Abstract
One of the clinical features of cystic fibrosis (CF) is a deep inflammatory process, which is characterized by production and release of cytokines and chemokines, among which interleukin 8 (IL-8) represents one of the most important. Accordingly, there is a growing interest in developing therapies against CF to reduce the excessive inflammatory response in the airways of CF patients. Since transcription factor NF-kappaB plays a critical role in IL-8 expression, the transcription factor decoy (TFD) strategy might be of interest. In order to demonstrate that TFD against NF-kappaB interferes with the NF-kappaB pathway we proved, by chromatin immunoprecipitation (ChIP) that treatment with TFD oligodeoxyribonucleotides of cystic fibrosis IB3–1 cells infected with Pseudomonas aeruginosa leads to a decrease occupancy of the Il-8 gene promoter by NF-kappaB factors. In order to develop more stable therapeutic molecules, peptide nucleic acids (PNAs) based agents were considered. In this respect PNA-DNA-PNA (PDP) chimeras are molecules of great interest from several points of view: (1) they can be complexed with liposomes and microspheres; (2) they are resistant to DNases, serum and cytoplasmic extracts; (3) they are potent decoy molecules. By using electrophoretic mobility shift assay and RT-PCR analysis we have demonstrated that (1) the effects of PDP/PDP NF-kappaB decoy chimera on accumulation of pro-inflammatory mRNAs in P.aeruginosa infected IB3–1 cells reproduce that of decoy oligonucleotides; in particular (2) the PDP/PDP chimera is a strong inhibitor of IL-8 gene expression; (3) the effect of PDP/PDP chimeras, unlike those of ODN-based decoys, are observed even in the absence of protection with lipofectamine. These informations are of great impact, in our opinion, for the development of stable molecules to be used in non-viral gene therapy of cystic fibrosis.
Keywords: NF-kappaB, transcription factor decoy, inflammation, Peptide Nucleic Acids, PNA-DNA chimeras
Introduction
Cystic fibrosis (CF) is a severe inherited disease caused by mutations of a gene encoding a chloride channel termed Cystic fibrosis transmembrane conductance regulator (CFTR).1 Although most of the CF patients are affected by multiple organ pathologies, lung disease is the major cause of morbidity and mortality in CF. In the lung of CF patients a hyper-inflammatory condition is established, which is characterized by predominant infiltrates of polymorphonuclear neutrophils (PMNs) in bronchial lumina and increased expression of pro-inflammatory cytokines and chemokines, in particular Interleukin-8 (IL-8).2-8 Recently, it has been established that excessive IL-8 release from lung epithelial cells may also play a role in bacterial proliferation and adhesion.9 Accordingly, IL-8 is presently considered a critical pharmacological target to reduce the excessive inflammation in CF lungs.10 We recently described an analysis of the transcription machinery of the IL-8 gene in human bronchial epithelial cells, and found NF-kappaB pathway very important.11,12 These results are in line with several reports indicating lung inflammation in CF related to an increased NF-kappaB signaling causing the induction of IL-8 gene expression.13
One of the possible strategies to inhibit IL-8 gene transcription is the so called “transcription factor decoy” (TFD) approach,14-17 based on oligodeoxynucleotides (ODNs) mimicking consensus sequences identified within the proximal promoter region of the IL-8 gene. In order to verify whether the TFD against NF-kappaB interferes with the NF-kappaB pathway we employed in this study chromatin immunoprecipitation (ChIP) on cystic fibrosis IB3–1 cells infected with Pseudomonas aeruginosa and treated with TFD ODNs against NF-kappaB factors.
In consideration of the fact that an important drawback of the decoy approach designed for the modulation of gene expression is the presence of intracellular and extracellular DNases,18 therefore, large amounts of DNA must be loaded into target cells in order to obtain biological responses leading to alteration of gene expression. On the other hand, modified oligonucleotides (either methylphosphonate or phosphorothioate) have the advantage in that they are resistant to DNase cleavage; unfortunately, these molecules are highly toxic.18
In this respect, in recent reports we presented the possible use of peptide nucleic acids (PNAs)19-21 as alternative reagents in experiments aimed at the control of gene expression involving the TFD approach. In PNAs, the pseudopeptide backbone is composed of N-(2-aminoethyl)glycine units.19 PNAs hybridize with high affinity to complementary sequences of single-stranded RNA and DNA, forming Watson-Crick double helices21 and are resistant to both nucleases and proteases.22 PNAs were found to be excellent candidates for antisense and antigéne therapies.23-26 Among PNA-based molecules, we found that PNA-DNA chimeras are of great potential in gene therapy being active as decoy molecules against the NF-kappaB and Sp1 transcription factors.23,27-29
Following these considerations, we determined the effect of PNA-DNA-PNA (PDP) chimeras mimicking the binding sites of NF-kappaB on the transcription of IL-8 gene in cystic fibrosis IB3–1 cells infected by Pseudomonas aeruginosa. We have described this system in several reports and reviews, pointing out that after Pseudomonas aeruginosa exposure, IB3–1 are induced to increase accumulation of IL-8 mRNA in respect to basal levels of uninfected cells. In addition to IL-8 mRNA, other sequences induced by PAO1 are GRO-γ, GRO-α, IL-6, IL-1β, ICAM-1.11,12 Therefore, IB3–1 cells infected by P.aeruginosa are an excellent system to verify (1) whether decoy molecules against NF-kappaB inhibit IL-8 gene transcription and (2) whether this effect is restricted to the IL-8 gene, or affects other genes containing NF-kappaB binding sites, such as GRO-γ, IL-6, IL-1β and ICAM-1.
