Abstract
Transposable elements are ubiquitous residents in eukaryotic genomes. Often considered to be genomic parasites, they can lead to dramatic changes in genome organization, gene expression, and gene evolution. The oomycete plant pathogen Phytophthora infestans has evolved a genome organization where core biology genes are predominantly located in genome regions that have relatively few resident transposons. In contrast, disease effector-encoding genes are most frequently located in rapidly evolving genomic regions that are rich in transposons. P. infestans, as a eukaryote, likely uses RNA silencing to minimize the activity of transposons. We have shown that fusion of a short interspersed element (SINE) to an effector gene in P. infestans leads to the silencing of both the introduced fusion and endogenous homologous sequences. This is also likely to occur naturally in the genome of P. infestans, as transcriptional inactivation of effectors is known to occur, and over half of the translocated “RXLR class” of effectors are located within 2 kb of transposon sequences in the P. infestans genome. In this commentary, we review the diverse transposon inventory of P. infestans, its control by RNA silencing, and consequences for expression modulation of nearby effector genes in this economically important plant pathogen.
Keywords: Phytophthora infestans, RXLR, effector, oomycete, silencing, small RNA, transposon
Oomycete Plant Pathogens and Their Effectors
Many eukaryotic plant pathogens exhibit variation in traits such as specific virulence and avirulence, and pathogenicity. The mechanisms underlying this variation have remained largely unaddressed. Much research is presently focused on identifying the molecules (proteins or metabolites) that act at the interface of pathogen and host. Outcomes from this research have led to the development of general models that describe interactions between plants and pathogens at the molecular level. Central to these evolutionary hypotheses, such as the “zig-zag” model, are pathogen molecules called effectors. Mechanistic details regarding plant immunity can be found in many outstanding reviews (for example, see refs. 1 and 2). In brief, all pathogens trigger defense responses in plants, due to detection of conserved molecules called pathogen associated molecular patterns (PAMPs). This detection triggers an array of immune responses, or PAMP triggered immunity (PTI). Pathogens may adapt and produce secreted effectors to suppress PTI in plants, leading to effector-triggered susceptibility (ETS). Effectors can also be recognized by specific plant host resistance (R) proteins, resulting in effector-triggered immunity (ETI). In this instance, the recognized effector is termed an avirulence (Avr) protein. It is postulated that the high numbers of R-genes in plant genomes and their large sequence diversity are essential evolutionary factors in the surveillance machinery for resisting pathogen attack. Plant R-genes evolve through duplication, unequal crossing over, recombination and diversification, leading to clusters of paralogous genes.3 In comparison, pathogens have evolved various ways to evade detection by these resistance proteins, such as variations in sequence, gene loss, or transcriptional inactivation (reviewed in refs. 4 and 5). Effectors are thus considered to define the host range of a pathogen, by adapting to specific host target proteins. The selection pressure on effectors has resulted in their placement among the most rapidly evolving proteins in pathogen genomes.
How expression of effector genes varies or is regulated in pathogen genomes is little understood, but it is likely that epigenetic mechanisms may have some influence. Epigenetic control of genes is well described in eukaryotes and its mechanism(s) frequently involves overlap with genome defense mechanisms such as RNA mediated silencing, DNA or histone methylation, and heterochromatin formation (reviewed in refs. 6 and 7). These latter mechanisms often have the endogenous role of restricting the deleterious impact of transposable element activity on their host genome (reviewed in ref. 8).
Oomycetes are a group of eukaryotes that superficially resemble fungi in their hyphal growth habit and formation of spores, but are only distantly related to fungi, being placed in the stramenopiles.9 Oomycetes encompass many extremely destructive plant pathogens such as the potato late blight agent, Phytophthora infestans. This pathogen precipitated the Irish potato famine in the mid-1800s and can still cause economically significant losses, thus making it a continuing threat to food security.10,11 Recent years have witnessed a renaissance in molecular biology research into oomycetes, culminating in the genome sequencing of at least seven plant pathogenic species and the discovery of vast numbers of disease promoting effector proteins.12-17 These effector proteins are grouped into two broad classes; those that act in the apoplast (outside the plasma membrane of plant cells), and those that are translocated into host cells to exert their action (reviewed in ref. 18). This latter group contains the intensely studied “RXLR” and “Crinkler” classes of effectors that are defined by specific amino acid motifs within their peptide sequences (reviewed in refs. 18 and 19).
