Abstract
Autophagy is a catabolic process involved in the turnover of organelles and macromolecules which, depending on conditions, may lead to cell death or preserve cell survival. We found that some lung cancer cell lines and tumor samples are characterized by increased levels of lipidated LC3. Inhibition of autophagy sensitized non-small cell lung carcinoma (NSCLC) cells to cisplatin-induced apoptosis; however, such response was attenuated in cells treated with etoposide. Inhibition of autophagy stimulated ROS formation and treatment with cisplatin had a synergistic effect on ROS accumulation. Using genetically encoded hydrogen peroxide probes directed to intracellular compartments we found that autophagy inhibition facilitated formation of hydrogen peroxide in the cytosol and mitochondria of cisplatin-treated cells. The enhancement of cell death under conditions of inhibited autophagy was partially dependent on caspases, however, antioxidant NAC or hydroxyl radical scavengers, but not the scavengers of superoxide or a MnSOD mimetic, reduced the release of cytochrome c and abolished the sensitization of the cells to cisplatin-induced apoptosis. Such inhibition of ROS prevented the processing and release of AIF (apoptosis-inducing factor) and HTRA2 from mitochondria. Furthermore, suppression of autophagy in NSCLC cells with active basal autophagy reduced their proliferation without significant effect on the cell-cycle distribution. Inhibition of cell proliferation delayed accumulation of cells in the S phase upon treatment with etoposide that could attenuate the execution stage of etoposide-induced apoptosis. These findings suggest that autophagy suppression leads to inhibition of NSCLC cell proliferation and sensitizes them to cisplatin-induced caspase-dependent and -independent apoptosis by stimulation of ROS formation.
Keywords: autophagy, apoptosis, ROS, hydroxyl radical, superoxide, NSCLC, caspase-independent cell death
Introduction
Autophagy is a catabolic process that regulates the degradation of intracellular components in lysosomes. Although autophagy maintains cell survival under different stress conditions, it has also been described as a form of non-apoptotic (type II) programmed cell death. Thus, whether autophagy preserves cell survival or drives cells to death is not fully understood. Activation of autophagy has been suggested to promote cell survival by ensuring metabolic supply or by removing protein aggregates and abnormal organelles. For instance, the turnover of mitochondria is thought to be important for the protection of cells from oxidative stress.1 Several molecular links between autophagy and apoptosis have been described. For instance, ATG5, an important autophagy-related protein, was shown to be cleaved by calpain, and the cleavable N-terminal product was able to switch the autophagic function of ATG5 to proapoptotic by triggering the release of cytochrome c from mitochondria.2 Similarly, the caspase-mediated cleaved form of the autophagy-related protein BECN1 can amplify the apoptotic signal by facilitating the permeabilization of mitochondria.3 Furthermore, proteins such as TP53, the BCL2 family proteins, and DAPk are involved in both apoptosis and autophagy.4
Various cytotoxic drugs induce autophagy. However, the inhibition of autophagy may differently regulate the sensitivity of cells to treatment. Depending on the nature of the inducing agent or the cell content, autophagy has been shown to promote survival or to sensitize cells to the death stimulus. For example, ATG5-deficient mouse embryonic fibroblasts (MEFs) were more resistant to cell death induced by H2O2 than wild-type fibroblasts. Nevertheless, the same autophagy-deficient MEFs were more sensitive to cell death triggered by TNFα.5 Interestingly, inhibiting autophagy was found to prevent apoptosis in sarcoma cells following TNFα treatment in combination with NFκB suppression,6 or apoptosis in nontransformed fibroblasts followed by ER stress.7
Reactive oxygen species (ROS) play an important role in different types of cell death. Several studies demonstrate the involvement of autophagy in the degradation and formation of ROS. For instance, autophagy activation leads to selective autophagic degradation of catalase, the enzyme involved in decomposition of hydrogen peroxide.8 Suppression of autophagy by silencing ATG7 and ATG8 also blocks ROS accumulation and inhibits caspase-independent cell death in macrophages.9 In contrast to these studies, it was suggested that loss of autophagy leads to an accumulation of SQSTM1, ER chaperones and damaged mitochondria that might be potential substrates of ROS, and suppresses tumorigenesis.10 Thus, autophagy either induces ROS formation or is involved in decomposition of ROS, suggesting that ROS might link autophagy with other cell death modalities.
Here, we addressed the question of how inhibition of autophagy affects cell death induced by the therapeutic drugs etoposide and cisplatin, used as the first line of treatment for NSCLC. Our results suggest that inhibition of autophagy facilitates stimulation of ROS formation, leading to sensitization of cells to cisplatin-mediated apoptosis. Although scavenging of superoxide did not affect the sensitivity of cells with inhibited autophagy to cisplatin treatment, apoptosis was completely blocked when scavengers of hydroxyl radicals or antioxidant N-acetyl cysteine (NAC) were used. This increase in ROS facilitated the permeabilization of the mitochondrial outer membrane, followed by the release of cytochrome c and HTRA2, which subsequently enhanced caspase-dependent apoptosis. The suppression of autophagy also accelerates the processing and release of apoptosis-inducing factor (AIF) from mitochondria, which contributes to caspase-independent cell death. Furthermore, inhibiting autophagy delayed proliferation of NSCLC cells and progression of cells through the cell cycle. Thus, inhibition of autophagy must be considered as a new therapeutic approach to reduce lung cancer proliferation and as a promising strategy for the sensitization of cells to drug treatment through facilitating formation of ROS.