Results
The experimental system and preliminary assays on the specificity of the decoy molecules
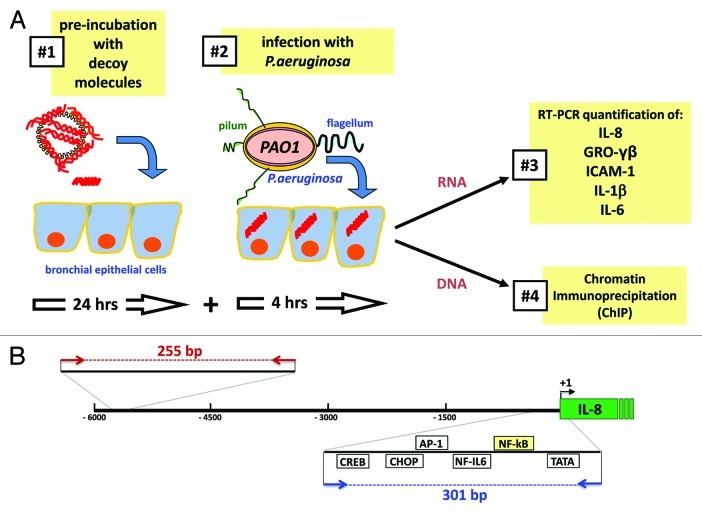
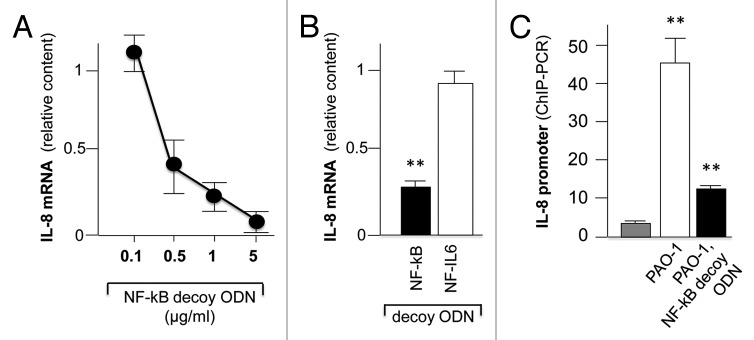
Figure 1 shows the experimental strategy followed in our study. Complexes of cationic liposomes with NF-kappaB ODNs or PDP/PDP chimeras have been pre-incubated with cystic fibrosis IB3–1 cells 24 h before exposure to the PAO1 laboratory strain of P.aeruginosa (100 CFU/cell) for a further four hour time period. A scrambled ODN was used as negative control (Table 1). After the treatment RNA was extracted and real-time quantitative RT-PCR performed; at the same time chromatin was purified from IB3–1 cells for chromatin immunoprecipitation assay (ChIP). In the first set of experiments, described in Figure 2, we provided evidences that the TF decoy approach interfere(s) with the NF-kappaB activity in vitro and in the IB3–1 cellular system. In order to verify the ability of this decoy ODNs to compete for the binding of NF-kappaB to the sequences contained in the promoter of IL-8 gene, the NF-kappaB decoy ODNs was incubated with nuclear extracts from IB3–1 cells in the presence of a radiolabeled probe 100% homologous with the NF-kappaB binding sequences and performed electrophoretic mobility shift assays (EMSA). Complete inhibition of interaction of the 32P-labeled probes with specific transcription factor proteins (NF-kappaB/DNA complexes) has been obtained, providing the proof of principle of the competition of this NF-kappaB decoy ODNs for the DNA consensus sequence contained in the promoter of the IL-8 gene. On the contrary, other ODN containing the binding sites for other transcription factors are inactive. ODN for CREB, NF-IL6, AP-1 and CHOP were selected because these transcription factors play a crucial role in the control of transcription of the IL-8 gene.11
Figure 1. (A) Experimental design followed for the analysis of the effects of the transcription factor decoy (TFD) strategy employing TF decoy ODNs against NF-kappaB and human bronchial epithelial cells. (#1) Human bronchial CF-derived respiratory epithelial IB3–1 cells were pre-incubated for 24 h with NF-kappaB ODNs or PDP/PDP chimeras before infection with the laboratory strain of P.aeruginosa PAO1 (#2). After 4 h post-infection, total RNA was extracted, reverse-transcribed to cDNA and analyzed by RT-qPCR (#3). In parallel, chromatin was purified by the IB3–1 cells for chromatin immunoprecipitation assay (ChIP) (#4). In (B) representation of the genomic region located:6 kb upstream of IL-8 gene. The location of primers used for IL-8 promoter amplification in the ChIP assay and the respective product length are indicated: PCR product obtained from IL-8 promoter amplification, containing NF-κappaB binding site (301bp, in blue); PCR product obtained using control primers flanking a genomic region:5 kb upstream of IL-8 promoter (255 bp, in red).
Table 1. Sequence of synthetic oligonucleotides used in this study.
| Double-stranded oligonucleotides used in gel shift assays and decoy transfectionsa | |||
|---|---|---|---|
| |
|
|
|
| NF-kappaB |
|
5'-AGAGGAATTTCCACGATT-3' |
|
| CREB |
|
5'-AAAACTTTCGTCATACTC-3' |
|
| NF-IL6 |
|
5'-CATCAGTTGCAAATCGTGG-3' |
|
| AP-1 |
|
5'-TGTGATGACTCAGGTTTG-3' |
|
| CHOP |
|
5'-CGCTGGTGTGATGCACGG-3' |
|
| SCRAMBLED |
|
5'-CACAAAGTGTAACAGTCT-3' |
|
| ChIP Q-PCR Primers | Amplified region | Sequences | |
|---|---|---|---|
| |
|
|
|
| IL-8 ChIP f |
IL-8 promoter region |
5'-TCACCAAATTGTGGAGCTTCAGTAT-3' |
|
| IL-8 ChIP r |
5'-GGCTCTTGTCCTAGAAGCTTGTGT-3' |
|
|
| |
|
|
|
| Neg ChIP f |
Negative control regionb |
5'-TCCCTAAGTCACTTTCTTCAAGTTGC-3' |
|
| Neg ChIP r |
5'-CGTGCATTTAATTGTGTCTTGTGG-3' |
|
|
| RT Q-PCR Primers | Amplified region | Sequences | Accession Number |
|---|---|---|---|
| |
|
|
|
| IL-8 f |
IL-8 transcripts |
5'-GACCACACTGCGCCAACA-3' |
AF385628.2 |
| IL-8 r |
5'-GCTCTCTTCCATCAGAAAGTTACATAATTT-3' |
||
| |
|
|
|
| ICAM-1 f |
ICAM-1 transcripts |
5'-TATGGCAACGACTCCTTCTCG-3' |
NM_000201 |
| ICAM-1 r |
5'-CTCTGCGGTCACACTGACTGA-3' |
||
| |
|
|
|
| GRO-γβ f |
GRO-γβ transcripts |
5'-CCGGACCCCACTGCG-3' |
M36821 |
| GRO-γβ r |
5'-TTCCCATTCTTGAGTGTGGCTA-3' |
||
| |
|
|
|
| IL-1-β f |
IL-1-β transcripts |
5'-CTCCACCTCCAGGGACAGGA-3' |
BT007213.1 |
| IL-1-β r |
5'-GGACATGGAGAACACCACTTGTT-3' |
||
| |
|
|
|
| IL-6 f |
IL-6 transcripts |
5'-CGGTACATCCTCGACGGC-3' |
NM_000600 |
| IL-6 r |
5'-CTTGTTACATGTCTCCTTTCTCAGG-3' |
||
| |
|
|
|
| GAPDH f |
GAPDH mRNA | 5'-GTGGAGTCCACTGGCGTCTT-3' |
NM_001404.3 |
| GAPDH r | 5'-GCAAATGAGCCCAGCCTTC-3' |
a Sequence of decoy ODNs based on IL-8 promoter regulatory elements; bRegion about 5 kb upstream of the IL-8 promoter, lacking NF-kappaB binding sites.
