Abstract
Mankind is confronted by the outbreaks of highly virulent and multi-drug resistant pathogens. The outbreak strains often belong to well-known diseases associated species such as Salmonella, Klebsiella and Mycobacterium, but even normally commensal and environmental microorganisms may suddenly acquire properties of virulent bacteria and cause nosocomial infections. The acquired virulence is often associated with lateral exchange of pathogenicity genomic islands containing drug and heavy metal resistance determinants. Metal ions are used by the immune system of macro-organisms against bactericidal agents. The ability to control heavy metal homeostasis is a factor that allows the survival of pathogenic microorganisms in macrophages. In this paper, we investigate the origin of heavy metal resistance operons in the recent outbreak strains and the possible routes which may lead to acquisitions of these genes by potentially new pathogens. We hypothesize that new outbreak microorganisms appear intermittently on an intersection of the non-specialized, genetically naïve strains of potential pathogens and virulence factor comprising vectors (plasmid and/or phages) newly generated in the environmental microflora. Global contamination of the environment and climate change may also have an effect toward the acceleration and appearance of new pathogens.
Keywords: heavy metal resistance, horizontal gene transfer, outbreak, virulence
Despite the availability of complete genome sequences of multiple pathogens and the profound knowledge we encompass on both epidemiological and molecular aspects of bacterial pathogenicity, the sudden outbreaks that arise from new highly virulent pathogens do not cease to take mankind by surprise. The last striking outbreak was caused by an enterohemorrhagic Escherichia coli in Germany in 2011, where hundreds of people were affected. The first outbreak strain TY2482 was isolated at Hamburg-Eppendorf University Medical Center on May 25th and sequenced at the Beijing Genomics Institute in Shenzhen on 2nd of June.1 Isolation and sequencing of the other outbreak associated strains followed and a number of genome comparative studies were published2-4 which served as a demonstration of an impressive increase in the level of sequencing technologies and their relevance to clinical studies. The studies showed a connection between the new outbreak E. coli and an earlier outbreak strain isolated in Africa ten years ago. However, the origin and mechanisms of emergence of the outbreak variants of the usually harmless E. coli remained generally unclear5-7 even upon the immense analysis.
A number of genomic islands (GIs) comprising of possible virulence determinants were identified in TY2482, and at least one of these possessed a plasmid associated mercury resistance operon.8 Heavy metal resistance genes are often found in pathogenicity genomic islands (PAIs) of different highly virulent microorganisms.9-14 These are utilized by virulence microorganisms for heavy metal sensing and removal which aid against the exploitation of a transition metal such as copper by mammalian immune defenses.15 Copper is known to be an essential toxicity component used by macrophages for killing pathogens within phagosomes.15-17 However, the most frequent heavy metal resistance genes reported to be associated with virulence GIs and plasmids are the ones that make up mercury resistance operons. Schottel et al. reported that in a collection of some 800 antibiotic-resistance plasmids isolated from clinical E. coli, 25% carried mercury resistance determinants.18 The latter serves as an indication that these determinants are frequent in plasmids and may also be associated with microbes which confer resistance to antibiotics. In fact, the mer operons are highly versatile19 and it has recently been demonstrated experimentally that the sensitivity of mercury-sensing regulators may be re-directed by mutagenesis to sense other heavy metal pollutants.20 The role of the mer operons in pathogenicity remains unclear, but their prevalence in PAIs suggests that they may be important factors which pathogens may use for alternative functions such as transport and detoxification of antibiotics and other detrimental compounds. Furthermore, the roles of mercury resistance genes in bacterial resistance toward clinical disinfectants have also been reported.21 In relation to the latter, an acquired antiseptic and disinfectant resistance of Acinetobacter baumannii was associated with the arsenic and mercury resistance operons14 which could possibly be of plasmid origin.
