Abstract
Heat shock factor 1 (HSF1) is essential for protecting cells from protein-damaging stress associated with misfolded proteins and regulates the insulin-signaling pathway and aging. Here, we show that human HSF1 is inducibly acetylated at a critical residue that negatively regulates DNA binding activity. Activation of the deacetylase and longevity factor SIRT1 prolonged HSF1 binding to the heat shock promoter Hsp70 by maintaining HSF1 in a deacetylated, DNA–binding competent state. Conversely, down-regulation of SIRT1 accelerated the attenuation of the heat shock response (HSR) and release of HSF1 from its cognate promoter elements. These results provide a mechanistic basis for the requirement of HSF1 in the regulation of life span and establish a role for SIRT1 in protein homeostasis and the HSR.
Transient activation of heat shock factor 1 (HSF1) by diverse environmental and physiological stress is a multistep process that involves constitutive expression of an inert HSF1 monomer, conversion of the monomer to a DNA–binding competent trimer, increased phosphorylation of HSF1 at serine residues, enhanced transcription, and attenuation of HSF1 DNA binding and transcriptional activity (1). HSF1 activates the transcription of a large number of genes that regulate protein homeostasis including the molecular chaperones heat shock proteins 70 and 90 (Hsp70 and Hsp90, respectively). These chaperones associate with HSF1 to initiate a negative-feedback loop and to inhibit HSF1 transcriptional activity (2). However, HSF1 is not released from its target promoter sites (3); this suggests that additional mechanisms must exist to complete the HSF1 cycle.
Stress resistance and metabolic state are intimately coupled to protein homeostasis and increased life span. In Caenorhabditis elegans, the protective effects of reduced insulin signaling require HSF1 and the FOXO transcription factor DAF-16 to prevent damage by protein misfolding and to promote longevity (4, 5). The beneficial effects of low caloric intake are mediated by the sirtuin family member Sir2, a deacetylase that is dependent on nicotinamide adenine dinucleotide (oxidized form) (NAD) and that is under metabolic control (6). The mammalian Sir2 homolog SIRT1 regulates the transcription factor FOXO3 among other cellular protective pathways (7). We therefore tested whether the sirtuins, specifically SIRT1, regulate HSF1 activity and thereby provide a direct link between these three longevity factors.
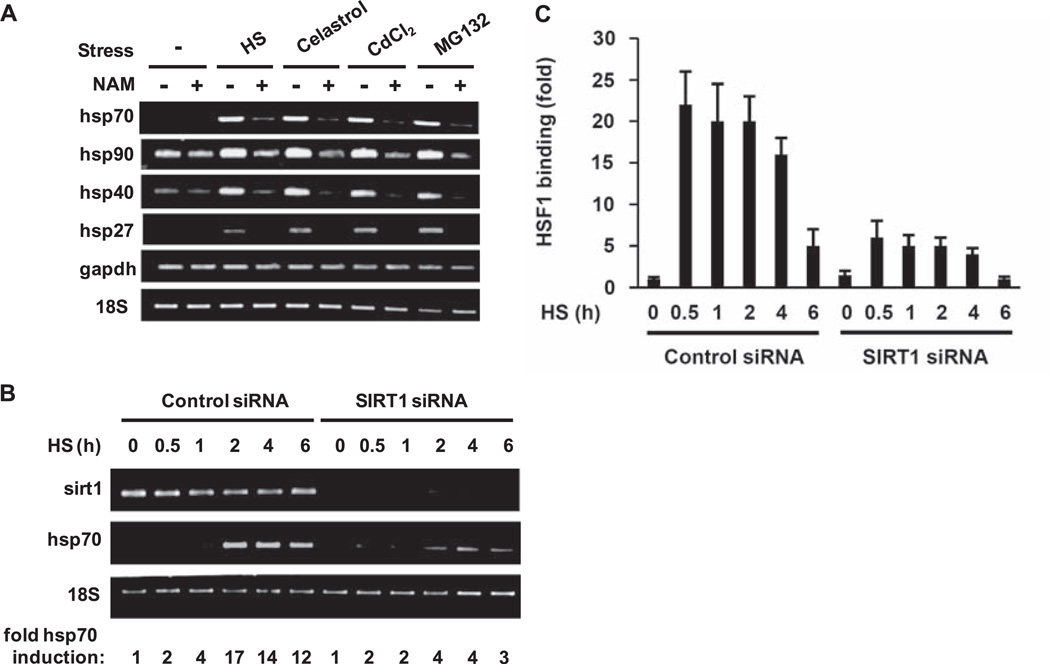
We treated HeLa cells with the sirtuin inhibitor nicotinamide (8) and then exposed the cells to various stresses known to induce the heat shock response (HSR) (9). Nicotinamide treatment decreased abundance of the stress-induced mRNAs from all major classes of heat shock genes (hsp70, hsp90, hsp40, and hsp27) (Fig. 1A), which indicated that the sirtuins are required for full induction of the HSR. Of three nuclear sirtuins, SIRT1 has well-characterized targets (10). We therefore investigated SIRT1 as a candidate for regulation of the HSR. When SIRT1 was depleted by small interfering RNA (siRNA), the amount of hsp70 mRNA produced during a 6-hour heat shock was one-fourth of that in cells transfected with control siRNA (Fig. 1B).
