whether NSC697923 inhibits the constitutive NF-κB pathway activity in ABC-DLBCL cells. We first examined the effect of this compound on the formation of the Ubc13∼Ub conjugate in these cells. Similar to what has been observed in vitro, NSC697923 inhibited the formation of the Ubc13∼Ub conjugate (Figure 5A) in OCI-Ly10 cells, an ABC-DLBCL cell line, suggesting that the compound also inhibits Ubc13 function in DLBCL cells. We next examined the effect of the compound on NF-κB activation. NSC697923 inhibited phosphorylation of IKK and IκBα in OCI-Ly10 cells (Figure 5B), indicating that the compound impeded NF-κB activation in these cells. Consistently, expression of p100 and Mcl-1, known NF-κB targets,42,43 was inhibited in OCI-Ly10 cells treated with the compound (Figure 5B). Therefore, the results indicate that NSC697923 inhibits the constitutive NF-κB signaling in ABC-DLBCL cells.
Figure 5.
NSC697923 inhibits constitutive NF-κB signaling, proliferation, and survival in ABC-DLBCL cells. (A) Inhibition of the Ubc13∼Ub conjugate formation in DLBCL cells by NSC697923. OCI-Ly10 cells were treated with the indicated concentrations of NSC697923 for 3 hours. The levels of Ubc13∼Ub conjugate were analyzed by Western blotting using a Ubc13-specific Ab as described in Figure 4E. In the figure, the leftmost lane was moved and placed next to the rest of the lanes on the same gel for easy comparison. (B) NSC697923 inhibits constitutive NF-κB activation in ABC-DLBCL cells. OCI-Ly10 cells were treated with the indicated concentrations of NSC697923 for 3.5 hours. The levels of the indicated proteins were analyzed by Western blotting with the respective Abs. (C) NSC697923 inhibits the proliferation and survival of ABC-DLBCL cells. The indicated ABC-DLBCL cells were seeded at 3 × 105 cells/mL in 6-well plates and cultured in the presence of DMSO (0.2%, control) or various concentrations of NSC697923 for 24 hours. The live and dead cells were counted using the trypan blue exclusion assay. Shown are the means from 3 separate experiments. The average number of live cells in the control was set at 100. (D) OCI-Ly10 cells were treated with the indicated concentrations of NSC697923 for 5 hours. The levels of the indicated proteins were analyzed by Western blotting. For the detection of caspase-3, a mixture of a caspase-3 Ab and an Ab specific for the cleaved form of this protein was used. (E) OCI-Ly10 cells were treated as in panel C. The apoptotic cells (annexin V–positive cells) were measured by flow cytometry as described in “Analyses of cell viability, apoptosis, and cell-cycle progression.” A representative result from 3 independent experiments is depicted. (F) OCI-Ly10 cells were treated with the indicated concentrations of NSC697923 for 4.5 hours. Bromodeoxyuridine was then added into the culture medium for 30 minutes before the cells were harvested for cell-cycle distribution analysis as described in “Analyses of cell viability, apoptosis, and cell-cycle progression.” Shown are the averages from 2 independent experiments.
NSC697923 inhibits the proliferation and survival of ABC-DLBCL cells
Because NSC697923 inhibits NF-κB activation in ABC-DLBCL cells, which depend on constitutive NF-κB activity for growth,9,16,17 we investigated the effect of this compound on the proliferation and survival of these cells. Consistent with the observation that ABC-DLBCL cells rely on constitutive NF-κB activity for proliferation and survival, NSC697923 induced proliferation arrest and cell death of ABC-DLBCL cells, including OCI-Ly3 cells, which carry a mutated CARMA1 (CARD11) and lacking PKCβ expression,12,33 as judged by a dramatic decrease of live cells and a marked increase of dead cells (Figure 5C). Treatment of OCI-Ly10 cells with the compound resulted in activation of caspase 3, cleavage of PARP, and an increase in annexin V–positive cells (Figure 5D-E), all hallmarks of apoptosis.34 Therefore, NSC697923 induces apoptosis in ABC-DLBCL cells.
