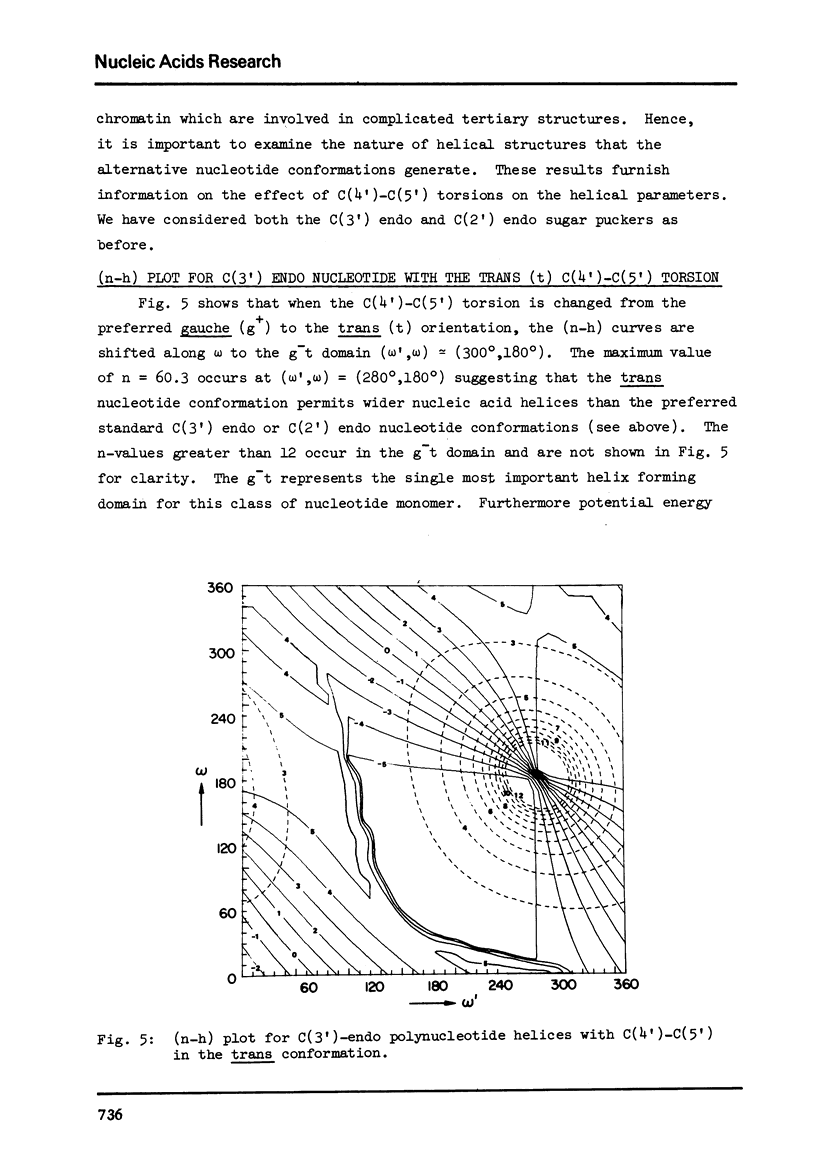
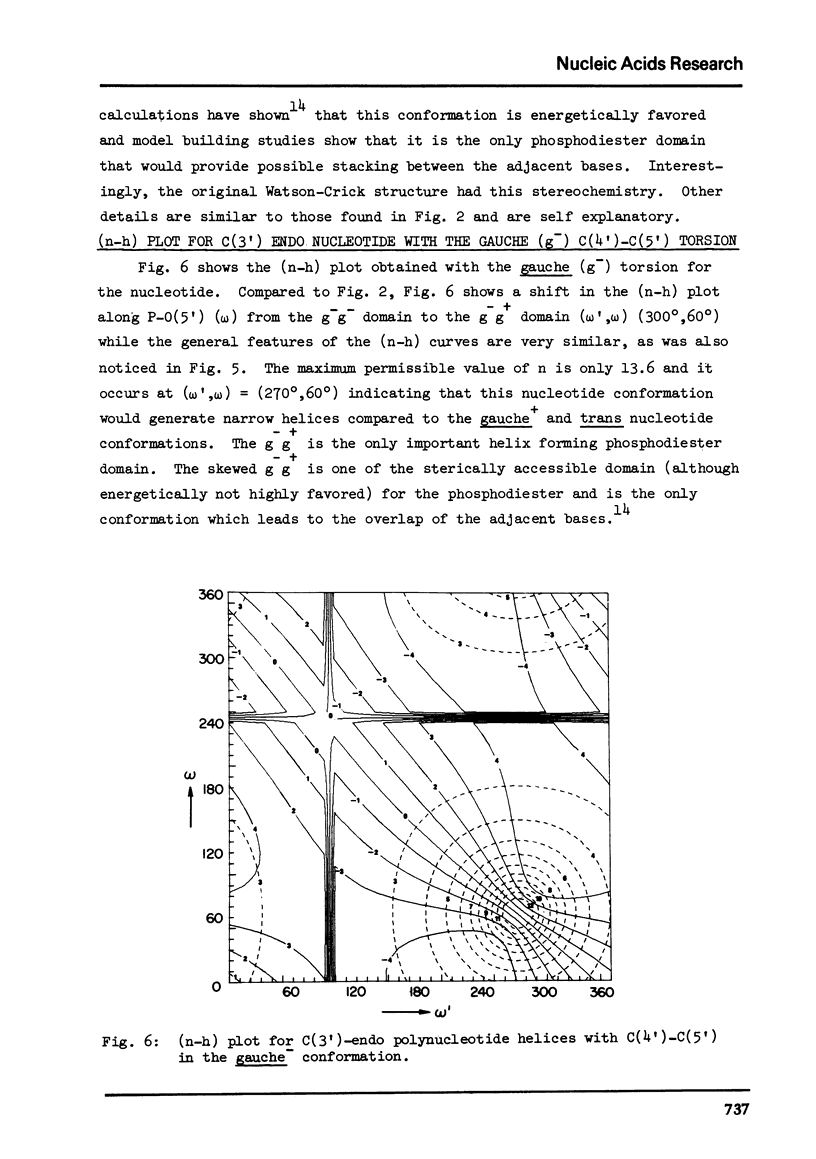
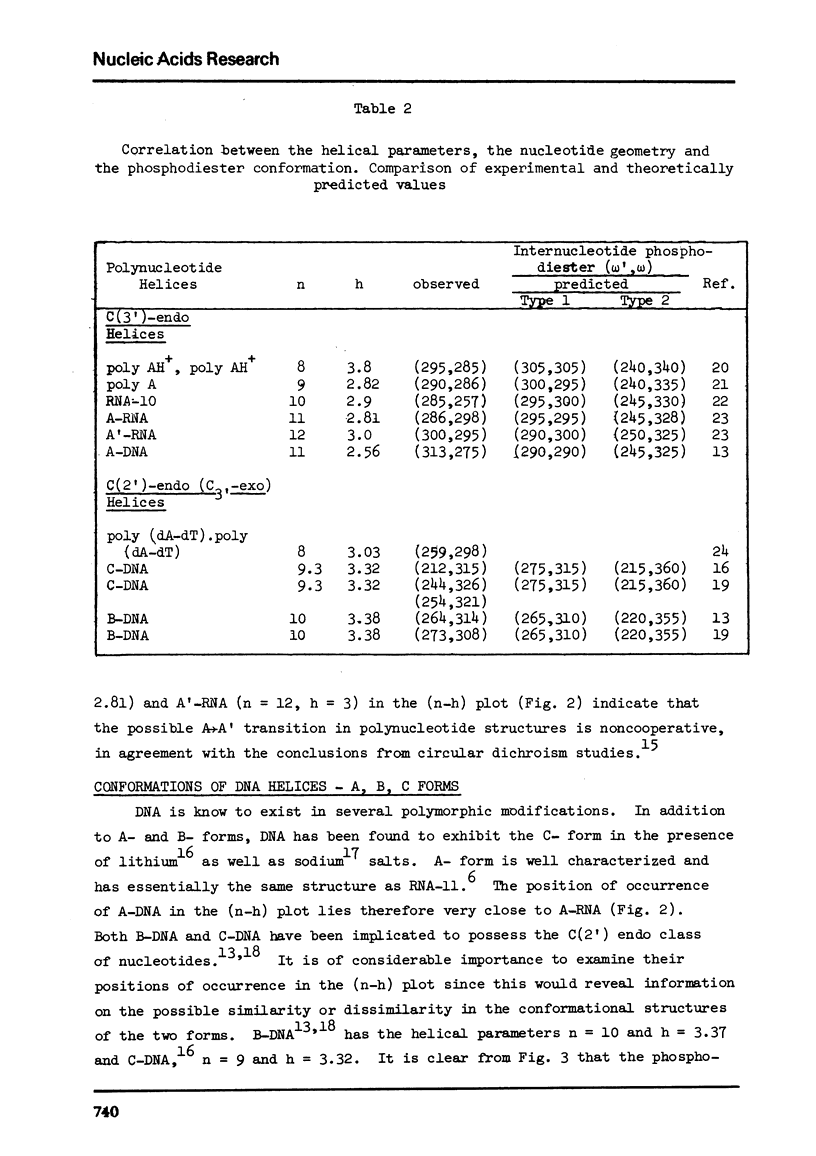
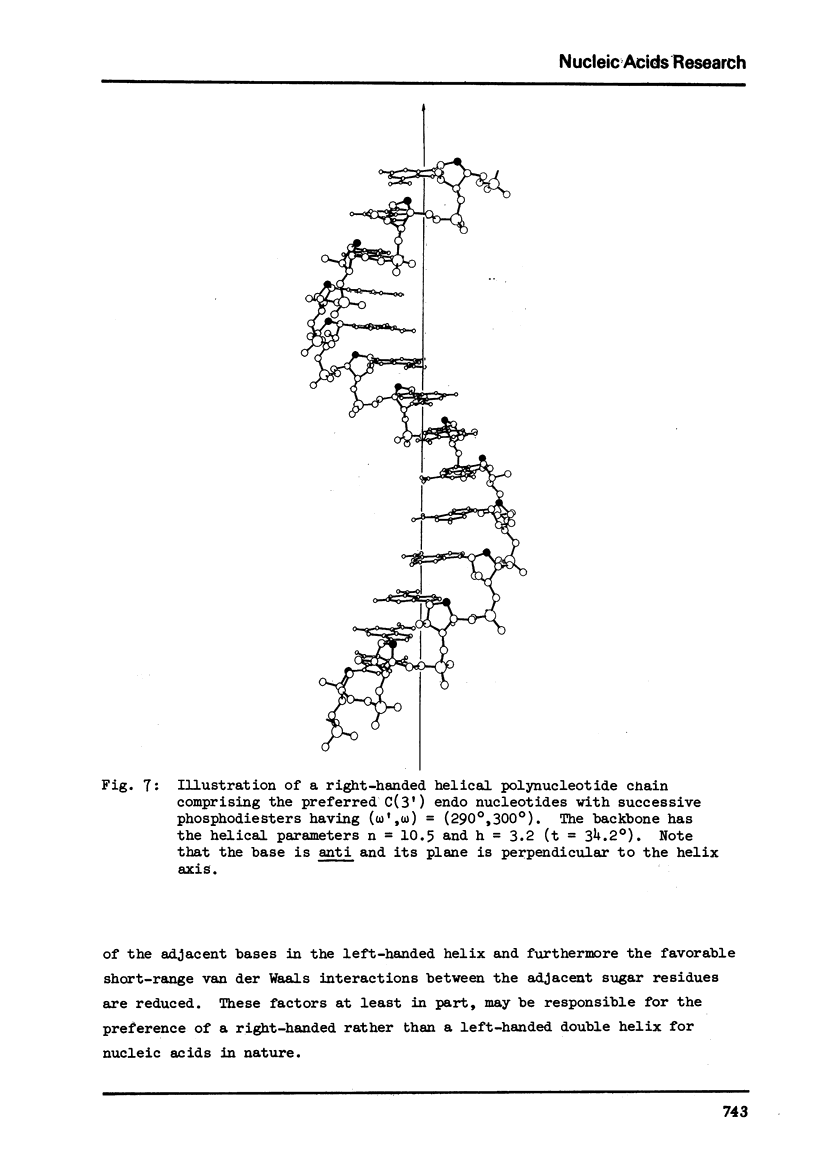
Abstract
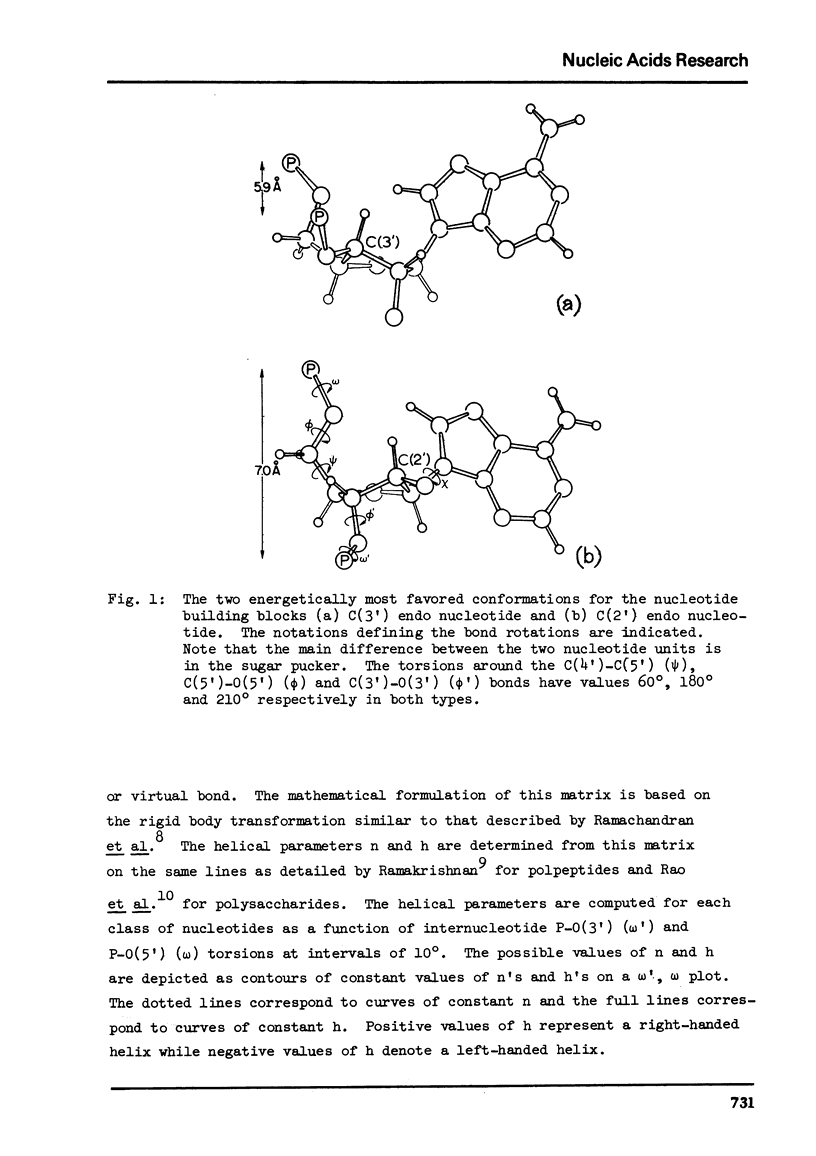
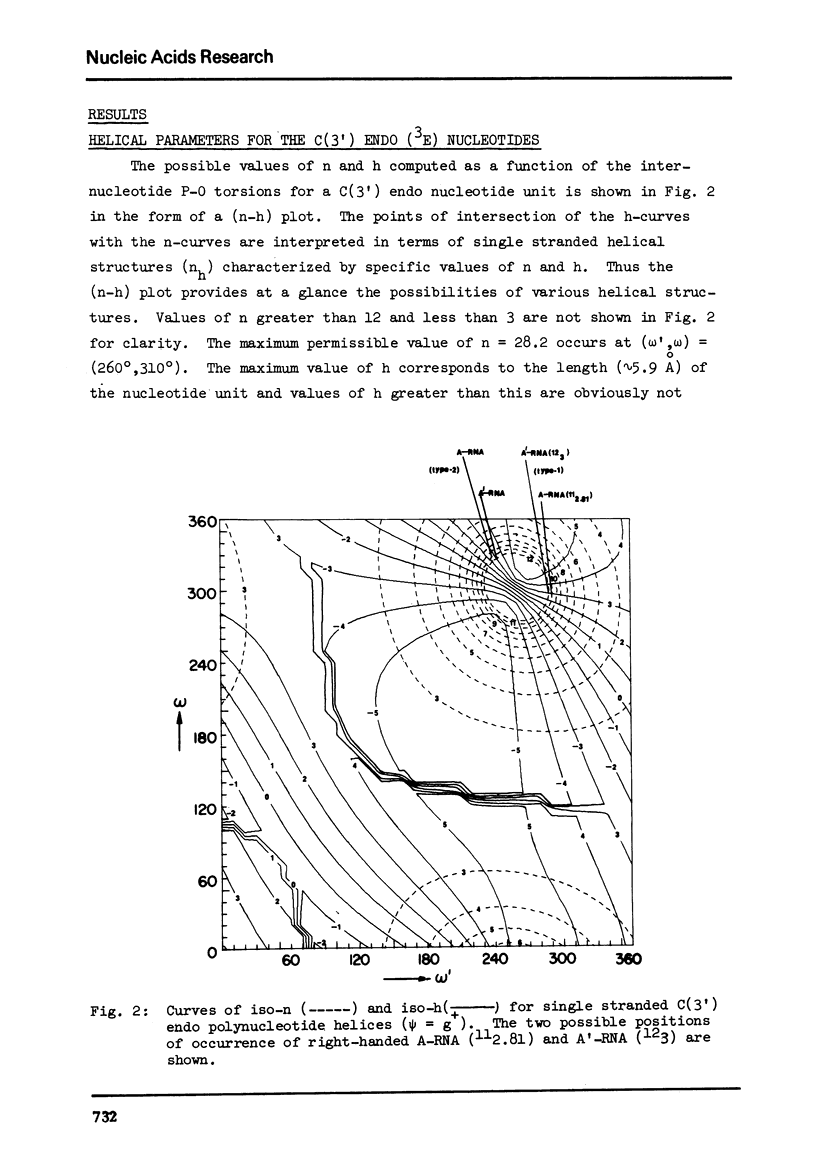
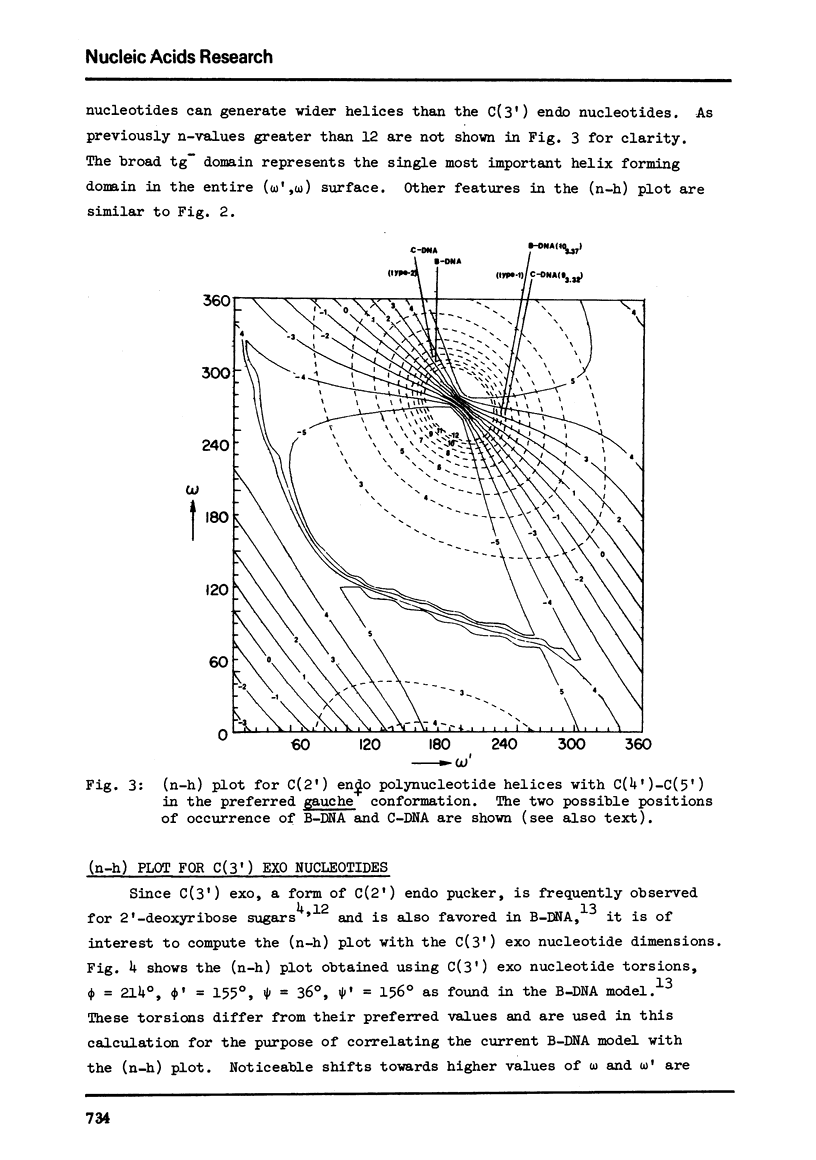
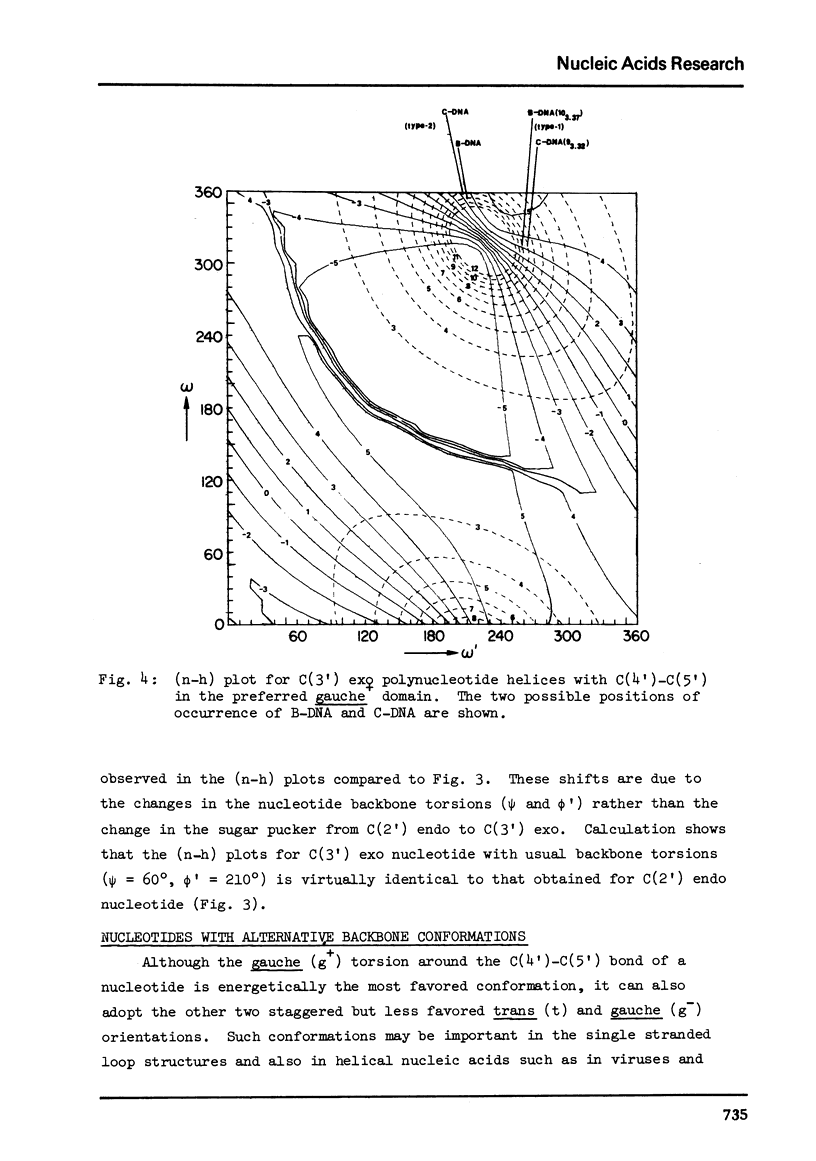
The two helical parameters n and h where n is the number of nucleotide residues per turn and h is the height per nucleotide residue have been evaluated for single stranded helical polynucleotide chains comprising C(3') -endo and C(2') endo class of nucleotides. The helical parameters are found to be especially sensitive to the C(4')-C(3') (sugar pucker) and