Oomycete Genomes, Transposons and RNA Silencing
Of the available oomycete genome sequences, the genome of P. infestans has been analyzed and annotated in the most detail. Bioinformatic analyses have revealed that the P. infestans genome is organized into gene-rich islands, separated by extensive stretches of gene-poor and highly repetitive DNA.12,13 The repetitive DNA is rich in transposable elements. Effector genes are preferentially located in the gene-poor and repeat (transposon)-rich genomic regions.13,20 This raises the possibility that, first, transposon activity may contribute to the evolution of effectors, and second that the proximity of transposons to effectors may influence their expression. The proximity of transposon sequences to active genes has been reported to influence their expression in numerous organisms.21,22
P. infestans has the largest known oomycete genome, at 240 Mb.13 It has been estimated that 74% of the genome comprises highly repeated sequences. The repetitive DNA of P. infestans encompasses a wide repertoire of transposons: short interspersed elements (SINEs), non-long-terminal repeat (non-LTR) long interspersed elements (LINEs), Copia and Gypsy LTR retrotransposons, Cryptons, Helitrons, DIRS-like, mini-transposable elements (MITEs), hATs, PiggyBACs, Mutators, Mariners, and a broad diversity of novel LTR and DNA transposons.13,23,24 Some of these transposons are presumably active, as their transcripts are present at high levels in some lifecycle stages.25 However, many are believed to represent ancient insertions into the P. infestans genome and are therefore now inactive.13,26
P. infestans has an active RNA silencing pathway,27 which has been exploited in RNA interference (RNAi) studies to determine the role(s) of specific genes (reviewed in refs. 28 and 29). The silencing pathway presumably acts, as in many other eukaryotes (reviewed in ref. 8), to restrict the activity of its heavy genomic load of transposons. A hallmark of silencing is the presence of small non-coding RNAs (sRNA) of 19–40 nt. The general processes and components involved in RNA silencing are reviewed elsewhere.6,7,28
Consistent with this, in a recent report we identified small non-coding RNAs of 40 nt that were homologous to a non-autonomous P. infestans SINE called infSINEm.30 The P. infestans genome contains 32 copies of infSINEm, and some copies are expressed at a low level, likely through its internal RNA polymerase III promoter. It was hypothesized that the identified 40 nt sRNAs were likely to be involved in silencing the expression of infSINEm, and that any P. infestans sequence transcriptionally fused to infSINEm, together with its endogenous copy, would also be subject to silencing (Fig. 1). An additional reason to examine the spread of silencing from a transposon to an endogenous gene was to exploit this to develop simpler vectors for targeted gene silencing in P. infestans. The PiAvr3a gene was selected to deliver a phenotypic readout of silencing spread from infSINEm, as this effector gene is essential for pathogenicity on potato leaves,30,31 and overexpression in the sense direction had not previously led to silencing.32 Transcriptional fusions of infSINEm in transgenic lines of P. infestans, under control of a strong constitutive promoter, initially yielded several lines that were partially silenced for PiAvr3a. This was most pronounced when infSINEm was fused to the 3′ end of PiAvr3a, although silencing was also observed for infSINEm-PiAvr3a fusions. However, over time the silencing was overcome or released in most transgenic lines; the expression of PiAvr3a and infSINEm returned to wild-type or higher levels. In a small number of lines, both PiAvr3a and infSINEm became fully silenced.
