Introduction
Buruli ulcer disease (BUD) caused by Mycobacterium ulcerans involves the skin and soft tissue. If left untreated, extensive destruction of tissue followed by scarring and contractures may lead to severe functional limitations. Following the introduction of standardized antimycobacterial chemotherapy with rifampicin and streptomycin, recurrence rates of less than 2% were reported. However, treatment failures occur and a variety of secondary lesions necessitating customized clinical management strategies have been reported. True recurrences by definition occur more than three months after completion of antibiotic treatment, are characterised by the presence of viable bacilli, and require a second course of antibiotics. “Non-healers” may harbour viable, possibly drug-resistant M. ulcerans strains and may benefit from surgical intervention. Early-onset immune-mediated paradoxical reactions emerging during or shortly after treatment do not contain viable bacilli and may heal under conventional wound care and/or minor surgery; late-onset secondary lesions presumably attributable to secondary infection foci may clear spontaneously through enhanced immune responses primed by initial treatment. None of the current diagnostic techniques is applicable to rapidly address the pivotal question of the presence of viable bacilli in non-healers and patients with secondary BUD lesions, and optimal time points for collection of follow-up samples have not yet been investigated. Therefore, to date treatment monitoring is mainly based on clinical observation [1]–[5]. Reverse transcriptase assays targeting 16S rRNA and mRNA were successfully applied for the rapid detection of viable mycobacteria in clinical samples from patients with tuberculosis and leprosy [6], [7]. To employ this technique for classification of BUD lesions and monitoring of treatment success we developed a M. ulcerans–specific RNA-based viability assay combining a 16S rRNA reverse transcriptase real-time PCR (RT-qPCR) to determine bacterial viability with an IS2404 quantitative real-time PCR (qPCR) for increased specificity and simultaneous quantification of bacilli.
Development and Validation
Ethical Approval
The study was approved by the Committee of Human Research Publication and Ethics, School of Medical Sciences, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana (CHRPE/28/09). Written informed consent was obtained from all study participants, or their legal representatives.
Bacterial Strains, DNA Extracts, and Clinical Samples
Technical validation of the assay was performed with 29 M. ulcerans strains originating from Cameroon [8] and Ghana (Table 1), as well as DNA extracts from 18 closely related human pathogenic mycobacterial species and five bacterial species frequently colonizing human skin (Table 2).
Table 1. M. ulcerans cultures subjected to the 16S rRNA RT/IS2404 qPCR assay.
| M. ulcerans Strain | Source | Origina | 16S rRNA RT-qPCRb | IS2404 qPCRc | IS2404 qPCR – Wipeout Controld |
| K4s-C1 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K4s-C2 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K4s-C3 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K5d-C1 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K5d-C2 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K5d-C1 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K5d-C2 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K5d-C3 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K5d-C4 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K5s-C1 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K5s-C2 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K5s-C3 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K5s-C4 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K5s-C5 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K7b-C1 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K7b-C2 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K7b-C3 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K7b-C4 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K7s-C1 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K7s-C2 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| K12S-C1 | DITM | Human isolate – Kamerun | Positive | Positive | Negative |
| 941328-C1 | DITM | Human isolate – Ghana | Positive | Positive | Negative |
| 07-C1 | DITM | Human isolate – Ghana | Positive | Positive | Negative |
| DS1-C1 | DITM | Human isolate – Ghana | Positive | Positive | Negative |
| 97680-C1 | DITM | Human isolate – Ghana | Positive | Positive | Negative |
| G.A.P.001-C1 | KCCR | Human isolate – Ghana | Positive | Positive | Negative |
| G.A.P.033-C1 | KCCR | Human isolate – Ghana | Positive | Positive | Negative |
| G.A.P.071-C1 | KCCR | Human isolate – Ghana | Positive | Positive | Negative |
| G.A.P.078-C1 | KCCR | Human isolate – Ghana | Positive | Positive | Negative |
Table 1 shows 29 M. ulcerans cultures that were available at the Department of Infectious Diseases and Tropical Medicine (DITM) and the Kumasi Centre for Collaborative Research (KCCR) for development and technical validation of the 16S rRNA RT/IS2404 qPCR viability assay and the corresponding test results. Sequence analysis of 16S rRNA genes from the listed strains revealed 100% nucleotide concordance of the corresponding genomic regions amplified by the 16S rRNA RT-qPCR; no SNPs or mutations were detected, suggesting a high selectivity of the assay. Sequencing primers are described in Table 3 [11].
