Abstract
Mitoribosome in mammalian cells is responsible for synthesis of 13 mtDNA-encoded proteins, which are integral parts of four mitochondrial respiratory chain complexes (I, III, IV and V). ERAL1 is a nuclear-encoded GTPase important for the formation of the 28S small mitoribosomal subunit. Here, we demonstrate that knockdown of ERAL1 by RNA interference inhibits mitochondrial protein synthesis and promotes reactive oxygen species (ROS) generation, leading to autophagic vacuolization in HeLa cells. Cells that lack ERAL1 expression showed a significant conversion of LC3-I to LC3-II and an enhanced accumulation of autophagic vacuoles carrying the LC3 marker, all of which were blocked by the autophagy inhibitor 3-MA as well as by the ROS scavenger NAC. Inhibition of mitochondrial protein synthesis either by ERAL1 siRNA or chloramphenicol (CAP), a specific inhibitor of mitoribosomes, induced autophagy in HTC-116 TP53+/+ cells, but not in HTC-116 TP53−/− cells, indicating that tumor protein 53 (TP53) is essential for the autophagy induction. The ROS elevation resulting from mitochondrial protein synthesis inhibition induced TP53 expression at transcriptional levels by enhancing TP53 promoter activity, and increased TP53 protein stability by suppressing TP53 ubiquitination through MAPK14/p38 MAPK-mediated TP53 phosphorylation. Upregulation of TP53 and its downstream target gene DRAM1, but not CDKN1A/p21, was required for the autophagy induction in ERAL1 siRNA or CAP-treated cells. Altogether, these data indicate that autophagy is induced through the ROS-TP53-DRAM1 pathway in response to mitochondrial protein synthesis inhibition.
Keywords: ERAL1, mitoribosome, autophagy, ROS, TP53, DRAM1, chloramphenicol
Introduction
Autophagy is one of the main mechanisms for maintaining cellular homeostasis. During autophagy, unused long-lived proteins, damaged organelles, and even invasive pathogens are sequestered into double-membrane vesicles, called autophagosomes. The autophagosome fuses with a lysosome to form an autolysosome where its contents are degraded via acidic lysosomal hydrolases.1-3 Autophagy can be activated in response to extra- or intracellular stress and signals such as starvation, growth factor deprivation, ER stress and pathogen infection.1 Growing evidence has shown that autophagy is closely associated with human disease and physiology, such as cancer, neurodegeneration, microbial infection and aging.2 Damaged and superfluous mitochondria are predominantly cleared by autophagy, while dysfunction of mitochondria also plays an important role in the activation of autophagy.4,5 Reactive oxygen species (ROS) are generally small, short-lived and highly reactive molecules produced by ionizing radiation of biological molecules. Accumulation of ROS, the byproduct of respiration in mitochondria, is an oxidative stress associated with various cellular processes, including autophagy.5
Human mitochondrial DNA (mtDNA) encodes two rRNAs, 22 tRNAs and 13 proteins. All of the 13 proteins are essential subunits of four mitochondrial respiratory chain complexes (I, III, IV and V). Complex II is the only respiratory chain complex without mtDNA-encoded subunits. Synthesis of these 13 proteins is performed by mitoribosomes, which are 55S particles composed of a small 28S and a large 39S subunit. All of the ~80 mitochondrial ribosomal proteins (MRPs) are the products of nuclear genes and are imported into the mitochondrial matrix.6,7 To assemble the mitochondrial ribosome, the rRNA synthesis by the mitochondrial transcription machinery needs to coordinate with the nuclear expression of MRPs. Components of the mitochondrial translational machinery (translational factors, elongation factors and mitoribosome) are distinct from eukaryotic ones in cytosol and generally resemble the bacterial counterparts.7,8
The homologs of Escherichia coli Ras-like protein (ERA) consist of a conserved GTPase superfamily. ERA was originally reported as a bacterial homolog of RAS, but it is distinguished from RAS by containing not only a GTPase domain but also an hnRNPK homology (KH) domain, which can bind to RNA.9 Almost all of the sequenced bacterial genomes have the gene encoding the ERA protein. Deletion of era is lethal in bacteria indicating that the era gene is essential. Bacterial ERA binds to the 3′ end of 16S rRNA as a chaperone for 16S rRNA processing and maturation.10 ERA also plays a role during the final stages of the 30S subunit assembly and inhibits the formation of a translation initiation complex on a prematurely assembled 30S subunit.11 DNA database searches and cDNA cloning studies have shown the existence of ERA homologs in eukaryotic species including human, mouse, chicken, Drosophila, Caenorhabditis elegans and Antirrhinum majus.12-14 ERG, the ERA homolog in Antirrhinum is required for embryonic viability.13 Deletion of chicken ERA (GdERA) in lymphoma B-cell line DT40 induces cell apoptosis, which is blocked by BCL2 expression.15 It has been reported recently that human ERA, termed ERAL1, locates in the mitochondria matrix and associates with the 28S small mitoribosomal subunit, where it acts as a chaperone for the 12S mt-rRNA. Depletion of ERAL1 leads to instability of 12S mt-rRNA and a consequent loss of newly synthesized 28S subunit, resulting in mitochondrial protein synthesis inhibition.16,17
In this study, we demonstrate that ERAL1 knockdown inhibits protein synthesis in mitochondria, leading to ROS accumulation and autophagy induction in mammalian cells. ERAL1 knockdown resulted in LC3-I to LC3-II conversion and autophagic vacuole formation, the hallmarkers of autophagy, all of which were blocked by the autophagy inhibitor 3-MA as well as by NAC, a specific scavenger of ROS. Moreover, inhibition of mitochondrial protein synthesis by the mitoribosome inhibitor CAP also induced autophagy in a ROS-dependent manner. ROS enhanced tumor protein 53 (TP53) expression at both transcriptional and post-translational levels to activate TP53-DRAM1 signaling, which was required for the autophagy induction. Therefore, mitochondrial protein synthesis inhibition induces autophagy through the ROS-TP53-DRAM1 pathway.