Fig. 1.
Regulation of the HSR by sirtuins. (A) Effect of the sirtuin inhibitor nicotinamide on chaperone gene expression. HeLa cells were treated with nicotinamide (NAM) before exposure to heat shock (HS), celastrol, CdCl2, or MG132, and reverse transcription (RT)–PCR analysis was performed with the indicated primers. (B) SIRT1 siRNA inhibits transcription of hsp70. HeLa cells transfected with siRNA against SIRT1 or a control siRNA were treated with heat shock for the indicated times. RT-PCR analysis was performed and fold increase in hsp70 mRNA abundance was determined by densitometry and normalized to 18S ribosomal RNA. (C) SIRT1 siRNA inhibits HSF1 binding to the hsp70 promoter. HeLa cells were treated as described above (B). ChIP analysis was performed using an HSF1 antibody and qPCR, and the results were normalized to reactions performed with 1% of input. Experiments were performed in triplicate, and error bars indicate ±SD.
To examine whether SIRT1 influences recruitment of HSF1 to the hsp70 promoter, we performed chromatin immunoprecipitation (ChIP) assays with cells transfected with control or SIRT1 siRNA before heat shock (Fig. 1C). In control siRNA-treated cells, binding of HSF1 to the hsp70 promoter occurs rapidly and begins to attenuate at 30 min of heat shock (11), with a gradual decline over a 6-hour period. However, in SIRT1 siRNA-transfected cells, about one-fourth as much HSF1 was associated with the promoter throughout the time course. These results support a role for SIRT1 as an in vivo regulator of HSF1 DNA binding activity and hsp70 expression.
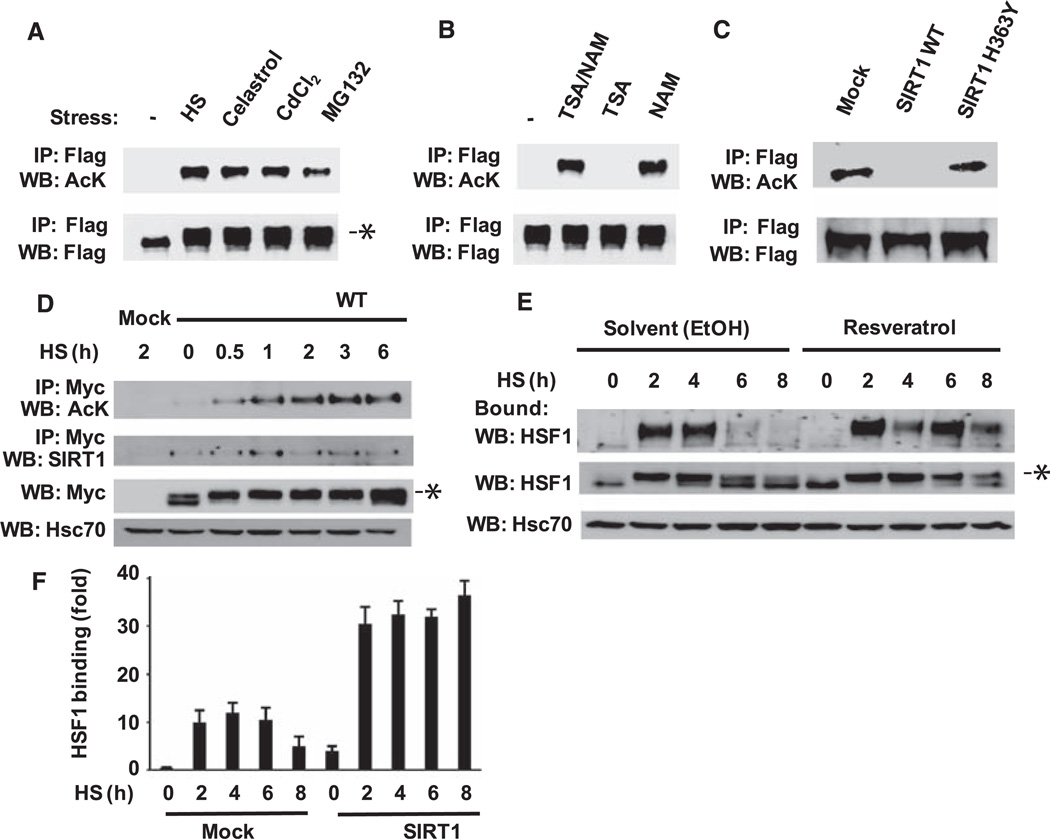
To determine whether HSF1 is a direct target of SIRT1, we examined the acetylation status of HSF1. We transfected 293T cells with vectors encoding a Flag-HSF1 fusion protein and p300 and exposed them to several HSR inducers. Immunoprecipitated HSF1 was analyzed by Western blotting with an antibody that binds acetylated lysines. Acetylated HSF1 was not detected in untreated cells but was present in cells exposed to various stress conditions (Fig. 2A). The endogenous acetyltransferase that regulates HSF1 acetylation may be p300/CBP [adenosine 