Abstract
Bacteria and their viruses (phages) are antagonists, yet have coexisted in nature for billions of years. Models proposed to explain the paradox of antagonistic coexistence generally reach two types of solutions: Arms race-like dynamics that lead to hosts and viruses with increasing resistance and infection ranges; and population fluctuations between diverse host and viral types due to a metabolic cost of resistance. Recently, we found that populations of the marine cyanobacterium, Prochlorococcus, consist of cells with extreme hypervariability in gene sequence and gene content in a viral susceptibility region of the genome. Furthermore, we found a novel cost of resistance where resistance to one set of viruses is accompanied by changes in infection dynamics by other viruses. In this combined mini-review and commentary paper we discuss these findings in the context of existing ecological, evolutionary and genetic models of host-virus coexistence. We suggest that this coexistence is governed mainly by fluctuations between microbial subpopulations with differing viral susceptibility regions and that these fluctuations are driven by both metabolic and enhanced infection costs of resistance. Furthermore, we suggest that enhanced infection leads to passive host-switching by viruses, preventing the development of hosts with universal resistance. These findings highlight the vital importance of community complexity for host-virus coexistence.
Keywords: antagonistic coexistence, arms race, fluctuating selection, kill the winner, matching-alleles, gene-for-gene, cost of resistance, virus, cyanobacteria, Prochlorococcus
The paradox of host-parasite coexistence despite their antagonistic relationships has long been of interest in ecological and evolutionary studies1 and has often focused on interactions between bacteria and the viruses (phages) that infect them.2-8 Viruses, being obligate parasites, depend on the infection of host cells for the production of progeny. Seeing as the end result of lytic infection is the death of the bacterial host, and the phage replication cycle is typically more rapid than that of their hosts, the question arises as to how it is that phages have not killed off their bacterial hosts. And, how is it that they, in the process, have not caused their own extinction? This paradox of host-virus coexistence became even more pronounced for the marine environment with the discovery that the oceans are teeming with large numbers of bacteria and viruses.9,10
Recently, we reported a high degree of gene sequence and gene content diversity in regions of the genome responsible for viral susceptibility among populations of the abundant marine cyanobacterium, Prochlorococcus.2 We further described a novel cost of resistance to viral infection, whereby resistance to one set of viruses led to enhanced infection dynamics by other viruses.2 In this combined mini-review and commentary paper, we discuss these recent findings in the context of existing evolutionary, ecological and genetic models developed to explain antagonistic coexistence. We first briefly review the main models and highlight some similarities and differences between them in light of the distinctive approaches, perspectives and assumptions used to build them, and discuss some of their limitations. For a more comprehensive review of the literature and other aspects of coexistence we refer the reader to Thompson,1 Bohannan and Lenski3 and Woolhouse et al.11 We conclude that the biological context in which organisms reside in nature needs to be incorporated into these models if we are to gain a better understanding of antagonistic coexistence between hosts and viruses in the oceans.
The Arms Race
One of the current evolutionary models put forward to explain antagonistic coexistence is the continual arms race between bacterial hosts and their viral parasites.3,4 In this model, mutations occur in the host that confer resistance to viruses and thus prevent infection and decimation of the host population. Subsequently, mutations occur in the virus that enables it to re-infect the recently emergent resistant population. Hence the coexistence of host and parasite is maintained through repeated cycles of host mutation and viral counter-mutation. Under this model selection is directional with a continuous increase in the breadth of resistance and infectivity of host and parasite, respectively,4 resulting in generalist hosts and viruses. Despite this directionality, these antagonists often remain in the same evolutionary position relative to each other in a Red Queen-like manner. However, it is important to point out that Red Queen dynamics do not necessarily lead to directional selection and therefore the terms “arms race” and “Red Queen dynamics” are not interchangeable.11
Despite the wide recognition of the arms race model in the scientific community, most coevolutionary experimental studies indicate that the arms race between bacteria and viruses does not continue indefinitely,3,5,12 often being limited to no more than a few cycles.3,6,12,13 There are likely to be two reasons for this based on existing experimental evidence. The first is due to genetic constraints on the phage for counter-mutation, whereby a resistant host emerges yet no subsequent phage acquires a mutation enabling it to reinfect this resistant strain.3,6,12,13 In such situations resistant bacteria would come to dominate the population with a concomitant decline in virus numbers3,6,12 that could even lead to viral extinction.6
The second factor likely to limit the arms race is related to metabolic constraints on the host that are associated with resistance.3,5,14 This can result, for example, if host nutrient acquisition proteins at the surface of the host also serve as viral receptors. Resistance-conferring mutations in such proteins would prevent viral attachment to the cell surface, but are also likely to impair nutrient uptake and utilization by the host (see Winter et al.15 and refs. within). This metabolic limitation is a cost of resistance that is manifested as a reduction in growth rate and thus lowers fitness of the mutants,2,5,14 making the resistant host less competitive than previous hosts.