To confirm that NSC697923 indeed inhibits proliferation, we examined the effect of NSC697923 on cell-cycle progression of OCI-Ly10 cells. Because long treatment of ABC-DLBCL cells with NSC697923 leads to massive cell death (Figure 5C-E) and may thus interfere with cell-cycle analysis, we carried out cell-cycle progression analysis with a short (5-hour) treatment. Treatment of OCI-Ly10 with the compound resulted in a marked decrease of cells in the S phase (Figure 5F), verifying that NSC697923 inhibits cell-cycle progression in ABC-DLBCL cells.
NSC697923 also inhibits proliferation and survival of GCB-DLBCL cells
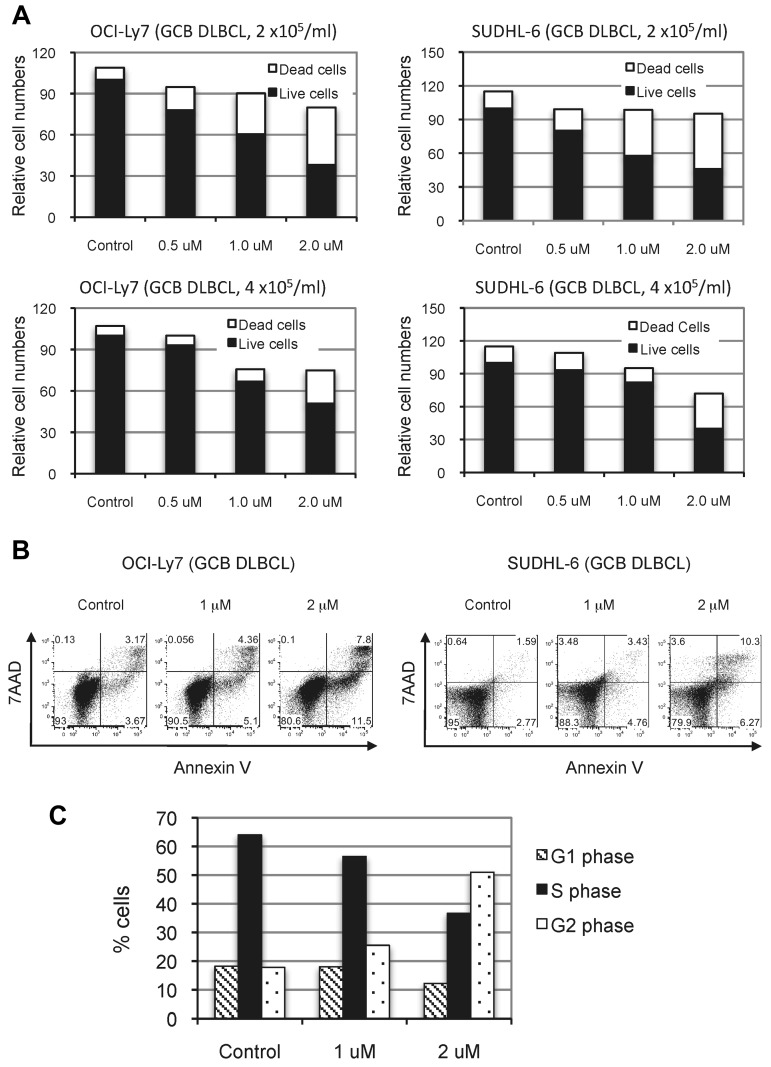
The observation that NSC697923 inhibited the proliferation and survival of ABC-DLBCL cells prompted us to investigate whether this compound has similar inhibitory effects on GCB-DLBCL cells, which are independent of NF-κB activity for growth.16,17 As shown in Figure 6, NSC697923 also inhibited proliferation and survival of OCI-Ly7 and SUDHL-6, 2 GCB-DLBCL cell lines. Therefore, NSC697923 apparently blocks an essential pathway other than NF-κB signaling in GCB-DLBCL cells, possibly reflecting an NF-κB–independent function of Ubc13-Uev1A in these cells.
