Abstract
The sorting nexins Atg20/Snx42 and Snx41 regulate membrane traffic and endosomal protein sorting and are essential for Cvt and/or pexophagy in yeast. Previously, we showed that macroautophagy is necessary for conidiation in the rice-blast fungus Magnaporthe oryzae. Here, we analyzed the physiological function(s) of selective autophagy in Magnaporthe through targeted deletion of MGG_12832, an ortholog of yeast SNX41 and ATG20/SNX42. Loss of MGG_12832 (hereafter SNX41) abolished conidia formation and pathogenesis in M. oryzae. Snx41-GFP localized as dynamic puncta or short tubules that are partially associated with autophagosomes and/or autophagic vacuoles. PX domain, but not macroautophagy per se, was required for such localization of Snx41-GFP in Magnaporthe. Although not required for nonselective autophagy, Snx41 was essential for pexophagy in Magnaporthe. We identified Oxp1, an ATP-dependent oxoprolinase in the gamma-glutamyl cycle, as a binding partner and potential retrieval target of Snx41-dependent protein sorting. The substrate of Oxp1, 5-oxoproline, could partially restore conidiation in the snx41Δ. Exogenous glutathione, a product of the gamma-glutamyl cycle, significantly restored pathogenicity in the snx41Δ mutant, likely through counteracting the oxidative stress imposed by the host. We propose that the gamma-glutamyl cycle and glutathione biosynthesis are subject to regulation by Snx41-dependent vesicular trafficking, and mediate antioxidant defense crucial for in planta growth and pathogenic differentiation of Magnaporthe at the onset of blast disease in rice.
Keywords: Atg20, Snx41, fungal virulence, Magnaporthe, antioxidant, GSH, sorting nexin
Introduction
Magnaporthe oryzae, a filamentous ascomycete, is the causal agent of the devastating blast disease in rice and several crops. Magnaporthe depends on its asexual spores, conidia, for disease establishment and propagation.1,2 Upon contact with a suitable host surface, the conidia differentiate into specialized dome-shaped structures called the appressoria to facilitate penetration, followed by in planta growth, and colonization of the host.3,4
Production of conidia occurs in response to light exposure in Magnaporthe.5,6 So far, not much is known about the genetic regulation of Magnaporthe conidiation.7,8 G-protein signaling acts as an upstream regulator of Magnaporthe conidiation.9 More recently, we showed that autophagy is induced during Magnaporthe conidiation, wherein it facilitates carbon catabolism and nutrient homeostasis.10,11
Autophagy is a bulk degradation process conserved in the eukaryotic cells. It was first identified by electron microscopy and considered as nonselective for its cytosolic cargos.12 It was later found that different types of selective autophagy exist, including glycogen autophagy (specific for glycogen degradation),13,14 mitophagy (mitochondrial degradation)15 and pexophagy (specific degradation of peroxisomes).16
The molecular basis of autophagy has been elucidated in yeast and mammals, with the identification of 35 ATG genes so far.17,18 Among them, ATG20 and ATG24 encode Phox (PX)- and BAR-domain containing proteins involved in specific cargo assembly.19 The localization of Atg20 and Atg24 to the PAS depends on the phospholipid/PtdIns3P content of the membrane and thus is subject to regulation through PtdIns 3-kinase complex I, composed of Atg6/Vps30, Atg14 and several other Vps proteins.20,21 On the other hand, emerging evidence suggests the requirement of Atg20 in pexophagy in Pichia pastoris.22 Peroxisomes represent important sites for lipid catabolism, which is essential for the formation of melanin layer in the appressorium and thus required for Magnaporthe pathogenicity.23
Besides ATG20, a subset of ATG and PEX genes is specifically required for pexophagy but not for nonselective autophagy. ATG30 encodes a 44 kDa protein with two coiled-coil domains and is required for both forms of pexophagy (macropexophagy and micropexophagy), but not for the Cvt and general autophagy pathways in Pichia.24 Atg26 was reported to be essential for pexophagy in P. pastoris25,26 and in Colletotrichum orbiculare.27 Magnaporthe lacks ATG30, however gene deletion analysis of ATG26 in Magnaporthe showed that the atg26Δ mutant is pathogenic,28 but it is unclear whether Atg26 is essential for pexophagy in Magnaporthe. PEX14 encodes a peroxisomal membrane protein implicated in both peroxisome biogenesis and pexophagy in Hansenula.29,30 In S. cerevisiae, Snx41 forms an endosomal-sorting complex with Atg20/Snx42 and Atg24/Snx4 to carry out autophagy-independent functions in protein sorting and traffic.31
The Gamma-glutamyl cycle is a metabolic pathway for transporting amino acids into cells,32 and for synthesis of an antioxidative tripeptide, reduced glutathione (GSH).33 Hereditary defects in the gamma-glutamyl cycle lead to metabolic acidosis, 5-oxoprolinuria, hemolytic anemia and progressive neurological symptoms.33,34 The investigation of the biological function and regulation of gamma-glutamyl cycle during fungal development, is, however, limited.
In this study, we reported the functional analysis and an essential role in conidiation and pathogenesis of an Snx41- and Atg20-related sorting nexin, MGG_12832, in Magnaporthe. We showed that Oxp1, an enzyme in the gamma-glutamyl cycle, physically interacts with Snx41 and likely serves as a cargo for its sorting function. Importantly, exogenous 5-oxoproline (the substrate of Oxp1) partially restored the conidiation in the snx41Δ mutant. Furthermore, exogenous GSH (an antioxidant produced by the gamma-glutamyl cycle) significantly increased invasive hyphal growth in planta. Our observations suggest that MGG_12832 protein shares overlapping functions with Snx41 and Atg20 and contributes to Magnaporthe conidiogenesis and pathogenesis by regulating the production of important intermediate(s) and/or product(s) from the gamma-glutamyl cycle. Lastly, we uncovered a hitherto uncharacterized role for this cycle in protecting Magnaporthe against the host-resistance response.
Results
Identification and gene deletion analysis of SNX41 in Magnaporthe
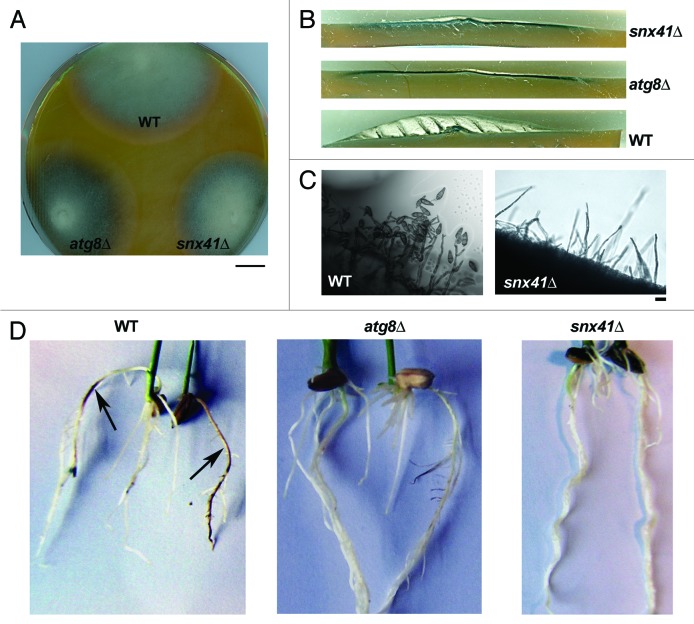
We identified Magnaporthe MGG_12832 as a gene encoding a sorting nexin-like protein (627 aa; GenBank EHA50783.1) significantly similar to S. cerevisiae Snx41 (41% over entire length) and Snx42/Atg20 (34%; Fig. S1). The predicted polypeptide encoded by MGG_12832 [designated MoSNX41; hereafter referred to as SNX41 (gene) or Snx41 (encoded protein)] showed the characteristic phosphoinositide-binding PX domain (N-terminal; aa 101–211) and 2 BAR domains (C-terminal half). MGG_12832/Snx41 showed limited homology (5%) to the Snx3 ortholog in Magnaporthe. An snx41Δ strain was created by replacing MGG_12832 with the hygromycin-resistance cassette (HPH1) in wild-type Magnaporthe strain B157 as well as in the GFP-SRL (as the peroxisome marker23) strain (Fig. S2A and S2B). As judged by colony morphology, the snx41Δ mutant exhibited reduced aerial hyphal growth compared with the wild type (Fig. 1A and B). The snx41Δ mutant showed a total loss of conidiation [Fig. 1C, WT with 97.5 ± 5.5 (×100 conidia/cm2), snx41Δ with 0 (×100 conidia/cm2)]. We had earlier reported that autophagy-assisted glycogen catabolism is necessary for Magnaporthe conidiation, but could be bypassed by exogenous supplementation with sucrose or glucose 6-phosphate (G6P) in the atg8Δ mutant.10 External addition of sucrose or G6P could also suppress the conidiation defects in another autophagy-deficient mutant, atg1Δ (Fig. S3A), further confirming the importance of glycogen autophagy in carbon homeostasis during Magnporthe conidiation. Compared with the atg8Δ mutant, the snx41Δ showed better aerial hyphal growth (Fig. 1A and B). However, the loss of conidiation could not be restored by addition of sucrose or G6P (Fig. S3A), implying that the conidiation defects in the snx41Δ were likely not linked to defects in glycogen hydrolysis and/or autophagy function therein. Furthermore, iodine staining confirmed that unlike atg8Δ, which accumulates high level of total glycogen under conditions conducive to conidiation, the snx41Δ contained comparable level of total glycogen as that of the WT (Fig. S3B). Thus, the conidiation defects in the snx41Δ mutant were not caused by anomalies in glycogen catabolism. Pathogenicity assays on barley leaf explants could not be performed with the snx41Δ mutant due to lack of conidia. Inoculation on wounded barley leaf explants with snx41Δ mycelial suspension caused no lesion on the leaf surface (data not shown). Infection assays of rice roots with mycelial plugs of the snx41Δ showed no symptoms on the root surface as opposed to the typical dark/brown lesions seen in the roots infected by WT Magnaporthe (Fig. 1D, denoted by arrows), suggesting that Snx41 plays a significant role in Magnaporthe virulence.