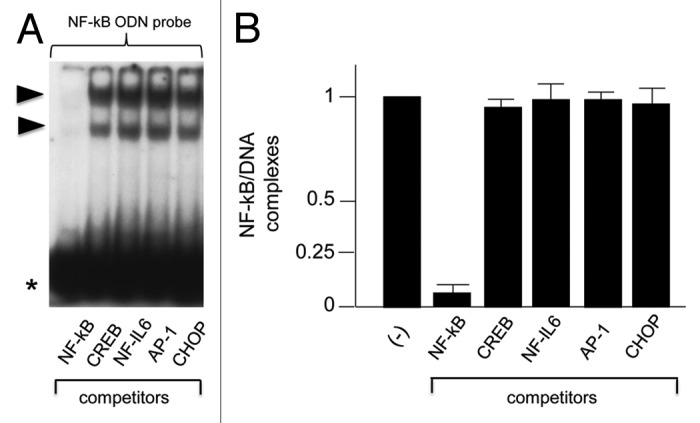
Figure 2. (A) Representative autoradiogram showing the effects of cold ODN competitors specific for NF-kappaB, CREB, NF-IL6, AP-1, and CHOP transcription factors on molecular interactions between nuclear factors from IB3–1 cells and 32P-labeled NF-kappaB ODN (►, NF-kappaB/DNA complexes; *, free probe). (B) Effects of the indicated ODN competitors on the NF-kappaB/DNA complexes. Data (average ± SD of three independent EMSA experiments) represent the amount of the complexes in respect to that found in control EMSA reaction conducted in the absence of any competitors (-).
Effects of decoy ODNs targeting NF-kappaB on gene expression of Pseudomonas aeruginosa infected IB3–1 cells
The results reported in Figure 3A show that treatment with 0.5 μg/ml of ODN NF-kappaB decoy is sufficient to obtain decreased expression of the IL-8 gene. However also the other signals from transcription factors present within the IL-8 gene promoter (Fig. 1) might play an important transcriptional role; this is supported by the observation that decoys for CREB, AP-1 and CHOP-1 also induce to some extent inhibition of IL-8 mRNA content in parallel experiments (data not shown). In any case, some decoy oligonucleotides are not active in inhibiting P. aeruginosa induced IL-8 gene increased expression, as outlined in Figure 3B, showing that treatment of PAO1 infected IB3–1 CF cells with NF-IL6 decoy does not lead to significant reduction of IL-8 mRNA. In order to clarify the effects of decoy molecules on the NF-kappaB intracellular dynamics, we studied by chromatin immunoprecipitation (ChIP) the possible effects of the treatment with decoy ODNs on the recruitment of NF-kappaB to the IL-8 gene promoters. Chromatin was isolated from uninfected IB3–1 cells, from cells infected with PAO1, and from cells infected with PAO1 and treated with 2 μg/ml of ODN NF-kappaB decoy. After immunoprecipitation (Fig. 1B), amplification of the IL-8 gene promoter segment containing the NF-kappaB binding sites was performed; as a negative control a parallel PCR was also performed using primers amplifying an upstream sequence of the IL-8 gene (located at - 5 Kb from the transcription start site) lacking NF-kappaB consensus elements. Figure 3B shows one important feature of our experimental cell systems, i.e., that NF-kappaB is recruited with high efficiency to the IL-8 gene promoter following infection with PAO1; second, the data clearly indicate that the NF-kappaB decoy ODN interferes with this recruitment. No changes in occupancy by NF-kappaB were detected when ChIP-DNA was amplified using PCR primers specific for the upstream sequence of the IL-8 gene lacking NF-kappaB consensus elements.
Figure 3. (A) Effect of NF-kappaB decoy ODNs on P.aeruginosa-dependent induction of IL-8 mRNA in IB3–1 cells. Cells were pre-incubated for 24 h with the indicated concentration of NF-kappaB decoy ODN, the same amounts of scrambled ODNs or medium alone before infection with P.aeruginosa, PAO1 strain. Total RNA was extracted 4 h post-infection and IL-8 mRNA was quantified as described in the Methods section. Data are mean ± SEM. Representative of 3 separate experiments. (B) Effects of 1 μg/ml of NF-kappaB and NF-IL6 decoy on IL-8 mRNA production in IB3–1 CF cells infected PAO-1. C. Chromatin Immunoprecipitation (ChIP) assay showing the effects of NF-kappaB ODN decoy on the recruitment of NF-kappaB transcription factor on the promoter of the IL-8 gene. Values represent the ratios between the IL-8 promoter Q-PCR performed on NF-kappaB specific ChIP and negative IgG ChIP. Each value was derived employing chromatin isolated from untreated IB3–1 cells (gray bar) or cells infected with PAO-1 in the absence (white bar) or in the presence (black bar) of pre-treatment with the NF-kappaB decoy ODN. Significance in Student’s t test between PAO-1 infected and uninfected cells and between PAO-1 infected, NF-kB decoy treated, and PAO-1 infected cells: ** p < 0.01.