3′,5′-monophosphate (cAMP) response element–binding protein], as overexpression of either p300 or CBP, but not p300/CBP-associated factor, resulted in acetylation of HSF1 (fig. S1A), and p300 was recruited to the hsp70 promoter after heat shock (fig. S1B). SIRT1 also binds to the hsp70 promoter under both basal and stress conditions (fig. S1C).
Fig. 2.
Regulation of stress-induced acetylation of HSF1 by SIRT1. (A) Acetylation of HSF1 in response to HSR inducers. 293T cells transfected with Flag-HSF1 and p300 were treated with heat shock (HS), celastrol, CdCl2 or MG132. Cell lysates were analyzed by acetylation assay using immunoprecipitation and Western blotting (9). Asterisk (*) indicates HSF1 that is slowly migrating because of increased phosphorylation (17). (B) Effects of nicotinamide and trichostatin A on HSF1 acetylation. 293T cells transfected with Flag-HSF1 and p300 were treated with trichostatin A (TSA), nicotinamide (NAM), or both, and exposed to heat shock, then cell lysates were analyzed by acetylation assay. (C) Wild-type SIRT1, but not a catalytic mutant, inhibits HSF1 acetylation. 293T cells were transfected with Flag-HSF1, p300, and either SIRT1 WT or a SIRT1 H363Y mutant before treatment with heat shock and analysis by acetylation assay. (D) HSF1 acetylation in response to heat shock. Cos7 cells transfected with Myc-HSF1 were treated with heat shock for the indicated times and analyzed by acetylation assay. SIRT1 and HSF1 were detected with an antibody to SIRT1 or a Myc-specific antibody. Heat-shock cognate protein 70 (Hsc70) was a loading control. (E) Effects of resveratrol. HeLa cells were treated with ethanol solvent (EtOH) or resveratrol before heat-shock treatment for the indicated times. Cell extracts were analyzed by oligonucleotide pull-down assay and Western blotting. Increased phosphorylation of HSF1 is indicated by an asterisk. (F) SIRT1 overexpression effect on HSF1 DNA binding. 293T cells were transfected with SIRT1 WT and then subjected to heat shock for the indicated times. ChIP analysis was performed using an HSF1 antibody and qPCR. Experiments in (A) to (G) were performed in triplicate, and error bars indicate ±SD.