Figure 1. Characterization of vegetative and pathogenic development in snx41Δmutant. (A) Colony and growth characteristics of the wild type (WT), atg8Δ and snx41Δ mutants. Colonies of the indicated strains were grown on PA medium for 7 d in the dark and photographed. Scale bar: 1 cm. (B) Loss of SNX41 leads to reduction in aerial hyphal growth. Photograph was taken with cross-section from 5d old colonies grown under constant illumination. (C) Loss of SNX41 leads to total loss of conidiation. Wild type (WT) or snx41Δ grown on PA medium containing lactose as the sole carbon source, were stained with acid fuchsin (for better visualization of hyphal outline) and analyzed by bright field microscopy at 24 h post photo-induction. Scale bar: 10 μm. (D) The snx41Δ mutant is incapable of infecting the rice roots. Surface-sterilized rice seeds were allowed to germinate and grow in direct contact with the fungal mycelial plugs from the wild type, atg8Δ or the snx41Δ. Disease symptoms (necrosis) were examined at 6 dpi. Arrows indicate necrotic lesions produced on the roots infected by WT.
Subcellular localization of Snx41-GFP in Magnaporthe
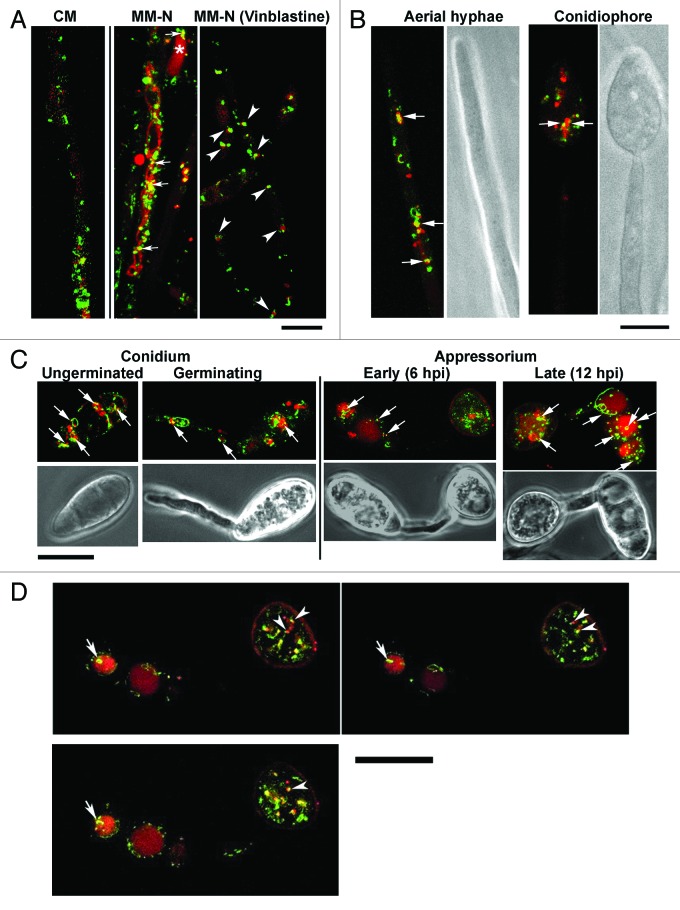
To gain further insight into the molecular function, subcellular localization of Snx41 was studied in a strain expressing Snx41-GFP fusion protein under native regulation. RFP-ATG8 was also expressed in the Snx41-GFP strain to examine the spatial relationship between autophagy and Snx41. RFP-Atg8 was found to be expressed at basal/uninduced levels in vegetative mycelia grown in rich medium, while Snx41-GFP was abundant as punctate or tubular structures, thus indicating an Atg8 (autophagy)-independent function for Snx41 (Fig. 2A). Upon nitrogen starvation, RFP-Atg8 was significantly induced and RFP-tagged vesicles or vacuoles (spherical and filamentous) became abundant and 27.6% of the Snx41-GFP vesicles overlapped with autophagosome/autophagic vacuoles (Fig. 2A; please refer to Materials and Methods for details on quantitation). About 22% of the Snx41-GFP puncta associated with the RFP-Atg8 tagged autophagosomes (Fig. 2A; vinblastine treatment to prevent autophagic vacuole formation35).
Figure 2. Snx41-GFP partially colocalizes with RFP-Atg8 (autophagosome and/or autophagic vacuole). (A) Confocal microscopic analysis of the Snx41-GFP/RFP-Atg8 strain grown in CM or in PMSF-containing MM-N (with or without Vinblastine). Arrows indicate Snx41-GFP association with autophagosomes and/or autophagic vacuoles. Asterisk denotes a spherical vacuole. Association between Snx41-GFP and autophagosomes is highlighted by arrowheads. Scale bar: 10 μm. (B) Snx41-GFP and RFP-Atg8 localization was examined in aerial hyphae (6–10 h post photo-induction) and in conidiophores (16–24 h post photo-induction). Arrows indicate Snx41-GFP association with autophagosomes and/or autophagic vacuoles. Scale bar: 5 μm. (C) Snx41-GFP and RFP-Atg8 localization during conidial germination (2–5 hpi), appressorium formation (6–12 hpi) and appressorium maturation (12–24 hpi). Arrows mark the association of Snx41-GFP with autophagosomes and/or autophagic vacuoles. Scale bar: 10 μm. (D) Montage of Movie 1 showing Snx41-GFP associated with autophagosomes and/or autophagic vacuoles during appressorium development. Arrowheads denote association between Snx41-GFP vesicles and autophagosomes labeled by RFP-Atg8. Arrows denote a ring-shaped tubular compartment labeled by Snx41-GFP at the surface of autophagic vacuoles. Scale bar: 10 μm.
During conidiation, Snx41-GFP partially associated with RFP-Atg8 in aerial hyphae (16.8%), conidiophores (22.7%) (Fig. 2B, denoted by arrows) and mature conidia (28.9%, Fig. 2C). During conidial germination Snx41-GFP puncta or short tubules associated with filamentous vacuoles labeled with RFP-Atg8 (18.9%, Fig. 2C). In nascent or mature appressorium, 12.6% or 28.3% of Snx41-GFP vesicles associated with RFP-Atg8 tagged autophagosome/autophagic vacuoles, respectively (Fig. 2C). Live imaging (Movie 1) showed RFP-Atg8 and Snx41-GFP puncta associate with each other and move together into the newly formed appressorium (Fig. 2D, arrowheads). In the conidium, RFP-Atg8 accumulated in larger vacuoles with dynamic tubular Snx41-GFP attached to or surrounding the vacuolar surface (Movie 1; Fig. 2D, arrows).