Fluctuating Selection
In light of the limitations described above, an alternative evolutionary model has been suggested to explain antagonistic coexistence between bacterial hosts and viral parasites, that of density dependent fluctuating selection.1,4,5,11 In this model, mutants emerge that lead to diverse bacterial and viral populations. Different to the arms race model, however, mutations do not need to be continuously produced and the emergent bacteria and viruses have different rather than greater resistance and infectivity ranges, respectively. Furthermore, while a metabolic cost of resistance is considered to limit the extent of the arms race, such a cost is a basic assumption of fluctuating selection. This cost of resistance prevents competitive exclusion of susceptible hosts within which viral populations propagate.
To illustrate the fluctuating nature of population dynamics in this model we will describe a simplified situation of a susceptible and a resistant host and a single parasite. As the abundance of the fast-growing susceptible host rises to dominate the population, so does the contact rate with its abundant parasite, resulting in increased host mortality and a subsequent decline in the susceptible host population. Resistant mutants are unaffected by the abundant virus and will subsequently increase in numbers despite their slower growth rate. Concomitantly, the abundance of the parasite decreases due to the obligatory requirement of susceptible hosts for parasite propagation. The ensuing low parasite abundance eliminates the advantage for resistant cells, and the faster growing susceptible hosts will now out-compete the resistant ones, and the cycle can begin again. In this manner neither resistant nor susceptible strains are driven to extinction, and the latter allow viral populations to also persist. Thus, abundances of resistant cells, susceptible hosts and parasites oscillate in the community over time due to the negative density dependent selection driven by alternating selection pressures: viral selection for resistant mutants and competitive selection for faster growing hosts.1,11
Kill the Winner
The “kill the winner” hypothesis is an ecological model similar to that of fluctuating selection that relates to oceanic communities.7 Similar to fluctuating selection, kill the winner assumes a metabolic cost for resistance, and is linked to resource utilization.7 The outcome of both models is fluctuations in population composition resulting in a diverse community.7,15 However, some other assumptions differ between the two models. For example, kill the winner assumes that a particular host can be infected by only one phage and that one phage can infect only a single host.7,15 Other major differences between these models relate to the ecological vs. evolutionary perspectives used to build them. For example the kill the winner hypothesis relates to abiotic conditions, particularly nutrient availability, together with differences in uptake rates by diverse bacteria and the ensuing competition that results.7,15
These ecological and evolutionary models of fluctuating selection cannot be easily reconciled with existing experimental data. First, they assume a tradeoff between viral resistance and fitness and generally relate this tradeoff to competition for resources that negatively impact the growth rate of the resistant bacterium. While resistance is often accompanied by a decrease in growth rate relative to the susceptible cells from which they were derived,2,3,5,14,16,17 it is also quite common that a growth rate cost of resistance is not detected.2,3,5,14,18 Therefore, it is generally assumed that the conditions under which a reduction in growth rate occur were simply not found.3,14,18 We will argue below that this is not necessarily the case. Furthermore, while mutations conferring resistance to infection are sometimes found in nutrient uptake associated genes,2,3,17 some are associated with lipopolysaccharide and cell-wall biosynthesis genes.2,3 Such mutants are not necessarily competitively inferior in their ability to utilize resources (unless they show a growth rate cost which would indirectly affect their capacity for resource utilization). Additionally, even when a growth rate cost of resistance does appear, compensatory mutations can evolve and thus decrease, if not eliminate the cost.16 Therefore the question remains: If a cell can develop resistance without paying a growth rate cost what prevents it from taking over the community?