M. ulcerans cultures were available from previous studies from Kamerun (n = 21) and Ghana (n = 4) at DITM [8] or were available at KCCR (n = 4) from the present study. All strains were of human origin (BUD patients) and confirmed by conventional IS2404 PCR and sequencing of rpoB- and rpsL-genes that revealed the M. ulcerans Agy99 wild-type sequences (GenBank accession no. CP000325.1) [11], [12].
Results of the 16S rRNA RT-qPCR of mycobacterial RNA extracts.
Results of the IS2404 qPCR of mycobacterial DNA extracts.
Results of the IS2404 qPCR of genomic DNA (gDNA) wipeout controls (see Protocols S2 and S3); a positive result indicates gDNA contamination of RNA extracts following DNAse digestions, and a negative result indicates RNA extracts free of gDNA.
Table 2. Specificity of 16S rRNA and IS2404 qPCR assays.
| Bacterial Species | Sourcea | Originb | 16S rRNAd | IS2404 e |
| M. abscessus | NRZ | Human isolatep | − | − |
| M. africanum | NRZ | Human isolatep | − | − |
| M. avium | NRZ | Human isolatep | − | − |
| M. bovis | NRZ | Cattle isolatep | − | − |
| M. chelonae | NRZ | Human isolatep | − | − |
| M. fortuitum | NRZ | Human isolatec | − | − |
| M. gordonae | NRZ | Human isolatec | − | − |
| M. gordonae | DITM | Human isolatec | − | − |
| M. kansasii | NRZ | Human isolatep | − | − |
| M. leprae | DITM | Human isolatep | − | − |
| M. malmoense | NRZ | Human isolatec | − | − |
| M. marinum | NRZ | Human isolatep | + | − |
| M. microti | NRZ | Mouse isolatep | − | − |
| M. scrofulaceum | NRZ | Human isolatep | − | − |
| M. smegmatis | NRZ | Human isolatep | − | − |
| M. szulgai | NRZ | Human isolatep | − | − |
| M. tuberculosis | NRZ | Human isolatep | − | − |
| M. ulcerans | DITM | Human isolatep | + | + |
| M. xenopi | NRZ | Human isolatec | − | − |
| E. coli | MVP | Human isolatec | − | − |
| P. acnes | MVP | Human isolatep | − | − |
| Staph. aureus | MVP | Human isolatec | − | − |
| Staph. epidermidis | MVP | Human isolatec | − | − |
| Str. pyogenes | MVP | Human isolatep | − | − |
Table 2 shows DNA extracts from closely related mycobacterial species and bacteria potentially contaminating the human skin subjected to the combined 16S rRNA RT/IS2404 qPCR viability assay and the corresponding test results. Mycobacterial species were selected according to their respective genetic contiguousness to M. ulcerans Agy99 (GenBank accession no. CP000325.1) within the 16S rRNA gene sequences as determined by BLASTN analysis (GenBank, NCBI) [13]. M., Mycobacterium; E., Escherichia; P., Propionibacterium; Staph., Staphylococcus; Str., Streptococcus. While in-silico analysis revealed that the combined 16S rRNA RT/IS2404 assay will also amplify mycolactone-producing mycobacteria (MPM) other than M. ulcerans (e.g., M. pseudoshottsii, M. liflandii, and the environmental M. marinum [GenBank accession No. NR_042988.1, AY500838.1, and AF456241.1, respectively]), these MPM species were not included in specificity testing.
DNA extracts that were not available at the DITM were provided by the National Reference Center (NRZ) for Mycobacteria, Borstel, Germany, and the Max von Pettenkofer-Institute (MVP), Ludwig-Maximilians University, Munich, Germany.
The respective primary patient isolates were considered as ppathogenic bacteria or as ccommensals/contaminants of clinical samples.
Results of the 16S rRNA RT-qPCR of DNA extracts; “+” indicates a positive and “–” a negative test result.
Results of the IS2404 qPCR of DNA extracts; “+” indicates a positive and “–” a negative test result.
Clinical validation was conducted on pre-treatment swab samples in PANTA (BD, Heidelberg, Germany) from 24 suspected BUD cases from Agogo Presbyterian Hospital (n = 14) and Tepa Government Hospital (n = 10), Ghana (Protocol S1). In addition, post-treatment swab samples from seven IS2404 PCR confirmed BUD patients with incomplete wound healing were collected at week nine (Figures 1 and 2).