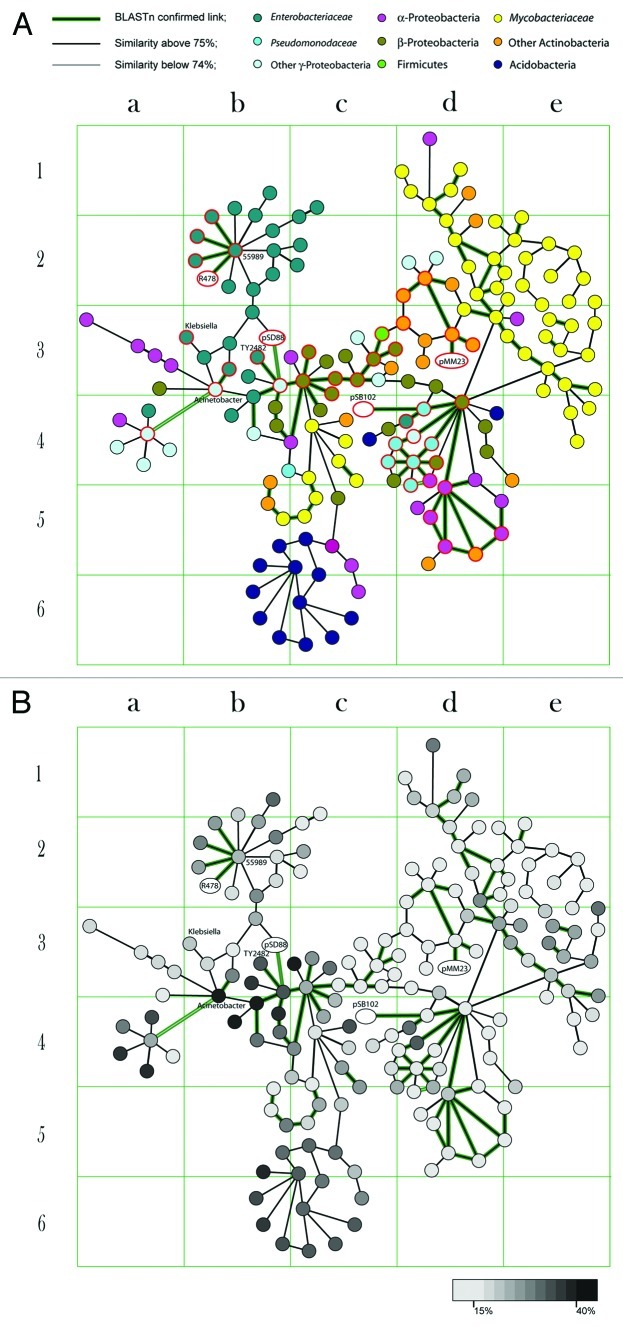
In this work we traced down the distribution of heavy metal resistance operons associated with mobile genomic regions. Initially, a total of 17,984 GIs were identified by SeqWord Sniffer program in 1,639 bacterial chromosomes and plasmids.8 The GIs that showed DNA sequence similarity with the virulence plasmids pSB102, R478 and pSD_889,10,22 comprising of multiple heavy metal resistance operons and also shared compositional similarity with GIs of enterohemorragic E. coli, were selected for a detailed analysis. From the analysis it was found that GIs sharing both sequence and compositional similarity were distributed among distant bacteria of different taxonomic orders (Fig. 1A). The latter finding implicates that these GIs are possibly associated with a wide range of plasmids. Plasmids may exchange transposable elements with the chromosomes or sometimes the whole plasmid may be integrated into a bacterial genome. Having been fixed on the chromosome, the mobile element undergoes amelioration, a process that smoothes out differences in oligonucleotide usage patterns of the host chromosome and that of the acquired element.23 Figure 1B shows the results of a stratigraphic analysis of GIs, represented as linked nodes. The stratigraphic method calculates the distances in OU patterns between the GIs and host chromosomes and determines their relative time of acquisition. In Figure 1B the nodes which are depicted by a lighter color show higher levels of compositional similarity to the host chromosomes than those depicted by a darker color, which probably still resemble the composition of their donor genomes. The colors of nodes are linked with the acquisition periods. Recent acquisitions are distant from the hosts in terms of composition, they therefore have a darker color. A color in this case means that these GIs have not lost their specific original composition yet. Overtime these GIs get affected by the mutational pressures of their new hosts, they thus start to ameliorate and resemble the patterns of the host organisms. The lighter GIs in Figure 1B are ancient acquisitions, which have been in the host chromosomes for longer, hence the resemblance of the patterns. As illustrated, the heavy metal resistance GI of the strain E. coli 55989 depicted as the central node in the cell b2 (Fig. 1B), appears to be an older insert as compared with the mercury resistance operon of E. coli TY2482 (Fig. 1B, cell b3). The former GI shares strong sequence and compositional similarity with the plasmid R478 isolated from Serratia marcescens in 1969,10 while the latter is similar to the fragment of the plasmid pSD_88 isolated from Salmonella enterica in 2011.22 E. coli TY2482 caused the outbreak in Germany in 2011 while E. coli 55989 appeared as the causative agent of a similar outbreak in Central African Republic in the late 1990s. Comparison of their genomes showed 99.8% identity and it was logical to conclude that TY2482 and 55989 shared a common ancestor.24 In contrast to commensal E. coli and pathogenic Shigella which share a variety of newly acquired virulence associated GIs, the genomes of TY2482 and 55989 comprise mostly of old uniform