Results
Autophagy in lung cancer cells and its activity upon treatment with cisplatin and etoposide
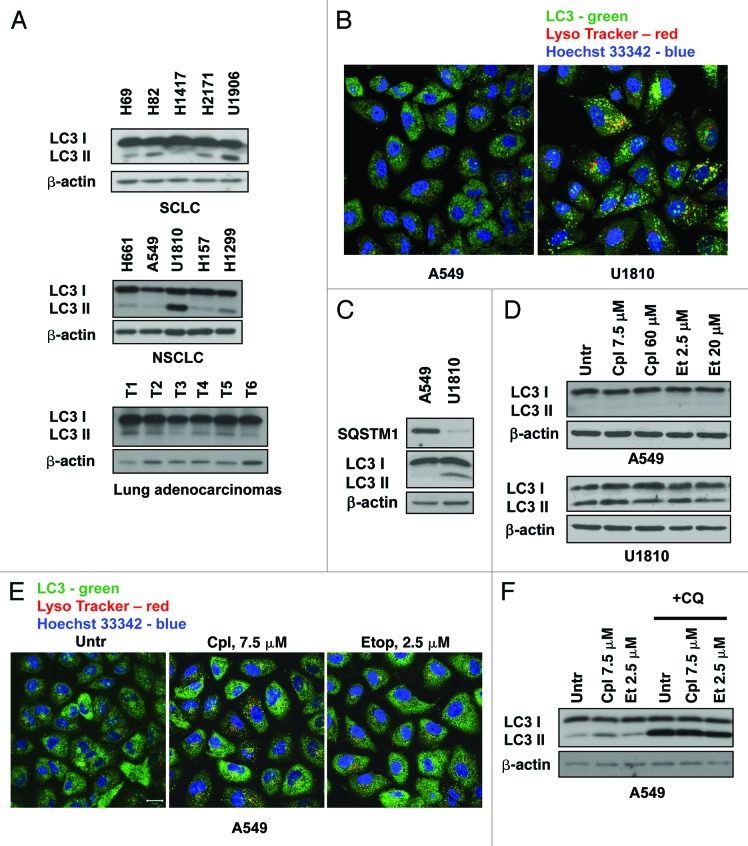
Our previous data, and the results presented in Figure 1A, have shown that lung cancer cell lines and human lung adenocarcinomas are characterized by different levels of basal autophagy activity.11 To explore the role of autophagy in the sensitivity of lung cancer cells to treatment, the present study was performed using two NSCLC cell lines, U1810 and A549, which respond differently to conventional therapeutic drugs. The basal level of autophagy in these cells was examined by confocal microscopy using immunostaining of autophagosomes with an LC3 antibody and lysosomes with Lysotracker Red. U1810 cells contain a significant number of autophagosomes; however, autophagosomes were not detected in A549 cells (Fig. 1B). The higher basal autophagy in U1810 cells was also confirmed biochemically by measuring the lipidation of autophagy-related LC3 protein and the expression of SQSTM1 protein in these cell lines (Fig. 1C). Two drugs, cisplatin and etoposide, which are used as the first line of therapy for lung cancer, were selected in order to understand the role of autophagy in the sensitivity of NSCLC cells to treatment. To determine the effect of cisplatin and etoposide on the activation of autophagy, cells were treated with different concentrations of these drugs. Twenty-four hours of treatment had a negligible effect on macroautophagy in both U1810 and A549 cells (Fig. 1D). Formation of autophagosomes in A549 cells was also not detectable by immunocytochemistry (Fig. 1E). However, inhibition of lysosomal degradation with chloroquine (50 μM, 3 h) in cells treated with cisplatin (7.5 μM) or etoposide (2.5 μM) for 36h showed accumulation of lipidated form of LC3, indicating that autophagic flux in A549 cells was active and comparable to basal level of autophagy in these cells (Fig. 1F) Thus, the level of basal autophagy was present and maintained active in cells upon treatment with etoposide or cisplatin. Therefore, we decided to investigate whether the inhibition of autophagy affects the sensitivity of NSCLC to treatment with these drugs.
Figure 1. Autophagy in lung cancer cell lines and its activity upon treatment with cisplatin and etoposide. (A) Different level of LC3 lipidation in lung carcinoma cell lines and human lung adenocarcinomas. (B) Co-staining with LC3 antibody and Lysotracker Red shows significant numbers of autophagosomes in U1810 cells fused with lysosomes. (C) Expression of SQSTM1 protein and lipidation of LC3 in A549 and U1810 cells detected by immunoblotting. (D) Expression of LC3 in U1810 and A549 cells treated (24 h) with cisplatin (7.5 or 60 μM) or etoposide (2.5 and 20 μM). (E) Immunocytochemical detection of autophagosome formation in A549 cells treated (24 h) with cisplatin (7.5 μM) or etoposide (2.5 μM). (F) Autophagic flux in A549 cells treated (36 h) with etoposide (2.5 μM) or cisplatin (7.5 μM). Cells were treated with chloroquine (CQ, 50 μM) 3 h before collecting the samples and accumulation of LC3-II was measured by immunoblotting.
Inhibition of autophagy sensitizes NSCLC to cisplatin-mediated cell death
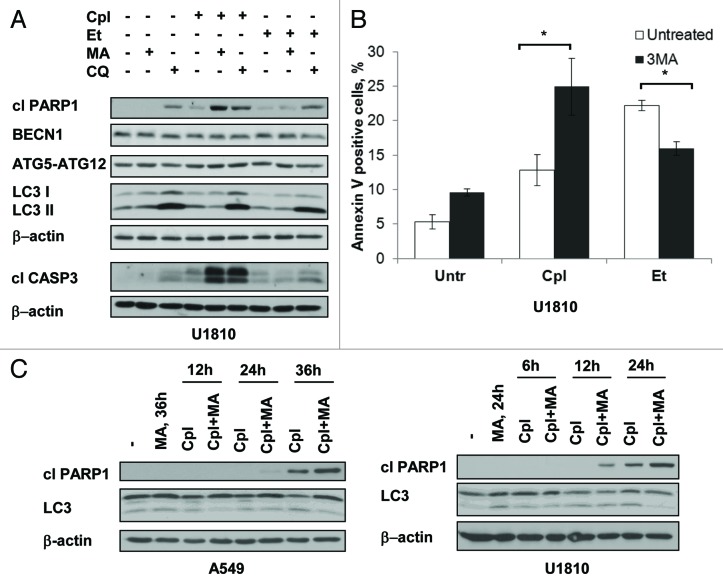
First, to examine whether the inhibition of autophagy sensitizes NSCLC cells to either etoposide or cisplatin, cells were treated with these compounds in combination with an inhibitor of autophagy: either 3-methyladenine or chloroquine (Fig. 2A). Treatments with these drugs, either alone or in combination, did not significantly affect the expression of the autophagy-related proteins ATG5–12, ATG7, BECN1 and LC3. An accumulation of LC3-II was observed upon treatment with chloroquine, indicating that autophagic flux was efficiently blocked. Interestingly, combining the treatment of U1810 cells with an autophagy inhibitor and cisplatin had a synergistic effect on activation of CASP3 and PARP1 cleavage, however, the inhibition of autophagy in the cells treated with etoposide did not result in marked changes in the level of active caspase-3 or cleaved PARP1. Importantly, the suppression of autophagy weakened the execution stages of apoptosis in another NSCLC cell line (A549), also treated with etoposide (Fig. S1A). To validate the observations that inhibition of autophagy differentially affects cell sensitivity to the studied drugs, we assessed the outcome of etoposide and cisplatin on U1810 cells, with basal or suppressed autophagy, using Annexin V–FITC and PI staining. The number of AV- and AV/PI-positive cells significantly increased in cisplatin-treated samples in which autophagy was suppressed. However, apoptotic cell death was attenuated when etoposide was used in combination with the inhibitor of autophagy (Fig. 2B; Fig. S1B and C). To examine the kinetics of the sensitization of cells to cisplatin-induced apoptosis, the cells were treated for different periods of time with the drugs in combination with the inhibitor of autophagy. The level of autophagy was analyzed by immunoblotting using antibodies to LC3 protein, and caspase-dependent apoptosis was evaluated by PARP1 cleavage (Fig. 2C). U1810 cells with high basal level of lipidated LC3 responded well to the combined treatment even at 12 h; however, inhibition of autophagy in A549 cells sensitized them to cisplatin after a longer time period. A significant effect due to autophagy inhibition on cisplatin-mediated apoptosis in A549 cells was observed even using a relatively low dose of the drug (7.5 μM) (Fig. S1D). Thus, inhibitors of autophagy sensitize NSCLC cells to cisplatin and to some extend attenuate the cell death induced by etoposide.