The NF-kappB decoy PNA-DNA-PNA chimeras are strong inhibitors of IL-8 gene expression and do not require transfection reagents
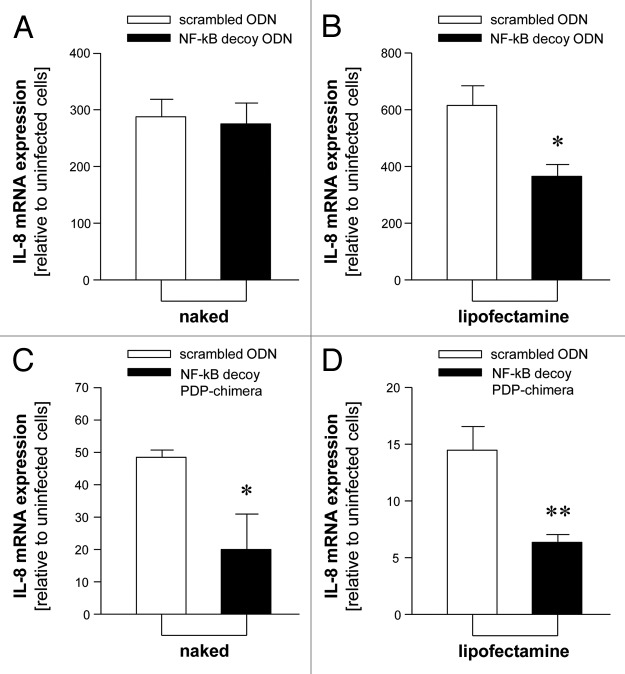
While it is firmly established that the entry of oligonucleotides into eukaryotic cells can occur through a nucleic acid channel,30 the complexation with lipofectamine (or other delivery systems) is required in vitro, since the degradation of ODNs exposed to fetal calf serum is an important drawback of these transfection approaches.31,32 On the contrary, we have elsewhere reported that, unlike ODN-based decoys, PDP/PDP chimeras are fully resistant to serum and cytoplasmic extracts.18,28 This information is of great impact, in our opinion, for the development of stable molecules to be used in non-viral gene therapy. Therefore, we compared the effects of ODN-based and PDP-based NF-kappaB decoys on P. aeruginosa infected cells in the presence or in the absence of the lipofectamine transfection reagents. In the first set of experiments we confirmed that the NF-kappB decoy PNA-DNA-PNA chimera (see Fig. 4A for molecular structure) reproduces the activity of the NF-kappaB decoy ODN on P.aeruginosa induced genes. In fact, the NF-kappaB PDP/PDP chimera exhibits differential effects on expression of PAO1 activated genes (Fig. 4B), such as ICAM-1, GRO-γ, IL-1β, IL-6, IL-8. The major inhibitory effect was found on IL-8 gene expression, confirming on one hand elsewhere reported studies on the role of NF-kappaB for IL-8 gene transcription and, on the other hand, that decoy for NF-kB might retain differential effects of genes regulated by promoters containing NF-kB signal sequences, such as ICAM-1, GRO-γ, IL-1β, IL-6. This suggest that other transcription factors are the master regulators of these genes (such as Sp1 for IL-6 gene).33 Despite the fact that this specific issue should be object of future investigations, the effects of the NF-kappaB PNA-DNA-PNA chimeras, as shown in the insert of Figure 4B, reproduce those obtained using NF-kappB ODNs. The results shown in Figure 5, while confirming that the NF-kappaB decoy ODN exhibit lipofectamine-dependent effects, provide evidence that the NF-kappaB decoy PDP interferes with the NF-kappaB activity without the need of lipofectamine.
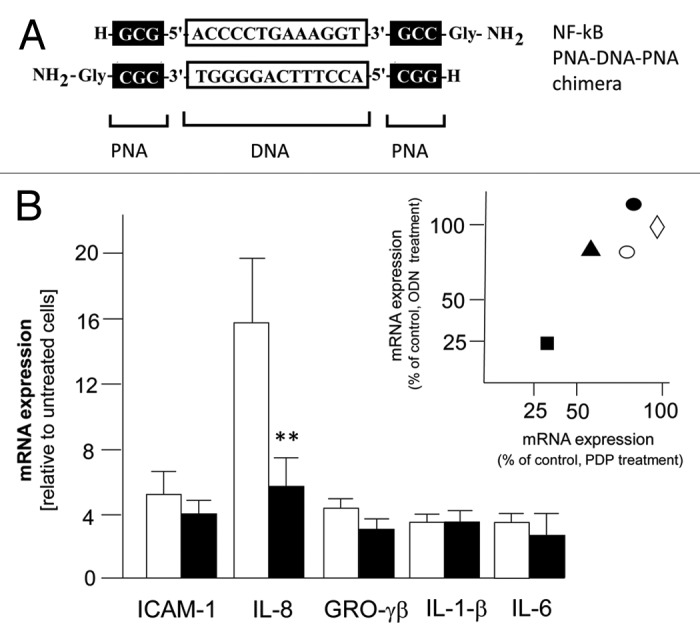
Figure 4. (A) Sequences of the double stranded NF-kappaB PNA-DNA-PNA chimeras. DNA sequences are within white boxes; PNA sequences are within black boxes. (B) Effects of PDP/PDP NF-kappaB decoy molecules on induction of different pro-inflammatory mRNAs. The PDP/PDP NF-kappaB decoy chimera was tested on transcription of ICAM-1, IL-8, GRO-γβ, IL-1β and IL-6 genes in IB3–1 bronchial cells after infection with P.aeruginosa. Total RNA was extracted and processed for quantification of transcripts as described in Materials and Methods. White bars: PAO-1 infected IB3–1 cells; black bars: IB3–1 cells infected with PAO-1 and treated with NF-kappaB decoy PDP/PDP chimeras. Values are mean ± SEM of four separate experiments. Significance in Student’s t test between each scrambled and decoy ODNs: ** p < 0.01. Insert. Relationship between the effects of PDP based and ODN-based NF-kappaB decoys on accumulation of IL-8 (■), GRO-γβ (▲), ICAM-1 (○), IL-1β (◊) and IL-6 (●) in PAO-1 infected IB3–1 cells. Data represent mRNA content in respect to control decoy-untreated PAO-1 infected cells.