the C(4')-C(5') torsions. The (n-h) plots display only one important helix forming domain for each class of nucleotides characterized by the sugar pucker and the C(4')-C(5') torsion. A correlation between the (n-h) plots and the known RNA (A,A') and DNA (A,B,C) helical forms has been established. It is found that all forms of helices except the C-DNA possess a favorable combination of P-O torsions. The analysis of the (n-h) plots suggests that C-DNA can have a conformation very similar to B-DNA. Although the (n-h) plots predict the stereochemical possibility of both right-handed and left-handed helices, nucleic acids apparently prefer right-handed conformation because of the energetics associated with the sugar-phosphate backbone and the base.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Hukins D. W., Smith P. J., Watts L. Structural details of double-helix observed for DNAs containing alternating purine and pyrimidine sequences. J Mol Biol. 1974 Sep 15;88(2):523–533. doi: 10.1016/0022-2836(74)90499-9. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Marttila C. M. Structures for polyinosinic acid and polyguanylic acid. Biochem J. 1974 Aug;141(2):537–543. doi: 10.1042/bj1410537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W., Dover S. D. Optimised parameters for RNA double-helices. Biochem Biophys Res Commun. 1972 Sep 26;48(6):1392–1399. doi: 10.1016/0006-291x(72)90867-4. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W. Optimised parameters for A-DNA and B-DNA. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1504–1509. doi: 10.1016/0006-291X(72)90243-4. [DOI] [PubMed] [Google Scholar]
- Arnott S., Selsing E. The conformation of C-DNA. J Mol Biol. 1975 Oct 15;98(1):265–269. doi: 10.1016/s0022-2836(75)80115-x. [DOI] [PubMed] [Google Scholar]
- Arnott S. The geometry of nucleic acids. Prog Biophys Mol Biol. 1970;21:265–319. doi: 10.1016/0079-6107(70)90027-1. [DOI] [PubMed] [Google Scholar]
- Brahms J., Pilet J., Phuong Lan T. T., Hill L. R. Direct evidence of the C-like form of sodium deoxyribonucleate. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3352–3355. doi: 10.1073/pnas.70.12.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller W., Hutchinson F., Spencer M., Wilkins M. H. Molecular and crystal structures of double-helical RNA. I. An x-ray diffraction study of fragmented yeast RNA and a preliminary double-helical RNA model. J Mol Biol. 1967 Aug 14;27(3):507–524. doi: 10.1016/0022-2836(67)90055-1. [DOI] [PubMed] [Google Scholar]
- Ivanov V. I., Minchenkova L. E., Schyolkina A. K., Poletayev A. I. Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers. 1973;12(1):89–110. doi: 10.1002/bip.1973.360120109. [DOI] [PubMed] [Google Scholar]
