A gratifying story is emerging on how the complementary strands of the DNA double helix are unlinked and partitioned after replication with astonishing accuracy as finished chromosomes to daughter cells. There are three key conclusions:
DNA condensation is a driving force for DNA unlinking and chromosome partitioning.
Condensation is achieved by supercoiling.
Supercoiling results from topological strain and the contortion of DNA by proteins, notably the nucleosomal histone octet and the structural maintenance of chromosomes (SMC) proteins.
The paper by Sawitzke and Austin (1) ties together all three conclusions. Their work concerns the Muk (from the Japanese mukaku, meaning anucleate) protein complex of Escherichia coli, which was discovered and characterized by Hiraga and coworkers (2). Mutations in any of the three muk genes, mukB, mukE, and mukF, disrupt chromosome segregation such that many progeny have missing or incomplete chromosomes that have been guillotined by septation. What links this fate of muk− bacteria to our story is that their chromosomes are decondensed (3) and that MukB is the structural and functional analogue of the ubiquitous SMC family of proteins (4). These huge molecules form coiled-coil dimers that, along with associated proteins, are thought to bind DNA segments separated by as much as 1,000 Å and then to contract the intervening DNA at the expense of ATP (5). Sawitzke and Austin show that the severity of the Muk phenotype can be controlled by changing the level of supercoiling in the cell. The harsh consequences of being Mukless are suppressed by just a modest increase in chromosomal supercoiling, and muk− cells are hypersensitive to gyrase inhibitors. The authors conclude that Muk and supercoiling cooperate in condensing DNA, which drives partitioning. These findings fit beautifully with those from biophysics to cell biology in a wide array of organisms.
DNA Condensation.
To put this work in context, we need first to review the properties of DNA condensation. DNA is vastly longer than the space assigned to it because of the shortsightedness of evolution in fashioning a linear genetic code. The needed condensation has been traditionally described one-dimensionally as the ratio of DNA length to the diameter of its container, a nucleus, or nucleoid.
There is a more biophysical way of quantifying the space crunch. The persistence length of DNA (a) is so small compared with its total length (L) that its natural low energy conformation is already substantially condensed [dimensions of (aL)1/2]. Moreover, DNA must be condensed not in one dimension but in three. Instead of linear compaction, a better way to judge condensation is the volume reduction of a random coil of free DNA to its final volume in a cell (refs. 6 and 7; Table 1). The amount of DNA compaction increases with genome size and is on the order of 103 for E. coli and 105 for humans.
Table 1.
Comparison of linear and volumetric condensation
| DNA | Sequence length, bp | Physical length, μm | Container* diameter, μm | Linear reduction† | Radius of gyration for free DNA, μm | Volume of free DNA, random coil, μm3 | Container volume, μm3 | Volume reduction‡ |
|---|---|---|---|---|---|---|---|---|
| E. coli | 4.6 × 106 | 1.6 × 103 | 1.0 | 1.6 × 103 | 3.6 | 1.9 × 102 | 0.52 | 3.7 × 102 |
| Yeast | 2.8 × 107 | 9.3 × 103 | 2.0 | 4.7 × 103 | 13 | 8.1 × 103 | 4.2 | 1.9 × 103 |
| Human | 6.0 × 109 | 2.0 × 106 | 10.0 | 2.0 × 105 | 1.8 × 102 | 2.6 × 107 | 5.2 × 102 | 4.9 × 104 |
*Nucleoid or nucleus.
†Linear reduction = physical length/container diameter.
‡Volume reduction = volume of free DNA container volume.