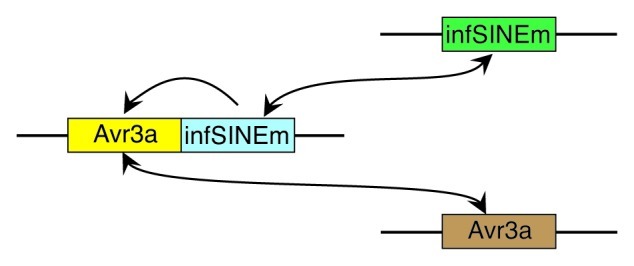
Figure 1. Model for silencing of an effector-encoding gene by transcriptional fusion to a SINE in P. infestans. Small RNAs derived from endogenous infSINEm (green) initiate silencing of the transgenic PiAvr3a-infSINEm fusion transcript (yellow-blue) through degradation by Argonaute (Ago). Secondary sRNAs are formed from the fusion transcript through the action of RNA dependent RNA polymerase (RdR) and Dicer-like (Dcl). Secondary sRNAs target the endogenous copies of both infSINEm and PiAvr3a (brown) to initiate (Ago) or reinforce silencing (RdR→Dcl→Ago). Arrows indicate the direction and reinforcement of silencing.
Evidence for Endogenous Silencing of Effectors in P. infestans through Proximity to Transposons
The biological significance of our findings is that a transposable element-derived sequence, silenced via sRNAs, can potentially also bi-directionally silence nearby sequences in P. infestans. Similar proliferation of transposons and gene repression has been reported from Drosophila melanogaster.33 However, in the P. infestans system, transposition rates and distance to a potential target gene remains to be determined.
In a study into the effects of silencing a series of P. infestans NIF transcription factors, it was demonstrated that silencing of these genes led to formation of heterochromatin at the affected locus.34 The formation of heterochromatin was also demonstrated to spread outwards from the silenced locus for approximately 300 bp, but was also detectable up to 600 bp.
Taken together with our results from infSINEm, it is possible that genes located near silenced transposons (within 300 bp) may be subject to reduction in expression due to heterochromatin formation (Fig. 2). This is particularly of interest when the genomic locations of the RXLR effectors are considered. These genes are preferentially located in genomic regions also heavily populated with transposons.13 Little is known of which effector genes are essential for infection, and which are dispensable. In P. infestans, to date PiAvr3a is the only effector that has been demonstrated to be essential for pathogenicity.30,31 Therefore, until many more RXLR encoding genes have been assessed for their effects on pathogenicity through silencing, it is difficult to definitively associate the genomic location of specific RXLR effectors and neighboring transposons with reduced pathogenicity. However, the recent report of PiAvr2 may provide some evidence that transposon-initiated silencing of effectors may also occur naturally.35
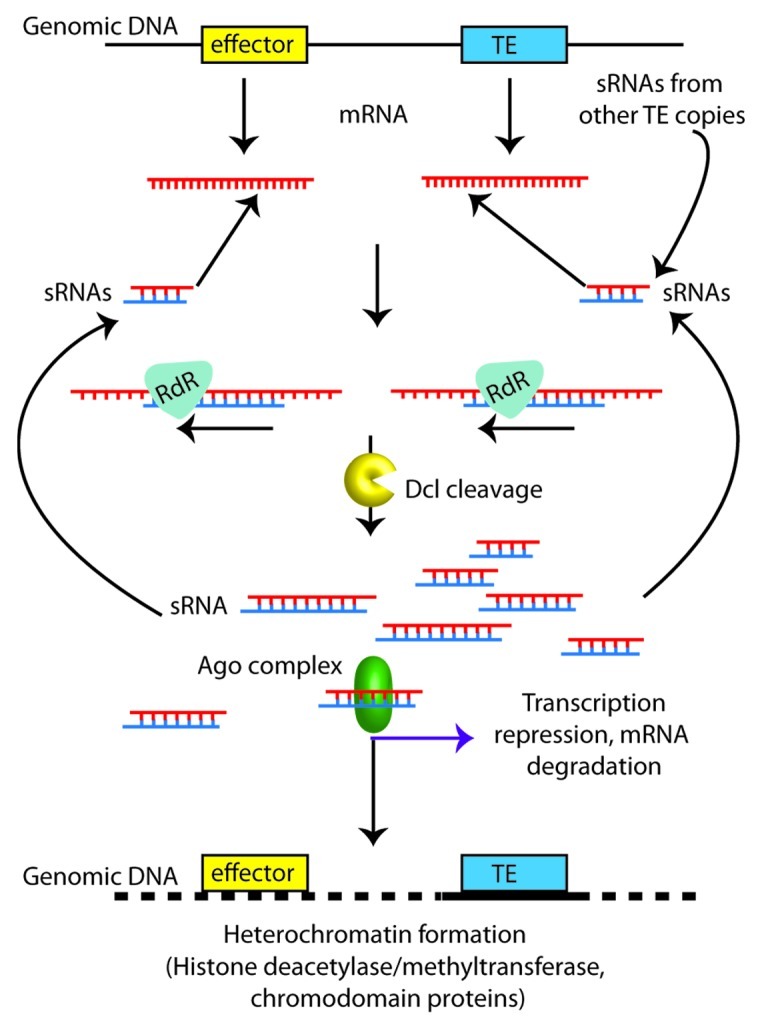