Figure 2. The inhibitors of autophagy sensitize NSCLC cells to cisplatin-induced apoptosis and attenuate the cell death induced by etoposide. (A) Activation of CASP3, cleavage of PARP1 and expression of BECN1, ATG7, ATG5–12 and LC3 in U1810 cells treated (24 h) with cisplatin (Cpl, 15 μM) or etoposide (Et, 2.5 μM) alone or in combination with either inhibitors of autophagy 3-methyladenine (3MA, 5 mM) or chloroquine (CQ, 50 μM). (B) Annexin V/ PI staining of U1810 cells treated (24 h) with cisplatin (15 μM) or etoposide (2.5 μM) alone, or in combination with 3-methyladenine. (C) Time-course analysis of PARP1 cleavage, expression of LC3 in A549 and U1810 cells treated with cisplatin (15 μM), 3-methyladenine (5 mM), or their combination. Statistical significance: *p < 0.05
Autophagy inhibition delays cell proliferation and the cell cycle progression
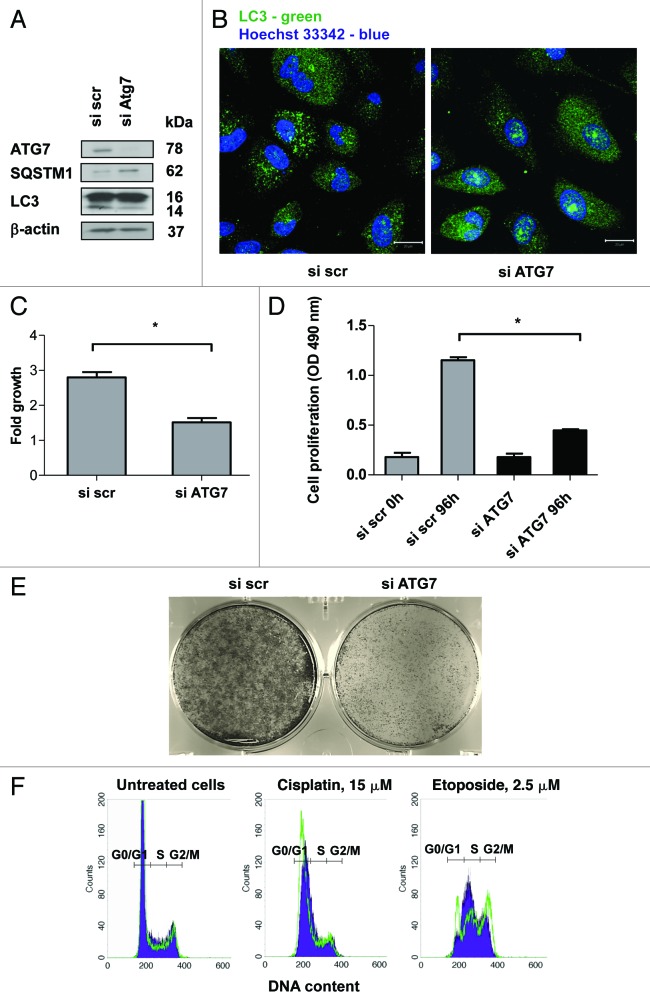
To explore the mechanism underlying the influence of autophagy inhibition on the cell death induced by etoposide and cisplatin, autophagy was suppressed via silencing of the autophagy-related gene ATG7, which is important for LC3 and ATG5-ATG12 ubiquitin-like conjugation systems. ATG7 expression was depleted by more than 80% in the siRNA-transfected cells (Fig. 3A). The efficiency of autophagy inhibition was confirmed by staining autophagosomes with an LC3 antibody (Fig. 3B). This inhibition was associated with a decrease in the level of lipidated LC3 protein and an increase in the expression of the autophagic substrate SQSTM1 in the cells with silenced ATG7 (Fig. 3A). Since the effect of some drugs is cell-cycle dependent and, considering that autophagy is a catabolic process, we suggest that inhibition of autophagy might affect cell proliferation, presumably by supplying nutrients and, therefore, facilitate the progression of cells through the cell cycle. Indeed, our data showed that the inhibition of autophagy affects proliferation of NSCLC cells (Fig. 3C and D). The effect of autophagy inhibition on cell growth was also confirmed using a clonogenic assay (Fig. 3E).
Figure 3. The inhibition of autophagy delays the growth and progression of cells through the cell cycle. (A) Inhibition of autophagy in U1810 cells transfected with siRNA targeting ATG7. Autophagy was detected by measurement of the expression level of SQSTM1 and conversion of LC3-I to LC3-II proteins, or (B) using immunostaining of autophagosomes with LC3 antibody. (C) Autophagy inhibition delays the growth of U1810 cells. The cells were incubated (24 h) with scrambled or ATG7-targeting siRNAs. The cells were plated in 6-well plates and counted 48h after seeding. The bars represent a fold increase in the number of seeded cells. (D) Suppression of autophagy inhibits cell proliferation. U1810 cells were plated in 96-well plates and MTS assay was performed 96h after seeding. (E) Suppression of autophagy inhibits colony formation in U1810 cells. Autophagy was suppressed using siRNA targeting ATG7. The cells were grown in 6-well plates and five days after fixed and stained with crystal violet. (F) The cell cycle analysis of U1810 cells with basal or suppressed autophagy was performed after treatment with cisplatin (15 μM), or etoposide (2.5 μM). The filled blue histogram represents the cells transfected with scrambled siRNA and the green histogram corresponds to the cells with suppressed autophagy. Statistical significance: *p < 0.05
To explain the difference in response of NSCLC cells to cisplatin and etoposide, we considered their targeting effects on cells. Both of these drugs interact with DNA; however, the precise molecular mechanism of their action on DNA differs. The cell death induced by etoposide is dependent on the cell cycle progression, since it inhibits the activity of topoisomerase II, an enzyme that is essential for progression through the S phase. Although detailing the molecular mechanisms concerning how autophagy inhibition influences cell proliferation requires further investigation, our results showed that inhibition of autophagy attenuates accumulation of cells in the S phase, leading to a reduction in cell death after treatment with etoposide (Fig. 3F; Fig. S2A and B). This suggests that due to delay in the entry of cells into the S phase, the effect of the S-phase-specific drug etoposide could be attenuated. However, no significant changes in the cell cycle distribution of U1810 cells with basal or suppressed autophagy treated with cisplatin were found (Fig. 3F). Thus, suppression of autophagy reduces cell proliferation and therefore can be proposed as a new therapeutic target for the treatment of lung carcinomas.