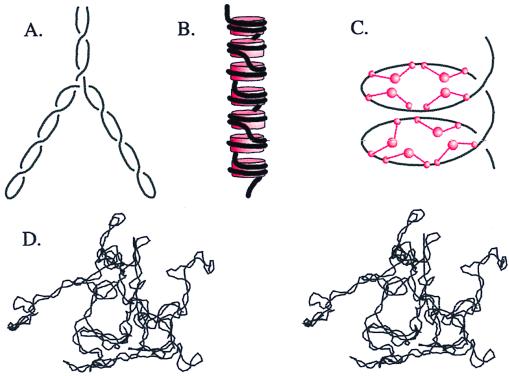
Three mechanisms of in vivo condensation are illustrated in Fig. 1. Two of these, (−) supercoiling of DNA free in solution (ref. 8; Fig. 1 A and D) and the constrained DNA supercoiling in nucleosomes (ref. 9; Fig. 1B) are well understood physically and physiologically. The third mechanism has been discovered only recently and is at the core of our story today. This is the (+) supercoiling buttressed by 13S condensin, the key SMC protein complex of frogs (ref. 10; Fig. 1C).
Figure 1.
Comparison of three types of DNA compaction by supercoiling. (A) Free (−) supercoils twist DNA into a right-handed plectonemic superhelix. The tight interwinding brings DNA an average of only 100 Å apart. Branching of the superhelical axis causes additional compaction. (B) Wrapping around the histone octamer (red cylinders) compacts DNA by forming left-handed solenoidal supercoils. (C) SMC proteins, such as Xenopus 13S condensin (schematized as red ball and stalk structures), effect global DNA writhe by forming large (+) solenoidal supercoils. (D) Stereo image of a 25-kilobase (kb) (−) supercoiled DNA generated by a Metropolis Monte Carlo simulation. A and C represent approximately 2 kb of DNA (700 nm) at 200,000-fold magnification, whereas B is only 1.5 kb of DNA (500 nm) but at 4-fold greater magnification. D is at 100,000-fold magnification.
Unconstrained (−) Supercoiling.
Because (−) supercoiling in bacteria arises from a topological misalignment and not a protein corset, it has the flexibility to do work. Its labors are so important that a slight reduction in supercoiling is lethal (11). Supercoiling has three essential roles.
First, (−) supercoiling promotes the unwinding of DNA and thereby the myriad processes that depend on helix opening (8). Whenever DNA is doing anything interesting, it is single-stranded, and (−) supercoils provide a vital sequence-independent assistance to denaturation.
The second essential role of supercoiling is in DNA replication. For replication to be completed, the linking number of the DNA, Lk, must be reduced from its vast (+) value to exactly zero. In bacteria, DNA gyrase introduces (−) supercoils and thereby removes parental Lk (11).
The third essential role of supercoiling is conformational (ref. 8; Fig. 1 A and D). DNA manifests the difference between the relaxed and naturally occurring values of Lk by winding up into supercoils. These supercoils condense DNA and promote the disentanglement of topological domains. This can be accomplished equally well by (−) or (+) supercoiling. It is this condensation role of supercoiling that directly concerns us now.
Key observations linking condensation by supercoiling to partitioning were made some time ago, but their significance was missed. In 1968, Hirota, Ryter, and Jacob (12) isolated conditionally lethal mutants of E. coli that had the dramatic chromosome partition defect (Par−) phenotype characterized by anucleate and guillotined cells. The very first mutation isolated, parA, mapped to a subunit of DNA gyrase (13).
For years, the message of these mutants was misunderstood; the conclusion was that gyrase was needed for partitioning, because it decatenated daughter chromosomes. The story unraveled further, however, with the discovery of a second bacterial type-2 topoisomerase, topo IV (15), that decatenates DNA orders of magnitude better than gyrase. Experiments in vivo have shown that topo IV, and not gyrase, is responsible for decatenation (16, 17). Topo IV mutants are richly represented in par collections (14, 18).
Why then are gyrase mutants defective in partitioning? Supercoiling draws DNA in on itself and thereby pulls it away from other DNAs. This molecularly antisocial behavior has two roots. First, (Fig. 1A) supercoiled DNA tightens into a plectonemic (interwound) superhelix whose diameter is only about five times as wide as the DNA itself (8). As a result, random strand passages by a topoisomerase are much more likely to decatenate. Indeed, supercoiling of a 7-kb plasmid promoted decatenation by three orders of magnitude (19). The promotion of decatenation by supercoiling has also been directly demonstrated in vivo (20).