Figure 2. Proposed model for transcriptional repression of effector genes due to proximity to transposon sequences. Transposon sequences (TE; blue box) are strongly targeted for silencing by abundant homologous small RNAs (sRNAs). Inactivation of transposons results from formation of heterochromatin, guided by sRNAs. The heterochromatic region may spread outwards along the genomic DNA (dotted line), and affect nearby effector gene sequences (yellow), either through degradation of mRNAs (Ago) or repression of transcription (histone methylation). The formation of additional sRNAs, maintaining or reinforcing the silenced state, can occur through the action of Rdr and Dcl on mRNAs from the transposon or effector gene.
The PiAvr2 effector is recognized by the R2 resistance protein in potato,35 and initiates a defense response called the hypersensitive response, a form of programmed cell death that restricts the growth of invading pathogens, including P. infestans. Genotypes of P. infestans that are virulent on potato plants with the R2 gene express a sequence variant called PiAvr2-like that is not recognized by R2. Virulent isolates are typically homozygous for PiAvr2-like, while avirulent isolates may be homozygous for PiAvr2 or heterozygous. However, a small number of virulent genotypes are heterozygous, but express only PiAvr2-like. Other heterozygous isolates may express predominantly one allele. These results demonstrate an allele-specific inactivation of expression at this effector locus. The sequence and organization of the genomic region encompassing the PiAvr2 locus is highly variable between virulent and avirulent alleles, and is bounded by transposable element-derived sequences. The nearest of these transposons (transposase-like) is 231 bp from the 3′ end of PiAvr2, which is within the range of heterochromatin formation determined experimentally.34 It remains to be determined if this proximity can influence PiAvr2 expression.
Transcriptional inactivation of avirulence effectors has also been observed in P. sojae, the soybean root rot pathogen. Genotypes have been identified that exhibit transcriptional inactivation of the PsAvr1a, 1b and 3a/5 avirulence genes.36-38 For PsAvr1a and 3a/5, transposable element sequences are located at the 3′ end, or in the promoter of these genes, respectively.37,38 Although it is intriguing that transposons are found nearby transcriptionally inactivated effector genes, it should be cautioned that ascribing effector silencing events to transposons is complicated by the nature of effector gene evolution in P. infestans. That is, many RXLR effectors are part of gene families, with members exhibiting very closely related gene sequences.13 Furthermore, effectors may also exhibit copy number polymorphism between isolates.37 Transcriptional inactivation by silencing mechanisms may lead to a loss, or reduced transcription, for the entire gene family. However, such a possibility remains to be demonstrated.