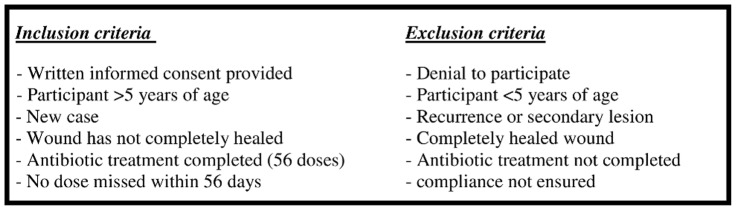
Figure 1. Enrolment criteria for the pre-treatment study population.
Figure 1 describes enrolment criteria for clinically suspected BUD patients presenting at Agogo Presbyterian Hospital (n = 14) and Tepa Governmental Hospital (n = 10), Ghana, respectively. None of the eligible study participants was excluded.
Figure 2. Enrolment criteria for the post-treatment study population.
Figure 2 describes enrolment criteria for IS2404 PCR confirmed BUD patients with incomplete wound healing (collection of swab samples feasible) who presented at Agogo Presbyterian Hospital, Ghana (n = 7), following completion of 56 doses of rifampicin and streptomycin administered within eight weeks. None of the eligible study participants was excluded.
All clinical samples were subjected to routine diagnostics (microscopy and IS2404 dry-reagent-based [DRB] PCR) at the Kumasi Centre for Collaborative Research (KCCR) [3].
Primers and Probes
Primers and a hydrolysis probe (TibMolBiol, Berlin, Germany) for specific amplification of M. ulcerans 16S rRNA were designed using DNAsis Max (MiraiBio, San Francisco, USA) by alignment of 16S rRNA gene sequences (GenBank, National Center for Biotechnology Information [NCBI]) from closely related mycobacteria and other bacteria potentially contaminating the human skin (Table 2).
For simultaneous quantification by IS2404 qPCR, the primers described by Fyfe et al. [9] were used in combination with a hydrolysis probe (Table 3) that was re-designed by DNAsis Max for thermodynamic reasons.
Table 3. Primers and probes.
| Primer/Probea | Sequence (5′–3′) | Target Geneb | Nucleotide Positionc | Amplicon Sized |
| MU16S TFMU16S TRMU16S TP | CGA TCT GCC CTG CAC TTC CCA CAC CGC AAA AGC TT6 FAM-CAC AGG ACA TGA ATC CCG TGG TC-BBQe | 16S rRNA | 4414800–44148174414718–44147344414740–4414762 | 100 bp |
| IS2404 TFIS2404 TRIS2404 TP2 | AAA GCA CCA CGC AGC ATC T AGC GAC CCC AGT GGA TTG6 FAM-CCG TCC AAC GCG ATC GGC A-BBQe | IS2404 | 96685–9666796627–9664496664–96646 | 59 bp |
| T13fT39f | TGC ACA CAG GCC ACA AGG GACG AAC GGG TGA GTA ACA CG | 16S rRNA | 4413906–44139254414822–4414840 | 935 bp |
Table 3 indicates primers and probes designed for the 16S rRNA RT-qPCR, the primers described by Fyfe et al., and a re-designed hydrolysis probe used for the amplification, detection, and quantification of IS2404 [9].
TF, forward primer; TR, reverse primer; TP2, hydrolysis probe (TibMolBiol, Berlin, Germany).
16S rRNA, gene for the ribosomal 16S RNA detected as 16S cDNA; IS2404, insertion sequence 2404.
Nucleotide positions are provided for the first (IS2404) or single (16S rRNA) copy of the respective amplicon in M. ulcerans Agy99 (GenBank accession no. CP000325.1) as determined by BLASTN analysis within GenBank (NCBI) [13].
bp, base pairs.
6 FAM, 6-Carboxyfluorescein fluorescent dye; BBQ, BlackBerry Quencher.
Primers T13 (forward) and T39 (reverse) were used for the amplification of a 935-bp region of the M. ulcerans 16S rRNA gene, encompassing the region amplified by qPCR primers MU16S TF and MU16S TR, to generate single copy replicates. Furthermore, these primers were used for sequencing of the M. ulcerans 16S rRNA gene (Table 1).
Combined RNA/DNA Extraction, Reverse Transcription, and Real-Time qPCR
Culture suspensions and swab samples were stabilized by RNA protect (Qiagen, Hilden, Germany) and subjected to AllPrep DNA/RNA extraction kit (Qiagen) (Protocols S1 and S2).