Second, the volume occupied by a supercoiled molecule is much smaller than that of a relaxed DNA. This difference in volume is due mostly to the formation of superhelical branches. Fig. 1D shows a 25-kb supercoiled DNA branching and bending itself into a ball. The decrease in chromosomal volume by supercoiling decreases the probability that the septum will pass through the chromosome during cell division.
Hence, the explanation for the Par phenotype of gyrase mutants is now clear. (−) Supercoiling by gyrase compacts the chromosomes such that random passages by topo IV disentangle them.
Supercoiling Around Core Histones in Nucleosomes.
The second type of condensation via supercoiling, that by core histones (Fig. 1B), is so well known (21) that we will limit ourselves to a few comments.
One could imagine several ways to compact DNA in a eukaryotic cell. The DNA could be crammed into the nucleus like dirty clothes in a teenager's closet. The result is compact but poorly reversible or searchable. A second alternative is that DNA is wrapped like a fishing line around a reel. This is orderly and reversible, but only the most superficial genes are readily accessible. Nature has apparently adopted a third alternative: DNA is compacted in independent successive stages such that the total compaction is the product of compaction in each stage. The first stage of this compaction is via solenoidal wrapping of DNA in the nucleosome. Although the compaction achieved is modest, the nucleosome provides a fundamental structure for genome organization and function.
The role of the (+) charge on histones is often misunderstood. Molecules with a (+) charge of three or more greatly condense DNA (6), but the high (+) charge on histones is not the basis of their ability to fold DNA. Polyvalent cations induce condensation by nonspecific DNA aggregation such that they exacerbate rather than relieve the problem of DNA entanglement. Whereas supercoiling greatly decreases the probability of catenation, polyvalent cations greatly increase it. The structure of a nucleosome reveals a scaffolding that forces the DNA to adopt ordered solenoidal supercoils (9).
Compaction by 13S Condensin-Mediated (+) Supercoiling.
The third type of compaction cum supercoiling, that by 13S condensin (Fig. 1C), is needed for the formation of mitotic chromosomes from the open interphase forms. In a dramatic experiment, Hirano and Mitchison (22) took demembranated sperm as a source of extended chromatin and added an extract of frog eggs and ATP. Mirabile dictu, the chromosomes condensed into forms very much like those seen in mitosis. Blocking experiments with antibodies demonstrated that 13S condensin was required for both the assembly and maintenance of these chromosomes.
This result gained significance from the genetic studies performed on homologues in more genetically amenable organisms. Mutants of SMC homologues in yeast have decondensed chromosomes and are defective in partitioning (4, 23). Mutations in Mix-1, a Caenorhabditis elegans SMC protein, result in mitotic segregation defects and decondensed chromosomes (24). This theme was repeated with SMC mutants of Bacillus subtilis (25), which have a classic Par phenotype.
The first important observation as to the mechanism of condensation by condensin was provided again by the Hirano laboratory. Purified mitotic 13S condensin, hydrolysis of ATP, and the action of a type-1 topoisomerase resulted in (+) supercoiling of plasmid DNA (10).
Two interpretations of these results were suggested (10). Condensin could locally overtwist DNA or, like nucleosomes, could have a tight external wrapping of DNA but with opposite handedness. Either would lead to compensatory (−) supercoils that were suggested as the basis of condensation. Unfortunately, neither possibility is an attractive explanation for the role of this protein in DNA condensation. The compensatory (−) supercoils would be relaxed by abundant eukaryotic topoisomerases. A local overwinding of DNA would have no effect on condensation; nor could a tight wrapping around condensin greatly compact DNA, because there is no more than one condensin molecule per 10 kb of DNA (26). Fortunately, there is a third possible explanation for the (+) supercoiling that is compatible with its physiological role. Condensin is so large, reaching out perhaps 1,000 Å, that it could torque the DNA between its reach. Thus, condensin could introduce (+) supercoiling by effecting global writhe, as schematized in Fig. 1C. Strong evidence for this was provided by the finding that incubation of 13S condensin and a type-2 topoisomerase with plasmid DNA forms chiral DNA knots (26). These knots were almost exclusively (+), as expected if condensin introduces a regular (+) writhe. This activity requires ATP hydrolysis and 13S condensin from mitotic cells. Condensin from interphase cells had knotting activity only after being phosphorylated in vitro by Cdc2 kinase.