Second, the “kill the winner” model predicts that under viral predation the bacterial population will be highly diverse and that the dominant species in the marine pelagic system will not exceed a density threshold of ~10,000 cells per ml.7 This is in direct contrast to the high densities of dominant bacterial groups present in the marine environment. For example, single ecotypes of the dominant photosynthetic cyanobacterium, Prochlorococcus, reach 200,000 cells per ml19,20 and that of the heterotrophic bacterium, Pelagibacter, can reach 125,000 cells per ml.21 Therefore other factors must be taken into consideration if we are to adequately explain long-term antagonistic coexistence.
Genetic Models of Host-Parasite Coevolution
At the genetic level, two models of infection-resistance specificity between hosts and parasites are commonly considered. The gene-for-gene model relates to the recognition between a specific host receptor and a matching pathogen elicitor molecule that leads to host resistance. A lack of a match between these molecules leads to infection. As in the arms race, this model predicts the appearance of a universally infective pathogen or resistant host.22 However, at the population level, this would lead to the fixation of the generalist, and seeing as this doesn’t occur in nature, these models invoke a metabolic cost associated with generalism that would lead to fluctuating selection.4,5,22
The matching-alleles model assumes that an exact match between a host and parasite is needed for infection and therefore each pathogen is specific to, and can only infect, a single host. Furthermore, in this model if mutations occur they cause a change in the range of resistance and infectivity, but a single parasite still only infects a single host (similar to the assumption used in the kill the winner hypothesis). Therefore this model does not permit universal infectivity or resistance by assuming a genetic constraint on directional selection of an arms race.4,22 Such populations are controlled by fluctuating selection between different genotypes and population diversity is maintained without a metabolic cost associated with resistance. Experimental data suggest that the gene-for-gene model is found in plant-pathogen systems22 and that matching-alleles-like interactions occur at some level in vertebrate immunity systems.23
These genetic models may actually be at either ends of a specificity continuum with specificity more realistically lying somewhere in the middle.22 While this is a purely theoretical combination of these two models, it predicts that a single parasite can both efficiently infect a specific host (as in the matching-alleles model) as well as infect other hosts with lower efficiency (as in the gene-for-gene model)22 in an imperfect lock and key manner.24
The above suggestion that genetic diversity within populations can be maintained without a metabolic cost of resistance is seldom incorporated into models of fluctuating selection (but see refs. 4 and 22) and has not been shown for systems other than vertebrate immunity. Moreover it is completely ignored in ecological models such as kill the winner.7,15 Below we describe our recent findings of hyperdiversity in viral susceptibility regions of cyanobacterial genomes and argue that such genetic microdiversity, together with mutations that lead to changes in infection dynamics, facilitates fluctuations at the subpopulation level, providing experimental evidence in line with the ideas outlined in Agrawal and Lively.22
Hyperdiversity in Viral Susceptibility Regions
We investigated bacterial resistance to viral infection using the cyanobacterium Prochlorococcus and its viruses as a model system, as it exemplifies a microbe that coexists with antagonistic viruses at high abundances in the marine environment. Using four ancestral laboratory strains belonging to two high-light adapted Prochlorococcus ecotypes, we isolated 77 substrains, each experimentally selected for resistance to one of 10 different viruses.2 The genomic characterization of these resistant substrains revealed mutations in a variety of predicted cell-surface genes. Only one of these 24 genes is known to encode for a protein involved in resource utilization—a subunit for phosphorus uptake. The majority of mutations were in genes that appear to be involved in lipopolysaccharide or cell wall biosynthesis and modification or are of unknown function.2
The resistance conferring mutations were in strain specific genes that localized primarily to a single hypervariable region of the genome, known as genomic island 4. This region is highly variable in terms of gene content and gene sequence diversity when comparing among strains from the same ecotype.25 This is in stark contrast to the high degree of gene synteny and gene sequence identity in most other regions of the genome.25 The strain specific nature of the genes in this genomic region, suggests that viral selection has promoted the loss of susceptibility genes accompanied by the lateral gain of new genes with similar cell-surface functions, but with different viral recognition determinants.2 The high diversity of this genome region in natural populations of Prochlorococcus suggests that it is constantly losing and gaining genes and accumulating other mutations. Therefore this genome region can be considered a viral susceptibility region that is prone to enhanced diversification, that has developed over long evolutionary scales, and that is continuing to develop today in response to viral selection pressure.2,26
The hypervariability resulting from this viral selection pressure has given rise to Prochlorococcus populations in the oceans that are composed of various subpopulations with diverse viral susceptibility regions. Since each subpopulation is susceptible to a different set of viruses no single viral type can cause the collapse of the entire Prochlorococcus population. We suggest that fluctuating selection acts at this subpopulation level facilitating long-term host-virus coexistence despite high densities of the overall host population.