GIs which they inherited from their common ancestor except for those comprising mercury resistance operons. Their ancestor is believed to have been a genetically naïve organism which managed to avoid the major fluxes of gene exchange between enterobacteria for a very long time.8 The virulent TY2482 which is thought to have evolved from this hypothetical ancestor acquired pathogenicity determinants such as Shiga-toxin encoding prophage; multi-drug and heavy metal resistance operons from both pathogenic enterobacteria24 and environmental bacteria.8 Figure 1 shows that similar and much older mercury resistance GIs are harboured by β-Proteobacteria of Acidovorax and Alicycliphilus genera, which very likely were the initial hosts of these mobile genetic elements as far as we could see. These mobilomes were at later stages disseminated to other distant microorganisms such as mycobacteria through Frankia/Nocardia (cells d1-e4 in Figure 1) or directly from β-Proteobacteria (c4-c5). These elements also dispersed to Pseudomonas (d4), thereafter to the soil α-Protobacteria of genera Ochrobacterium, Methylobacterium, Arthrobacter, Xanthobacter, Oligotropha and Acidiphilum (d5); and also Acidobacteria (b5-c6). It is an intriguing question whether the distribution of these GIs played a role in the evolution of pathogenic Mycobacterium or in the pathogenicity development of opportunistic nosocomial pathogens of Ochrobacterium and Arthrobacter. The most recent insertions of these GIs were found in virulent E. coli TY2482, 55989 and UMNO26; Salmonella enterica Typhi, Dublin and Newport; Acinetobacter baumannii AYE; in a clinical isolate Pseudomonas aeruginosa UCBPP-PA14 and a fish pathogen Aeromonas salmonicida A449. It is remarkable that the metal resistance operons are more frequently identified in recently acquired GIs of this group (Fig. 1). This observation is in consistence with the discovery by Ramaiah and De that in a period of 5 y (1997 to 2003) the number of microorganisms isolated from sea water which were tolerant to 10 ppm of HgCl2 increased from none to 75% or even 95% of CFU in the polluted zones of Mumbai.25 The general environmental pollution may be behind the activation of these dormant GIs and making the genes which increase tolerance to heavy metals and other pollutants to be on demand by a great range of environmental bacteria. Having reached potentially pathogenic bacteria, these genes get adjusted toward resistance against drugs and antibacterial immune responses.18,26
Figure 1. The figure illustrates the grouping of GIs which share similarities in both composition and sequence. Each node corresponds to one GI. Nodes sharing compositional similarity of 75% and above are linked by edges. As it was shown previously, this level of compositional similarity implies common ancestry between mobile genomic elements.8 Plasmid genomes are depicted by ovals. More details about these GIs are available on an interactive map at www.bi.up.ac.za/SeqWord/maps/map.html. (A) Nodes are colored according to the taxonomic belonging of the host microorganisms. (B) Grey gradient colors depict divergence of GI pattern from the tetranucleotide usage pattern of the host chromosome.