During blast disease initiation, punctate or tubular Snx41-GFP signal, similar to those seen in mycelia or conidia, were observed in penetration hyphae in rice leaf sheath (Fig. 3A). In some cases, we adjudged the Snx41-GFP to be proximal/peripheral to vacuoles (based on DIC images; Fig. 3A, arrowheads). Snx41-GFP localization in planta further supports a functional requirement during Magnaporthe pathogenesis.
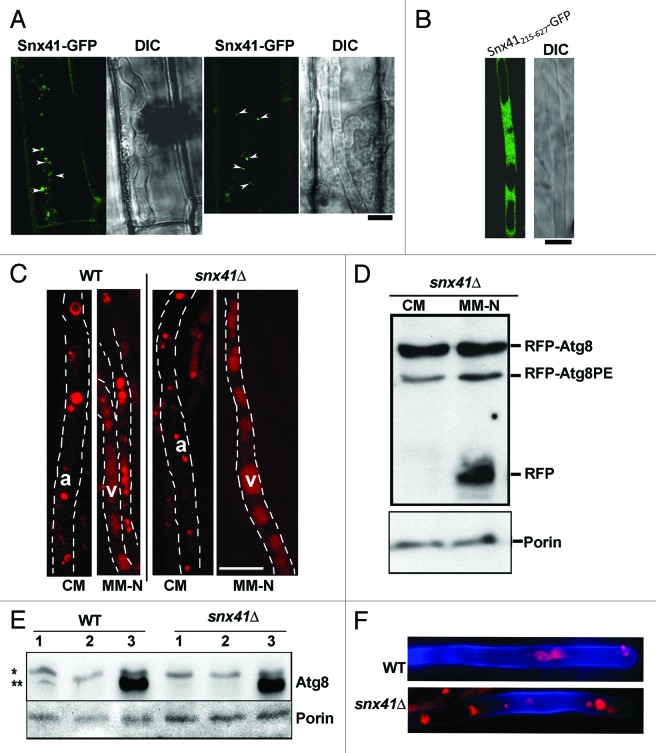
Figure 3. Subcellular localization of Snx41-GFP depends on the PX domain, and is dispensable for the autophagy pathway. (A) Subcellular localization of Snx41-GFP during in planta growth. A conidial suspension of the Snx41-GFP strain was inoculated on the surface of rice leaf sheath. Confocal microscopy was carried at 27–32 hpi. Arrowheads denote punctate Snx41-GFP signal at the margin of vacuoles in the invasive hyphae. Scale bar: 5 μm. (B) The PX domain is essential for vesicular localization of Snx41. Snx41215–627-GFP was localized uniformly in the cytosol in vegetative mycelia. Scale bar: 5 μm. (C) Mycelia of snx41Δ mutant expressing RFP-Atg8 were cultured in CM and MM-N (with 2mM PMSF, for 6 h) and visualized by confocal microscopy. RFP-Atg8 was barely detectable in CM-cultured mycelia, and it was significantly induced by nitrogen starvation. a, autophagosome; v, (autophagic) vacuole. Scale bar: 10 μm. (D) Post-translational processing of Atg8 is normal in snx41Δ. Immunoblot analysis with total lysates from CM- and MM-N- cultured snx41Δ (in RFP-ATG8 background), using anti-RFP antibody or anti-porin antibody. (E) Snx41 is not required for nonselective autophagy. Total lysates from the WT or the snx41Δ, grown in three different conditions, were analyzed by immunoblots with anti-Atg8 or anti-Porin antibody. “*” denotes full-length Atg8 and “**” marks the Atg8–PE. 1. CM culture; 2. MM-N without PMSF; 3. MM-N with PMSF. (F) Autophagy is naturally induced in the aerial hyphae of the wild type and the snx41Δ mutant. Confocal microscopy with RFP-Atg8 expressing snx41Δ, was performed at 6–12 h post photo-induction. Calcofluor White was co-stained to detect the cell wall. Scale bar: 5 μm.
Next, we verified whether the PtdIns(3)P-interacting PX domain of Snx41 contributes to (vesicular) membrane association/localization. We created an snx41ΔPX strain through targeted deletion of exons 1–3 of the SNX41 locus, which encode the PX domain, leaving exons 4–5 intact (amino acid 215–627) and fused in-frame with GFP, as the sole variant of Snx41 protein. The resultant Snx41215–627-GFP was predominantly cytosolic (Fig. 3B) in contrast to the full-length Snx41-GFP. Lack of the PX motif and/or the consequential mislocalization of the Snx41215–627-GFP to the cytosol (Fig. 3B) led to conidiation and pathogenicity defects similar to snx41Δ mutant. We conclude that the PX domain-dependent localization of Snx41 is essential for its proper function during Magnaporthe conidiation and pathogenicity.
Next, the interdependence between autophagy and Snx41-mediated vesicle trafficking was investigated. Localization of RFP-Atg8 was initially assessed in the snx41Δ mutant. In rich medium (CM), RFP-Atg8 remained uninduced and was sporadically seen as punctate structures, likely representing autophagosomes. Upon nitrogen starvation in the presence of PMSF (to inhibit vacuolar proteinase B36), RFP-Atg8 accumulated inside the vacuoles (Fig. 3C). The observation of vacuolar RFP signal indicates the successful delivery and fusion of autophagosome to the vacuole in the snx41Δ (Fig. 3C). Immunoblot analysis using anti-RFP antibody, with CM or MM-N cultured mycelia from the RFP-Atg8 expressing snx41Δ detected RFP-Atg8–PE as well as free RFP, judged by the mobility in SDS-PAGE (Fig. 3D). The N-terminal RFP tag was likely cleaved from the RFP-Atg8 fusion protein and accumulated in the vacuoles upon nitrogen starvation,10 indicating proper fusion of autophagosomes with the vacuole in snx41Δ. This result further supported the conclusion that autophagy was active and functional in the snx41Δ mutant. Therefore, Snx41 is not required for the induction or completion of autophagy. Likewise, the requisite increase in Atg8–PE levels upon nitrogen starvation in the presence of PMSF confirmed a functional (nonselective) autophagy in snx41Δ mutant (Fig. 3E). Furthermore, in the RFP-Atg8 expressing snx41Δ mutant, induction of autophagy and localization of autophagic vesicles in aerial hyphae was comparable to that seen in the wild-type strain (Fig. 3F). We conclude that lack of conidiation in the snx41Δ mutant is not due to any perturbation in autophagy per se.