Thus, the hypothesis for condensation in vivo is that condensin regularly supercoils interphase chromatin. The loops formed are so large that even 30-nm fibers could be compacted in this way.
Muk, Supercoiling, Condensation, and Chromosome Structure.
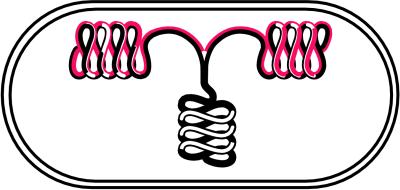
The paper by Sawitzke and Austin (1) fits well into this general picture and advances it. Muk was originally thought to be a classical motor protein that, like proteins that act along spindle fibers, would move the replicated bacterial chromosomes from the middle of the cell to the 1/4 and 3/4 positions before septation. But how can suppression of muk− by increased supercoiling be reconciled with chromosomes stuck in the middle of the cell? The authors instead adopt the model put forth by Grossman's group (27) that parental DNA in the center of the cell is decondensed, forced through a stationary replication factory, and then recondensed toward the 1/4 or 3/4 positions (Fig. 2). Muk does not act like a motor to transport chromosomes actively but instead pulls the newly replicated DNA toward the poles by supercoiling it into a more condensed form.
Figure 2.
Model for flow of DNA during replication. Condensed parental DNA (black-black) in the center of the cell is decondensed before replication and pulled through a central replication factory. The newly replicated daughters (black-red) are quickly condensed again.
Consistent with this interpretation is that the suppression is reversed by mild inhibition of DNA gyrase. The validity of this conclusion is strengthened, not spoiled, by the hypersensitivity of mukB mutants by themselves to gyrase inhibitors. This latter result was also obtained independently (3). The explanation is that supercoiling by both Muk and gyrase is needed to condense daughter DNAs away from each other and their parent. In the absence of the Muk-driven condensation, the cell is particularly vulnerable to diminution of gyrase.
We cannot yet conclude that Muk supercoils DNA in the same fashion as Xenopus 13S condensin. This reasonable hypothesis cannot be tested until a functional Muk complex is purified. Even if Muk and 13S condensin do have identical mechanisms, the path of free supercoiled DNA would be radically different from that of SMC-condensed DNA (Fig. 1 A and C). Although both have right-handed superhelices, the former is (−), interwound, and branched. The latter is (+), solenoidal, unbranched, and structured. The suppression implies that the only essential feature of Muk action is DNA condensation. Finally, it is surprising that a number of mutations can partially rescue muk− cells. It has been reported that mutations in seqA or dam suppress the Muk deficiency (3). Perhaps disruption of cell cycle control allows more time for DNA to be compacted. High copy number suppressors of camphor sensitivity also suppress muk− (8). Because camphor decondenses the nucleoid, the latter suppressors may represent yet another mechanism of DNA condensation.
Although Sawitzke and Austin (1) are the first to link chromosomal condensation, supercoiling, and SMC proteins, a relationship between plasmid supercoiling and partitioning had already been widely established. Cohen's group (28, 29) showed that topA10 mutations suppressed partition defects for pSC101 and mini F plasmids. Suppression by topA− was also found for partition mutants of P1 (30). The segregation of P1 and F was worse than random in the absence of the partitioning complex but improved to random in topA mutants. This is now easy to explain. Supercoiling would condense the plasmid such that it would likely avoid guillotining by septation even if randomly localized in the cell. The repeated suppression by (−) supercoiling of defects in disparate partitioning systems implies that the conclusions of Sawitzke and Austin have broad significance.