Metagenomic analyses revealed genomic islands enriched with uncommon cell-surface genes in other marine Bacterial and Archaeal genomes.26 Thus this genome mechanism of diversification of viral susceptibility regions, both through nucleotide polymorphisms as well as loss and gain of cell-surface genes, is potentially widespread among a variety of host-virus systems. Therefore, when addressing antagonistic host-virus coexistence in abundant populations in the environment, the microdiversity in viral susceptibility regions of these populations needs to betaken into consideration. Indeed this microdiversity has been addressed in recent discussions of subpopulation oscillations in aquatic environments.15,26 The challenge now will be to determine what constitutes a discrete subpopulation and whether these can be discerned from the standpoint of host-virus interactions.
Enhanced Infection, a Novel Cost of Resistance
Many of the mutants in our study exhibited a reduced growth rate, including those with a mutation in the phosphorus uptake gene and others with mutations in genes involved in lipopolysaccharide and cell wall biosynthesis and modification.2 However, as discussed above, many other mutants displayed no detectable growth rate cost associated with resistance.2,3,5,14,18 We found that a number of these strains exhibited a novel type of cost of resistance, that of enhanced susceptibility to other viruses.2 In these strains, the exact same mutations that conferred resistance to one set of viruses brought about enhanced infection dynamics by other viruses. In at least two cases this was due to more rapid adsorption by these other viruses to the resistant mutant relative to the ancestral strain.2 Additional situations were also observed, whereby resistant strains were infected more slowly by other viruses. Moreover, studies by others have shown that mutations that conferred resistance to one virus enabled infection by another virus that was incapable of infecting the ancestral bacterium from which the resistant strain was derived.14,27 Therefore, a bacterial mutation can be under varying levels of selection pressures due to different degrees of infectivity by distinct viruses, including those belonging to different viral families.2 These findings indicate that infection is not a binary off or on trait, but rather that infectivity can be considered a continuum that ranges from complete resistance, through varying infection efficiencies, to highly infective. This was also suggested by Holmfeldt et al.28 who showed drastic differences in infection efficiency by the same virus for different strains of Flavobacteria isolated from the marine environment.