M. ulcerans whole transcriptome RNA from cultures and swab samples was transcribed to cDNA by QuantiTect Reverse Transcription Kit (Qiagen) including genomic DNA (gDNA) wipeout (Protocol S2). DNA and cDNA were subjected to IS2404 qPCR and 16S rRNA RT-qPCR, respectively, with corresponding controls (Table 4, Protocols S3 and S4).
Table 4. Controls applied in 16S rRNA RT/IS2404 qPCR.
| Control | Purpose | Material | |
| 16S rRNA RT-qPCRa | IS2404 qPCRb | ||
| gDNA wipeout controlc | To exclude DNA contamination of RNA extracts | Aliquot of each RNA extract following gDNA wipeout before reverse transcription | NA |
| Internal positive control | To exclude false negative results due to inhibition | TaqMan exogenous internal positive control (IPC)d | TaqMan exogenous internal positive control (IPC)d |
| Positive run control | To ensure adequate performance of PCR | M. ulcerans cDNAe | Cloned IS2404 standard |
| Negative no template control | To exclude contamination during PCR set up | H2O | H2O |
| Negative extraction control | To exclude contamination during extraction procedure | NA | Extract treated in the same way as samples |
Table 4 indicates controls applied in 16S rRNA RT/IS2404 qPCR. NA, not applicable.
16S rRNA RT PCR, reverse transcriptase real-time PCR targeting the 16S ribosomal RNA of M. ulcerans.
IS2404 qPCR, quantitative real-time PCR targeting the insertion sequence (IS) 2404 of M. ulcerans.
gDNA, genomic DNA wipeout was conducted using DNAses provided in the QuantiTect Reverse Transcription Kit (Qiagen).
TaqMan exogenous internal positive control (Applied Biosystems, Carlsbad, CA).
cDNA, complementary DNA obtained through reverse transcription of M. ulcerans RNA by QuantiTect Reverse Transcription Kit (Qiagen).
Intra- and Inter-Assay Variability
Intra- and inter-assay variability was assessed by testing of each sample in quadruplicate within one 96-well plate, repeated on three different days (Table 5).
Table 5. Intra- and inter-assay variability of the 16S rRNA RT/IS2404 qPCR assay.
| qPCR Targeta | Standard No. | Run No. 1 | Run No. 2 | Run No. 3 | Intra-Assay Variability | Inter-Assay Variability | |||||||
| Ct-rangeb | CVc | Ct-rangeb | CVc | Ct-rangeb | CVc | ΔCt max.d | CV max.e | Ct-rangef | CVg | ΔCt max.h | CV max.i | ||
| 16S rRNA | 12345 | 0.230.090.120.150.07 | 0.500.190.180.220.10 | 0.120.160.060.170.15 | 0.480.300.200.250.20 | 0.170.190.200.120.16 | 0.420.350.320.220.20 | 0.23 | 0.49 | 0.550.240.310.750.71 | 1.330.530.551.150.92 | 0.75 | 1.33 |
| IS2404 | 1234567 | 0.120.180.020.180.310.150.35 | 0.530.650.070.390.580.230.48 | 0.130.150.230.140.250.310.15 | 0.540.480.600.280.420.470.33 | 0.100.180.110.100.220.200.08 | 0.420.570.280.220.380.320.29 | 0.35 | 0.65 | 0.610.710.800.800.580.310.74 | 2.672.352.131.761.090.581.10 | 0.80 | 2.66 |
Table 5 shows intra- and inter-assay variability of the 16S rRNA RT/IS2404 qPCR assay.
16S rRNA RT-qPCR: 16S rRNA gene standards (935 bp) were generated by conventional PCR according to Talaat et al. [12]. Quantification of PCR products was conducted by Picogreen fluorometry (Invitrogen) and copy numbers were calculated based on the known mass of one amplicon. Serial standards were prepared from PCR products in 5 Log dilutions ranging from 3E+6 (standard no. 1) to 3E+2 copies (standard no. 5) of the 16S rRNA amplicon (PCR template: 2 µl) and were subjected to the 16S rRNA RT-qPCR in quadruplicate on one 96-well plate to assess intra-assay variability. The runs were repeated on three days to determine the inter-assay variability between runs 1 through 3. The intra- and inter-assay variability of the 16S rRNA RT-qPCR was low with maximum coefficients of variation (CV) of 0.49 (intra-assay) and 1.33 (inter-assay).