Figure 6.
NSC697923 inhibits the proliferation and survival of GCB-DLBCL cells. (A) GCB-DLBCL cells were plated at the indicated cell concentrations and treated with various concentrations of NSC697923 for 24 hours. The effects of NSC697923 on the growth of these cells were analyzed as described in Figure 5C. (B) The cells were plated at a concentration of 4 × 105 cells/mL and treated with NSC697923 as in panel A and the apoptotic cells were analyzed as described in Figure 5E. (C) Cell-cycle distribution of OCI-Ly7 cells treated with the indicated concentrations of NSC697923 for 24 hours. Shown are the average results from 2 independent experiments.
NSC697923 reduces the viability of primary DLBCL cells
In addition to the DLBCL cell lines, we tested the effect of NSC697923 on the survival of primary DLBCL cells. We obtained primary DLBCL cells from 2 of the tested DLBCL patient samples that were able to grow for more than 7 days in an in vitro culture condition. Treatment of these primary DLBCL cells with the compound resulted in an increase in annexin V–positive (apoptotic) cells (Figure 7), indicating that NSC697923 also induces apoptosis of primary DLBCL cells.
Figure 7.
NSC697923 induces cell death of primary DLBCL cells. Primary cancer cells prepared from tumor specimens of 2 DLBCL patients were seeded at 4.5 × 105 cells/mL in RPMI medium supplemented with 20% FBS and treated with the indicated concentrations of NSC697923 for 24 hours. The apoptotic cells were analyzed as described in Figure 5E. Shown are the averages from 2 independent experiments.
Knockdown of Ubc13 inhibits constitutive NF-κB signaling and cell survival of ABC-DLBCL cells
Our studies on NSC697923 suggested that Ubc13 plays a crucial role in the proliferation and survival of DLBCL cells. To test this possibility further, we examined whether Ubc13 expression is required for constitutive NF-κB signaling and the survival of ABC-DLBCL cells. We used the miRNA-derived shRNA from a lentiviral vector to knock down the expression of Ubc13 (Figure 8A). Consistent with the results of NSC697923 treatment, suppression of Ubc13 expression led to inhibition of IKK activation and NF-κB target expression (Figure 8B), as well as a dramatic increase in the cell death of OCI-Ly10 cells (Figure 8C-D), supporting the notion that Ubc13 is essential for constitutive NF-κB activation and survival of ABC-DLBCL cells.
Figure 8.
Suppression of Ubc13 expression inhibits constitutive NF-κB signaling and cell survival of ABC-DLBCL cell. (A) Analysis of Ubc13 expression. OCI-Ly10 cells were infected with lentiviruses that express either shControl or shUbc13. The levels of Ubc13 protein in the infected cells were analyzed 7 days after infection by Western blotting with a Ubc13-specific Ab. The amount of total protein lysates loaded in each lane is indicated. (B) Effect of Ubc13 knockdown on constitutive NF-κB signaling in OCI-Ly10 cells. Expression of the indicated proteins was analyzed by Western blotting. (C) Viability of the infected OCI-Ly10 cells was measured by trypan blue exclusion assay at the indicated times after infection. (D) Apoptosis of the infected OCI-Ly10 cells at 9 days after infection was analyzed as described in “Analyses of cell viability, apoptosis, and cell-cycle progression.”
We also explored the effect of Ubc13 knockdown on the growth of GCB-DLBCL cells. Despite our repeated efforts, only a partial (approximately 40%-50%) suppression of Ubc13 expression in GCB-DLBCL cells was achieved with the available shRNA constructs (supplemental Figure 2A and data not shown). Consistent with the idea that Ubc13 is crucial for the proliferation of DLBCL cells, OCI-Ly7 cells with decreased Ubc13 expression displayed a reduced growth rate (supplemental Figure 2B).