Based on these findings, we propose that the enhanced infection cost of resistance can serve as the tradeoff between viral resistance and fitness and drive fluctuating selection. This novel cost of resistance can, therefore, explain antagonistic coexistence, even when there is no metabolic cost associated with it. In this scenario, oscillations in population sizes are still density dependent, but the selective force is always viral pressure, alternating between pressure from one virus and pressure from another virus rather than between viral pressure and competitive selection. However, host resistance or susceptibility and the degree of that susceptibility in any given interaction, will vary depending on the virus encountered. The continuum of infection efficiencies resulting from enhanced infection, that would lead to the generation of fluctuating selection between diverse genotypes without the requirement for a metabolic cost of resistance, provides empirical evidence for the combined theoretical model proposed by Agrawal and Lively (described above).22
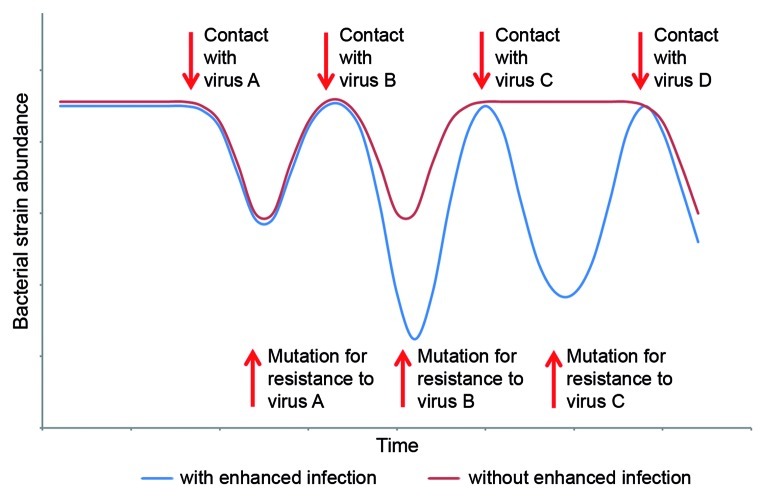
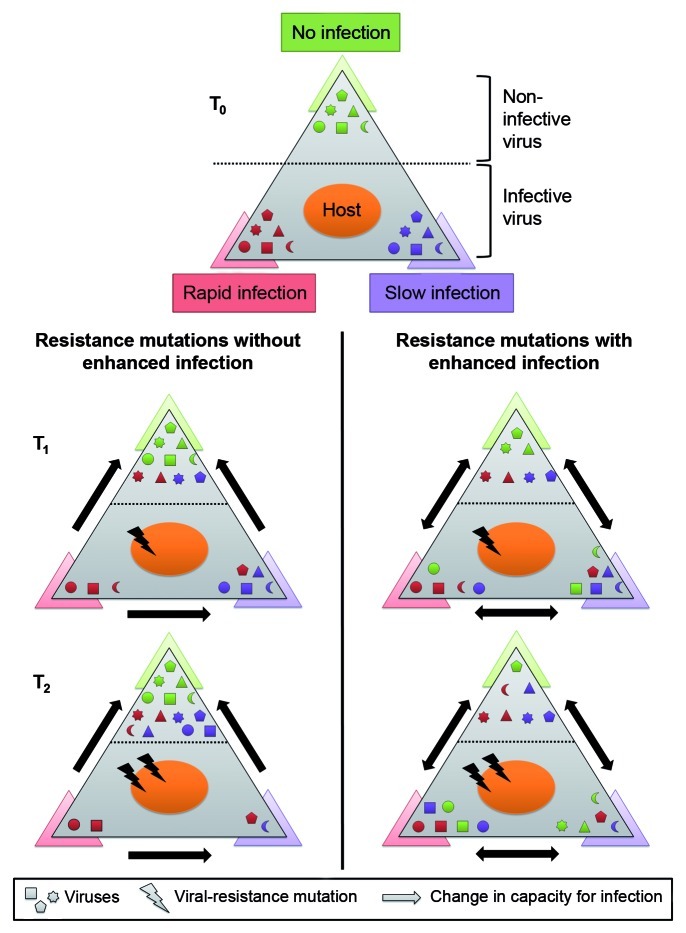
It should be noted that fluctuations between host subpopulations can be driven by viruses even when new mutations conferring resistance have neither type of cost, as long as a particular bacterium can be infected by more than one virus and that each resistance mutation affects the ability of only one of them to infect the host. However, these fluctuations will be much more common and considerably more pronounced when resistance is accompanied with the enhanced infection cost (Fig. 1). This is because mutation-induced enhanced infection and susceptibility to a changing set of viruses dramatically increases the probability for infection relative to mutations that confer resistance alone. This is due both to a greater chance for the mutant to meet infectious viruses (Fig. 2) and the rapidity of the ensuing infection once these viruses are encountered. Furthermore, enhanced infection would prevent the development of generalist hosts with a wide range of resistance to co-occurring viruses, and would thus preclude the directional selection expected from a continuous arms race (Fig. 2). Enhanced infection can, however, support Red Queen dynamics, in which mutations serve to enhance diversity within host and virus populations without any directionality of selection.
Figure 1. Schematic representation of bacterial population dynamics with and without enhanced infection. In this simplified depiction, a single ancestral host and 4 viruses are present. The ancestral host is susceptible to viruses A, B and D each of which infect the host with equal efficiency. This ancestral host is not initially susceptible to virus C. In the absence of enhanced infection, declines in the abundance of the host (red line) result from contact with viruses A, B and D but not with virus C and reduce the population to the same abundance. With the existence of enhanced infection, declines in the abundance of the host (blue line) result from interactions with all 4 viruses and are to different lower bounds. In this latter scenario, the mutation conferring resistance to virus A increased the infection efficiency of virus B for the emergent resistant strain, and the mutation conferring resistance to virus A or B made it susceptible to virus C. This depiction assumes that there is no growth rate cost to mutations and no virus counter-mutations occur.