Inhibition of autophagy sensitizes NSCLC cells to caspase-dependent and -independent apoptosis
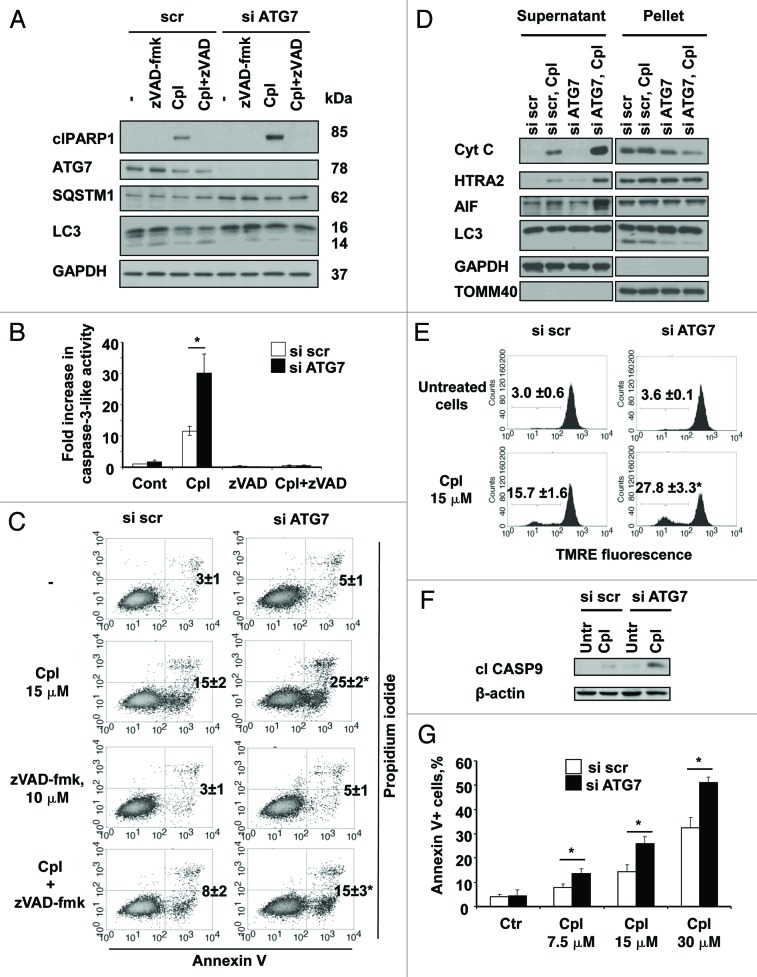
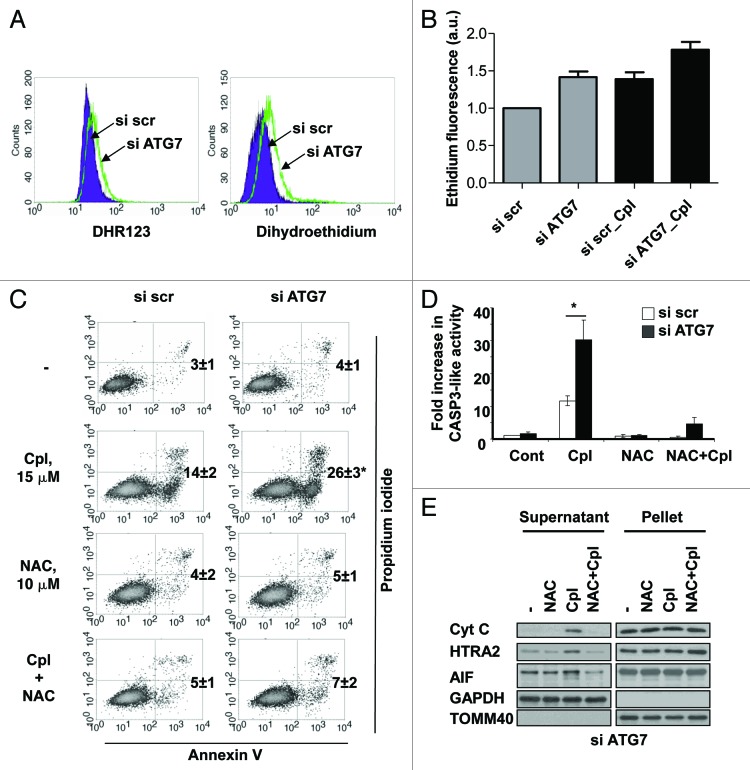
The effect of autophagy inhibition on sensitization of NSCLC cells to cisplatin-mediated apoptosis was studied in cells in which autophagy was suppressed by silencing ATG7 (Fig. 4A). Caspase-3-like activity in the cisplatin-treated cells with inhibited autophagy was significantly higher than in cells treated with cisplatin alone. This activity was also validated by an increase in the level of cleaved PARP1 (Fig. 4A). To explore whether cell death is dependent on caspases, the activity of these enzymes was blocked using the pan-caspase inhibitor, zVAD-fmk (10 μM). At this concentration caspase-3-like activity and specific cleavage of its substrate PARP1 were completely blocked (Fig. 4A and B). However, apoptotic cell death measured by Annexin V/PI staining was only partially inhibited (Fig. 4C). Previous studies from our group showed that the activation of caspases was not sufficient to trigger apoptotic cell death in NSCLCs and, instead, suggested the importance of caspase-independent apoptosis driven by AIF.12 Therefore, the impact of inhibited autophagy on the cisplatin-mediated release of AIF was investigated using cell fractionation. The results we obtained revealed that inhibition of autophagy in cisplatin-treated cells led to a drastic increase in the accumulation of cytochrome c, HTRA2 and AIF in the cytosol (Fig. 4D). Silencing of AIF reduced the number of Annexin V-positive cells (Fig. S3A), indicating the contribution of AIF to this type of cell death. The effect on mitochondria was also confirmed by a drop in mitochondrial membrane potential and activation of caspase-9 (Fig. 4E and F). In addition, we found that the effect of autophagy inhibition on apoptotic cell death was more pronounced when the concentration of cisplatin increased (Fig. 4G).
Figure 4. The inhibition of autophagy sensitizes the cells to caspase-dependent and -independent apoptotic cell death induced by cisplatin. (A) U1810 cells with basal or siRNA-mediated suppressed autophagy were pretreated (1 h) with DMSO or pan-caspase inhibitor zVAD-fmk (10 μM) and were then treated with cisplatin (15 μM). The level of autophagy was measured by the detection of expression of SQSTM1 and LC3 proteins by western blot and the efficiency of effector caspases inhibition was proved by the detection of PARP1 cleavage and (B) a caspase activity assay. (C) U1810 cells were transfected with scrambled or siRNA-targeting ATG7, and were then pretreated (1 h) with zVAD-fmk (10 μM) followed by cisplatin administration (15 μM, 24 h). Cell death was measured by annexin V/PI-staining. The number of apoptotic/secondary necrotic cells is shown as the sum of annexin V-positive and annexin V/PI double-positive cells. (D) The effect of autophagy inhibition on the distribution of cytochrome c, HTRA2 and AIF in the cytosolic and membrane fractions of U1810 cells treated with cisplatin. (E) The effect of autophagy inhibition on mitochondrial membrane potential in U1810 cells treated with cisplatin. The cells were treated (24 h) with cisplatin and MMP was assessed by TMRE staining. (F) Activation of CASP9 in cells with basal and suppressed autophagy. U1810 cells were treated (24 h) with cisplatin and the active form of CASP9 was detected by immunoblotting. (G) The effect of autophagy inhibition on cell death induced by different concentrations of cisplatin (7.5–30 μM). U1810 cells were treated (24 h) with cisplatin and cell death was measured by Annexin V/PI-staining. Statistical significance: *p < 0.05–untreated or treated cells transfected with scrambled siRNA in comparison to the untreated control (or same cell treatment) transfected with siRNA targeting ATG7.