Discussion
Despite recent advances in treatment, a significant proportion of DLBCL patients, especially ABC-DLBCL patients, still die of this malignancy.3,4,44 The development of new therapies targeting pathways essential for the proliferation and survival of both the ABC subgroup and the GCB subtype may improve clinical outcome. In the present study, we report the identification of a small-molecule compound inhibitor of the Ub-conjugating (E2) enzyme Ubc13-Uev1A and demonstrate that this compound, NSC697923, induces proliferation arrest and apoptosis in both ABC-DLBCL and GCB-DLBCL cell lines, as well as apoptosis in primary DLBCL cells. These results, together with the observation that Ubc13 knockdown inhibits the proliferation and survival of DLBCL cells, indicate that Ubc13 is crucial for the proliferation and survival of DLBCL cells. Therefore, Ubc13/Uev1A may represent a potential therapeutic target for DLBCL.
Our studies herein demonstrate that NSC697923 is a specific inhibitor of Ubc13-Uev1A, which acts through inhibiting the formation of the Ubc13∼Ub conjugate. Structure-activity relationship analysis suggested that the nitrofuran group is important, but not sufficient, for the inhibition of Ubc13 activity. Interestingly, several nitrofuran-containing pyrazolidine compounds were shown to inhibit the activity of the Ub-activating enzyme (E1), but not E2, by blocking the formation of the E1∼Ub conjugate.45,46 Therefore, it may be possible that the nitrofuran moiety of these pyrazolidine compounds and NSC697923 interfere with the thioester bound formation between the Ub molecule and the active-site cysteine residue of an E1 or E2 enzyme, whereas the remaining structures of these compounds direct them to a specific ubiquitination enzyme.
NSC697923, through the inhibition of the Ubc13-Uev1A enzyme, may impede cell proliferation and survival of DLBCL cells by several distinct mechanisms. The cytotoxic effect of NSC697923 on ABC-DLBCL cells likely results in part from the inhibition of NF-κB signaling, which is known to be essential for the proliferation and survival of these cells.11,13,16,17 Because Ubc13 negatively regulates nuclear localization of p53,26 which can inhibit both cell proliferation and survival,47 NSC697923 may also exert its effect through p53 activation. Indeed, we observed that NSC697923 treatment increased the nuclear level of p53 protein and p21 expression in OCI-Ly10 cells (supplemental Figure 3A). Consistently, knockdown of Ubc13 expression in OCI-Ly10 cells resulted in increased expression of p21 and GADD45 (supplemental Figure 3B), known p53 targets, supporting the idea that inhibition of Ubc13 induces p53 activation in ABC-DLBCL cells. NSC697923 is also toxic to GCB-DLBCL cells (Figure 6) such as OCI-Ly7 cells, which are independent of NF-κB activity for proliferation and survival16,17 and lack functional p53.48 This observation indicates that NSC697923 may inhibit the proliferation and survival of GCB-DLBCL cells through a mechanism independent of NF-κB and p53 signaling. One potential target of NSC697923 in these cells may be the MAPK pathway(s), which depends on Ubc13 for activation induced by several stimuli in mouse B cells.25 Therefore, inhibition of Ubc13 may affect multiple cellular processes, in addition to NF-κB signaling, in DLBCL cells that are essential for the proliferation and survival of these cells.
Our observations indicate that therapeutic targeting of Ubc13 may provide a promising treatment for DLBCL. NSC697923 inhibited the proliferation and survival of OCL-Ly3 cells (Figure 5C), suggesting that inhibition of Ubc13 may also offer an effective treatment for DLBCLs with oncogenic mutations in the CBM complex and those resistant to PKCβ inhibition.10,12,15,33 Therefore, inhibition of Ubc13 may provide a potential advantage over targeting components upstream of the CBM complex. Moreover, because Ubc13 plays a critical role in DNA double-strand break repair,24 inhibition of Ubc13 may sensitize cancer cells to radiation therapy or chemotherapy. Indeed, we found that NSC697923 inhibited recruitment of DNA-repair proteins to the sites of DNA double-strand breaks in mammalian cells (supplemental Figure 4) and exhibited an additive cytotoxic effect on DLBCL cells with CHOP (cyclophosphamide, hydroxydaunorubicin, oncovin, and prednisone/prednisolone) agents (supplemental Figure 5). Inhibitors of Ubc13 can thus be potentially used either as a single agent or in combination with CHOP agents in DLBCL treatment. The development of therapeutic strategies targeting Ubc13 may provide clinical benefits for DLBCL patients. Our studies on NSC697923 may be explored for this purpose. Other B-cell diseases such as autoimmune diseases may also rely on active NF-κB signaling49 and inhibitors of Ubc13 may be beneficial for the treatment of these diseases as well.