Figure 2. Enhanced infection leads to shifts in the phages exerting population control. In a given environment a host population (large oval shape) encounters diverse viral types (small geometric symbols) with different capacities for infecting the host. Non-infective viruses are positioned at the top corner, viruses with high infectivity (rapid) are at the bottom left corner and viruses with low infectivity (slow) are at the bottom right corner of each infectivity triangle. Over time the host acquires mutations that change the ability of viruses to infect it, thus changing their position in the infectivity triangle. To aid traceability the color of the viruses represents their initial capacity for infection in the T0 triangle while their position at subsequent time points (T1 and T2) represents their current capacity for infection. In the left panel host mutations conferring resistance to viruses are not associated with enhanced infection by other viruses and therefore viral shifts in infection capacity are unidirectional. In this case, the accumulation of resistance mutations leads to a generalist bacterium that, in the absence of a metabolic cost of resistance or a counter-mutation in the virus, would come to dominate the population. In the right panel host mutations conferring resistance to viruses are associated with enhanced infection leading to bidirectional changes in infection capabilities by other viruses. In this case, the accumulation of resistance mutations results in shifts in the viruses capable of infecting the host rather than a reduction in the number of viruses that can infect it. Therefore enhanced infection prevents a single bacterial strain from taking over the population by continuous shifts in the virus exerting control, and thus helps maintain bacterial diversity.
Therefore resistance costs, whether caused by a metabolic or enhanced infection cost, can drive fluctuating selection. While metabolic costs are linked to environmental conditions through competition for resource availability,7,15 the enhanced infection cost is manifested in the context of the community. Therefore, fluctuating selection will be driven by enhanced infection costs in both resource deplete and replete conditions. As such, enhanced infection is likely to be an important driver of fluctuating population dynamics facilitating long-term host-parasite coexistence in nature and should be taken into consideration in future ecological models of fluctuating selection.
Enhanced Infection Leads to Passive Host-Switching in Viruses
In our discussions so far we have focused our attention on the host. Let us now consider the matter from the parasite's perspective. As discussed above, bacterial microdiversity in genomic regions responsible for viral susceptibility is expected to significantly reduce host population abundance and availability to any given virus.2,26 This raises the question of what enables persistence of viruses if only a small fraction of the potential host population is actually available for infection? This is especially acute both because viruses constantly lose some hosts due to mutations that provide resistance to it,2-5,14 and because of the low rate of counter-mutation for the reinfection of these hosts.3,6,13 One possible explanation is mutations in viruses that enable them to switch hosts after the original host gained resistance.1 Tail fiber diversification due to point mutations29 and domain swapping,30 are well known phenomena that can lead to a change in host-range for the phage.
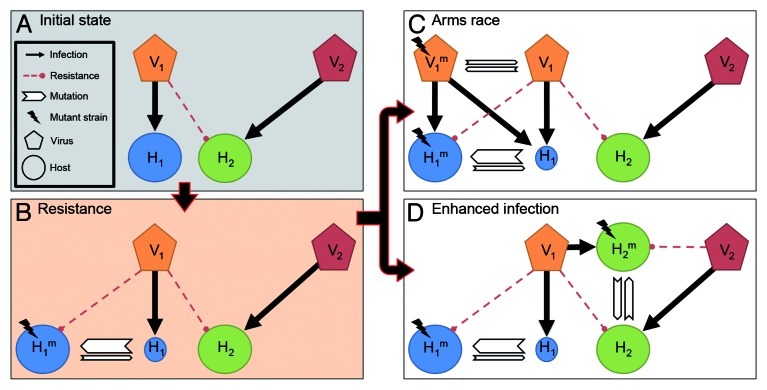
Another possibility is that enhanced infection supplies a virus with new infection opportunities without any change occurring in the virus. Indeed, the same resistance mutation in a host that caused a narrowing of host range for one virus can sometimes provide other viruses with new infection capabilities.2,14,27 Taken from the perspective of a single virus, a virus that constantly loses some hosts due to resistance, profits from interactions between other host-virus pairs: resistance mutations and selection for resistance in these other interactions, can provide the first virus with greater infectivity for some hosts2 and even afford it with additional hosts14,27 (Fig. 3). In this light we should consider the host range of viruses as a dynamic set of bacterial strains, as its host range includes bacteria which are just one mutation away from serving as hosts (Fig. 3).