Figure 5. Comparison of the effects on IL-8 mRNA expression of ODN (A and B) and PDP-based decoys (C and D) in the absence (naked molecules) or in the presence of lipofectamine. Values are mean ± SD of three separate experiments. Significance in Student’s t test between each scrambled and decoy ODNs: * p < 0.05; ** p < 0.01.
Discussion
Recent studies have indicated that the transcription factor decoy (TFD) strategy targeting NF-kappaB might be of interest to develop anti-inflammatory approaches for cystic fibrosis.12,13 The first set of data of the present study demonstrate that decoy ODNs against NF-kappaB interfere with the NF-kappaB pathway in cystic fibrosis cells infected with Pseudomonas aeruginosa. This was demonstrated by chromatin immunoprecipitation (ChIP) using as a model system cystic fibrosis IB3–1 cells infected with Pseudomonas aeruginosa. Treatment with ODN decoys against NF-kappaB led to a decreased occupancy of the IL-8 gene promoter by NF-kappaB factors.
In order to develop more stable therapeutic molecules, peptide nucleic acids (PNAs) based agents were considered. PNAs are DNA mimicking molecules in which the pseudopeptide backbone is composed of N-(2-aminoethyl)glycine units.19-21 PNAs are resistant to both nucleases and proteases22 and, more importantly, hybridize with high affinity to complementary sequences of single-stranded RNA and DNA, forming Watson-Crick double helices.21 For these reasons, PNAs were found to be excellent candidates for antisense and antigéne therapies.23-26 In recent studies, PNA-DNA chimeras have been described as reagents for the transcription factor decoy approach.27-29 PNA-DNA chimeras are PNA-DNA covalently bonded hybrids and were designed on one hand to improve the poor cellular uptake and solubility of PNAs, on the other hand to exhibit biological properties typical of DNA, such as the ability to stimulate RNaseH activity and to act as substrate for cellular enzymes (for instance DNA polymerases). The results published by Romanelli et al.29 Borgatti et al.27,28 and Moggio et al.34 firmly demonstrate that decoy molecules based on PNA-DNA chimeras are powerful decoy molecules. In respect to experiments aimed at pharmacological modification of gene expression, PNA-DNA-PNA chimeras are molecules of interest for several points of view: (1) unlike PNAs, they can be complexed with liposomes and microspheres;28 (2) unlike ODNs, they are resistant to DNases, serum and cytoplasmic extracts;22 (3) unlike PNA/PNA and PNA/DNA hybrids,35 they are potent decoy molecules.27-29
The second set of results presented in this paper (Figs. Four and 5) demonstrated that PDP/PDP chimeras targeting NF-kappaB are strong inhibitors of IL-8 gene expression even in the absence of protection with lipofectamine (Fig. 5) and mimick the biological activity of ODN-based lipofectamine-delivered decoys (insert of Fig. 4B). The extent of inhibition of IL-8 gene expression using naked NF-kappaB decoy ODN (Fig. 5C) approaches that found in lipofectamine-delivered NF-kappaB decoy ODN (Fig. 5B). This is the first research article from our group reporting this information, which is of great impact, in our opinion, for the development of stable molecules to be used in non-viral gene therapy. Despite the fact that these results should be considered as a proof-of-principle that PNA-DNA chimeras targeting NF-kB retain selective biological functions, we like to remark that no translation to clinical settings can be proposed based only on data reported in the present paper. On the other hand, we like to underline that IL-8 is one of the master genes in pro-inflammatory processes affecting cystic fibrosis. Accordingly, inhibition of its functions might have a clinical relevance and research efforts on this issue deserve great attention.
Materials and Methods
Synthetic oligonucleotides and peptide nucleic acids
The synthetic oligonucleotides used in this study were purchased from Pharmacia.
PNA-DNA-PNA oligomer assembly
Chimeras were assembled on solid phase, by sequential elongation of the PNA fragment, to whom DNA first and PNA were attached. The chimeras were obtained using Mmt(Bz)PNA monomers, as reported in previously published papers.29,36 ESI-MS for gcg-ACC CCT GAA AGG T-gcc: [M+4H]4+: 1438.2, [M+5H]5+ 1150.5, calculated. for C193H245N90O95P13 5748.26. ESI-MS for cgc-TGG GGA CTT TCC A-cgg: [M+4H]4+: 1443.6, [M+5H]5+: 1154.9, calculated for C194H247N86O99P13 5770.26.
Cell cultures and bacteria
IB3–1 cells (LGC Promochem) are human bronchial epithelial cells immortalized with adeno12/SV40, derived from a CF patient with a mutant F508del/W1282X genotype.37 Cells were grown in LHC-8 basal medium (Biofluids) supplemented with 5% fetal bovine serum (FBS). All culture flasks and plates were coated with a solution containing 35 μg/ml bovine collagen (Becton-Dickinson), 1μg/ml BSA (Sigma-Aldrich) and 1 μg/ml human fibronectin (Becton-Dickinson). P.aeruginosa, PAO1 laboratory strain, was kindly provided by A. Prince (Columbia University). Bacteria were grown in trypticase soy broth (TSB) or agar (TSA) (Difco).
Electrophoretic mobility shift assays
The double-stranded oligonucleotides (ODN) used in the EMSA are reported in Table 1. ODN (3 pmol each) were 32P-labeled using 10 U T4 polinucleotide kinase (MBI Fermentas) annealed to an excess of complementary ODN and purified from [γ-32P]ATP (Perkin Elmer). Binding reactions were performed by incubating 2.5 µg of nuclear extract, obtained from IB3 cells as previously described,12 and 16 fmol of 32P-labeled double-stranded ODN, with or without competitor in a final volume of 20 µL of binding buffer (20 mM TRIS-HCl, pH 7.5, 50 mM KCl, 1 mM MgCl2, 0.2 mM EDTA, 5% glycerol, 1 mM dithiothreitol, 0.01% TritonX100, 0.05 µg•µL-1 of poly dI-dC, 0.05 µg•µL-1 of a single-stranded ODN). Competitor (100-fold excess of unlabeled ODNs) and nuclear extract mixture were incubated for 15 min and then probe was added to the reaction. After a further incubation of 30 min at room temperature samples were immediately loaded onto a 6% nondenaturing polyacrylamide gel containing 0.25 x Tris/borate/EDTA (22.5 mM Tris, 22.5 mM boric acid, 0.5 mM EDTA, pH 8) buffer. Electrophoresis was performed at 200 V. Gels were vacuum heat-dried and subjected to autoradiography.