- MARVIN D. A., SPENCER M., WILKINS M. H., HAMILTON L. D. The molecular configuration of deoxyribonucleic acid. III. X-ray diffraction study of the C form of the lithium salt. J Mol Biol. 1961 Oct;3:547–565. doi: 10.1016/s0022-2836(61)80021-1. [DOI] [PubMed] [Google Scholar]
- RICH A., DAVIES D. R., CRICK F. H., WATSON J. D. The molecular structure of polyadenylic acid. J Mol Biol. 1961 Feb;3:71–86. doi: 10.1016/s0022-2836(61)80009-0. [DOI] [PubMed] [Google Scholar]
- Ramachandran G. N., Venkatachalam C. M., Krimm S. Stereochemical criteria for polypeptide and protein chain conformations. 3. Helical and hydrogen-bonded polypeptide chains. Biophys J. 1966 Nov;6(6):849–872. doi: 10.1016/s0006-3495(66)86699-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger W., Riecke J., Suck D. A structural model for the polyadenylic acid single helix. J Mol Biol. 1975 Apr 25;93(4):529–534. doi: 10.1016/0022-2836(75)90244-2. [DOI] [PubMed] [Google Scholar]
- Yathindra N., Sundaralingam M. Backbone conformations in secondary and tertiary structural units of nucleic acids. Constraint in the phosphodiester conformation. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3325–3328. doi: 10.1073/pnas.71.9.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S. B., Cohen G. H., Davies D. R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J Mol Biol. 1975 Feb 25;92(2):181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]