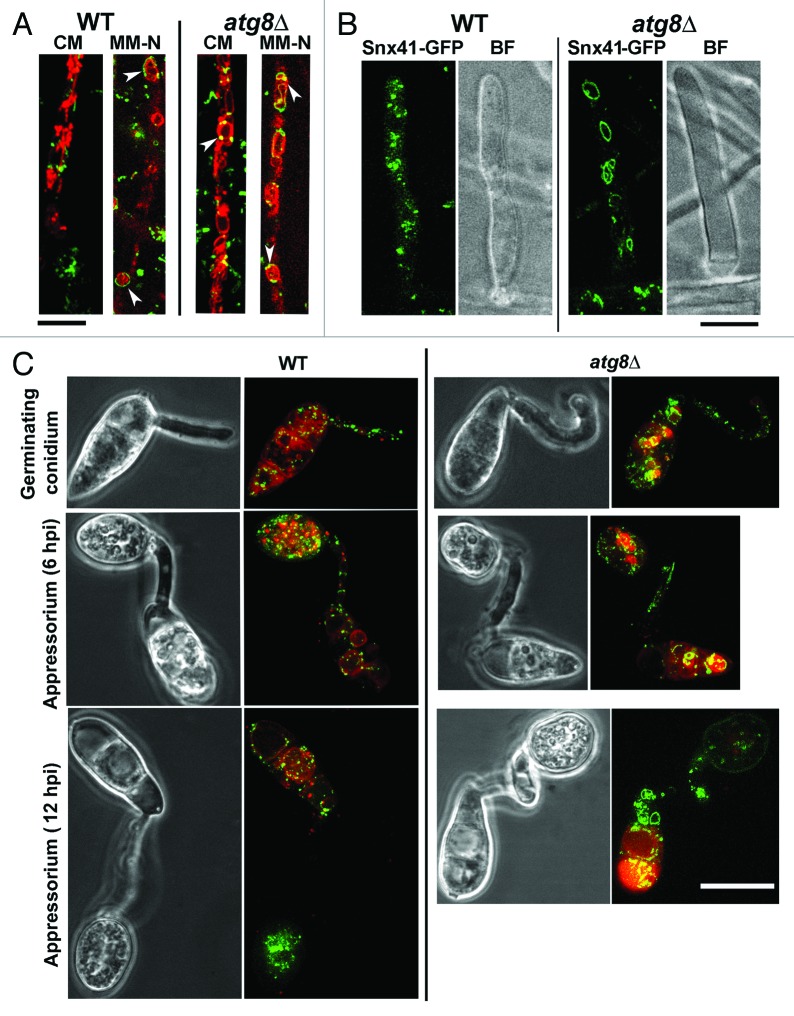
On the other hand, Snx41-GFP localization is likely independent of the autophagy pathway, since Snx41-GFP was ubiquitously expressed in vegetative hyphae as well as during nitrogen starvation (MM-N, with 2 mM PMSF; Fig. 4A), and in the aerial hyphae (10–12 h post photo-induction; Fig. 4B) in both the wild type and the atg8Δ mutant. Loss of autophagy did not appear to affect the vacuolar (surface or lumen) localization of Snx41-GFP in vegetative mycelia (cultured in CM or MM-N, and co-stained with FM4–64; Fig. 4A, arrowheads), aerial hyphae (Fig. 4B), or developing conidia (co-stained with Lysotracker Red DND99; Fig. 4C). We conclude that Snx41-GFP is present as puncta or short tubules that partially associate with autophagosomes or distribute at the surface of the vacuoles during vegetative and pathogenic development in Magnaporthe. Furthermore, the localization of Snx41-GFP was inferred to be independent of the autophagy pathway.
Figure 4. Subcellular localization of Snx41-GFP and its dependence on autophagy. (A) Snx41-GFP localizes to puncta or short tubules that associate with the vacuolar membrane. Snx41-GFP localization was assessed in WT or atg8Δ co-stained with FM4–64 to visualize the vacuolar compartment. Arrowheads indicate Snx41-GFP associated with autophagosomes and/or autophagic vacuoles. Scale bar: 10 μm. (B) Snx41-GFP localization was observed in aerial hyphae of WT or atg8Δ after 6–10 h post photo-induction. Scale bar: 5 μm. (C) Snx41-GFP localization during conidial germination (2–5 hpi), appressorium formation (6–12 hpi) and appressorium maturation (12–24 hpi). Lysotracker Red was used to visualize the vacuolar compartment. Scale bar: 10 μm.
Snx41 is required for pexophagy in Magnaporthe
Atg20/Snx42 was reported to be essential for pexophagy in Pichia,22 therefore we investigated whether the related Snx41 contributes to pexophagy in Magnaporthe too. Pexophagy was examined by immunoblot assay using antithiolase antibody in the wild type and the snx41Δ grown in BM + OL (olive oil; peroxisome biogenesis) or CM (glucose; pexophagy induction).25 As expected, thiolase accumulated in the WT in response to peroxisomal fatty acid metabolism, but was undetectable when glucose was present as a carbon source (Fig. S2C). Atg8 is known to be required for pexophagy in yeast37 and here we showed that Magnaporthe atg8Δ was similarly blocked in pexophagy too (Fig. S2C). Densitometry analysis revealed that thiolase reduction was 60% in the wild type upon pexophagy induction compared with peroxisome-biogenesis conditions (Fig. S2C), while in the atg8Δ mutant the reduction was 18% (Fig. S2C), indicating a significant delay/block in pexophagy. Similar pexophagy failure was evident in snx41Δ mutant, judged by the reduction of thiolase (20%) in response to pexophagy induction (Fig. S2C). Taken together, our data revealed that Snx41 plays an important role during pexophagy in Magnaporthe. We infer that Magnaporthe Snx41 likely shares the pexophagy function attributed to Atg20/Snx42 in budding yeast.
Snx41 cooperates with the gamma-glutamyl cycle during Magnaporthe conidiation
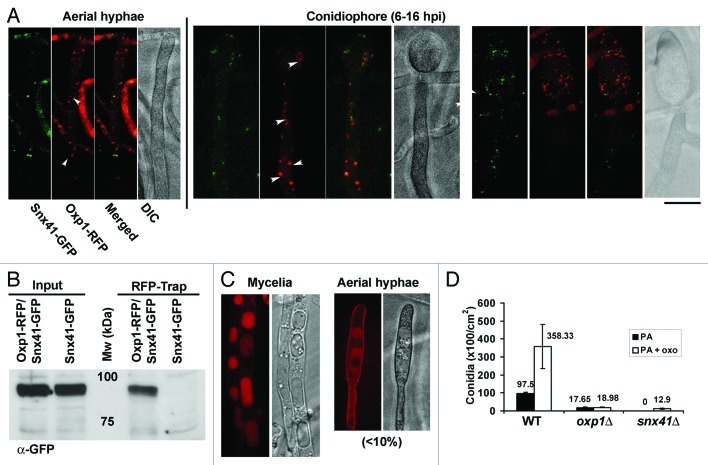
Next, we investigated whether retrieval trafficking or other vesicular trafficking pathway dependent on Snx41 is required for Magnaporthe conidiation. Using GFP-TrapA kit (Chromotek) and mass spectrometry, we identified Oxp1 as a protein that physically associates with Snx41-GFP in Magnaporthe. Oxp1 is an ATP-dependent oxoprolinase that catalyzes conversion of 5-oxoproline to glutamic acid in the gamma-glutamyl cycle.38 To determine whether Oxp1 and/or gamma-glutamyl cycle is involved in Magnaporthe conidiogenesis, we first generated a strain expressing Oxp1-RFP and Snx41-GFP and examined the interaction between Snx41 and Oxp1 by confocal microscopy. During conidiation, Oxp1-RFP localized to some punctate structures in the cytoplasm, and associated with 29 - 43% of Snx41-GFP localized puncta/tubules, in the aerial hyphae and conidiophores (Fig. 5A, arrowheads). Using RFP-Trap (Chromotek, RFP-Trap A kit) and the aforementioned Magnaporthe strain, we successfully pulled down Snx41-GFP and thus confirmed the physical interaction between Snx41 and Oxp1 (Fig. 5B). In contrast to the wild type, Oxp1-RFP lost punctate localization but appeared to be vacuolar in vegetative mycelia and occasionally in the aerial hyphae, in the snx41Δ mutant (Fig. 5C). Oxp1-RFP signal was weak or undetectable in snx41Δ aerial hyphae exposed to light for 6–16 h. Based on cell biology data, we conclude that Oxp1 depends on Snx41-based sorting and retrieval from the vacuoles, for proper localization and function in the cytoplasm. We construe that loss of Snx41 renders Oxp1 vacuolar.
Figure 5. The role of gamma-glutamyl cycle during Magnaporthe conidiation. (A) Oxp1-RFP and Snx41-GFP colocalize in aerial hyphae and conidiophore during conidiogenesis. Confocal microscopy was performed with conidiating culture at 6–16 h post photo-induction. Arrowheads denote colocalization of Oxp1-GFP and Snx41-GFP vesicles. Scale bar: 5 μm. (B) Snx41 physically interacts with Oxp1. Affinity purification with RFP-Trap beads using total protein lysates from Magnaporthe strains expressing Oxp1-RFP and Snx41-GFP, or Snx41-GFP alone, Immuno-blotting was performed with anti-GFP. (C) Oxp1-RFP is predominantly vacuolar in the snx41Δ mutant. In vegetative mycelia of snx41Δ, Oxp1-RFP was frequently seen inside the enlarged spherical vacuoles. In the aerial hyphae of snx41Δ, Oxp1-RFP was barely detectable, except for occasional vacuolar localization. (D) Bar chart depicting quantification of conidiation in the wild type (WT), oxp1Δ and snx41Δ grown on PA medium supplemented with 5-oxoproline. Mean values (± S.E.) presented as number of conidia were derived from three independent experiments (n = 30 colonies for each sample). Total conidia counts were performed five days post photo-induction.