IB3–1 transfection with decoy ODNs and PDP/PDP chimeras
IB3–1 cells were seeded in 24-wells plates at a density of 30,000 cells/cm2 and transfected with ODNs using cationic liposome vector Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Lipofectamine 2000 (4 μl) was diluted in 1 ml of serum-free LHC-8 basal medium (Biofluids) and double-stranded decoy or PDP/PDP chimeras were added and incubated for 10 min to generate Liposome:DNA complexes. In parallel, a scrambled ODN was employed as negative control (Table 1). Then, the complexes were added to IB3–1 cells and incubated for 6 h. After this time of incubation cells were washed twice with serum-free culture medium and left at 37°C and 5% CO2 for 20–24 h before pro-inflammatory challenge with P.aeruginosa (100 CFU/ml), IL-1β (10 ng/ml) or TNF-α (50 ng/ml) for a further 4 h.
RNA extraction and quantitative RT-PCR
Total RNA from IB3–1 cells was purified using High Pure RNA Isolation Kit (Roche) and 2.5 μg RNA were reverse-transcribed to cDNA using the High Capacity cDNA Archive Kit and random primers (Applied Biosystems) in a final reaction volume of 100 μl. cDNA (5 μl) and primers (15 nM each) (Table 1) were mixed with Fast SYBR Green Master Mix (Applied Biosystems). GAPDH gene expression was determined as normalizer gene. Primer sets were purchased from Sigma-Genosys. Real Time PCR was performed in duplicate for both target and normalizer genes using the ABI PRISM 7900 HT Fast Real Time PCR System (Applied Biosystems) as follows: 50°C for 2 min, 95°C for 20 sec and 40 two-step cycles: 95°C for 1 sec and 60°C for 20 sec. Results were collected with SDS 2.3 Software (Applied Biosystems) and relative quantification was performed using the comparative threshold (Ct) method. Data were analyzed with RQ Manager Software 1.2 (Applied Biosystems). Changes in mRNA expression levels were calculated after normalization to calibrator gene. The ratios obtained after normalization are expressed as -fold change over untreated samples.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation assays were performed by using the Chromatin Immunoprecipitation Assay Kit (Upstate Biotechnology). A total of 5 x 106 IB3 cells were treated, for 10 min at room temperature, with 1% formaldehyde culture medium. Cells were washed in phosphate-buffered saline, and then glycine was added to a final concentration of 0.125 M. The cells were then suspended in 0.5 ml of lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-Cl, pH 8.1) plus protease inhibitors (1 μg/ml pepstatin A, 1 μg/ml leupeptin, 1 μg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride) and the chromatin subjected to sonication (using a Sonics Vibracell VC130 sonicator with a 2 mm probe). Fifteen 15 sec sonication pulses at 30% amplitude were required to shear chromatin to 200–1000 bp fragments. 0.2 ml aliquots of chromatin were diluted to 2 ml in ChIP dilution buffer containing protease inhibitors and then cleared with 75 μl of salmon sperm DNA/protein A-agarose 50% gel slurry (Upstate Biotechnology) for 1 h at 4°C before incubation on a rocking platform with either 10 μg of NF-kappaB p65 specific antiserum (sc-372X, Santa Cruz Biotechnology) or normal rabbit serum as negative control IgG (Upstate Biotechnology). Twenty μl of diluted chromatin was saved and stored for later PCR analysis as 1% of the input extract. Incubations occurred overnight at 4°C and continued an additional 1 h after the addition of 60 μl protein A-agarose slurry. Thereafter the agarose pellets were washed consecutively with low salt, high salt and LiCl buffers. DNA/protein complexes were recovered from the pellets with elution buffer (0.1 M NaHCO3 with 1% SDS), and cross-links were reversed by incubating overnight at 65°C with 0.2 M NaCl. The samples were treated with RNase A and proteinase K, extracted with phenol/chloroform and ethanol-precipitated. The pelletted DNAs were washed with 70% ethanol and dissolved in 40 μl of Tris/EDTA. Two μl aliquots were used for each real-time PCR reaction to quantitate immunoprecipitated promoter fragments. For quantitative real-time PCR reaction, a pair of primers that amplify a 301 bp region on the IL-8 promoter, containing the NF-kappaB binding site, was designed (IL-8 ChIP f, IL-8 ChIP r, Table 1). PCR reactions were also performed using negative control primers that amplify a 255 bp genomic region about 5 kb upstream of the IL-8 promoter, lacking NF-kappaB binding sites (Neg ChIP f, Neg ChIP r, Table 1). Each Real-time PCR reactions were performed in 25 μl of final volume, using 2 μl of template DNA (from chromatin immunoprecipitations), 10 pmol of primers and 1 x iQ™ SYBR® Green Supermix (Bio-Rad) for a total of 45 cycles (96°C for 15 sec, 66°C for 30 sec, and 72°C for 20 sec) using an iCycler IQ® (Bio-Rad). The relative proportions of immunoprecipitated promoter fragments were determined based on the threshold cycle (Tc) value for each PCR reaction. Real time PCR data analysis were obtained using the comparative cycle threshold method.38 The data represent the ratios between the IL-8 promoter Q-PCR performed on NF-kappaB specific ChIP and negative IgG (preimmune rabbit serum) ChIP. Each value was derived employing chromatin isolated from untreated IB3–1 cells or cells infected with PAO-1 in the absence or in the presence of pre-treatment with the NF-kappaB decoy ODN. Each sample was performed in duplicate on at least three separate experiments.
Statistics
Results are expressed as mean ± standard error of the mean (SEM). Comparisons between groups were made by using paired Student's t test and a one-way analysis of variance (ANOVA). Statistical significance was defined with p < 0.05 (*, significant) and p < 0.01 (**, highly significant).
Acknowledgments
This work was supported by grants from the Italian Cystic Fibrosis Research Foundation (grants # 15/2004 and # 13/2007 to R.G. and G.C.). R.G. is granted by Fondazione Cariparo, by MUR COFIN-2005, by TELETHON (GGP10124), by C.I.B. and by Associazione Veneta per la Lotta alla Talassemia. V.B. was fellow of the “Fondazione Cariverona.”