Figure 3. The dynamic host range of a virus. Initially (A) two virus-host pairs (V1-H1 and V2-H2) are present in a common environment, but do not cross infect each other. Following a mutation, H1 gains resistance to V1 giving rise to H1m (B). In this scenario we assume that the emergent host has no growth rate cost of resistance. Thus the population size of H1m will increase while that of H1 will decline due to viral infection (depicted by a smaller circle). The survival of V1 now depends on its ability to gain a new host. To keep up in an arms race (C) a counter-mutation in V1 gives rise to a host range mutant (V1m) which can reinfect H1m, thereby enabling the recoupling of the virus-host pair. Alternatively, passive host switching occurs through enhanced infection (D), whereby V1 is provided with a new host (H2m) due to a mutation in H2 that conferred resistance to, and was selected by, another virus, V2 (D). Thus an independent interaction between H2 and V2 provided the V1 population with another host without any mutation in V1. In summation, the arms race and enhanced infection lead to a dynamic host range for V1. Despite the fact that this virus lost its immediate host (H1) through a resistance mutation, its host range can be considered to include both H1m and H2. These bacterial types are only one mutation away from serving as its immediate hosts, as a single mutation in these bacteria or in V1 can lead to productive infection.
We propose that due to this constant “fresh serving” of potential hosts, the virus can sustain its population at times when the abundance of its initial host is extremely low and the chance of contact is minimal. This continuous, passive supply of new hosts is thus likely to compensate at least partially for the asymmetry in counter-mutation in phages. This would enable viral persistence in the face of changes in host availability and abundance that result from mutation induced resistance and fluctuating host dynamics, respectively.
Summary
In summary, natural communities are complex systems whose members are constantly interacting with each other. The diversity of the players in this system are the outcome of millions of years of large-scale coevolution between viruses and their various hosts, during which host mutations have led to hypervariability in gene sequence and gene content in viral susceptibility regions of microbial genomes. Enhanced infection can serve as a driver of fluctuating selection among these diverse subpopulations which would ultimately prevent any single subpopulation from taking over the entire population. Enhanced infection also leads to passive host-switching in viruses that provides a constant supply of alternative hosts to the virus. This is especially important for viral persistence during periods of low abundances, and thus low contact rates, with preexisting susceptible hosts and likely compensates for the inherent asymmetry in the degree of phage counter-mutations. Moreover, this passive switching is also expected to prevent arms race-like super-resistant hosts from developing. These features of the system illustrate the importance of the biotic context in which an organism lives. Simple experimental set-ups that are detached from this biotic complexity neglect this aspect of antagonistic coexistence. Therefore, efforts are needed to develop meaningful experimental systems and models that take this complexity into account. This will be no easy task.
Acknowledgments
We thank Hywel Williams, an anonymous reviewer and Lindell lab members for comments on the manuscript. The researchers were funded by European Research Council (ERC) Starting Grant 203406 (to D.L.). D.L. is a Shillman fellow.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/20031
References
- 1.Thompson JN. The Coevolutionary Process. Chicago, IL: University of Chicago Press, 1994:253-75. [Google Scholar]
- 2.Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature. 2011;474:604–8. doi: 10.1038/nature10172. [DOI] [PubMed] [Google Scholar]
- 3.Bohannan BJM, Lenski RE. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol Lett. 2000;3:362–77. doi: 10.1046/j.1461-0248.2000.00161.x. [DOI] [Google Scholar]
- 4.Buckling A, Rainey PB. Antagonistic coevolution between a bacterium and a bacteriophage. Proc Biol Sci. 2002;269:931–6. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall AR, Scanlan PD, Morgan AD, Buckling A. Host-parasite coevolutionary arms races give way to fluctuating selection. Ecol Lett. 2011;14:635–42. doi: 10.1111/j.1461-0248.2011.01624.x. [DOI] [PubMed] [Google Scholar]
- 6.Lenski RE, Levin BR. Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am Nat. 1985;125:585–602. doi: 10.1086/284364. [DOI] [Google Scholar]
- 7.Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic ecosystems. Limnol Oceanogr. 2000;45:1320–8. doi: 10.4319/lo.2000.45.6.1320. [DOI] [Google Scholar]