IS2404 qPCR: Cloned IS2404 replicates (1,047 bp, complete sequence; M. ulcerans Agy99) were used as standards. Quantification of IS2404 templates was conducted by Picogreen fluorometry (Invitrogen) and copy numbers were calculated based on the known mass of one template. Serial standards were prepared in 7 Log dilutions ranging from 2E+8 to 2E+2 copies of the IS2404 (PCR template: 2 µl) and were subjected to the IS2404 qPCR in quadruplicate on one 96-well plate to assess intra-assay variability. The runs were repeated on three days to determine the inter-assay variability between runs 1 through 3. The intra- and inter-assay variability of the IS2404 qPCR was low with maximum CV of 0.65 (intra-assay) and 2.66 (inter-assay).
16S rRNA, target of the 16S rRNA RT-qPCR; IS2404, target of the IS2404 qPCR.
Ct-range, range of Ct-values of samples tested in the same dilution.
CV, coefficient of variation of copy numbers from samples tested in quadruplicate of the same dilution.
ΔCt max., maximum Ct-variation of all samples tested within one run.
CV max., maximum CV of all samples tested within one run.
Ct-range, range of Ct-values of samples tested in the same dilution within three runs.
CV of samples in the same dilution tested within three runs.
ΔCt max. of all samples tested within three runs.
CV max. of all samples tested within three runs.
Sensitivity
The analytical sensitivity was determined as lower limit of detection (LOD, lowest template concentration rendering amplification of 95% of samples) [10] for both qPCR components using 10-fold serial dilutions of cloned IS2404 templates (GenExpress, Berlin, Germany) with known copy numbers (IS2404 qPCR) and exactly quantified M. ulcerans whole genome DNA extracts from cultures (16S rRNA RT-qPCR). The LOD was two (IS2404) and six templates (16S rRNA gene), respectively (Figures 3 and 4).
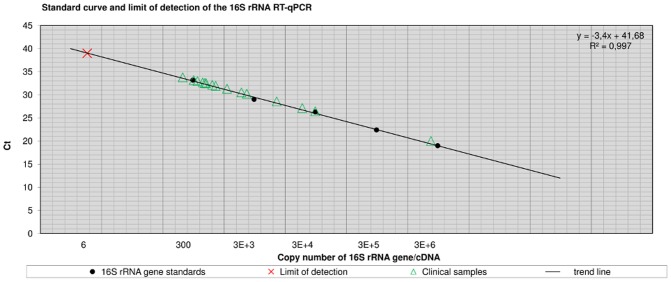
Figure 3. Standard curve and limit of detection of the 16S rRNA RT-qPCR.
Figure 3 shows Ct-values of clinical samples plotted versus quantified 16S rRNA copy numbers. Standards for the 16S rRNA RT-qPCR were generated by conventional PCR amplification (Table 5). Log 10 fold serial dilutions (n = 5) were prepared ranging from 3E+6 to 300 copies of the 16S rRNA gene (PCR template: 2 µl) and were subjected to the assay in quadruplicate to generate a calibration curve. The regression line was y = −3.4x+41.68 with a coefficient of correlation >0.99 and the efficiency was E = 0.97. M. ulcerans whole genome extracts were quantified by means of IS2404 qPCR and the analytical sensitivity was determined as limit of detection (LOD) by subjecting 10 aliquots of a dilution series containing 30, 15, 10, 8, 6, 3, or 2 copies of the 16S rRNA gene to the assay. The LOD was 6 copies of the target sequence. The copy number (n = 1) of the 16S rRNA gene per M. ulcerans genome was determined by copy number variation assay (unpublished data).
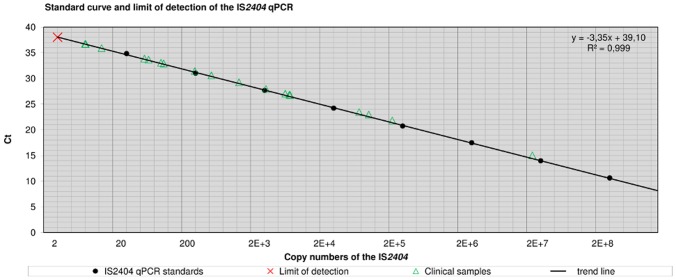
Figure 4. Standard curve and limit of detection of the IS2404 qPCR.