Glossary
Abbreviations:
- ODN
oligodeoxyribonucleitides
- PNA
peptide nucleic acids
- PDP
PNA-DNA-PNA chimeras
- TFD
transcription factor decoy
- NF-kappaB
NF-kappaB, nuclear factor kappa B
- I-kB
I-kappaB, inhibitor of NF-kappaB
- EMSA
electrophoretic mobility shift assay
- CF
cystic fibrosis
- CFTR
Cystic Fibrosis Transmembrane conductance Regulator
- PAO-1
Pseudomonas aeruginosa, strain O1
- IL-8
Interleukin 8
- PCR
polymerase chain reaction
- RT-PCR
reverse transcription PCR
- ChIP
chromatin immunoprecipitation
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/artificialdna/article/21061
References
- 1.Boucher RC. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur Respir J. 2004;23:146–58. doi: 10.1183/09031936.03.00057003. [DOI] [PubMed] [Google Scholar]
- 2.Verkman AS, Song Y, Thiagarajah JR. Role of airway surface liquid and submucosal glands in cystic fibrosis lung disease. Am J Physiol Cell Physiol. 2003;284:C2–15. doi: 10.1152/ajpcell.00417.2002. [DOI] [PubMed] [Google Scholar]
- 3.Machen TE. Innate immune response in CF airway epithelia: hyperinflammatory? Am J Physiol Cell Physiol. 2006;291:C218–30. doi: 10.1152/ajpcell.00605.2005. [DOI] [PubMed] [Google Scholar]
- 4.Puchelle E, De Bentzmann S, Hubeau C, Jacquot J, Gaillard D. Mechanisms involved in cystic fibrosis airway inflammation. Pediatr Pulmonol. 2001;(Suppl 23):143–5. [PubMed] [Google Scholar]
- 5.Bonfield TL, Panuska JR, Konstan MW, Hilliard KA, Hilliard JB, Ghnaim H, et al. Inflammatory cytokines in cystic fibrosis lungs. Am J Respir Crit Care Med. 1995;152:2111–8. doi: 10.1164/ajrccm.152.6.8520783. [DOI] [PubMed] [Google Scholar]
- 6.Jones AM, Martin L, Bright-Thomas RJ, Dodd ME, McDowell A, Moffitt KL, et al. Inflammatory markers in cystic fibrosis patients with transmissible Pseudomonas aeruginosa. Eur Respir J. 2003;22:503–6. doi: 10.1183/09031936.03.00004503. [DOI] [PubMed] [Google Scholar]
- 7.Becker MN, Sauer MS, Muhlebach MS, Hirsh AJ, Wu Q, Verghese MW, et al. Cytokine secretion by cystic fibrosis airway epithelial cells. Am J Respir Crit Care Med. 2004;169:645–53. doi: 10.1164/rccm.200207-765OC. [DOI] [PubMed] [Google Scholar]
- 8.Koehler DR, Downey GP, Sweezey NB, Tanswell AK, Hu J. Lung inflammation as a therapeutic target in cystic fibrosis. Am J Respir Cell Mol Biol. 2004;31:377–81. doi: 10.1165/rcmb.2004-0124TR. [DOI] [PubMed] [Google Scholar]
- 9.Groux-Degroote S, Krzewinski-Recchi MA, Cazet A, Vincent A, Lehoux S, Lafitte JJ, et al. IL-6 and IL-8 increase the expression of glycosyltransferases and sulfotransferases involved in the biosynthesis of sialylated and/or sulfated Lewisx epitopes in the human bronchial mucosa. Biochem J. 2008;410:213–23. doi: 10.1042/BJ20070958. [DOI] [PubMed] [Google Scholar]
- 10.Griesenbach U, Scheid P, Hillery E, de Martin R, Huang L, Geddes DM, et al. Anti-inflammatory gene therapy directed at the airway epithelium. Gene Ther. 2000;7:306–13. doi: 10.1038/sj.gt.3301078. [DOI] [PubMed] [Google Scholar]
- 11.Bezzerri V, Borgatti M, Finotti A, Tamanini A, Gambari R, Cabrini G. Mapping the transcriptional machinery of the IL-8 gene in human bronchial epithelial cells. J Immunol. 2011;187:6069–81. doi: 10.4049/jimmunol.1100821. [DOI] [PubMed] [Google Scholar]
- 12.Bezzerri V, Borgatti M, Nicolis E, Lampronti I, Dechecchi MC, Mancini I, et al. Transcription factor oligodeoxynucleotides to NF-kappaB inhibit transcription of IL-8 in bronchial cells. Am J Respir Cell Mol Biol. 2008;39:86–96. doi: 10.1165/rcmb.2007-0176OC. [DOI] [PubMed] [Google Scholar]
- 13.Cooper JA, Jr., Parks JM, Carcelen R, Kahlon SS, Sheffield M, Culbreth R. Attenuation of interleukin-8 production by inhibiting nuclear factor-kappaB translocation using decoy oligonucleotides. Biochem Pharmacol. 2000;59:605–13. doi: 10.1016/S0006-2952(99)00375-5. [DOI] [PubMed] [Google Scholar]
- 14.Mann MJ. Transcription factor decoys: a new model for disease intervention. Ann N Y Acad Sci. 2005;1058:128–39. doi: 10.1196/annals.1359.021. [DOI] [PubMed] [Google Scholar]
- 15.Mann MJ, Dzau VJ. Therapeutic applications of transcription factor decoy oligonucleotides. J Clin Invest. 2000;106:1071–5. doi: 10.1172/JCI11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gambari R. Biological activity and delivery of peptide nucleic acids (PNA)-DNA chimeras for transcription factor decoy (TFD) pharmacotherapy. Curr Med Chem. 2004;11:1253–63. doi: 10.2174/0929867043365242. [DOI] [PubMed] [Google Scholar]
- 17.Dzau VJ. Transcription factor decoy. Circ Res. 2002;90:1234–6. doi: 10.1161/01.RES.0000025209.24283.73. [DOI] [PubMed] [Google Scholar]
- 18.Borgatti M, Romanelli A, Saviano M, Pedone C, Lampronti I, Breda L, et al. Resistance of decoy PNA-DNA chimeras to enzymatic degradation in cellular extracts and serum. Oncol Res. 2003;13:279–87. doi: 10.3727/096504003108748339. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 20.Borgatti M, Finotti A, Romanelli A, Saviano M, Bianchi N, Lampronti I, et al. Peptide nucleic acids (PNA)-DNA chimeras targeting transcription factors as a tool to modify gene expression. Curr Drug Targets. 2004;5:735–44. doi: 10.2174/1389450043345155. [DOI] [PubMed] [Google Scholar]