Because the constitutive NF-κB activity in ABC-DLBCL cells is primarily controlled by chronic BCR signaling,11 the observation that NSC697923 inhibited NF-κB activation in these cells (Figure 5B) suggests that Ubc13-Uev1A may play a role in this signaling pathway. Consistent with this view, NSC697923 inhibited NF-κB activation induced by PMA (Figures 1 and 3), an activator of PKCβ that is essential for BCR-induced NF-κB activation.33 In addition, a previous study reported that Ubc13 deficiency impairs PMA-induced NF-κB activation.50 However, the conditional deletion of Ubc13 in normal mouse B cells apparently had little effect on BCR-induced NF-κB activation.25 Therefore, Ubc13 may have acquired a lymphoma-specific role in chronic BCR signaling. Alternatively, the reported lack of effect of Ubc13 deficiency on NF-κB activation in mouse B cells might be because of the incomplete deletion of Ubc13 in these cells,50 given that Ubc13 was shown to be required for NF-κB activation induced by TCR signaling, which shares extensive similarities with the BCR signaling pathway.50
One concern for targeting Ubc13 are the potential side effects that it may generate, because Ubc13 is involved in multiple signaling pathways and has been shown to play crucial roles in mouse embryo development and hematopoiesis.25,51 Interestingly, NSC697923 (and knockdown of Ubc13) has much less toxic effects on the proliferation and survival of several other types of cells, such as breast cancer and osteosarcoma cells, compared with its effect on DLBCL cells (supplemental Figure 6 and data not shown). It thus appears that not all cells depend on Ubc13 for survival, thereby providing a potential opportunity to target this E2 enzyme in DLBCL with tolerable side effects. Nevertheless, future research, including in vivo studies, is required to comprehensively address the feasibility of targeting Ubc13 as a therapeutic strategy for DLBCL.
Supplementary Material
Acknowledgments
The authors thank Hartmut Land and Dirk Bohmann for helpful discussions and critical reading of the manuscript; Craig Jordan for suggestions; Richard Fisher for comments on the manuscript; Eric Davis for HBL-1 and OCI-Ly10 cells; Edward Schwarz for RAW264.7 cells; Bryant Darnay for PGX-TRAF6 plasmid; Ed Harlow for GST Ab; the flow cytometry core of University of Rochester Medical Center for help with cell sorting and flow cytometric analysis; and the National Cancer Institute Developmental Therapeutic Program for the Mechanistic Set compound library, compound NSC697923, and its analogs.
This work was supported by a developmental research program grant from the NCI lymphoma SPORE (1P50CA130805) at the James P. Wilmot Cancer Center of University of Rochester. J.Z. and L.C are also supported by NIH R01 CA127530, and I.S. is supported by NIH U19-AI56390, P01 AI078907, and CA130805-02S2.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.P. designed the experiments, performed the research, analyzed and interpreted the data, and wrote the manuscript; Y.L., D.O., and M.D. performed the research and analyzed the data; E.V.P. performed the research; J.S. and I.S. provided resources and critically reviewed the manuscript; L.C. designed the research, analyzed and interpreted the data, and critically reviewed the manuscript; J.Z. conceived the research, designed the experiments, analyzed and interpreted the data, and wrote the manuscript; and all authors read and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.D. is Cutaneous Biology Research Center, Massachusetts General Hospital, Harvard Medical School, Charlestown, MA. The current affiliation for I.S. is Division of Rheumatology, Lowance Center for Human Immunology, Emory University School of Medicine, Atlanta, GA.