Deacetylases are grouped into three families, with the class I and II histone deacetylase (HDAC) families inhibited by trichostatin A (12) and the nicotinamide adenine dinucleotide (NAD+)–dependent class III sirtuin family inhibited by nicotinamide (8). Trichostatin A had no effect on deacetylation of HSF1, whereas nicotinamide inhibited deacetylation alone or in the presence of trichostatin A (Fig. 2B). Overexpression of SIRT1 WT, but not a point mutant with impaired NAD-dependent deacetylase activity [SIRT1 H363Y in which histidine at position 363 was replaced by tyrosine (13, 14)], inhibited HSF1 acetylation (Fig. 2C), which supported a role for SIRT1 in HSF1 function. HSF1 acetylation was not cell type–specific, as it was detected in 293T and Cos7 cells, and although HSF1 acetylation was enhanced by p300 overexpression, it did not require p300 overexpression (Fig. 2, A and D). The kinetics of HSF1 acetylation do not match the kinetics of HSF1 activation. Acetylation is delayed and persists during the period when HSF1 activity and DNA binding have attenuated (11). In addition, an HSF1 in which 10 potentially phosphorylated serines were replaced with alanines remained competent for acetylation, which suggests that phosphorylation of HSF1 is not a prerequisite for acetylation (fig. S2).
The persistence of HSF1 acetylation during later time points of the HSR and the coimmunoprecipitation of SIRT1, together with HSF1 (Fig. 2D), led us to investigate whether SIRT1 has a role in attenuation of HSF1 activity. We treated HeLa cells with resveratrol, a small-molecule inducer of SIRT1 activity (15), and assayed HSF1 DNA binding activity in an oligonucleotide-based pull-down assay (16) (Fig. 2E). In cells treated with heat shock and vehicle alone, HSF1 DNA binding was induced within 2 hours and attenuated after 6 hours. The transient activation of HSF1 was reflected by altered mobility on SDS–polyacrylamide gel electrophoresis (SDS-PAGE), which detects the stress-induced phosphorylated state of HSF1 (17). In contrast, HSF1 in resveratrol-treated cells persisted in a DNA–binding competent and phosphorylated state even after 8 hours of continuous heat shock. In cells overexpressing SIRT1, HSF1 DNA binding was enhanced, and attenuation was suppressed as measured by ChIP experiments (Fig. 2F). These results suggest that changes in the abundance and activity of SIRT1 regulate the attenuation of the HSR.
To elucidate the mechanism by which acetylation regulates HSF1 DNA binding, we identified the sites of acetylation on HSF1 by mass spectrometry of peptides from Flag-HSF1 purified from 293T cells. At least nine lysines in HSF1 were acetylated in response to stress (fig. S3) of which K80, located in the DNA binding domain, was particularly intriguing because mutations of the corresponding lysine of yeast HSF cause a loss-of-function phenotype (18, 19). Furthermore, analysis of the crystal structure of Kluyveromyces lactis HSF indicated that the lysine corresponding to human HSF1 K80 is located in a short domain that connects the main DNA binding helix to a flexible and solvent-exposed loop and forms a hydrogen bond with the DNA phosphate backbone (20). Comparative protein modeling of the HSF-HSE (HSF–heat shock element) crystal structure showed that human HSF1 K80 is in close contact with the DNA backbone (fig. S4), which suggests that neutralizing the positive charge of lysine by acetylation should interfere with DNA binding.