Inhibition of autophagy stimulates ROS formation, which facilitates mitochondrial membrane permeabilization and is required for cisplatin-mediated apoptosis in NSCLC cells
Earlier work by our group demonstrated the importance of ROS in the release of AIF from mitochondria.13 Therefore, we decided to analyze whether inhibition of autophagy could stimulate the generation of ROS. Autophagy was suppressed in cells by siRNAs targeting ATG7 and BECN1 and ROS formation was measured using dihydroethidine and dihydrorhodamine123 staining (Fig. 5A; Fig. S3B). We found that inhibition of autophagy stimulated the formation of ROS and treatment of the cells with cisplatin had a synergistic effect on the generation of ROS when autophagy was suppressed (Fig. 5B). Since the level of ROS was elevated in cells with suppressed autophagy, we further analyzed whether the inhibition of ROS influenced cisplatin-mediated cell death. For this purpose, ROS formation was inhibited by N-acetyl cysteine and apoptosis was measured using the Annexin V-binding assay (Fig. 5C). This experiment showed that sensitization of the cells to cisplatin-induced cell death was completely blocked by preventing ROS formation. Finally, we investigated whether the inhibition of ROS affected caspase activity and influenced the release of AIF. Indeed, scavenging of ROS in cells with genetically inhibited autophagy prevented the release of cytochrome c, HTRA2 and the processing and release of AIF from mitochondria, and rescued them from cisplatin-mediated apoptosis (Fig. 5D and E).
Figure 5. The inhibition of autophagy stimulates ROS formation which is required for sensitization of NSCLC cells to cisplatin-induced apoptosis. (A) ROS formation in U1810 cells transfected with scrambled or ATG7-targeting siRNA. Forty-eight hours after transfection ROS formation was measured using dihydrorodamine 123 and dihydroethidium probes, as described in the Materials and Methods. (B) The effect of autophagy inhibition on ROS formation in U1810 cells. Cells were transfected with scrambled or ATG7-targeting siRNAs followed by treatment with cisplatin (15 μM, 24 h). (C) U1810 cells with basal or suppressed autophagy were pretreated (1 h) with N-acetyl-cysteine (10 mM) and then incubated with cisplatin (15 μM, 24 h). Cell death was measured by Annexin V/PI staining and (D) the CASP3-like activity assay was performed as described in the Materials and Methods. (E) The effect of the antioxidant NAC on cisplatin-mediated release of cytochrome c and AIF from mitochondria in U1810 cells with suppressed autophagy. Cells were treated as described in (C). Statistical significance: *p < 0.05.
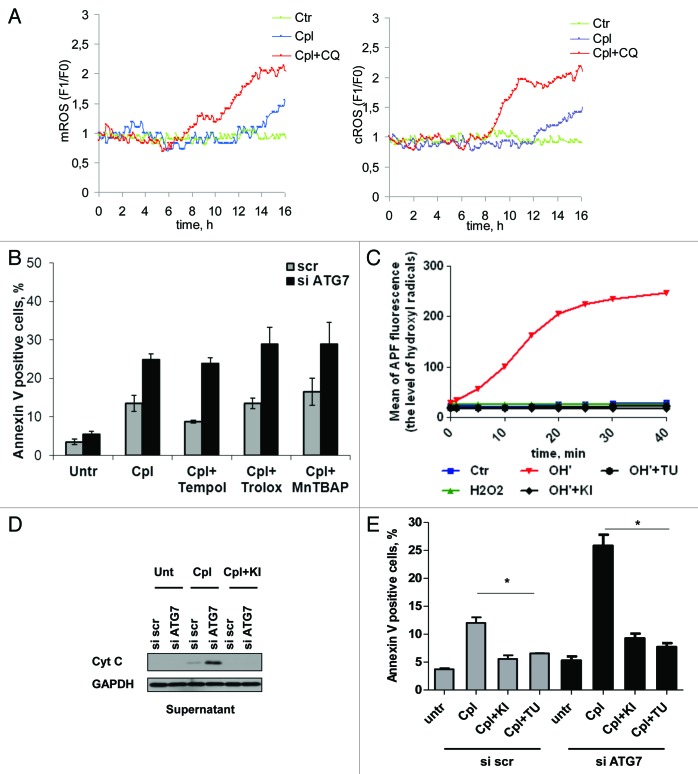
To further explore the source of ROS that mediates cisplatin-induced cell death, we studied formation of H2O2 in different intracellular compartments using genetically encoded fluorescent indicators directed to cytosol and mitochondria. Inhibition of autophagy in combination with cisplatin treatment leads to an increase in cytosolic and mitochondrial H2O2 within 8–16 h (Fig. 6A; Movies 1 and 2). Next, to determine which type of ROS is important, we utilized scavengers of superoxide and hydroxyl radicals. Since previously it was suggested that accumulation of damaged mitochondria might be a source of ROS in cells with suppressed autophagy, two inhibitors of mitochondrial respiration, rotenone and antimycin A, were used to stimulate formation of superoxide by mitochondrial complexes I and III (Fig. S4A). An increase in superoxide formation was detected already within 3 h and observed up to 24 h after cell treatment. However, such ROS stimulation was not capable of sensitizing cells to cisplatin (Fig. S4B). Furthermore, scavengers of superoxide or the MnSOD mimetic MnTBAP did not prevent cisplatin-mediated cell death (Fig. 6B). Therefore, in the next set of experiments we utilized thiourea and potassium iodide, two compounds that might potentially be used as hydroxyl radical scavengers. The results we obtained showed that these reagents efficiently scavenge the hydroxyl radicals induced in vitro (Fig. 6C). Pretreatment of cells with scavengers of hydroxyl radicals prevented the outer mitochondria membrane permeabilization (Fig. 6D), and efficiently blocked apoptosis, suggesting a critical role for hydroxyl radicals in sensitization of cells to cisplatin under conditions of suppressed autophagy (Fig. 6E). Thus, such increases in mitochondrial and cytosolic H2O2, a source of hydroxyl radicals, is proposed as the mechanism which allows autophagy inhibition to contribute to drug-mediated apoptotic cell death.
Figure 6. The scavengers of hydroxyl radicals prevent cisplatin-mediated cell death in NSCLC cells with suppressed autophagy. (A) Detection of H2O2 formation in cytosolic and mitochondrial compartments of U1810 cells treated with cisplatin (15 μM) alone or in combination with chloroquine (50 μM). Representative single cells that responded to treatment from different live cell imaging experiments are shown. (B) The effect of the MnSOD mimetic MnTBAP and scavengers of superoxide Tempol and Trolox on cisplatin-mediated cell death in U1810 cells with suppressed autophagy. Autophagy was suppressed in U1810 cells using siRNA targeting ATG7 and 48 h after transfection cells were pretreated for 1 h with Trolox (200 μM), Tempol (0.5 mM) or MnTBAP (25 μM) followed by treatment with cipslatin (15 μM, 24 h). (C) Thiourea and potassium iodide efficiently scavenge hydroxyl radicals in vitro. (D) The scavengers of hydroxyl radical potassium iodide (5 mM) and thiourea (1 mM) prevent the release of cytochrome c and processing of AIF in U1810 cells treated with cisplatin (15 μM, 24 h). Cells were pretreated for 1 h with the scavengers of hydroxyl radicals followed by treatment with cisplatin. (E) The effect of hydroxyl radical scavengers on cell death induced by cisplatin in U1810 cells with basal or suppressed autophagy. Cells were treated as described in (D). Statistical significance: *p < 0.05.