Figure 4 shows mean Ct-values of calibration standards and clinical samples plotted versus the quantified copy number of IS2404. Cloned IS2404 templates were used as standards (Table 5). Log 10 fold serial dilutions (n = 8) were prepared ranging from 2E+8 to 20 copies of the IS2404 (PCR template: 2 µl) and were subjected to the IS2404 qPCR in quadruplicate to generate a calibration curve. The regression line was y = −3.35x+39.10 with a coefficient of correlation >0.99 and the efficiency was E = 0.97. The analytical sensitivity was determined as limit of detection (LOD) by subjecting 10 aliquots of a dilution series containing 10, 5, 4, 3, 2, or 1 copy of the IS2404 to the assay. The LOD was 2 copies of the target sequence.
M. ulcerans DNA and rRNA was detected in all culture extracts. Out of 24 pre-treatment swab samples, 18 (75.0%; 95%-CI: 57.7%–92.3%) had a positive IS2404 qPCR result, 12 out of those were also positive in routine DRB PCR, and rRNA was detected in 15 out of these 18 samples (83.3%; 95%-CI: 66.1%–100%); quantification of the three negative samples revealed a bacillary load below the LOD of the 16S rRNA RT-qPCR (Table 6).
Table 6. Study participants, clinical information, and diagnostic results.
| Clinical Data | Molecular Viability Assaya | Routine Diagnosticsb | ||||||
| No.c | BUD Patientd | Duration (Weeks)e | Category of Lesionf | IS2404 [Ct]g | Bacillary Loadh | 16S rRNAi | MICk | PCRl |
| 1 | No | NA | NA | Neg [NA] | NA | Neg | 0 | Neg |
| 2 | Yes | 6 | III | Pos [15,04] | >1000 | Pos | +1 | Pos |
| 3 | Yes | 4 | III | Pos [26,80] | 584 | Pos | +1 | Pos |
| 4 | Yes | 9 | III | Pos [32,93] | 6–10 | Pos | 0 | Neg |
| 5 | Yes | 4 | I | Pos [35,94] | 1–5 | Neg | 0 | Neg |
| 6 | Yes | 8 | II | Pos [36,72] | 1–5 | Neg | 0 | Neg |
| 7 | Yes | 2 | I | Pos [36,74] | 1–5 | Neg | 0 | Neg |
| 8 | Yes | 10 | I | Pos [27,05] | 497 | Pos | +1 | Pos |
| 9 | No | NA | NA | Neg [NA] | NA | Neg | 0 | Neg |
| 10 | Yes | 3 | I | Pos [30,61] | 42 | Pos | +1 | Pos |
| 11 | Yes | 8 | II | Pos [33,89] | 6–10 | Pos | 0 | Neg |
| 12 | Yes | 9 | I | Pos [33,68] | 6–10 | Pos | 0 | Neg |
| 13 | Yes | 3 | III | Pos [29,27] | 106 | Pos | +1 | Pos |
| 14 | Yes | 3 | I | Pos [27,98] | 261 | Pos | +1 | Pos |
| 15 | Yes | 1 | I | Pos [26,85] | 571 | Pos | +1 | Pos |
| 16 | Yes | 2 | I | Pos [33,07] | 6–10 | Pos | 0 | Pos |
| 17 | Yes | 2 | II | Pos [31,44] | 24 | Pos | +1 | Pos |
| 18 | Yes | 3 | II | Pos [21,85] | >1000 | Pos | +2 | Pos |
| 19 | Yes | 4 | III | Pos [22,98] | >1000 | Pos | +1 | Pos |
| 20 | Yes | 3 | I | Pos [23,47] | >1000 | Pos | +2 | Pos |
| 21 | No | NA | NA | Neg [NA] | NA | Neg | 0 | Neg |
| 22 | No | NA | NA | Neg [NA] | NA | Neg | 0 | Neg |
| 23 | No | NA | NA | Neg [NA] | NA | Neg | 0 | Neg |
| 24 | No | NA | NA | Neg [NA] | NA | Neg | 0 | Neg |
Table 6 shows suspected BUD cases with ulcerative lesions enrolled in the pre-treatment cohort (Figure 1), clinical information, and diagnostic results. Swab samples from 24 suspected BUD cases were subjected to 16S rRNA RT/IS2404 qPCR viability assay (swab 1 in PANTA), microscopic examination and enumeration of acid fast bacilli (AFB) following Ziehl-Neelsen staining (swab 2, direct smear), and conventional IS2404 dry-reagent-based (DRB) PCR (swab 3 in Cell Lysis Solution [Qiagen]). 18 patients were laboratory confirmed by IS2404 qPCR and 15 out of those were RNA positive; the quantification by IS2404 qPCR revealed a bacillary load (1–2 bacilli per sample) below the lower limit of detection of the RNA assay for samples from three RNA negative patients. All samples from six IS2404 qPCR negative study participants were also RNA negative. Direct correlation of AFB enumeration with IS2404 qPCR quantification is not feasible due to inhomogeneous distribution of M. ulcerans in different clinical samples. NA, not applicable; Neg, negative test result; Pos, positive test result.