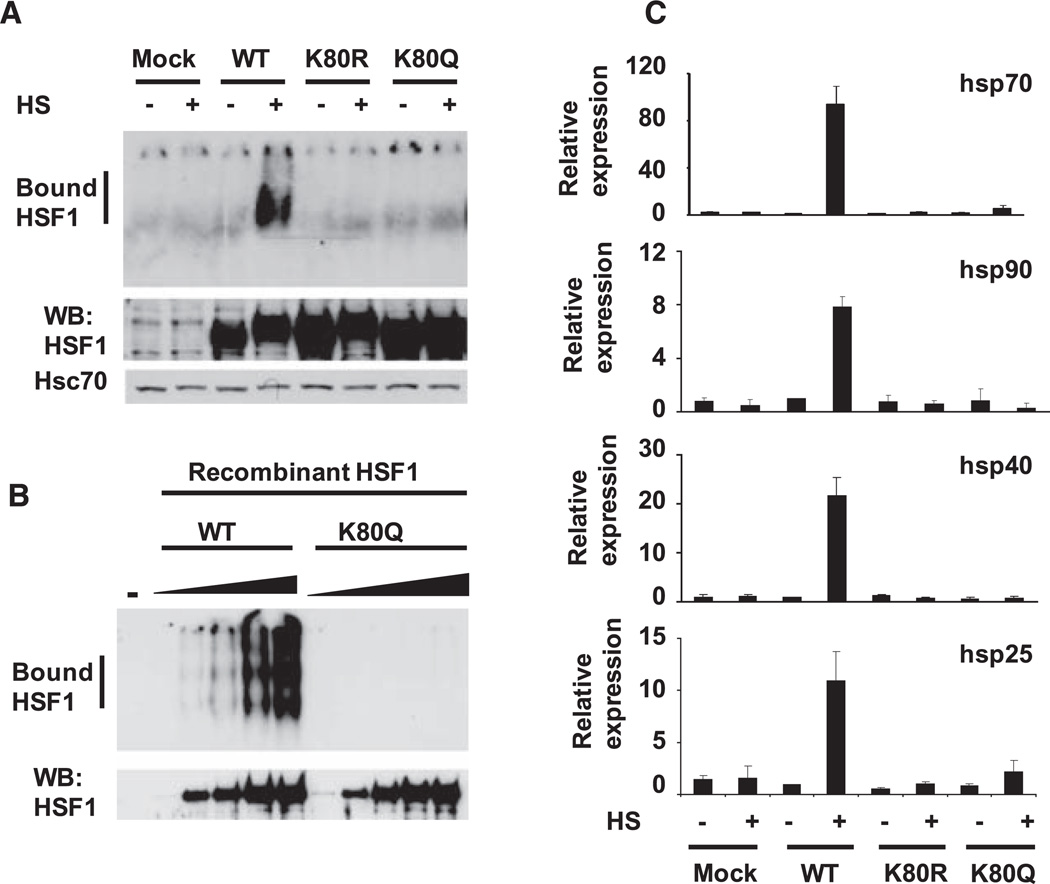
We therefore replaced K80 with a glutamine to mimic constitutive acetylation. In extracts from hsf1−/− fibroblasts (21) transfected with HSF1 wild-type (WT) and HSF1 K80Q expression constructs, the mutant protein failed to bind DNA in an electrophoretic mobility shift assay (EMSA) (Fig. 3A). The K80Q mutant, however, still assembled into heat shock–induced trimers, a hallmark of the DNA-bound state (fig. S5). Substitution of other amino acids at K80 [K80R, -A, -H, -N, and -T (14)] also resulted in defective DNA binding (Fig. 3A, fig. S6). In vitro, recombinant nonacetylated WT HSF1 readily bound to a synthetic HSE, but the K80Q mutant protein did not (Fig. 3B). We introduced WT and the K80 mutants into hsf1−/− fibroblasts and analyzed the heat shock–induced expression of HSF1 target genes by quantitative polymerase chain reaction (qPCR). Although WT HSF1 induced expression of heat shock protein mRNAs, HSF1 K80 mutants were nonfunctional (Fig. 3C). The mutants localized to the nucleus upon heat shock but were impaired in the relocalization into nuclear stress bodies that occurs in heat-shocked human cells (figs. S7 and S8) (22). An unmodified lysine side chain at residue 80 appears to be required for HSF1-HSE binding ability, relocalization into nuclear stress bodies, and expression of target genes. Therefore, we propose that acetylation of HSF1 K80 causes the regulated release of the HSF1 trimers from DNA and thus represents a regulatory step in the attenuation of the HSR (fig. S9).
Fig. 3.
Mutation of HSF1 K80 inhibits the HSR. (A) Mutation of HSF1 at K80 disrupts DNA binding activity. EMSA reactions were performed with extracts from hsf1−/− cells transfected with the indicated HSF1 constructs treated with or without heat shock (HS) (top). The EMSA probe contains the proximal HSE from the human hsp70 promoter. Western blot analysis was performed on the same samples to show HSF1 and Hsc70 levels. (B) Mutation of recombinant HSF1 at K80 disrupts DNA binding ability. EMSA reactions with increasing amounts (5, 20, 40, 80, or 120 ng) of recombinant WT HSF1 or HSF1 K80Q and a probe containing an HSE are shown (top). A sample without (−) HSF1 protein was a control. Western blot analysis was performed on the same samples to show HSF1 expression levels. (C) Failure of HSF1 mutated at K80 to rescue the HSR in hsf1−/− cells. hsf1−/− cells were transfected with the indicated versions of human HSF1 and treated with or without heat shock. RNA was quantified using qPCR with primers for the indicated genes. Data are normalized to values obtained for glyceraldehyde 3-phosphate dehydrogenase and are relative to the abundance of each mRNA in WT HSF1 cells treated without heat shock (value set as 1). Experiments in (A) to (C) were performed in triplicate, and error bars indicate ±SD.