Discussion
The high level of basal autophagy in some parts of tumors, or the large-scale activation of autophagy in tumor cells upon chemo- and radio-therapy, makes this area of research attractive to those attempting to modulate autophagy in order to increase the sensitivity of cancer cells to treatment. In tumors, autophagy often localizes to hypoxic regions prior to angiogenesis where it supports tumor cell survival.14 Since cancer cells frequently carry defects in their apoptotic machinery, autophagy can sustain tumor survival for weeks in conditions of deprivation.15 Interestingly, although autophagy supports tumor cell survival, some tumors have a mono-allelic loss of chromosome 17q21, which is where BECN1 is located. As a result, some tumor types, such as breast and ovarian carcinomas, are characterized by suppressed autophagy.16 Here, we focused on investigating the role of autophagy in the sensitivity of NSCLCs to conventional therapeutic drugs. Since the level of autophagy was relatively high in some NSCLC cell lines, and autophagy was still active in cells treated with cisplatin and etoposide, we were interested in understanding the effect of inhibiting autophagy on drug sensitivity.
The results obtained demonstrate that, depending on the treatment, inhibition of autophagy could enhance the pro-apoptotic effect of cisplatin and to some extent attenuate the cytotoxic effect of etoposide. It is known that one of the main molecular targets of these drugs is DNA. However, the mechanism of their interaction with DNA differs. For instance, cisplatin induces the formation of DNA adducts and is believed to kill cells in different phases of the cell cycle. In contrast, etoposide intercalates into the DNA molecule and induces DNA double-strand breaks (DSBs) by inhibiting topoisomerase II, the enzyme that forms transient DNA DSBs during the replication of DNA.17 Bearing this in mind, we supposed that the different effects of autophagy inhibition on drug-mediated apoptosis might be related to the influence of the former on the cell cycle progression. In previous studies it has been suggested that autophagy contributes to the cell cycle by regulation of the expression of proteins involved in specific cell cycle checkpoints,18 or perhaps by providing the energy and nutrients that are essential for cell growth and division.19 We showed that inhibiting autophagy in NSCLCs could influence their proliferation and thus delay the progression of cells through the cell cycle, suggesting that inhibition of cell proliferation affects the sensitivity of the cells to cell cycle-specific drugs. For instance, inhibition of autophagy in etoposide-treated NSCLC cells delayed their entry into the S phase and therefore the pro-apoptotic effect of this drug was weakened. Interestingly, although siRNA-mediated inhibition of autophagy delayed NSCLC cell growth, it did not significantly affect the cell cycle distribution. Our previous observations demonstrated the presence of autophagy in all stages of the cell cycle.11 Given this, it might be speculated that autophagy is an important process which is required for the progression of cells through all phases of the cell cycle. Since U1810 cells, similar to a number of other lung cancer cells, do not express the cell cycle regulator p53, we suggest that short-term suppression of autophagy delays progression through the cell cycle, rather than arresting them in a specific phase of the cell cycle. Since autophagy is a catabolic process, involving degradation of macromolecules including proteins and lipids, it has an important role as a supplier of the substrates essential for NSCLC cell growth. Thus, the suppression of autophagy affecting NSCLC cell proliferation may be a useful strategy for the treatment of lung carcinomas.
Activators of autophagy, such as lithium, or inhibitors of autophagy, such as the antimalarial drug chloroquine, are used in the clinical setting. As a cytoprotective mechanism, autophagy was shown to be involved in the degradation of damaged organelles or protein aggregates,20 and modulation of autophagy was reported to enhance the efficiency of chemotherapeutic drugs. Our data demonstrate that inhibition of autophagy sensitizes NSCLC cells to cisplatin-induced apoptosis, which is in line with previous findings showing that the inhibition of autophagy might enhance the ability of the drugs to induce apoptosis in tumor cells. Pharmacological inhibitors of autophagy have been shown to sensitize cells to a wide range of cytotoxic stimuli including radiation therapy,21 TRAIL,22 the tyrosine kinase receptor inhibitor imatinib,23 and a DNA-damaging drug temozolomide.24 However, the molecular mechanisms of this sensitization to treatment were not studied. Interestingly, activation of autophagy followed by blocking of lysosomal degradation (late stages of autophagy) was shown to be detrimental to cancer cell survival and provides a rational therapeutic approach to enhancing the anticancer efficacy of PtdIns3K–Akt pathway inhibition.25 The inhibition of autophagy significantly enhanced tumor regression and delayed the recurrence of a Myc-induced model of lymphoma following administration of cytotoxic chemotherapy, providing evidence supporting the use of autophagy inhibitors in combination with conventional therapy to induce apoptosis in human cancers.26 Thus, combining agents that disrupt autophagy with HDAC inhibitors was suggested as a promising approach to treat imatinib-refractory patients who find conventional therapy ineffective.
What is the mechanism of sensitization of NSCLC to apoptosis induced by cisplatin? It has previously been shown that the inhibition of autophagy could block ROS accumulation in L929 mouse fibroblasts by the selective degradation of catalase.8 In contrast to these findings, our results demonstrate that inhibition of autophagy leads to an increase in ROS formation and that treatment with cisplatin has a synergistic effect on ROS accumulation in NSCLC cells. Although the level of ROS induced by the suppression of autophagy did not kill cells per se, this level of ROS was required for the sensitization of NSCLC cells to cisplatin-induced apoptosis. Indeed, scavenging of ROS rescued the cells from the harmful effect of autophagy inhibition on cisplatin administration. The involvement of autophagy in the oxidative stress response remains incompletely established, however, the accumulation of protein aggregates in cells with suppressed autophagy could contribute to ROS production.10 Another possibility involves the inhibition of elimination of damaged mitochondria. It has been suggested that the presence of mutant Parkin, a multiprotein E3 ubiquitin ligase complex, which in turn is part of the ubiquitin-proteasome system which mediates the targeting of proteins for degradation, increases oxidative stress and sensitizes cells to death induced by different insults.27 Recent publications have shown a direct link between Parkin and mitochondria. Parkin is specifically recruited to impaired mitochondria and activates the process of mitophagy.1,28,29 Our studies demonstrate that neither stimulation of mitochondrial superoxide nor scavengers of superoxide affect the sensitivity of NSCLC cells to cisplatin. Previously, an increase in mitochondrial superoxide has been proposed as a mechanism involved in cell sensitization to autophagy inhibition,30 however, we have shown that a mitochondrial SOD mimetic or inhibitors of mitochondrial complex I and III (stimulation of mitochondrial superoxide), did not affect sensitivity of NSCLC cells to cisplatin, indicating the role of other forms or sources of ROS. Our data suggest that the accumulation of cytosolic and mitochondrial H2O2 is induced by combined treatment with cisplatin and inhibitors of autophagy, and that hydroxyl radicals play an important role in this sensitization process. It is likely that cytosolic ROS may facilitate activation of proapoptotic proteins, such as BAX, which subsequently leads to permeabilization of the mitochondrial membrane.