Acknowledgments
We thank A. Vologodskii for helpful discussions. V.H. is supported by a Howard Hughes Medical Institute Predoctoral Fellowship.
Footnotes
See companion article at www.pnas.org/cgi/doi/10.1073/pnas.030528397.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040576797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040576797
References
- 1.Sawitzke, J. A. & Austin, S. (January 31, 2000) Proc. Natl. Acad. Sci. USA, 10.1073/pnas.030528397. http://www.pnas.org/cgi/doi/10.1073/pnas.030528397
- 2.Hiraga S. Annu Rev Biochem. 1992;61:283–306. doi: 10.1146/annurev.bi.61.070192.001435. [DOI] [PubMed] [Google Scholar]
- 3.Weitao T, Nordstrom K, Dasgupta S. Mol Microbiol. 1999;34:157–168. doi: 10.1046/j.1365-2958.1999.01589.x. [DOI] [PubMed] [Google Scholar]
- 4.Koshland D, Strunnikov A. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- 5.Melby T E, Ciampaglio C N, Briscoe G, Erickson H P. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloomfield V. Biopolymers. 1998;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 7.Marko J, Trun N. Am Soc Microbiol News. 1998;64:276–283. [Google Scholar]
- 8.Vologodskii A V, Cozzarelli N R. Annu Rev Biophys Biomol Struct. 1994;23:609–643. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- 9.Luger K, Mäder A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 10.Kimura K, Hirano T. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- 11.Wang J C. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- 12.Hirota Y, Ryter A, Jacob F. Cold Spring Harbor Symp Quant Biol. 1968;33:1496–1505. doi: 10.1101/sqb.1968.033.01.077. [DOI] [PubMed] [Google Scholar]
- 13.Kato J, Nishimura Y, Suzuki H. Mol Gen Genet. 1989;217:178–181. doi: 10.1007/BF00330959. [DOI] [PubMed] [Google Scholar]
- 14.Luttinger A L, Springer A L, Schmid M B. New Biol. 1991;3:687–697. [PubMed] [Google Scholar]
- 15.Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Susuki H. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 16.Adams D E, Shekhtman E M, Zechiedrich E L, Schmid M B, Cozzarelli N R. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 17.Zechiedrich E L, Cozzarelli N R. Genes Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- 18.Kato J, Nishimura Y, Yamada M, Suzuki H, Hirota Y. J Bacteriol. 1988;170:3967–3977. doi: 10.1128/jb.170.9.3967-3977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rybenkov V V, Vologodskii A V, Cozzarelli N R. J Mol Biol. 1997;267:312–323. doi: 10.1006/jmbi.1996.0877. [DOI] [PubMed] [Google Scholar]
- 20.Zechiedrich E L, Khodursky A B, Cozzarelli N R. Genes Dev. 1997;11:2580–2592. doi: 10.1101/gad.11.19.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Widom J. Annu Rev Biophys Biomol Struct. 1998;27:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 22.Hirano T, Mitchison T J. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 23.Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieb J D, Albrecht M R, Chuang P T, Meyer B J. Cell. 1998;92:265–277. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- 25.Britton R A, Lin D C, Grossman A D. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura K, Rybenkov V, Crisona N, Hirano T, Cozzarelli N R. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- 27.Lemon K, Grossman A. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 28.Biek D, Cohen S. J Bacteriol. 1989;12:2056–2065. doi: 10.1128/jb.171.4.2056-2065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller C A, Beaucage S L, Cohen S N. Cell. 1990;62:127–133. doi: 10.1016/0092-8674(90)90246-b. [DOI] [PubMed] [Google Scholar]
- 30.Austin S J, Eichorn B G. J Bacteriol. 1992;174:5190–5195. doi: 10.1128/jb.174.16.5190-5195.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]