To investigate the function of Oxp1 and/or the gamma-glutamyl cycle during Magnaporthe conidiation, we generated an oxp1Δ mutant and found that production of conidia was significantly reduced (Fig. 5D). Interestingly, exogenous addition of 5-oxoproline could significantly promote conidiation in both the wild type and the snx41Δ (p < 0.01), but not in the oxp1Δ (Fig. 5D). We propose that Snx41 associates with and likely retrieves vacuolar Oxp1, which participates in the gamma-glutamyl cycle in the cytoplasm and is necessary for proper conidiation in Magnaporthe.
Snx41-dependent regulation of antioxidant defense during in planta growth of Magnaporthe
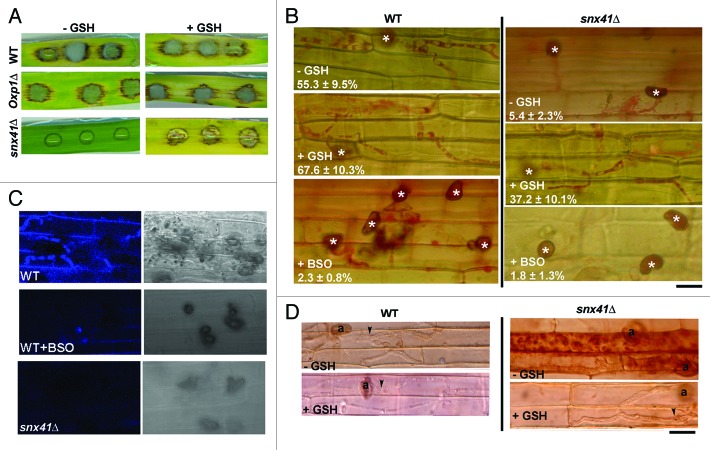
Using snx41Δ conidia obtained from 5-oxoproline-supplemented PA medium we performed barley leaf infection assays and found that snx41Δ mutant was nonpathogenic (Fig. 6A), which is consistent with the results of rice root infection assay (Fig. 1C). Microscopic observation showed retarded development of invasive hyphae, which were unable to cross the host cell walls and thus incapable of spread from the primary infection site to the neighboring cells (48 hpi; Fig. 6B).
Figure 6. The role of gamma-glutamyl cycle during Magnaporthe infection. (A) Barley leaf infection assay in the presence or absence of GSH, with conidia from the wild type, oxp1Δ (both PA grown) or the snx41Δ (5-oxoproline supplemented PA grown). (B) Microscopy observation of snx41Δ invasive hyphae developing in rice leaf sheath at 48 hpi, stained with acid fuschin for better visualization (mean values ± SE presented as percentage of appressoria differentiating invasive hyphae). BSO was used to pre-treat conidia for 16 h before inoculation on rice leaf sheath. “*” denotes appressorium. Arrowheads or arrows denote the invasive hyphae incapable or capable of crossing host cell wall respectively. Scale bar: 10 μm. (C) Monochlorobimane staining for glutathione during rice leaf sheath infection, at 27–30 hpi. (D) DAB staining indicated the ROS accumulation inside the infected cells of the rice leaf sheath by the snx41Δ mutants, but not by the wild type. (n = 50 appressoria examined in each instance). Scale bar: 10 μm.
Pathogenic microbes elicit innate immunity or defense response in plants, which is manifested through rapid accumulation of reactive oxygen species (ROS) and cell death at the site of pathogen invasion.39 Considering the connection between Snx41 and the gamma-glutamyl cycle, which produces reduced glutathione (GSH), an antioxidant, we hypothesized that the reduced infectivity of the snx41Δ might be due to inadequate production of GSH and the resultant weaker resistance against oxidative stress encountered in planta. To test whether the blocked invasive hyphal development of the snx41Δ was a consequence of host ROS, we added GSH (5 mM) to the snx41Δ conidial suspension on barley leaves at 24 hpi. Remarkably, GSH treatment led to a significant improvement in invasive growth in snx41Δ and elicited prominent blast symptoms in the host leaves (Fig. 6A). Invasive hyphae formation and spread was also significantly improved in rice leaf sheath infection assay at 48 hpi (Fig. 6B). Glutathione depletion using buthionine sulfoximine (BSO), a selective inhibitor of GSH biosynthesis, mimics the defective invasive hyphal development as seen in snx41Δ (Fig. 6B). We visualized GSH by monochlorobimane staining, which forms a fluorescent complex detectable by confocal microscopy. In the wild type, prominent GSH accumulation was evident in invasive hyphae and also diffused into the infected host cells; whereas GSH was absent or undetectable in the snx41Δ or BSO pre-treated wild-type infections (Fig. 6C). DAB staining showed that high ROS accumulated in the rice leaf sheath inoculated with the snx41Δ mutant, but not in the host cells infected by the wild type (Fig. 6D). When GSH was added to the snx41Δ conidial suspension, ROS levels were significantly reduced or undetectable in the infected rice leaf sheath (Fig. 6D). The atg8Δ mutant was reported as being nonpathogenic.40 Different from the snx41Δ mutant, the atg8Δ appressoria are unable to penetrate the rice sheath cells, and thus fail to trigger the HR reaction in the host (data not shown). Addition of GSH failed to suppress the penetration defects of the atg8Δ mutant (data not shown). Therefore the molecular function of Snx41, likely the regulation of redox signaling via retrieval trafficking, is different from that of Atg8 (autophagy) during Magnaporthe infection. An important observation was that the snx41Δ conidia (inoculated on barley leaf explant in the presence of GSH) were capable of initiating conidiation in planta (Fig. S2E).
We propose that Snx41, together with the gamma-glutamyl cycle, regulates amino acid metabolism to produce GSH, and is thus crucial for Magnaporthe response to counteract oxidative stress during biotrophic growth. Evidence supporting this hypothesis is that compared with the wild type and the atg8Δ mutant, the snx41Δ showed enhanced sensitivity toward menadione and paraquat, both of which are specific GSH-repressible oxidants (Fig. S4). In contrast, hydrogen peroxide is not a specific oxidant for GSH and the snx41Δ showed comparable sensitivity to it as the wild type and the atg8Δ (Fig. S4).
We concluded that the gamma-glutamyl cycle likely plays an important role in Magnaporthe conidiation and pathogenicity, by producing at least two important intermediate products, 5-oxoproline and GSH. Oxp1, a key enzyme of the gamma-glutamyl cycle, depends on Snx41-mediated retrieval trafficking for its function in Magnaporthe development.
Discussion
Distinct functions of Snx41 and Atg8
The conidiation and pathogenicity defects in the snx41Δ and the atg8Δ mutant are distinct: (1) Conidiation in the atg8Δ mutant could be restored through glucose or G6P supplementation, but not by 5-oxoproline. In contrast, 5-oxoproline but not glucose or G6P partially reinstated conidiation in snx41Δ. (2) The atg8Δ appressoria were defective in host penetration and thus incapable of triggering ROS build-up in the host. In contrast, the snx41Δ appressoria were capable of penetrating the host but could not develop invasive hyphae properly in planta, likely due to the inability to cope with host-derived ROS. Addition of GSH could promote snx41Δ invasive hyphal growth. Therefore, we propose that unlike Atg8, Snx41 carries out a different function (other than glycogen catabolism) during conidiation. In addition, and based on terminal phenotypic defects of the respective mutants, Snx41 acts at the host invasion step during Magnaporthe infection, likely after Atg8-dependent autophagic cell death of conidial cells post appressorium formation.40 (3) Subcellular localization of RFP-Atg8 and Snx41-GFP were shown to be distinct and independent from each other, but with a partial overlap. All these observations suggest that Snx41 may participate in a molecular pathway that is distinct and independent of bulk macroautophagy.