Thus, here we show that impaired autophagy stimulates ROS formation upon treatment with cisplatin and scavengers of hydroxyl radicals, or antioxidant NAC, significantly reduces apoptosis, suggesting that accumulation of ROS is an important mechanism in the sensitization of cells to apoptosis under conditions of suppressed autophagy. Furthermore, the apoptotic response to cisplatin treatment in NSCLC cells with impaired autophagy is, to some extent, dependent on caspases and also involves the activation of the caspase-independent pathway. Both these death signaling pathways implicate mitochondria. Inhibiting autophagy facilitates the release of cytochrome c, HTRA2 and the processing and release of AIF from mitochondria. Thus, it can be concluded that inhibition of autophagy could enhance the killing efficiency of the drug by increasing the formation of ROS that eventually facilitates the release of cytochrome c, HTRA2 and AIF from mitochondria and, subsequently, activates cell death.
Materials and Methods
Cell culture and treatments
Human lung carcinoma A549 (ATCC, CCL-185), H661 (ATCC, HTB-183), H157 (ATCC, CRL-5802), H1299 (ATCC, CRL-5803), H69 (ATCC, HTB-119), H82 (ATCC, HTB-175), H1417 (ATCC, CRL-5869), H2171 (ATCC, CRL-5929) and U1906 and U1810 cell lines (both from the collection at the Uppsala University, Sweden), were cultured in RPMI 1640 (Sigma, R0883) with 10% fetal calf serum (FCS; GIBCO, 10270), 2 mM glutamine (GIBCO, 25030), penicillin (100 μg/ml)/ streptomycin (100 μg/ml) (GIBCO, 15140). Cells were seeded 24 h before treatment for the indicated time period with 2.5–20 μM etoposide or 7.5–60 μM cisplatin (both from Bristol Myers). The inhibitors of autophagy chloroquine (50 μM; Sigma, C-6628) or 3-methyladenine (5 mM, Sigma; M9281) were added simultaneously with the drugs. The pan-caspase inhibitor z-VAD-fmk (10 μM, Enzyme Systems Products, FK109), ROS scavengers N-acetyl cysteine (10 mM, Sigma, A9165), Trolox (Sigma, 238813), Tempol (Sigma, T7263) were added 1 h prior to treatment.
Clinical material
Surgically resected specimens were collected from patients with lung adenocarcinomas at the Clinical Oncology Research Institute, N.N. Blokhin Russian Cancer Research Center during the period 2007–2009. After surgical removal the tumor specimens were frozen and stored in liquid nitrogen. All patients signed informed consent forms according to the legal institutional guidelines and ethical permission. The tumor clinic morphological stages were determined according to the standard tumor TNM classification systems of the International Union Against Cancer (edition 7).
Immunoblotting
Cells were lysed in RIPA buffer (25 mM Tris•HCl pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), supplemented with protease inhibitor cocktail (Roche, 04693159001). For preparation of the cytosolic fraction, cells were incubated for 5 min in a buffer containing 250 mM sucrose, 70 mM KCl and 100 μg/ml digitonin (Calbiochem, 300410) in PBS (GIBCO, 20012), and then pelleted for 5 min at 7000 × g. The supernatant that contained the cytosolic fraction was collected. The pellet was washed in the same buffer and lysed in RIPA buffer (mitochondrial fraction). The protein concentration was determined using the BCA protein assay (Pierce, 23223). The samples were mixed with Laemmli buffer, boiled for 5min, subjected to SDS-PAGE and blotted onto nitrocellulose membrane (Bio-Rad, 162-0115), which was blocked for 1 h with 5% non-fat milk in PBS and probed with primary antibodies diluted in PBS containing 2% BSA (Sigma, A3059) and 0.05% Tween-20 (Sigma, P1379). The following antibodies were used: mouse anti- SQSTM1 (Santa Cruz Biotechnology, 28359), goat anti-AIF (Santa Cruz Biotechnology, 9416), rabbit anti-TOMM40 (Santa Cruz Biotechnology, 11414), rabbit anti-ATG7 (Cell Signaling Technology, 2631), rabbit anti-BECN1 (Cell Signaling Technology, 3738), rabbit anti-ATG5 (Cell Signaling Technology, 2630), mouse anti-cleaved PARP1 (Cell Signaling Technology, 9546), rabbit anti-GAPDH (Trevigen, 2275-PC-100), rabbit anti-actin (Sigma, A2066), rabbit anti-HTRA2 (R&D Systems, AF1458), mouse anti-cytochrome c (BD PharMingen, 559027) and rabbit anti-LC3 (MBL, PM036). The recognized proteins were detected using horseradish peroxidase-labeled secondary antibodies: anti-goat IG, anti-mouse IgG (Pierce, 31430), anti-rabbit IgG (Pierce, 31460) and an enhanced chemiluminescence kit (Amersham, RPN2209).
Caspase activity assay
Cells were washed with ice-cold PBS, resuspended in 25 μl of PBS and after lysis in liquid nitrogen were loaded onto a microtiter plate. CASP3 substrate DEVD-AMC (50 μM, Peptide Institute, Osaka) was added and fluorescence was detected in a Fluoroscan II plate reader (Labsystems) using 355 nm excitation and 460 nm emission wavelengths. Fluorescent units were converted to pmols of released AMC and, subsequently, related to the amount of protein in each sample. Finally, caspase activity was expressed as a fold-increase compared with the appropriate control.
Annexin V/ PI staining
Annexin V/PI double staining was performed using an Annexin-V-FLUOS Staining Kit (Roche Applied Science, 11988549001) according to the manufacturer’s protocol. The cells were analyzed by flow cytometry (FACScan, Becton Dickinson) and the data were evaluated using Cell Quest software.
Mitochondrial membrane potential
Cells were washed in PBS, incubated for 20 min at 37°C with 25 nM of tetramethylrhodamine ethyl ester perchlorate (TMRE, Molecular Probes, T-669) in PBS and subsequently analyzed by flow cytometry (FACScan, Becton Dickinson).
Measurement of ROS generation
Cells were trypsinized, resuspended in PBS and stained with 10 µM dihydroethidium (10 min at 37°C) (Molecular probes, D-1168) or 10 μM dihydrorodamine 123 (10 min at 37°C) (Molecular probes, D-23806). Ethidium or rhodamine 123 fluorescence intensity resulting from dihydroethidium and dihydrorodamine 123 oxidation were measured using a FACScan flow cytometer (Becton-Dickinson). FL2 (ethidium) or FL1 (rhodamine 123) fluorescence was collected in each experiment by gating and acquiring the cell population.
The hydroxyl radical scavenging properties of thiourea and potassium iodide were studied using aminophenyl fluorescein (1–10 μM, APF, Cayman Chemical Company, 10157) probe. For this purpose, hydroxyl radicals were induced in a chemical reaction in which iron (II) sulfate (50 μM) interacts with hydrogen peroxide (50 μM). Thiourea (50 μM) or potassium iodide (1 mM) was applied to the reaction mixture to scavenge the hydroxyl radicals. The fluorescence of oxidized APF probe was collected in a Fluoroscan II plate reader (Labsystems) using 485 nm excitation and 538 nm emission wavelengths.
Cell cycle analysis
Cells were washed twice in PBS, fixed in 70% ice-cold ethanol for at least 12 h and stored at +4°C until analysis. Fixed cells were washed twice in PBS, incubated for 1 h in PBS containing 50 μg/ml RNase A (Boehringer, 109169) and then stained for 20 min with 50 μg/ml PI-solution (Sigma, P4170) in PBS. Analysis was performed using a flow cytometer (FACScan, Becton Dickinson) and the data were evaluated using ModFit and Cell Quest software.