Correspondence: Jiyong Zhao, Department of Biomedical Genetics, University of Rochester Medical Center, 601 Elmwood Ave, Rochester, NY 14642; e-mail: jiyong_zhao@urmc.rochester.edu.
References
- 1.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010;362(15):1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staudt LM, Dave S. The biology of human lymphoid malignancies revealed by gene expression profiling. Adv Immunol. 2005;87:163–208. doi: 10.1016/S0065-2776(05)87005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedberg JW, Fisher RI. Diffuse large B-cell lymphoma. Hematol Oncol Clin North Am. 2008;22(5):941–952. doi: 10.1016/j.hoc.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Häcker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;2006(357):re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- 6.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12(8):715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 8.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1(5):a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2(6):a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenz G, Davis RE, Ngo VN, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319(5870):1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 13.Ngo VN, Young RM, Schmitz R, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honma K, Tsuzuki S, Nakagawa M, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood. 2009;114(12):2467–2475. doi: 10.1182/blood-2008-12-194852. [DOI] [PubMed] [Google Scholar]
- 15.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194(12):1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam LT, Davis RE, Pierce J, et al. Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin Cancer Res. 2005;11(1):28–40. [PubMed] [Google Scholar]
- 18.Rui L, Schmitz R, Ceribelli M, Staudt LM. Malignant pirates of the immune system. Nat Immunol. 2011;12(10):933–940. doi: 10.1038/ni.2094. [DOI] [PubMed] [Google Scholar]
- 19.Weissman AM, Shabek N, Ciechanover A. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat Rev Mol Cell Biol. 2011;12(9):605–620. doi: 10.1038/nrm3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochem J. 2011;433(1):31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, Chen ZJ. Expanding role of ubiquitination in NF-kappaB signaling. Cell Res. 2011;21(1):6–21. doi: 10.1038/cr.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wertz IE, Dixit VM. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb Perspect Biol. 2010;2(3):a003350. doi: 10.1101/cshperspect.a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10(11):755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panier S, Durocher D. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair. 2009;8(4):436–443. doi: 10.1016/j.dnarep.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto M, Okamoto T, Takeda K, et al. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat Immunol. 2006;7(9):962–970. doi: 10.1038/ni1367. [DOI] [PubMed] [Google Scholar]
- 26.Laine A, Topisirovic I, Zhai D, Reed JC, Borden KL, Ronai Z. Regulation of p53 localization and activity by Ubc13. Mol Cell Biol. 2006;26(23):8901–8913. doi: 10.1128/MCB.01156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsukamoto S, Takeuchi T, Rotinsulu H, et al. Leucettamol A: a new inhibitor of Ubc13-Uev1A interaction isolated from a marine sponge, Leucetta aff. microrhaphis. Bioorg Med Chem Lett. 2008;18(24):6319–6320. doi: 10.1016/j.bmcl.2008.10.110. [DOI] [PubMed] [Google Scholar]
- 28.Scheper J, Guerra-Rebollo M, Sanclimens G, et al. Protein-protein interaction antagonists as novel inhibitors of non-canonical polyubiquitylation. PloS One. 2010;5(6):e11403. doi: 10.1371/journal.pone.0011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madiraju C, Welsh K, Cuddy MP, et al. TR-FRET-based high-throughput screening assay for identification of UBC13 inhibitors. J Biomol Screen. 2012;17(2):163–176. doi: 10.1177/1087057111423417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol. 2010;2(9):a003004. doi: 10.1101/cshperspect.a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siebenlist U, Brown K, Claudio E. Control of lymphocyte development by nuclear factor-kappaB. Nat Rev Immunol. 2005;5(6):435–445. doi: 10.1038/nri1629. [DOI] [PubMed] [Google Scholar]