Snx41 functions in selective autophagy and autophagy-independent pathways during Magnaporthe development
Autophagy is required for proper conidiation in Magnaporthe, wherein it assists carbohydrate metabolism to meet the energy requirements during cellular differentiation.10 Magnaporthe snx41Δ mutant displayed more severe defects in conidiation compared with the autophagy-deficient atg8Δ, and such defects could not be restored by exogenous glucose, indicating that Snx41 may play a different function than autophagy-assisted nutrient catabolism.10,11 By systematic characterization, we have shown that the nonselective autophagy was normal in the snx41Δ mutant. However, pexophagy, the selective autophagic degradation of peroxisomes, was delayed/blocked in the snx41Δ mutant. After obtaining snx41Δ conidia, via exogenous 5-oxoproline, we further assessed the pexophagy defects therein at the developmental stages when pexophagy is expected to occur in the wild type (Fig. S2D, arrowhead). During appressorium maturation in the snx41Δ (20–22 hpi), numerous GFP-SRL puncta/vesicles, corresponding to the peroxisomes, were observed outside the vacuoles in the conidium (Fig. S2D, arrow), indicating a reduction/loss of pexophagy.
Apart from pexophagy, Snx41 carries out autophagy-independent function in Magnaporthe. Evidence supporting this conclusion includes the partial association of Snx41-tagged vesicles with autophagic vesicles, Atg8-independent localization of Snx41, and the identification of a potential cargo of Snx41-dependent vesicle trafficking, Oxp1. Such varied and additional functions of Magnaporthe Snx41 are likely performed by separate proteins in yeast, viz Snx42/Atg20 (pexophagy) andSnx41 (endosomal sorting). Meijer et al. (2007) failed to identify a true ortholog of Atg20 in Magnaporthe and other filamentous fungi. However, we believe that Magnaporthe, MGG_12832 likely serves a dual role given its extensive relatedness to Atg20 and Snx41.
Peroxisomal lipid metabolism is essential for melanin layer formation and thus for appressorium function in Magnaporthe, but the turnover/degradation of peroxisomes via pexophagy, is likely dispensable. Thus, it remains to be seen whether pexophagy is required for conidiation and pathogenesis in Magnaporthe. Besides the snx41Δ mutant, other pexophagy-deficient mutants would be needed to investigate specific requirement (if any) of pexophagy in Magnaporthe. In contrast, Snx41-mediated trafficking of Oxp1 (and the resultant regulation of gamma-glutamyl cycle), on the other hand, may be essential for proper conidiation and in planta growth.
Snx41-mediated retrieval of Oxp1 during Magnaporthe conidiation
Affinity purification indicated a physical interaction between Snx41 and Oxp1, an ATP-dependent oxoprolinase that catalyzes conversion of 5-oxoproline to glutamic acid in the gamma-glutamyl cycle. Mislocalization of Oxp1-RFP to the vacuoles in the snx41Δ suggests that Snx41-dependent active sorting is necessary to retain Oxp1 on vesicles in the cytoplasm. It is possible that such vacuolar retention of Oxp1 in the snx41Δ mutant causes negative feedback inhibition of the gamma-glutamyl cycle, leading to reduction (or block) in synthesis of 5-oxoproline. This implies that the snx41Δ mutant possesses limited amounts of 5-oxoproline and likely explains the paradox that addition of substrate (but not the product) of Oxp1 suppresses the conidiation defects in the snx41Δ mutant. Alternately, a high level of 5-oxoproline accumulates in the snx41Δ mutant and triggers a functionally redundant, albeit a lower affinity enzyme, that bypasses the requirement of Oxp1 for the completion of the gamma-glutamyl cycle. However, the latter possibility may not be well supported by the observation that conidiation in 5-oxoproline supplemented oxp1Δ is still very limited even at higher concentrations. Addition of 5-oxoproline to the wild type promotes excessive conidiation, indicating that availability/biosynthesis of 5-oxoproline is otherwise a likely rate-limiting step for conidia formation in the presence of wild-type Oxp1 function. Direct measurement of the intracellular levels of 5-oxoproline and glutathione in wild type, snx41Δ and oxp1Δ would certainly help resolve such paradoxical issues pertaining to 5-oxoproline synthesis and usage in Magnaporthe. In the future, we hope to elucidate the exact function of the gamma-glutamyl cycle, and its intrinsic regulation by Snx41, during asexual development in Magnaporthe. As for overall differences in the level of conidiation defects in snx41Δ and oxp1Δ mutants, there may be other as yet unidentified targets of Snx41-mediated trafficking that are functionally redundant with Oxp1 and contribute to conidia formation in Magnaporthe.
ROS scavenging and survival of Magnaporthe in host plants
Gamma-glutamyl cycle produces two important intermediates serving important physiological functions. 5-oxoproline restores conidiation in the snx41Δ mutant, although the exact mechanism remains elusive and would be the subject of future research. GSH, the antioxidant, is involved in counteracting host resistance response during invasive growth. Gamma-glutamyl cycle regulates absorption and metabolism of three amino acids: glutamic acid, cysteine and glycine.41 None of these amino acids (individually or combinations therein) were able to restore pathogenesis in the snx41Δ (data not shown), leading us to propose that GSH may be actively synthesized (and probably secreted into the host42) to act as an ROS scavenger, and thus efficiently suppress host response to ensure invasive hyphal development during Magnaporthe infections. Regulation of GSH synthesis and/or trafficking by Snx41 (if any) would form the basis of future studies in this project.
ROS seems to play a major role in host resistance during incompatible interactions between WT (B157) Magnaporthe and the blast-resistant rice strain NIL127 (Pi-9). Brown aggregates (probably due to ROS-induced cell death) were seen in the primary host cell invaded by WT appressoria (Fig. S2F, upper panel), and none or very few invasive hyphae were formed inside the resistant host tissue. Successful invasive hyphae (that lacked the brown aggregates) spreading to the neighboring cells were frequently observed when GSH was included in the inoculation mix (Fig. S2F, lower panel). Thus, the incompatible interaction between Magnaporthe B157 and and NIL127 is mediated in part by ROS generation leading to host cell death at the biotrophy interface. This underscores the importance of GSH-mediated counter-defense employed by Magnaporthe; and future research would aim to elucidate the exact role and mechanism of Snx41 in such redox regulation during invasive growth of an important fungal pathogen of cereal crops.
Materials and Methods
Fungal strains and growth conditions
Magnaporthe wild-type strain B157 (Field isolate, mat1-2) was obtained from the Directorate of Rice Research (Hyderabad). Magnaporthe strains were propagated on prune-agar (PA) medium or complete medium (CM) as described.23,43 Sucrose-, G6P- or 5-oxoproline- supplemented prune-agar medium for assessing conidiation in atg8Δ and snx41Δ was the PA medium with lactose 5 g/L, and either sucrose at 10 g/L, or G6P at 5mM, or 5-oxoproline (L-Pyroglutamic acid, Sigma-Aldrich, 83160) at 2.5 mM. The composition of MM and MM-N (used for nitrogen starvation) was as reported earlier.44 Two-day old liquid CM-grown mycelia were ground in liquid nitrogen for the isolation of nucleic acids. To assess the growth and colony characteristics, Magnaporthe isolates were cultivated on PA medium, at 28°C for a week. Mycelia used for total protein extraction was obtained by growing the relevant strains in liquid CM for 2–3 d, with gentle shaking, followed by inoculation in MM or MM-N for about 16 h. To induce peroxisome biogenesis or pexophagy, vegetative hyphae of respective strains grown in CM liquid medium for 3 d, were shifted to basal medium containing olive oil as the sole carbon source (BM+OL; for peroxisome biogenesis), and then were shifted back to CM (to induce pexophagy).
For quantitative analysis of conidiation, colonies were cultivated on PA medium in the dark for 2 d, followed by a 5-d growth cycle under constant illumination at room temperature. The surface of the colonies was then scraped with inoculation loops in the presence of water and the fungal biomass was harvested in Falcon™ Conical Tubes (BD Biosciences). The suspension was vortexed thoroughly to ensure maximum detachment of conidia from mycelia, and then filtered through two layers of Miracloth (Calbiochem). Conidia thus collected were washed twice with and finally resuspended in sterile water. The radius of the colonies was initially measured to calculate the surface area of the colonies (area = πr2). Conidia production in a given colony was quantified using a hemocytometer and reported as the total number of conidia present per unit area of the colony [conidia (×100/cm2)].