Results of the 16S rRNA RT/IS2404 qPCR viability assay. Clinical swab samples in PANTA were directly processed at KCCR, and M. ulcerans DNA and cDNA were transported to DITM and subjected to qPCR.
Routine diagnostics were conducted following standardized procedures at KCCR [3].
No., consecutive number of study participants.
Yes, IS2404 qPCR confirmed BUD patients; No, IS2404 negative study participants.
Duration of disease before presentation of study participants in weeks.
Category of lesion according to the World Health Organization's clinical criteria [1].
Results of the IS2404 qPCR with corresponding cycle threshold (Ct)-values.
The bacillary load in the respective swab samples (No. 2) was estimated on the basis of IS2404 quantification given an IS2404 copy number of 209 copies per M. ulcerans genome [9]. For bacterial numbers <10 ranges were estimated.
Results of the 16S rRNA RT-qPCR.
MIC, microscopic detection and enumeration of AFB was conducted at KCCR including external quality assurance by DITM. The following scale was applied: 0 = negative, +1 = 10–99 AFB/100 fields, +2 = 1–10 AFB/1 field, +3 = more than 10 AFB/1 field.
PCR, conventional, single target gel-based IS2404 DRB PCR.
All seven post-treatment swab samples were IS2404 qPCR positive and 16S rRNA negative.
Specificity
Analysis of DNA extracts revealed 100% specificity for the combined assay. M. marinum (human isolate) was amplified by 16S rRNA RT-qPCR; however, simultaneous IS2404 qPCR was negative (Table 2).
Bacillary Survival Times
To investigate the effect of sample transport on bacillary survival, mycobacteriological transport media (PANTA and LTM) [3] were spiked with viable M. ulcerans and stored at 4°C and 31°C. RNA was detectable in both media for >4 weeks (4°C and 31°C).
After heat-inactivation of M. ulcerans–spiked PANTA-samples, RNA positivity decreased significantly within 12 h, whereas DNA was still detectable after seven days.
Future Application
The assay will support clinicians in classification of secondary lesions and selection of adequate clinical management strategies and provides a powerful tool for clinical research evaluating novel treatment regimens (Box 1).
Box 1. Advantages and Disadvantages of the Molecular Viability Assay
Advantages
Provides a rapid, sensitive, and specific tool to detect viable bacilli in clinical samples of BUD patients, thus offering an alternative to cultures.
Supports classification of secondary BUD lesions and monitoring of treatment success.
Disadvantages
Current test format requires well equipped laboratory with real-time PCR facilities.
Costs per test (approximately 14 €) may limit the applicability.
Through analysis of sequential samples collected during antimycobacterial treatment, the assay will be employed to determine the proportional decrease of bacterial viability over time and to establish laboratory-based evidence for optimal time-points to collect follow-up samples for treatment monitoring.
Whereas the current format of the assay is restricted to reference laboratories, sample collection on FTA cards in combination with isothermal dry-reagent-based reverse transcription and amplification formats would facilitate processing of samples also at a peripheral level and at lower costs.
Conclusions
The novel combined 16S rRNA RT/IS2404 qPCR assay proved to be highly sensitive, specific, and efficient in detecting viable M. ulcerans in clinical samples under field conditions. The assay is applicable for classification of secondary lesions and monitoring of treatment success and provides a powerful tool for clinical research.
GenBank Accession Numbers
Genes or DNA sequences of mycobacterial strains used in this study were retrieved from GenBank (NCBI) [13]. The respective sequences and accession numbers are summarized in Table S1.
Supporting Information
Preparation of PANTA transport medium and stabilisation of M. ulcerans RNA/DNA in swab samples and culture suspensions.
(PDF)
Simultaneous RNA/DNA extraction from swab samples and reverse transcription of whole transcriptome RNA from M. ulcerans.
(PDF)
Combined 16S rRNA RT/IS2404 qPCR assay.
(PDF)
16S rRNA RT/IS2404 qPCR run protocol.