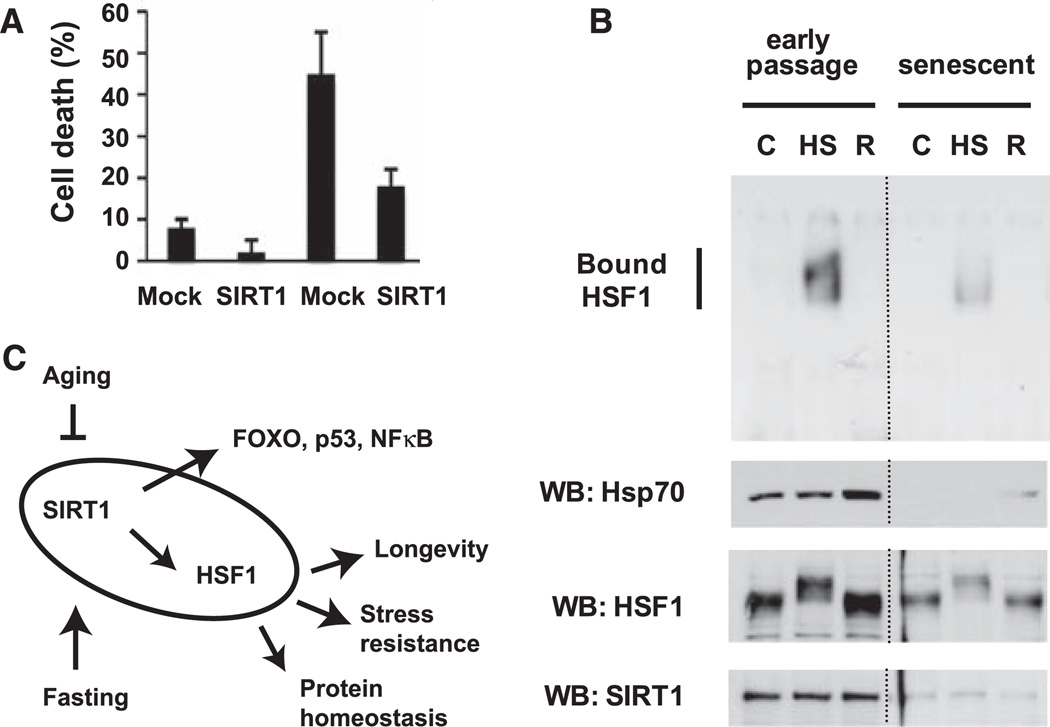
To verify the biological significance of the regulation of the HSR by SIRT1, we used an assay of stress resistance in which the expression of chaperones confers increased thermotolerance (23). 293T cells were transfected with or without SIRT1, exposed to a 45°C heat shock for 20 or 30 min, allowed to recover for 24 hours, and analyzed for cell death. As expected, the 45°C heat shock resulted in cell death that increased with treatment time (Fig. 4A). At both time points, the cells overexpressing SIRT1 had about one-third as many cells undergo cell death (Fig. 4A). To examine whether age-regulated changes in SIRT1 affect HSF1 activity and the HSR, we used human WI-38 fibroblasts that have been widely used in studies on molecular changes in the aging process. When comparing early and late passage numbers, we found that aging resulted in a decreased HSR and reduced activation of HSF1 DNA binding activity that correlated with the reduced abundance of SIRT1 (Fig. 4B).
Fig. 4.
Biological effects of SIRT1 on the HSR. (A) Protection of cells from severe stress by overexpressed SIRT1. 293T cells were transfected with empty vector (mock) or SIRT1 and treated with a 45°C heat shock for the indicated times, followed by recovery at 37°C. After 24 hours, cell death was determined by trypan blue uptake. The experiment was performed three times in triplicate, and error bars indicate ±SD. (B) Correlation of the age-dependent decline in the HSR with decreased abundance of SIRT1. Cell extracts from early passage (passage 21) and senescent (passage 44) WI-38 fibroblasts were treated with heat shock (HS) or heat shock followed by a 3-hour recovery at 37°C (R) and analyzed by EMSA with a probe containing an HSE (top). Western blot analysis was done on the same samples to show Hsp70, HSF1, and SIRT1 expression levels (bottom). (C) Model of the HSF1-SIRT1 regulatory network. The regulation of SIRT1 by aging and cellular metabolic state affects the activity of a network of transcription factors, including HSF1, to result in increased longevity and stress resistance.