- 8.Waterbury JB, Valois FW. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl Environ Microbiol. 1993;59:3393–9. doi: 10.1128/aem.59.10.3393-3399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergh Ø, Børsheim KY, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–8. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 10.Proctor LM, Fuhrman JA. Viral mortality of marine bacteria and cyanobacteria. Nature. 1990;343:60–2. doi: 10.1038/343060a0. [DOI] [Google Scholar]
- 11.Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32:569–77. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- 12.Middelboe M, Hagström A, Blackburn N, Sinn B, Fischer U, Borch NH, et al. Effects of bacteriophages on the population dynamics of four strains of pelagic marine bacteria. Microb Ecol. 2001;42:395–406. doi: 10.1007/s00248-001-0012-1. [DOI] [PubMed] [Google Scholar]
- 13.Lennon JT, Martiny JBH. Rapid evolution buffers ecosystem impacts of viruses in a microbial food web. Ecol Lett. 2008;11:1178–88. doi: 10.1111/j.1461-0248.2008.01225.x. [DOI] [PubMed] [Google Scholar]
- 14.Lennon JT, Khatana SAM, Marston MF, Martiny JBH. Is there a cost of virus resistance in marine cyanobacteria? ISME J. 2007;1:300–12. doi: 10.1038/ismej.2007.37. [DOI] [PubMed] [Google Scholar]
- 15.Winter C, Bouvier T, Weinbauer MG, Thingstad TF. Trade-offs between competition and defense specialists among unicellular planktonic organisms: the “killing the winner” hypothesis revisited. Microbiol Mol Biol Rev. 2010;74:42–57. doi: 10.1128/MMBR.00034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenski RE. Experimental studies of pleiotropy and epistasis in Escherichia coli. II. Compensation for maldaptive effects associated with resistance to virus T4. Evolution. 1988;42:433–40. doi: 10.2307/2409029. [DOI] [PubMed] [Google Scholar]
- 17.Middelboe M, Holmfeldt K, Riemann L, Nybroe O, Haaber J. Bacteriophages drive strain diversification in a marine Flavobacterium: implications for phage resistance and physiological properties. Environ Microbiol. 2009;11:1971–82. doi: 10.1111/j.1462-2920.2009.01920.x. [DOI] [PubMed] [Google Scholar]
- 18.Meyer JR, Agrawal AA, Quick RT, Dobias DT, Schneider D, Lenski RE. Parallel changes in host resistance to viral infection during 45,000 generations of relaxed selection. Evolution. 2010;64:3024–34. doi: 10.1111/j.1558-5646.2010.01049.x. [DOI] [PubMed] [Google Scholar]
- 19.Bouman HA, Ulloa O, Scanlan DJ, Zwirglmaier K, Li WKW, Platt T, et al. Oceanographic basis of the global surface distribution of Prochlorococcus ecotypes. Science. 2006;312:918–21. doi: 10.1126/science.1122692. [DOI] [PubMed] [Google Scholar]
- 20.Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EM, Chisholm SW. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–40. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 21.Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 2009;3:283–95. doi: 10.1038/ismej.2008.117. [DOI] [PubMed] [Google Scholar]
- 22.Agrawal A, Lively C. Infection genetics: gene-for-gene versus matching-alleles models and all points in between. Evol Ecol Res. 2002;4:1–12. [Google Scholar]
- 23.McClelland EE, Penn DJ, Potts WK. Major histocompatibility complex heterozygote superiority during coinfection. Infect Immun. 2003;71:2079–86. doi: 10.1128/IAI.71.4.2079-2086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weitz JS, Hartman H, Levin SA. Coevolutionary arms races between bacteria and bacteriophage. Proc Natl Acad Sci U S A. 2005;102:9535–40. doi: 10.1073/pnas.0504062102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF, et al. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–70. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasić L, Thingstad TF, Rohwer F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–36. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 27.Scanlan PD, Hall AR, Lopez-Pascua LD, Buckling A. Genetic basis of infectivity evolution in a bacteriophage. Mol Ecol. 2011;20:981–9. doi: 10.1111/j.1365-294X.2010.04903.x. [DOI] [PubMed] [Google Scholar]
- 28.Holmfeldt K, Middelboe M, Nybroe O, Riemann L. Large variabilities in host strain susceptibility and phage host range govern interactions between lytic marine phages and their Flavobacterium hosts. Appl Environ Microbiol. 2007;73:6730–9. doi: 10.1128/AEM.01399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paterson S, Vogwill T, Buckling A, Benmayor R, Spiers AJ, Thomson NR, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464:275–8. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abe M, Izumoji Y, Tanji Y. Phenotypic transformation including host-range transition through superinfection of T-even phages. FEMS Microbiol Lett. 2007;269:145–52. doi: 10.1111/j.1574-6968.2006.00615.x. [DOI] [PubMed] [Google Scholar]