(XLS)
GenBank accession numbers.
(DOC)
Acknowledgments
The authors thank Erna Fleischmann, Carolin Mengele and Kerstin Helfrich (DITM), and Mabel Peprah and Michael Frimpong (KCCR) for excellent technical assistance. The authors thank Dr. Sabine Rüsch-Gerdes and Dr. Elvira Richter (National Reference Center for Mycobacteria, Borstel, Germany), as well as Dr. Soeren Schubert (Max von Pettenkofer-Institute, Ludwig-Maximilians University, Munich, Germany) for providing (myco-) bacterial DNA extracts. The manuscript contains parts of the doctoral thesis of Dominik Symank.
Funding Statement
The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007–2013) under grant agreement N° 241500. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. World Health Organization (2008) Buruli ulcer: progress report, 2004–2008. Wkly Epidemiol Rec 83: 145–156. [PubMed] [Google Scholar]
- 2.World Health Organization (2008) Meeting of the WHO Technical Advisory Group on Buruli ulcer, 3 April 2008, Geneva, summary report. Geneva: World Health Organization.
- 3. Beissner M, Herbinger KH, Bretzel G (2010) Laboratory diagnosis of Buruli ulcer disease. Future Microbiol 5 (3) 363–370. [DOI] [PubMed] [Google Scholar]
- 4. Ruf MT, Chauty A, Adeye A, Ardant MF, Koussemou H, et al. (2011) Secondary Buruli ulcer skin lesions emerging several months after completion of chemotherapy: paradoxical reaction or evidence for immune protection? PLoS Negl Trop Dis 5 (8) e1252 doi:10.1371/journal.pntd.0001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nienhuis WA, Stienstra Y, Abass KM, Tuah W, Thompson WA, et al. (2011) Paradoxical responses after start of antimicrobial treatment in Mycobacterium ulcerans infection. Clin Infect Dis 54 (4) 519–526. [DOI] [PubMed] [Google Scholar]
- 6. Martinez AN, Lahiri R, Pittman TL, Scollard D, Truman R, et al. (2009) Molecular determination of Mycobacterium leprae viability by use of real-time PCR. J Clin Microbiol 47 (7) 2124–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Desjardin LE, Perkins MD, Wolski K, Haun S, Teixeira L, et al. (1999) Measurement of sputum Mycobacterium tuberculosis messenger RNA as a surrogate for response to chemotherapy. Am J Respir Crit Care Med 160: 203–210. [DOI] [PubMed] [Google Scholar]
- 8. Bretzel G, Siegmund V, Racz P, van Vloten F, Ngos F, et al. (2005) Post-surgical assessment of excised tissue from patients with Buruli ulcer disease: progression of infection in macroscopically healthy tissue. Trop Med Int Health 10 (11) 1199–1206. [DOI] [PubMed] [Google Scholar]
- 9. Fyfe JA, Lavender CJ, Johnson PD, Globan M, Sievers A, et al. (2007) Development and application of two multiplex real-time PCR assays for the detection of Mycobacterium ulcerans in clinical and environmental samples. Appl Environ Microbiol 73 (15) 4733–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taylor S, Wakem M, Dijkman G, Alsarraj M, Nguyen M (2010) A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 50 (4) S1–S5. [DOI] [PubMed] [Google Scholar]
- 11. Beissner M, Awua-Boateng NY, Thompson W, Nienhuis WA, Klutse E, et al. (2010) A genotypic approach for detection, identification, and characterization of drug resistance in Mycobacterium ulcerans in clinical samples and isolates from Ghana. Am J Trop Med Hyg 83 (5) 1059–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Talaat AM, Reimschuessel R, Trucksis M (1997) Identification of mycobacteria infecting fish to the species level using polymerase chain reaction and restriction enzyme analysis. Vet Microbiol 58 (2–4) 229–237. [DOI] [PubMed] [Google Scholar]
- 13. Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2008) GenBank. Nucleic Acids Res 36 (Database issue) D25–D30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Preparation of PANTA transport medium and stabilisation of M. ulcerans RNA/DNA in swab samples and culture suspensions.
(PDF)
Simultaneous RNA/DNA extraction from swab samples and reverse transcription of whole transcriptome RNA from M. ulcerans.
(PDF)
Combined 16S rRNA RT/IS2404 qPCR assay.
(PDF)
16S rRNA RT/IS2404 qPCR run protocol.
(XLS)
GenBank accession numbers.
(DOC)