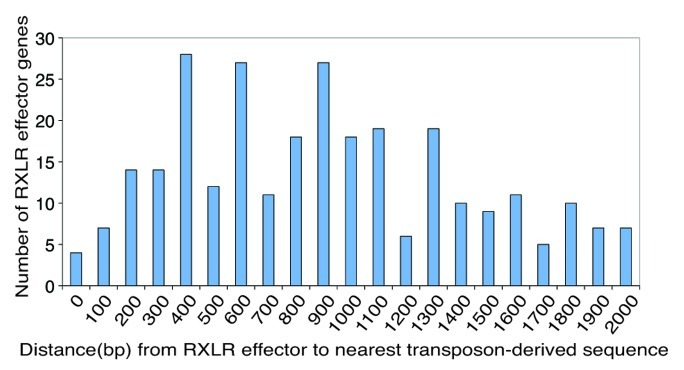
Of the 563 RXLR effectors predicted from the P. infestans genome, a total of 283 are located within 2 kb of a transposon-derived sequence (Fig. 3). Of these, four contain transposon insertions, a further 35 are located within 300 bp of a transposon-derived sequence, and a total of 106 within 600 bp. PiAvr2, together with PiAvr4, PiAvrBlb1, and PiAvrBlb2 (reviewed in ref. 5) are found within 2 kb of transposon-derived sequences.
Figure 3. RXLR effector gene proximity to transposon derived sequences in the P. infestans genome. The Y-axis represents the number of RXLR effector genes in each group. The X-axis represents the distance from RXLR effector to nearest transposon-derived sequence (100 bp window) up to a maximum of 2 kb. The PiAvr2 effector is located in the "300" window, PiAvr4 is in the “1200” window, and PiAvrBlb1 is in the “1500” window. The RXLR encoding gene family encompassing PiAvrBlb2 has three paralogs in each of the “400” and “900” windows.
The possibility that P. infestans can vary the expression of a large proportion of pathogenicity effectors may contribute to its adaptability when confronted with resistant host plants. In addition to the sequence variation present within populations, it is possible that it can also use epigenetic mechanisms to alter its specific virulence and overcome plant resistance, and vary pathogenicity.
Conclusions and Prospects
Transposable elements are often called selfish or junk DNA, but they have had a profound influence on the evolution of the genomes, and likely the biology, of many fungal and oomycete plant pathogens. This is exemplified by P. infestans and closely related species that have greatly expanded genomes,39 assumed to be due to extensive transposon amplification. It has been proposed that the location of the majority of disease effector genes in transposon-rich, rapidly evolving, genomic regions is likely to have had an impact on adaptability to new host plants throughout evolution.39 The next step in understanding the influence that transposons have on the biology of P. infestans will be to determine the extent of influence from heterochomatin formation in close proximity to transposons. Small RNAs are considered to be centrally involved in many aspects of gene silencing, either post-transcriptional or transcriptional. High-throughput deep-sequencing of sRNAs will reveal if these sRNAs map to, and thus silence, the regions of genome spanning effectors and transposons. Such studies are presently comparatively rare for plant pathogens,40 but hold promise in determining the role(s) of transposons in influencing the expression and evolution of effector genes, and thus host range of plant pathogens.
Acknowledgments
S.C.W. and A.O.A. were supported by the Scottish Government Rural and Environment Science and Analytical Services (RESAS), and the UK Biotechnology and Biological Sciences Research Council (BBSRC). R.R.V. and C.D. were supported by grants from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS), the Swedish Farmers Foundation for Agricultural Research (SLF), the Swedish Board of Agriculture (SJV), and the Swedish University of Agriculture (SLU).