The finding that SIRT1 regulates HSF1 complements previous observations on the role of HSF1 in regulating life span (4, 5). HSF1 appears to be at the hub of a regulatory network in which cell nutrition, stress, and life span are linked. Many SIRT1-regulated transcription factors, including FOXO3, p53, and nuclear factor–κB, have important roles in cellular stress responses (7, 24, 25). The addition of HSF1 to this stress regulatory network emphasizes the central role of protein homeostasis in SIRT1-mediated cellular protection (Fig. 4C) and may link the molecular response of the HSR to metabolic demands. A consistent observation in cell-based and animal studies has been the aging-related decline of the HSR (23), which may result, at least in part, from SIRT1 control of HSF1 activity. At the organismal level, we expect that regulation of HSF1 target genes may be influenced by diet and nutrition.
Supplementary Material
Acknowledgments
We thank N. Denslow, S. McClung, A. Schilling, and the Chicago Biomedical Consortium for assistance with the mass spectrometry; A. Mondragon for assistance with the molecular modeling; S. Raju for technical assistance; I. Benjamin for the hsf1−/− fibroblasts; and J. Brickner and members of the Morimoto laboratory for critical reading of the manuscript. This work was supported by an NIH training grant (S.D.W); Turku Graduate School of Biomedical Sciences (J.A.); Academy of Finland, Sigrid Juselius Foundation, and Åbo Akademi University (L.S.); and the National Institute for General Medical Science, the National Institute for Aging, and the Rice Institute for Biomedical Research (R.I.M.).
References and Notes
- 1.Anckar J, Sistonen L. Adv. Exp. Med. Biol. 2007;594:78. doi: 10.1007/978-0-387-39975-1_8. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y, Mosser DD, Morimoto RI. Genes Dev. 1998;12:654. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabindran SK, Wisniewski J, Li L, Li GC, Wu C. Mol. Cell. Biol. 1994;14:6552. doi: 10.1128/mcb.14.10.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu AL, Murphy CT, Kenyon C. Science. 2003;300:1142. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 5.Morley JF, Morimoto RI. Mol. Biol. Cell. 2004;15:657. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Nature. 2000;403:795. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 7.Brunet A, et al. Science. 2004;303:2011. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 8.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. J. Biol. Chem. 2002;277:45099. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 9.Materials and methods are available as supporting material on Science Online.
- 10.Dali-Youcef N, et al. Ann. Med. 2007;39:335. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- 11.Kline MP, Morimoto RI. Mol. Cell. Biol. 1997;17:2107. doi: 10.1128/mcb.17.4.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida M, Kijima M, Akita M, Beppu T. J. Biol. Chem. 1990;265:17174. [PubMed] [Google Scholar]
- 13.Langley E, et al. EMBO J. 2002;21:2383. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
- 15.Howitz KT, et al. Nature. 2003;425:191. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 16.Anckar J, et al. Mol. Cell. Biol. 2006;26:955. doi: 10.1128/MCB.26.3.955-964.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarge KD, Murphy SP, Morimoto RI. Mol. Cell. Biol. 1993;13:1392. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubl ST, Owens JC, Nelson HC. Nat. Struct. Biol. 1994;1:615. doi: 10.1038/nsb0994-615. [DOI] [PubMed] [Google Scholar]
- 19.Torres FA, Bonner JJ. Mol. Cell. Biol. 1995;15:5063. doi: 10.1128/mcb.15.9.5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Littlefield O, Nelson HC. Nat. Struct. Biol. 1999;6:464. doi: 10.1038/8269. [DOI] [PubMed] [Google Scholar]
- 21.McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. J. Biol. Chem. 1998;273:7523. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- 22.Jolly C, et al. J. Cell Biol. 2002;156:775. doi: 10.1083/jcb.200109018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kregel KC. J. Appl. Physiol. 2002;92:2177. doi: 10.1152/japplphysiol.01267.2001. [DOI] [PubMed] [Google Scholar]
- 24.Vaziri H, et al. Cell. 2001;107:149. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 25.Yeung F, et al. EMBO J. 2004;23:2369. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.