Infection assays
For rice root infection, surface-sterilized rice seeds were allowed to germinate and grow in direct contact with the fungal mycelial plugs. Resultant rice roots were then washed with water and evaluated for the black or browning lesions as disease symptoms after two weeks. For barley leaf explants infection, 20 microliter droplets of conidial suspension (containing 2000 or 1000 or 500 conidia per droplet) were inoculated onto the leaf surface and incubated in a growth chamber (22°C, 90% humidity and 16 h illumination/d). Disease symptoms were assessed 7–10 dpi. Rice leaf sheath infection was performed with conidial suspension at concentration of 105/ml. The inoculated rice leaf sheath was kept in growth chamber at 20°C and 90% humidity. Acid fuchsin (1%, Sigma-Aldrich, F8129) in ethanol was used to stain (37°C for 2 h) the infection hyphae in rice leaf sheath.
Nucleic acid and protein-related manipulation
Standard molecular manipulations were performed as described.45 Fungal genomic DNA was extracted with the MasterPure™ Yeast DNA Purification Kit (Epicenter, MPY80200) following the manufacturer’s instructions. For plasmid extraction from E. coli, the high-speed mini kit from Geneaid (PD300) was used. The recombinant DNA was examined by nucleotide sequencing using the ABI Prism Big Dye terminator method (PE-Applied Biosystems). Homology searches of DNA/protein sequences were performed using BLAST.46 The primers used for gene deletion and RFP or GFP tagging are listed in Table 1. Underlined text represents the restriction enzyme site introduced for cloning purposes.
Table 1. Oligonucleotide primers used in this study. Restriction enzyme sites introduced for cloning purposes are underlined.
| Gene (Locus) | Description | Enzyme sites | Primer sequence (5′–3′) |
|---|---|---|---|
|
MoSNX41 (MGG_12832) |
Deletion construct |
HindIII PstI BamHI SacI |
GAGAGTGTTAAGCTTGCCGGGGCAACATCTTGTTGCG GAGACTGTTCTGCAGGATGGAGCCGCACGGAGCTTAG GAGAGTGAGGATCCGTACCGCCGCTACGTGAGAGCG GAGTGAGAGCTCCCGGTACAGTCCGGGCGGGTTC |
| |
C-terminal tagging with GFP |
PstI NcoI EcoRI EcoRI |
GAGACTGTTCTGCAGGTCATCACTGGCGGCAGATCTT GAGTGACCATGGGAGACTCGTCGATCTTGTTGATC GAGTGAGAATTCGTACCGCCGCTACGTGAGAGCG GAGTGAGAATTCCTGCCGGCGATGATTGCGCTGG |
|
OXP1(MGG_00771) |
Deletion construct |
SacI BamHI HindIII HindIII |
GAGTGAGAGCTCCCGATGTCAAACTTCCTAGGTT GAGAGTGGATCCCTCTCTGGCTGCTCGGTTCAGCG GAGAGTAAGCTTGGGATGATATGAAGTACGCCGAG GAGAGTAAGCTTGTTATTTCTCCTCAGAGGCGGG |
| C-terminal tagging with RFP RFP: |
XhoI NcoI NcoI SacI |
GAGTGACTCGAGTGCCCCTGAGGCGGTTACTTAC GAGTGACCATGGGTGCCTGCAACGCCATCTCCTCCC GAGTGACCCCATGGACAACACCGAGGACGTCATCAAG GAGTGACCGAGCTCCTACTGGGAGCCGGAGTGGCGGG |
For total protein extractions, MM+N or MM-N grown mycelia were ground to a fine powder in liquid nitrogen and re-suspended in 0.3 ml of extraction buffer (10 mM Na2HPO4 pH 7.0, 0.5% SDS, 1mM DTT and 1 mM EDTA). Lysates were cleared by centrifugation at 12000 g for 30 min at 4°C. Protein samples from each extract (100 mg) were fractionated by SDS-PAGE, transferred onto a PVDF membrane (Millipore) and immunoblotted with appropriate primary antibody (anti-GFP: Invitrogen-Molecular Probes, A6455; anti-DsRed: Clontech, 632496; anti-Atg8: Abcam, ab4753-250; anti-ApeI: a gift from Suresh Subramani’s lab; anti-Thiolase and anti-Porin: a gift from the Jedd lab) at recommended dilutions. Secondary antibody conjugated to horseradish peroxidase was used at 1:20000 dilution. The Amersham ECL™ Kit (GE, RPN2135) was used to detect the chemiluminescent signals in western and Southern blotting experiments.
Generation of deletion mutants or epitope tagged strains
Fog gene deletion of SNX41 and OXP1, genomic DNA fragments (about 1 kb each) representing the 5′ and 3′ UTR of each gene were amplified by PCR, ligated sequentially so as to flank the HPH1 cassette. cDNAs were generated with AMV reversed transcriptase (Roche, 11495062001) and amplified by standard PCR. For Snx41-GFP and Oxp1-RFP constructs, the 1 Kb fragment just proximal to the translation stop codon in each gene was amplified and ligated with PCR-amplified GFP or RFP ORF respectively, in one step into the vectors (pFGL385 with the ILV1 cassette for Snx41-GFP, and pFGL97 with the BAR cassette for Oxp1-RFP). The 1 Kb SNX41 or OXP1 fragment immediately downstream of the stop codon was amplified and ligated into the corresponding plasmids so as to flank the fungal selection marker. The deletion construct or epitope-tagging construct for each of abovementioned genes was introduced into Magnaporthe strain to specifically replace the target genes with fungal selection markers, or with the GFP/RFP-tagged allele.
Staining protocols and microscopy
GFP epifluorescence was observed using a Zeiss LSM510 inverted confocal microscope (Carl Zeiss, Inc.) equipped with a 30 mW argon laser. The objective used was a 100 × Achromat (n.a. 1.25) oil immersion lens. EGFP was imaged with 488 nm wavelength laser excitation, using a 505–530 nm band pass emission filter or a 505 nm long-pass emission filter, while RFP imaging used 543 nm laser and a 560 nm long-pass emission filter.
To label acidified autophagic vacuoles, stained with 50 nM LysoTracker Green DND99 solution (Invitrogen-Molecular Probes, L7528) for 5 min at 37°C. Staining with FM4-64 (Invitrogen-Molecular Probes, T-13320) was performed with CM or MM-N cultured fungal mycelia, with FM4–64 added to a final concentration of 2 mM, and incubated for 30 min at 37°C, followed by a thorough wash with sterile water and additional 45 min incubation at 37°C before confocal microscopy.
Mycelial growth, treatment, collection and processing for TEM analysis of nitrogen starved wild type or the snx41Δ mutant was as described10 and samples examined using a JEM1400 electron microscope (Jeol) at 120 kV. Images were captured with a GATAN Orius SC200 camera and processed with Adobe Photoshop 7.0.1.
Image processing and analysis
For quantification of colocalization between RFP-Atg8 and Snx41-GFP or between Snx41-GFP and Oxp1-RFP, colocalization regions were selected by ImageJ’s Plug-in “colocalization test,” with the threshold for RFP and GFP channel set at 30. The region of colocalization was divided by the region of GFP (Snx41 vesicles) to give the percentage of Snx41 that colocalized with RFP-Atg8 (autophagosome/autophagic vacuole) or with Oxp1-RFP.
For densitometry analysis of immunoblot with anti-thiolase, signal density of lane3 (pexophagy-inducing condition without PMSF) and of lane1 (peroxisome biogenesis condition) from each strain were measured with ImageJ Measure. Percentage reduction of thiolase was calculated by 1-densitylane3/densitylane1.
Supplementary Material
Acknowledgments
We thank Meredith Calvert, Xuezhi Ouyang (TLL) and Kay En Low (Jeol Asia) for excellent EM support. We thank the Fungal Patho-biology group and Suresh Subramani for helpful discussion and suggestions. Y.Z.D acknowledges support from the Singapore Millennium Foundation. This work was funded by intramural grants from the Temasek Life Sciences Laboratory, Singapore.