The map of relations between GIs in Figure 1 illustrates possible paths for the distribution of PAIs supported by the compositional similarity between GIs and sharing of similar DNA sequences identified by blastn. It was reported that when compositional similarity reaches 75% and above the likelihood to find homologous DNA sequences in a pair of GIs rapidly increases. The latter statement is in support of the common ancestry hypothesis.8 However, it has to be noted that GIs which show strong compositional similarity may possess different fragments, only few of these contain matching sequences.
The figure suggests that Klebsiella (cell b3) could possibly acquire GIs from Salmonella and Acinetobacter. It could probably be that the latter had truly happened, as an outbreak of multidrug resistant K. pneumonia was reported in Rotterdam hospital and several other places27 in 2011 and it followed the spread of drug-resistant A. baumannii.14 The analysis of previous fluxes of GIs also implicates a possible transfer of new virulence genes to Brucella (Fig. 1, cell a4), Mycobacterium and Nocardia (cells d2–3), which may pose a serious impact on human health. There are many GIs in Mycobacterium, which most likely originated from β-Proteobacteria as discussed above (Fig. 1). All these are quite old and void of heavy metal resistance operons, but this does not rule out the possibility for new PAIs to be acquired by mycobacteria in future. An unexpected high frequency of mercury-resistant strains showing also an increased tolerance to gentamicin, streptomycin and D-cycloserine has been reported among the clinical nontuberculous mycobacteria isolates of the species Mycobacterium avium, M. intracellulare, and M. scrofulaceum.28 The genome of a fish pathogen Mycobacterium marinum, which sometimes causes opportunistic infections in humans, comprises a 23-kb mercury-resistance plasmid pMM23.29 Blastn and oligonucleotide composition analysis showed that this plasmid together with the mercury resistance operon which it possesses originate from either Nocardia or Pesudonocardia (Fig. 1). It is very likely that this plasmid has been acquired by M. marinum quite recently as it shows very strong sequence and pattern similarities to Nocardia. These newly acquired genes may be behind the reported increased drug resistance of M. marinum isolates. A mercury resistance plasmid similar to that of M. marinum together with multiple GIs of Pseudomonas and Actinobacteria origin were identified in Mycobacterium abscessus, a pseudotuberculous lung disease causing microbe.30
Drug and heavy metal resistance genes are abundant in the bacterial world. It is likely that these genes started being abundant in microorganisms soon after life began, when the world was heavily polluted with volcanic activity.19 A recent study has revealed that at least 30,000 y ago the major antibiotic resistance genes were already present, as these were identified in a permafrost sediment DNA sample.31 In spite of the abundance of potential virulence factors in nature, there is a clear periodicity in the occurrence of outbreaks. In our previous publication8 we hypothesized that this periodicity may reflect the oscillation of genetic vector activity. Bacteria develop resistance against foreign invasions initiated by existing vectors (plasmids and phages) and the efficiency of horizontal gene exchange gradually decreases. Thereafter, new vectors evolve by undergoing several mutations which allow them to evade host resistance mechanisms32 and get incorporated into host chromosomes. These bring new virulence genes or simply activate the gene exchange between potentially pathogenic microorganisms that eventually lead to the achievement of effective combination of pathogenicity determinants in new virulence GIs.8 Mutations which arise from these vectors in order to gain access to host organisms allow the interchange of various genomic fragments between bacteria. The interchanges may include fragments from both plasmids and phages. The latter could be the result of hybrid genomic islands which have been reported to possess elements of both: temperate phages and conjugative plasmids.33,34
There is a general belief that the infection outbreaks may possibly be caused by former commensal strains after their acquisitions of PAIs from pathogenic bacteria or by the change of a host niche.35 Analysis of the distribution of GIs showed that commensal E. coli and pathogenic Shigella share mobile genetic elements,8 but this did not turn either commensal E. coli to pathogenicity or pathogenic Shigella to commensalism. These microorganisms are highly specialized and saturated with horizontally acquired elements. One more acquisition will not be sufficient to change their lifestyles. Acquisition of a GI may have more profound effects on genetically naïve microorganisms which were initially not exposed to the horizontal exchange events due to geographic isolation. This was the case with the latest outbreak caused by a naïve E. coli TY2482 after it acquired a number of GIs from a general pool of PAIs of enterobacteria, and from environmental micro-flora. Climate change and general pollution may mix bacterial populations and activate previously dormant vectors for which all current pathogens will be “genetically naïve,” i.e., not exposed to these types of genetic vectors and thus become more vulnerable. Again this probably was the case with E. coli TY2482, after it acquired mercury resistance operons from microorganisms which are not known to be the usual sources of PAIs for enterobacteria.8 Our further work will be aimed at monitoring unusual recent GI inserts in potentially pathogenic microorganism and tracking down organisms which are likely to be probable donors and recipients of such entities. Potentially, this monitoring system will allow the likelihood estimate of new deadly outbreaks in future.