- 32.Dupire S, Coiffier B. Targeted treatment and new agents in diffuse large B cell lymphoma. Int J Hematol. 2010;92(1):12–24. doi: 10.1007/s12185-010-0609-6. [DOI] [PubMed] [Google Scholar]
- 33.Su TT, Guo B, Kawakami Y, et al. PKC-beta controls I kappa B kinase lipid raft recruitment and activation in response to BCR signaling. Nat Immunol. 2002;3(8):780–786. doi: 10.1038/ni823. [DOI] [PubMed] [Google Scholar]
- 34.Kim SW, Oleksyn DW, Rossi RM, et al. Protein kinase C-associated kinase is required for NF-kappaB signaling and survival in diffuse large B-cell lymphoma cells. Blood. 2008;111(3):1644–1653. doi: 10.1182/blood-2007-05-088591. [DOI] [PubMed] [Google Scholar]
- 35.Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J Biol Chem. 2007;282(6):4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeRan M, Pulvino M, Greene E, Su C, Zhao J. Transcriptional activation of histone genes requires NPAT-dependent recruitment of TRRAP-Tip60 complex to histone promoters during the G1/S phase transition. Mol Cell Biol. 2008;28(1):435–447. doi: 10.1128/MCB.00607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeRan M, Pulvino M, Zhao J. Assessing G1-to-S-phase progression after genotoxic stress. Methods Mol Biol. 2011;782:221–230. doi: 10.1007/978-1-61779-273-1_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baeuerle PA, Henkel T. Function and activation of NF-kappa B in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 39.Shi R, Re D, Dudl E, et al. Chemical biology strategy reveals pathway-selective inhibitor of NF-kappaB activation induced by protein kinase C. ACS Chem Biol. 2010;5(3):287–299. doi: 10.1021/cb9003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia ZP, Sun L, Chen X, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461(7260):114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruneda JN, Stoll KE, Bolton LJ, Brzovic PS, Klevit RE. Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme approximately ubiquitin conjugate. Biochemistry. 2011;50(10):1624–1633. doi: 10.1021/bi101913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lombardi L, Ciana P, Cappellini C, et al. Structural and functional characterization of the promoter regions of the NFKB2 gene. Nucleic Acids Res. 1995;23(12):2328–2336. doi: 10.1093/nar/23.12.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shetty S, Graham BA, Brown JG, et al. Transcription factor NF-kappaB differentially regulates death receptor 5 expression involving histone deacetylase 1. Mol Cell Biol. 2005;25(13):5404–5416. doi: 10.1128/MCB.25.13.5404-5416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahadevan D, Fisher RI. Novel therapeutics for aggressive non-Hodgkin's lymphoma. J Clin Oncol. 2011;29(14):1876–1884. doi: 10.1200/JCO.2010.32.7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu GW, Ali M, Wood TE, et al. The ubiquitin-activating enzyme E1 as a therapeutic target for the treatment of leukemia and multiple myeloma. Blood. 2010;115(11):2251–2259. doi: 10.1182/blood-2009-07-231191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, Kitagaki J, Dai RM, et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67(19):9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 47.Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2(12):a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farrugia MM, Duan LJ, Reis MD, Ngan BY, Berinstein NL. Alterations of the p53 tumor suppressor gene in diffuse large cell lymphomas with translocations of the c-MYC and BCL-2 proto-oncogenes. Blood. 1994;83(1):191–198. [PubMed] [Google Scholar]
- 49.Sanz I, Lee FE. B cells as therapeutic targets in SLE. Nat Rev Rheumatol. 2010;6(6):326–337. doi: 10.1038/nrrheum.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto M, Sato S, Saitoh T, et al. Cutting Edge: Pivotal function of Ubc13 in thymocyte TCR signaling. J Immunol. 2006;177(11):7520–7524. doi: 10.4049/jimmunol.177.11.7520. [DOI] [PubMed] [Google Scholar]
- 51.Wu X, Yamamoto M, Akira S, Sun SC. Regulation of hematopoiesis by the K63-specific ubiquitin-conjugating enzyme Ubc13. Proc Natl Acad Sci U S A. 2009;106(49):20836–20841. doi: 10.1073/pnas.0906547106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.