RNA interference
Cells were seeded in 100 mm Petri dishes at final density 1.5 × 106 cells in growth media without antibiotics. Non-targeting pool (ON-TARGETplus Non-targeting Pool, D-001810-10-05, Dharmacon), anti- BECN1 (ON-TARGETplus SMARTpool, L-034478-00-0005, Dharmacon), anti-ATG7 (ON-TARGETplus SMARTpool, L-020112-00-0005 Dharmacon) or anti-AIF (ON-TARGETplus SMARTpool, L-011912-00-0005, Dharmacon) siRNAs were diluted in 200 μl of OPTI-MEM (Gibco, 51985) and mixed with 15 μl of INTERFERin siRNA Transfection Reagent (Polyplus-transfection, 409-10). After 10 min incubation the complexes were added to the cells. The final concentration of siRNA in the medium was 50 nM. Twenty-four hours after transfection the cells were trypsinized and seeded in 6-well plates or 60 mm dishes. The desired treatments were administered 24 h after cell seeding.
Immunofluorescence microscopy
Cells were grown on coverslips. LysoTracker (Molecular Probes, L7528) was added for 20 min to a final concentration of 100 nM and the cells were then washed with PBS and fixed for 20 min with 4% paraformaldehyde (Sigma, P-6148) at RT. The cells were permeabilized for 15 min in digitonin (100 μg/ml) and incubated for 1 h at RT with primary rabbit anti-LC3 (MBL, PM036, 1:2000) antibody diluted in PBS supplemented with 1% BSA. Then the cells were rinsed with PBS and incubated with AlexaFluor488-conjugated donkey anti-rabbit IgG secondary antibodies (Molecular Probes, A21206, 1:500) for 60 min at RT. Nuclei were counterstained with Hoechst 33342 (10 μg/ml, Molecular Probes, H3570) by 5 min incubation at RT. After washing with PBS, slides were mounted in Vectashield mounting medium (Vector Laboratories, H-1000) and examined under a Zeiss LSM 510 META confocal laser scanner microscope (Zeiss).
Live cell imaging
Live cell imaging was performed according to the protocol described by Belousov et al.31 Briefly, cells were transfected with pHyPer-dMito and pHyPer-cyto fluorescent probes directed to mitochondria and cytosol (kindly provided by Dr. Belousov, Moscow, Russia), two days before desired treatments were applied. The oxidized form of the HyPer probes was exited using the 488-nm argon laser and the green fluorescent signal was detected at 505–550 nm using a Zeiss LSM 510 META confocal laser scanner microscope with the time series speed of one frame every 10 min.
Statistical analysis
All the results are expressed as means ± SE. Statistical evaluation was performed using nonpaired t-tests.
Supplementary Material
Acknowledgments
We thank Dr. Vsevolod V. Belousov (Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, Russia) for the generous gift of pHyPer-dMito and pHyPer-cyto plasmids. This study was supported by grants from the Swedish and Stockholm Cancer Societies, the Swedish Research Foundation, the Swedish Childhood Cancer Society, the European Union (Chemores and Apo-Sys) and the Russian Ministry of High Education and Science (11.G34.31.0006). V.K. was supported by a fellowship from the Swedish Institute and Karolinska Institutet.
Glossary
Abbreviations:
- NSCLC
non-small cell lung carcinoma
- PARP1
poly(ADP)ribose polymerase 1
- FCS
fetal calf serum
- NAC
N-acetylcysteine
- 3MA
3-methyladenine
- CQ
chloroquine
- AIF
apoptosis-inducing factor
- SOD
superoxide dismutase
- MMP
mitochondrial membrane potential
Disclosure of Potential Conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/20123
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/20123
References
- 1.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–32. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 3.Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis 2010; 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–75. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 5.Pyo JO, Nah J, Kim HJ, Lee HJ, Heo J, Lee H, et al. Compensatory activation of ERK1/2 in Atg5-deficient mouse embryo fibroblasts suppresses oxidative stress-induced cell death. Autophagy. 2008;4:315–21. doi: 10.4161/auto.5525. [DOI] [PubMed] [Google Scholar]
- 6.Djavaheri-Mergny M, Amelotti M, Mathieu J, Besançon F, Bauvy C, Souquère S, et al. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem. 2006;281:30373–82. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- 7.Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, et al. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282:4702–10. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 8.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A. 2006;103:4952–7. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y, Kim SO, Li Y, Han J. Autophagy contributes to caspase-independent macrophage cell death. J Biol Chem. 2006;281:19179–87. doi: 10.1074/jbc.M513377200. [DOI] [PubMed] [Google Scholar]
- 10.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaminskyy V, Abdi A, Zhivotovsky B. A quantitative assay for the monitoring of autophagosome accumulation in different phases of the cell cycle. Autophagy. 2011;7:83–90. doi: 10.4161/auto.7.1.13893. [DOI] [PubMed] [Google Scholar]
- 12.Gallego MA, Joseph B, Hemström TH, Tamiji S, Mortier L, Kroemer G, et al. Apoptosis-inducing factor determines the chemoresistance of non-small-cell lung carcinomas. Oncogene. 2004;23:6282–91. doi: 10.1038/sj.onc.1207835. [DOI] [PubMed] [Google Scholar]
- 13.Norberg E, Gogvadze V, Vakifahmetoglu H, Orrenius S, Zhivotovsky B. Oxidative modification sensitizes mitochondrial apoptosis-inducing factor to calpain-mediated processing. Free Radic Biol Med. 2010;48:791–7. doi: 10.1016/j.freeradbiomed.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 14.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–16. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 17.Burden DA, Kingma PS, Froelich-Ammon SJ, Bjornsti MA, Patchan MW, Thompson RB, et al. Topoisomerase II.etoposide interactions direct the formation of drug-induced enzyme-DNA cleavage complexes. J Biol Chem. 1996;271:29238–44. doi: 10.1074/jbc.271.46.29238. [DOI] [PubMed] [Google Scholar]
- 18.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 20.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–94. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 22.Han J, Hou W, Goldstein LA, Lu C, Stolz DB, Yin XM, et al. Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. J Biol Chem. 2008;283:19665–77. doi: 10.1074/jbc.M710169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shingu T, Fujiwara K, Bögler O, Akiyama Y, Moritake K, Shinojima N, et al. Inhibition of autophagy at a late stage enhances imatinib-induced cytotoxicity in human malignant glioma cells. Int J Cancer. 2009;124:1060–71. doi: 10.1002/ijc.24030. [DOI] [PubMed] [Google Scholar]
- 24.Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11:448–57. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 25.Degtyarev M, De Mazière A, Orr C, Lin J, Lee BB, Tien JY, et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183:101–16. doi: 10.1083/jcb.200801099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyun DH, Lee M, Hattori N, Kubo S, Mizuno Y, Halliwell B, et al. Effect of wild-type or mutant Parkin on oxidative damage, nitric oxide, antioxidant defenses, and the proteasome. J Biol Chem. 2002;277:28572–7. doi: 10.1074/jbc.M200666200. [DOI] [PubMed] [Google Scholar]
- 28.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–83. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–21. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–6. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.