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/20265
References
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–9. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–48. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 3.McDowell JM, Simon SA. Recent insights into R gene evolution. Mol Plant Pathol. 2006;7:437–48. doi: 10.1111/j.1364-3703.2006.00342.x. [DOI] [PubMed] [Google Scholar]
- 4.De Wit PJ, Mehrabi R, Van den Burg HA, Stergiopoulos I. Fungal effector proteins: past, present and future. Mol Plant Pathol. 2009;10:735–47. doi: 10.1111/j.1364-3703.2009.00591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whisson SC, Avrova AO, Boevink PC, Armstrong MR, Seman ZA, Hein I, et al. Exploiting knowledge of pathogen effectors to enhance late blight resistance in potato. Potato Res. 2011;54:325–40. doi: 10.1007/s11540-011-9197-y. [DOI] [Google Scholar]
- 6.Ketting RF. The many faces of RNAi. Dev Cell. 2011;20:148–61. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Goto DB, Nakayama JI. RNA and epigenetic silencing: Insight from fission yeast. Dev Growth Differ. 2011 doi: 10.1111/j.1440-169X.2011.01310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–68. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burki F, Shalchian-Tabrizi K, Minge M, Skjaeveland A, Nikolaev SI, Jakobsen KS, et al. Phylogenomics reshuffles the eukaryotic supergroups. PLoS One. 2007;2:e790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fry W. Phytophthora infestans: the plant (and R gene) destroyer. Mol Plant Pathol. 2008;9:385–402. doi: 10.1111/j.1364-3703.2007.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haverkort AJ, Boonekamp PM, Hutten R, Jacobsen E, Lotz LAP, Kessel GJT, et al. Societal costs of late blight in potato and prospects of durable resistance through cisgenic modification. Potato Res. 2008;51:47–57. doi: 10.1007/s11540-008-9089-y. [DOI] [Google Scholar]
- 12.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–6. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 13.Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–8. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 14.Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, Kemen E, et al. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science. 2010;330:1549–51. doi: 10.1126/science.1195203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lévesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, et al. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 2010;11:R73. doi: 10.1186/gb-2010-11-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kemen E, Gardiner A, Schultz-Larsen T, Kemen AC, Balmuth AL, Robert-Seilaniantz A, et al. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 2011;9:e1001094. doi: 10.1371/journal.pbio.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Links MG, Holub E, Jiang RH, Sharpe AG, Hegedus D, Beynon E, et al. De novo sequence assembly of Albugo candida reveals a small genome relative to other biotrophic oomycetes. BMC Genomics. 2011;12:503. doi: 10.1186/1471-2164-12-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schornack S, Huitema E, Cano LM, Bozkurt TO, Oliva R, Van Damme M, et al. Ten things to know about oomycete effectors. Mol Plant Pathol. 2009;10:795–803. doi: 10.1111/j.1364-3703.2009.00593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stassen JH, Van den Ackerveken G. How do oomycete effectors interfere with plant life? Curr Opin Plant Biol. 2011;14:407–14. doi: 10.1016/j.pbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Raffaele S, Win J, Cano LM, Kamoun S. Analyses of genome architecture and gene expression reveal novel candidate virulence factors in the secretome of Phytophthora infestans. BMC Genomics. 2010;11:637. doi: 10.1186/1471-2164-11-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anbar M, Bracha R, Nuchamowitz Y, Li Y, Florentin A, Mirelman D. Involvement of a short interspersed element in epigenetic transcriptional silencing of the amoebapore gene in Entamoeba histolytica. Eukaryot Cell. 2005;4:1775–84. doi: 10.1128/EC.4.11.1775-1784.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerat E, Sémon M. Influence of the transposable element neighborhood on human gene expression in normal and tumor tissues. Gene. 2007;396:303–11. doi: 10.1016/j.gene.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Jiang RH, Dawe AL, Weide R, van Staveren M, Peters S, Nuss DL, et al. Elicitin genes in Phytophthora infestans are clustered and interspersed with various transposon-like elements. Mol Genet Genomics. 2005;273:20–32. doi: 10.1007/s00438-005-1114-0. [DOI] [PubMed] [Google Scholar]