Glossary
Abbreviations:
- Cvt
cytoplasm to vacuole targeting
- dpi
days post inoculation
- GSH
reduced glutathione
- Hpi
hours post inoculation
- HR
hypersensitive response
- LC3
microtubule-associated protein 1 light chain 3
- ORF
open reading frame
- PAS
phagophore assembly site
- PtdIns3P
phosphatidylinositol-3-phosphate
- ROS
reactive oxygen species
- Snx
sorting nexin
- Vps
vacuolar protein sorting
- WT
wild type
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/20217
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/20217
References
- 1.Ou SH. Rice Diseases. Surrey, UK, 1985. [Google Scholar]
- 2.Teng PS, Klein-Gebbinck HW. H P. An analysis of the blast pathosystem to guide modeling and forecasting. Rice Blast Modeling and Forecasting. Manila, Philippines: International Rice Research Institute, 1991:1-30. [Google Scholar]
- 3.Lee YH, Dean RA. cAMP Regulates Infection Structure Formation in the Plant Pathogenic Fungus Magnaporthe grisea. Plant Cell. 1993;5:693–700. doi: 10.1105/tpc.5.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert RD, Johnson AM, Dean RA. Chemical signals responsible for appressorium formation in the rice blast fungus Magnaporthe grisea. Physiol Mol Plant Pathol. 1996;48:335–46. doi: 10.1006/pmpp.1996.0027. [DOI] [Google Scholar]
- 5.Cole GT. Models of cell differentiation in conidial fungi. Microbiol Rev. 1986;50:95–132. doi: 10.1128/mr.50.2.95-132.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee K, Singh P, Chung WC, Ash J, Kim TS, Hang L, et al. Light regulation of asexual development in the rice blast fungus, Magnaporthe oryzae. Fungal Genet Biol. 2006;43:694–706. doi: 10.1016/j.fgb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Shi Z, Leung H. Genetic Analysis of Sporulation in Magnaporthe grisea by Chemical and Insertional Mutagenesis. Mol Plant Microbe Interact. 1995;8:949–59. doi: 10.1094/MPMI-8-0949. [DOI] [Google Scholar]
- 8.Odenbach D, Breth B, Thines E, Weber RW, Anke H, Foster AJ. The transcription factor Con7p is a central regulator of infection-related morphogenesis in the rice blast fungus Magnaporthe grisea. Mol Microbiol. 2007;64:293–307. doi: 10.1111/j.1365-2958.2007.05643.x. [DOI] [PubMed] [Google Scholar]
- 9.Liu H, Suresh A, Willard FS, Siderovski DP, Lu S, Naqvi NI. Rgs1 regulates multiple Galpha subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. EMBO J. 2007;26:690–700. doi: 10.1038/sj.emboj.7601536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng YZ, Ramos-Pamplona M, Naqvi NI. Autophagy-assisted glycogen catabolism regulates asexual differentiation in Magnaporthe oryzae. Autophagy. 2009;5:33–43. doi: 10.4161/auto.5.1.7175. [DOI] [PubMed] [Google Scholar]
- 11.Deng YZ, Naqvi NI. A vacuolar glucoamylase, Sga1, participates in glycogen autophagy for proper asexual differentiation in Magnaporthe oryzae. Autophagy. 2010;6:455–61. doi: 10.4161/auto.6.4.11736. [DOI] [PubMed] [Google Scholar]
- 12.Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol Res Pract. 2006;202:631–8. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy. Microsc Res Tech. 2004;64:10–20. doi: 10.1002/jemt.20046. [DOI] [PubMed] [Google Scholar]
- 15.Tolkovsky AM. Mitophagy. Biochim Biophys Acta. 2009;1793:1508–15. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Sakai Y, Oku M, van der Klei IJ, Kiel JA. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta. 2006;1763:1767–75. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 17.Klionsky DJ, Cregg JM, Dunn WA, Jr., Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–45. doi: 10.1016/S1534-5807(03)00296-X. [DOI] [PubMed] [Google Scholar]
- 18.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem. 2002;277:30198–207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan J, Stromhaug PE, George MD, Habibzadegah-Tari P, Bevan A, Dunn WA, Jr., et al. Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol Biol Cell. 2001;12:3821–38. doi: 10.1091/mbc.12.12.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–30. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ano Y, Hattori T, Oku M, Mukaiyama H, Baba M, Ohsumi Y, et al. A sorting nexin PpAtg24 regulates vacuolar membrane dynamics during pexophagy via binding to phosphatidylinositol-3-phosphate. Mol Biol Cell. 2005;16:446–57. doi: 10.1091/mbc.E04-09-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramos-Pamplona M, Naqvi NI. Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol Microbiol. 2006;61:61–75. doi: 10.1111/j.1365-2958.2006.05194.x. [DOI] [PubMed] [Google Scholar]
- 24.Farré JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–76. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nazarko TY, Polupanov AS, Manjithaya RR, Subramani S, Sibirny AA. The requirement of sterol glucoside for pexophagy in yeast is dependent on the species and nature of peroxisome inducers. Mol Biol Cell. 2007;18:106–18. doi: 10.1091/mbc.E06-06-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nazarko TY, Farré JC, Subramani S. Peroxisome size provides insights into the function of autophagy-related proteins. Mol Biol Cell. 2009;20:3828–39. doi: 10.1091/mbc.E09-03-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takano Y, Asakura M, Sakai Y. Atg26-mediated pexophagy and fungal phytopathogenicity. Autophagy. 2009;5:1041–2. doi: 10.4161/auto.5.7.9316. [DOI] [PubMed] [Google Scholar]
- 28.Kershaw MJ, Talbot NJ. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc Natl Acad Sci U S A. 2009;106:15967–72. doi: 10.1073/pnas.0901477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellu AR, Komori M, van der Klei IJ, Kiel JA, Veenhuis M. Peroxisome biogenesis and selective degradation converge at Pex14p. J Biol Chem. 2001;276:44570–4. doi: 10.1074/jbc.M107599200. [DOI] [PubMed] [Google Scholar]
- 30.Zutphen T, Veenhuis M, van der Klei IJ. Pex14 is the sole component of the peroxisomal translocon that is required for pexophagy. Autophagy. 2008;4:63–6. doi: 10.4161/auto.5076. [DOI] [PubMed] [Google Scholar]
- 31.Hettema EH, Lewis MJ, Black MW, Pelham HR. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 2003;22:548–57. doi: 10.1093/emboj/cdg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orlowski M, Meister A. The gamma-glutamyl cycle: a possible transport system for amino acids. Proc Natl Acad Sci U S A. 1970;67:1248–55. doi: 10.1073/pnas.67.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Njålsson R, Norgren S. Physiological and pathological aspects of GSH metabolism. Acta Paediatr. 2005;94:132–7. doi: 10.1080/08035250410025285. [DOI] [PubMed] [Google Scholar]
- 34.Ristoff E, Larsson A. Patients with genetic defects in the gamma-glutamyl cycle. Chem Biol Interact. 1998;111-112:113–21. doi: 10.1016/S0009-2797(97)00155-5. [DOI] [PubMed] [Google Scholar]
- 35.Gordon PB, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun. 1988;151:40–7. doi: 10.1016/0006-291X(88)90556-6. [DOI] [PubMed] [Google Scholar]
- 36.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–11. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manjithaya R, Nazarko TY, Farré JC, Subramani S. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 2010;584:1367–73. doi: 10.1016/j.febslet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar A, Bachhawat AK. OXP1/YKL215c encodes an ATP-dependent 5-oxoprolinase in Saccharomyces cerevisiae: functional characterization, domain structure and identification of actin-like ATP-binding motifs in eukaryotic 5-oxoprolinases. FEMS Yeast Res. 2010;10:394–401. doi: 10.1111/j.1567-1364.2010.00619.x. [DOI] [PubMed] [Google Scholar]
- 39.Apostol I, Heinstein PF, Low PS. Rapid Stimulation of an Oxidative Burst during Elicitation of Cultured Plant Cells : Role in Defense and Signal Transduction. Plant Physiol. 1989;90:109–16. doi: 10.1104/pp.90.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312:580–3. doi: 10.1126/science.1124550. [DOI] [PubMed] [Google Scholar]
- 41.Osuji GO. Glutathione turnover and amino acid uptake in yeast: evidence for the participation of the gamma-glutamyl cycle in amino acid transport. FEBS Lett. 1979;105:283–5. doi: 10.1016/0014-5793(79)80630-4. [DOI] [PubMed] [Google Scholar]
- 42.Perrone GG, Grant CM, Dawes IW. Genetic and environmental factors influencing glutathione homeostasis in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:218–30. doi: 10.1091/mbc.E04-07-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soundararajan S, Jedd G, Li X, Ramos-Pamploña M, Chua NH, Naqvi NI. Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell. 2004;16:1564–74. doi: 10.1105/tpc.020677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–90. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1989. [Google Scholar]
- 46.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.