Acknowledgments
This work was funded by the National Research Foundation of South Africa grant NRF 71261.
Glossary
Abbreviations:
- GI
genomic island
- PAI
pathogenicity island
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/19963
References
- 1.Kupferschmidt K. Germany. Scientists rush to study genome of lethal E. coli. Science. 2011;332:1249–50. doi: 10.1126/science.332.6035.1249. [DOI] [PubMed] [Google Scholar]
- 2.Brzuszkiewicz E, Thürmer A, Schuldes J, Leimbach A, Liesegang H, Meyer FD, et al. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC) Arch Microbiol. 2011;193:883–91. doi: 10.1007/s00203-011-0725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manrique M, Pareja-Tobes P, Pareja-Tobes E, Pareja E, Tobes R. Escherichia coli EHEC Germany outbreak preliminary functional annotation using BG7 system. Nature Precedings. 2011 doi: 10.1038/npre.2011.6001.1. [DOI] [Google Scholar]
- 4.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One. 2011;6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709–17. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Struelens MJ, Palm D, Takkinen J. Enteroaggregative, Shiga toxin-producing Escherichia coli O104:H4 outbreak: new microbiological findings boost coordinated investigations by European public health laboratories. Euro Surveill. 2011;16:2–4. doi: 10.2807/ese.16.24.19890-en. [DOI] [PubMed] [Google Scholar]
- 7.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bezuidt O, Pierneef R, Mncube K, Lima-Mendez G, Reva ON. Mainstreams of horizontal gene exchange in enterobacteria: consideration of the outbreak of enterohemorrhagic E. coli O104:H4 in Germany in 2011. PLoS One. 2011;6:e25702. doi: 10.1371/journal.pone.0025702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneiker S, Keller M, Dröge M, Lanka E, Pühler A, Selbitschka W. The genetic organization and evolution of the broad host range mercury resistance plasmid pSB102 isolated from a microbial population residing in the rhizosphere of alfalfa. Nucleic Acids Res. 2001;29:5169–81. doi: 10.1093/nar/29.24.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilmour MW, Thomson NR, Sanders M, Parkhill J, Taylor DE. The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid. 2004;52:182–202. doi: 10.1016/j.plasmid.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Kamachi K, Sota M, Tamai Y, Nagata N, Konda T, Inoue T, et al. Plasmid pBP136 from Bordetella pertussis represents an ancestral form of IncP-1β plasmids without accessory mobile elements. Microbiology. 2006;152:3477–84. doi: 10.1099/mic.0.29056-0. [DOI] [PubMed] [Google Scholar]
- 12.Levings RS, Partridge SR, Djordjevic SP, Hall RM. SGI1-K, a variant of the SGI1 genomic island carrying a mercury resistance region, in Salmonella enterica serovar Kentucky. Antimicrob Agents Chemother. 2007;51:317–23. doi: 10.1128/AAC.01229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reith ME, Singh RK, Curtis B, Boyd JM, Bouevitch A, Kimball J, et al. The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics. 2008;9:427. doi: 10.1186/1471-2164-9-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durante-Mangoni E, Zarrilli R. Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol. 2011;6:407–22. doi: 10.2217/fmb.11.23. [DOI] [PubMed] [Google Scholar]
- 15.Osman D, Cavet JS. Metal sensing in Salmonella: implications for pathogenesis. Adv Microb Physiol. 2011;58:175–232. doi: 10.1016/B978-0-12-381043-4.00005-2. [DOI] [PubMed] [Google Scholar]
- 16.Percival SS. Copper and immunity. Am J Clin Nutr. 1998;67(Suppl):1064S–8S. doi: 10.1093/ajcn/67.5.1064S. [DOI] [PubMed] [Google Scholar]