- 24.Whisson SC, Avrova AO, Lavrova O, Pritchard L. Families of short interspersed elements in the genome of the oomycete plant pathogen, Phytophthora infestans. Fungal Genet Biol. 2005;42:351–65. doi: 10.1016/j.fgb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Judelson HS, Ah-Fong AM, Aux G, Avrova AO, Bruce C, Cakir C, et al. Gene expression profiling during asexual development of the late blight pathogen Phytophthora infestans reveals a highly dynamic transcriptome. Mol Plant Microbe Interact. 2008;21:433–47. doi: 10.1094/MPMI-21-4-0433. [DOI] [PubMed] [Google Scholar]
- 26.Judelson HS. Sequence variation and genomic amplification of a family of Gypsy-like elements in the oomycete genus Phytophthora. Mol Biol Evol. 2002;19:1313–22. doi: 10.1093/oxfordjournals.molbev.a004192. [DOI] [PubMed] [Google Scholar]
- 27.Vetukuri RR, Avrova AO, Grenville-Briggs LJ, Van West P, Söderbom F, Savenkov EI, et al. Evidence for involvement of Dicer-like, Argonaute and histone deacetylase proteins in gene silencing in Phytophthora infestans. Mol Plant Pathol. 2011;12:772–85. doi: 10.1111/j.1364-3703.2011.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nunes CC, Dean RA. Host-induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol Plant Pathol. 2011 doi: 10.1111/j.1364-3703.2011.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whisson SC, Avrova AO, Grenville-Briggs LJ, Van West P. Mechanisms and application of gene silencing in oomycetes. In: Lamour K, Kamoun S, eds. Oomycete Genetics and Genomics: Diversity, Interactions, and Research Tools. Hoboken, NJ, USA: Wiley-Blackwell, 2009: 493-516. [Google Scholar]
- 30.Vetukuri RR, Tian Z, Avrova AO, Savenkov EI, Dixelius C, Whisson SC. Silencing of the PiAvr3a effector-encoding gene from Phytophthora infestans by transcriptional fusion to a short interspersed element. Fungal Biol. 2011;115:1225–33. doi: 10.1016/j.funbio.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Bos JIB, Armstrong MR, Gilroy EM, Boevink PC, Hein I, Taylor RM, et al. Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc Natl Acad Sci U S A. 2010;107:9909–14. doi: 10.1073/pnas.0914408107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–8. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 33.Blumenstiel JP. Evolutionary dynamics of transposable elements in a small RNA world. Trends Genet. 2011;27:23–31. doi: 10.1016/j.tig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Judelson HS, Tani S. Transgene-induced silencing of the zoosporogenesis-specific NIFC gene cluster of Phytophthora infestans involves chromatin alterations. Eukaryot Cell. 2007;6:1200–9. doi: 10.1128/EC.00311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilroy EM, Breen S, Whisson SC, Squires J, Hein I, Kaczmarek M, et al. Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 2011;191:763–76. doi: 10.1111/j.1469-8137.2011.03736.x. [DOI] [PubMed] [Google Scholar]
- 36.Shan W, Cao M, Leung D, Tyler BM. The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol Plant Microbe Interact. 2004;17:394–403. doi: 10.1094/MPMI.2004.17.4.394. [DOI] [PubMed] [Google Scholar]
- 37.Qutob D, Tedman-Jones J, Dong S, Kuflu K, Pham H, Wang Y, et al. Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a. PLoS One. 2009;4:e5066. doi: 10.1371/journal.pone.0005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong S, Yu D, Cui L, Qutob D, Tedman-Jones J, Kale SD, et al. Sequence variants of the Phytophthora sojae RXLR effector Avr3a/5 are differentially recognized by Rps3a and Rps5 in soybean. PLoS One. 2011;6:e20172. doi: 10.1371/journal.pone.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raffaele S, Farrer RA, Cano LM, Studholme DJ, MacLean D, Thines M, et al. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science. 2010;330:1540–3. doi: 10.1126/science.1193070. [DOI] [PubMed] [Google Scholar]
- 40.Nunes CC, Gowda M, Sailsbery J, Xue M, Chen F, Brown DE, et al. Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae. BMC Genomics. 2011;12:288. doi: 10.1186/1471-2164-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]