- 21.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, et al. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–8. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 22.Demidov VV, Potaman VN, Frank-Kamenetskii MD, Egholm M, Buchard O, Sönnichsen SH, et al. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem Pharmacol. 1994;48:1310–3. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 23.Borgatti M, Boyd DD, Lampronti I, Bianchi N, Fabbri E, Saviano M, et al. Decoy molecules based on PNA-DNA chimeras and targeting Sp1 transcription factors inhibit the activity of urokinase-type plasminogen activator receptor (uPAR) promoter. Oncol Res. 2005;15:373–83. doi: 10.3727/096504005776449734. [DOI] [PubMed] [Google Scholar]
- 24.Penolazzi L, Borgatti M, Lambertini E, Mischiati C, Finotti A, Romanelli A, et al. Peptide nucleic acid-DNA decoy chimeras targeting NF-kappaB transcription factors: Induction of apoptosis in human primary osteoclasts. Int J Mol Med. 2004;14:145–52. [PubMed] [Google Scholar]
- 25.Gambari R. Peptide-nucleic acids (PNAs): a tool for the development of gene expression modifiers. Curr Pharm Des. 2001;7:1839–62. doi: 10.2174/1381612013397087. [DOI] [PubMed] [Google Scholar]
- 26.Uhlmann E. Peptide nucleic acids (PNA) and PNA-DNA chimeras: from high binding affinity towards biological function. Biol Chem. 1998;379:1045–52. [PubMed] [Google Scholar]
- 27.Borgatti M, Lampronti I, Romanelli A, Pedone C, Saviano M, Bianchi N, et al. Transcription factor decoy molecules based on a peptide nucleic acid (PNA)-DNA chimera mimicking Sp1 binding sites. J Biol Chem. 2003;278:7500–9. doi: 10.1074/jbc.M206780200. [DOI] [PubMed] [Google Scholar]
- 28.Borgatti M, Breda L, Cortesi R, Nastruzzi C, Romanelli A, Saviano M, et al. Cationic liposomes as delivery systems for double-stranded PNA-DNA chimeras exhibiting decoy activity against NF-kappaB transcription factors. Biochem Pharmacol. 2002;64:609–16. doi: 10.1016/S0006-2952(02)01188-7. [DOI] [PubMed] [Google Scholar]
- 29.Romanelli A, Pedone C, Saviano M, Bianchi N, Borgatti M, Mischiati C, et al. Molecular interactions with nuclear factor kappaB (NF-kappaB) transcription factors of a PNA-DNA chimera mimicking NF-kappaB binding sites. Eur J Biochem. 2001;268:6066–75. doi: 10.1046/j.0014-2956.2001.02549.x. [DOI] [PubMed] [Google Scholar]
- 30.Shi F, Gounko NV, Wang X, Ronken E, Hoekstra D. In situ entry of oligonucleotides into brain cells can occur through a nucleic acid channel. Oligonucleotides. 2007;17:122–33. doi: 10.1089/oli.2007.0034. [DOI] [PubMed] [Google Scholar]
- 31.Weyermann J, Lochmann D, Zimmer A. Comparison of antisense oligonucleotide drug delivery systems. J Control Release. 2004;100:411–23. doi: 10.1016/j.jconrel.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 32.Shi F, Hoekstra D. Effective intracellular delivery of oligonucleotides in order to make sense of antisense. J Control Release. 2004;97:189–209. doi: 10.1016/j.jconrel.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Borgatti M, Bezzerri V, Mancini I, Nicolis E, Dechecchi MC, Lampronti I, et al. Induction of IL-6 gene expression in a CF bronchial epithelial cell line by Pseudomonas aeruginosa is dependent on transcription factors belonging to the Sp1 superfamily. Biochem Biophys Res Commun. 2007;357:977–83. doi: 10.1016/j.bbrc.2007.04.081. [DOI] [PubMed] [Google Scholar]
- 34.Moggio L, Romanelli A, Gambari R, Bianchi N, Borgatti M, Fabbri E, et al. Alternate PNA-DNA chimeras (PNA-DNA)(n): synthesis, binding properties and biological activity. Biopolymers. 2007;88:815–22. doi: 10.1002/bip.20857. [DOI] [PubMed] [Google Scholar]
- 35.Saviano M, Romanelli A, Bucci E, Pedone C, Mischiati C, Bianchi N, et al. Computational procedures to explain the different biological activity of DNA/DNA, DNA/PNA and PNA/PNA hybrid molecules mimicking NF-kappaB binding sites. J Biomol Struct Dyn. 2000;18:353–62. doi: 10.1080/07391102.2000.10506672. [DOI] [PubMed] [Google Scholar]
- 36.Will DW, Breipohl G, Langner D, Knolle J, Uhlmann E. The synthesis of polyamide nucleic acids using a novel monomethoxytrityl protecting-group strategy. Tetrahedron. 1995;51:12069–82. doi: 10.1016/0040-4020(95)00766-2. [DOI] [Google Scholar]
- 37.Dechecchi MC, Nicolis E, Bezzerri V, Vella A, Colombatti M, Assael BM, et al. MPB-07 reduces the inflammatory response to Pseudomonas aeruginosa in cystic fibrosis bronchial cells. Am J Respir Cell Mol Biol. 2007;36:615–24. doi: 10.1165/rcmb.2006-0200OC. [DOI] [PubMed] [Google Scholar]
- 38.Finotti A, Treves S, Zorzato F, Gambari R, Feriotto G. Upstream stimulatory factors are involved in the P1 promoter directed transcription of the A beta H-J-J locus. BMC Mol Biol. 2008;16:9–110. doi: 10.1186/1471-2199-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]