- 17.Gold B, Deng H, Bryk R, Vargas D, Eliezer D, Roberts J, et al. Identification of a copper-binding metallothionein in pathogenic mycobacteria. Nat Chem Biol. 2008;4:609–16. doi: 10.1038/nchembio.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schottel J, Mandal A, Clark D, Silver S, Hedges RW. Volatilisation of mercury and organomercurials determined by inducible R-factor systems in enteric bacteria. Nature. 1974;251:335–7. doi: 10.1038/251335a0. [DOI] [PubMed] [Google Scholar]
- 19.Silver S, Phung LT. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50:753–89. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 20.Hakkila KM, Nikander PA, Junttila SM, Lamminmäki UJ, Virta MP. Cd-specific mutants of mercury-sensing regulatory protein MerR, generated by directed evolution. Appl Environ Microbiol. 2011;77:6215–24. doi: 10.1128/AEM.00662-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell AD. Bacterial resistance to disinfectants: present knowledge and future problems. J Hosp Infect. 1999;43(Suppl):S57–68. doi: 10.1016/S0195-6701(99)90066-X. [DOI] [PubMed] [Google Scholar]
- 22.Han J, Lynne AM, David DE, Nayak R, Foley SL. Sequencing of plasmids from a multi-antimicrobial resistant Salmonella enterica serovar Dublin strain. Food Res Int. 2012;45:931–4. doi: 10.1016/j.foodres.2011.04.016. [DOI] [Google Scholar]
- 23.Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–97. doi: 10.1007/PL00006158. [DOI] [PubMed] [Google Scholar]
- 24.Rohde H, Qin J, Cui Y, Li D, Loman NJ, Hentschke M, et al. E. coli O104:H4 Genome Analysis Crowd-Sourcing Consortium Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med. 2011;365:718–24. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- 25.Ramaiah N, De J. Unusual rise in mercury-resistant bacteria in coastal environs. Microb Ecol. 2003;45:444–54. doi: 10.1007/s00248-001-1068-7. [DOI] [PubMed] [Google Scholar]
- 26.Calomiris JJ, Armstrong JL, Seidler RJ. Association of metal tolerance with multiple antibiotic resistance of bacteria isolated from drinking water. Appl Environ Microbiol. 1984;47:1238–42. doi: 10.1128/aem.47.6.1238-1242.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potron A, Kalpoe J, Poirel L, Nordmann P. European dissemination of a single OXA-48-producing Klebsiella pneumoniae clone. Clin Microbiol Infect. 2011;17:E24–6. doi: 10.1111/j.1469-0691.2011.03669.x. [DOI] [PubMed] [Google Scholar]
- 28.Fry KL, Meissner PS, Falkinham JO., 3rd Epidemiology of infection by nontuberculous mycobacteria. VI. Identification and use of epidemiologic markers for studies of Mycobacterium avium, M. intracellulare, and M. scrofulaceum. Am Rev Respir Dis. 1986;134:39–43. doi: 10.1164/arrd.1986.134.1.39. [DOI] [PubMed] [Google Scholar]
- 29.Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008;18:729–41. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, et al. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One. 2009;4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–61. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 32.Heidelberg JF, Nelson WC, Schoenfeld T, Bhaya D. Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes. PLoS One. 2009;4:e4169. doi: 10.1371/journal.pone.0004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GS, Mavrodi DV, et al. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol. 2005;23:873–8. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson RW, Vinatzer B, Arnold DL, Dorus S, Murillo J. The influence of the accessory genome on bacterial pathogen evolution. Mob Genet Elements. 2011;1:55–65. doi: 10.4161/mge.1.1.16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hacker J, Hentschel U, Dobrindt U. Prokaryotic chromosomes and disease. Science. 2003;301:790–3. doi: 10.1126/science.1086802. [DOI] [PubMed] [Google Scholar]