Results
ERAL1 knockdown induces autophagy in HeLa cells
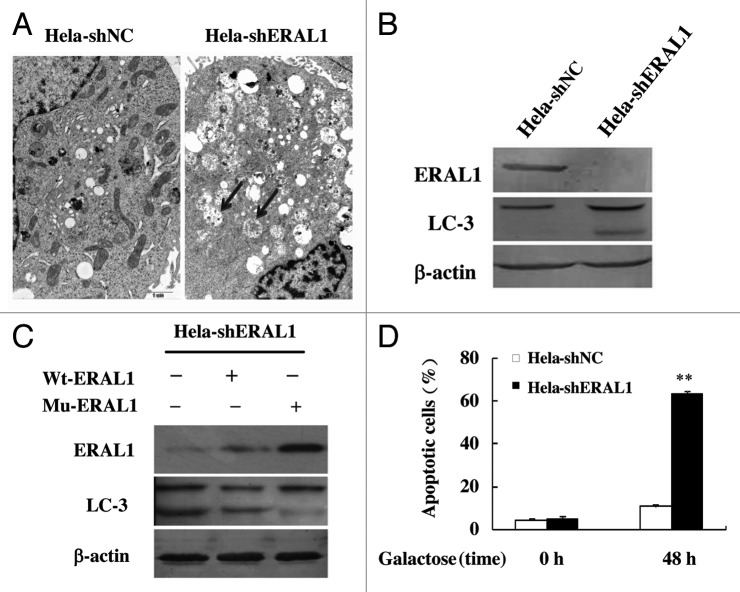
Human ERAL1, a member of the conserved ERA protein family, has been reported to locate in the mitochondria matrix as a novel nuclear-encoded mitoribosome assembly factor associated with mitochondrial 12S rRNA and playing an important role in the formation of 28S mitoribosomal small subunit.16,17 We generated a HeLa cell line with stable ERAL1 knockdown by expressing ERAL1-specific shRNA, termed HeLa-shERAL1, and a control HeLa cell line expressing scramble shRNA, termed HeLa-shNC. The growth rate of HeLa-shERAL1 cells was slightly slower than that of HeLa-shNC cells, but there was no significant difference (data not shown). When the two types of cells were subjected to electron microscopy, it was found that there were plenty of autophagosomes/autolysosomes, the characteristic feature of autophagy, in HeLa-shERAL1 but not in HeLa-shNC cells (Fig. 1A). Furthermore, the conversion from LC3-I to LC3-II was detected in HeLa-shERAL1 cells, while LC3-II formation was not found in HeLa-shNC cells (Fig. 1B). To confirm the effects of ERAL1 knockdown on autophagy activation, we constructed a plasmid expressing ERAL1 from its wild-type cDNA (Wt-ERAL1) and another plasmid expressing ERAL1 from its cDNA with silent mutations in the shRNA-targeting sequence (Mu-ERAL1). HeLa-shERAL1 cells were transfected with the plasmid expressing wt-ERAL1 or Mu-ERAL1 respectively, and then subjected to western blotting to detect the LC3-I to LC3-II conversion. Compared with wt-ERAL1, Mu-ERAL1, whose expression is resistant to shRNA inhibition, significantly suppressed the LC3-I to LC3-II conversion in HeLa-shERAL1 cells (Fig. 1C). These results indicate that autophagy is modulated by ERAL1 knockdown. With the significant autophagic phenomenon, HeLa-shERAL1 cells did not show obvious apoptosis when cultured in normal glucose medium. However, significant apoptosis was detected in HeLa-shERAL1 but not in HeLa-shNC cells after the cells were transferred into a glucose-free medium supplemented with galactose (Fig. 1D), suggesting that ERAL1 knockdown affected mitochondrial oxidative phosphorylation, which is required for ATP production in galactose medium. The mitochondrial dysfunction resulting from ERAL1 knockdown could be the reason for autophagy in HeLa-shERAL1 cells cultured in normal glucose medium.
Figure 1. Autophagy is induced by ERAL1 knockdown in HeLa cells. (A) Electron microscopy pictures were taken of HeLa cells with stable expression of ERAL1-shRNA (HeLa-shERAL1) or scramble shRNA (HeLa-shNC). Arrows represent autophagic vacuoles. (B) LC3-I to LC3-II conversion was induced in HeLa-shERAL1 cells. ERAL1 and LC3 in HeLa-shERAL1 and HeLa-shNC cells were detected by western blotting. (C) The LC3-I to LC3-II conversion in HeLa-shERAL1 was suppressed by Mu-ERAL1. Western blotting was performed to detect ERAL1 and LC3 in HeLa-shERAL1 cells transfected with the plasmid expressing ERAL1 from wild-type cDNA (wt-ERAL1) or from the cDNA with silent mutations in the ERAL1 shRNA targeting sequence (Mu-ERAL1). (D) ERAL1 knockdown induced apoptosis in HeLa cells when cultured in a galactose medium. HeLa-shERAL1 and HeLa-shNC cells were cultured in a glucose medium and then transferred into a galactose medium. Apoptotic cell death rates were detected before the medium change (0 h) and after being cultured in galactose medium for 48 h. The p value derived from a Student’s t-test is **p < 0.001.
ERAL1 knockdown increases ROS generation leading to autophagy
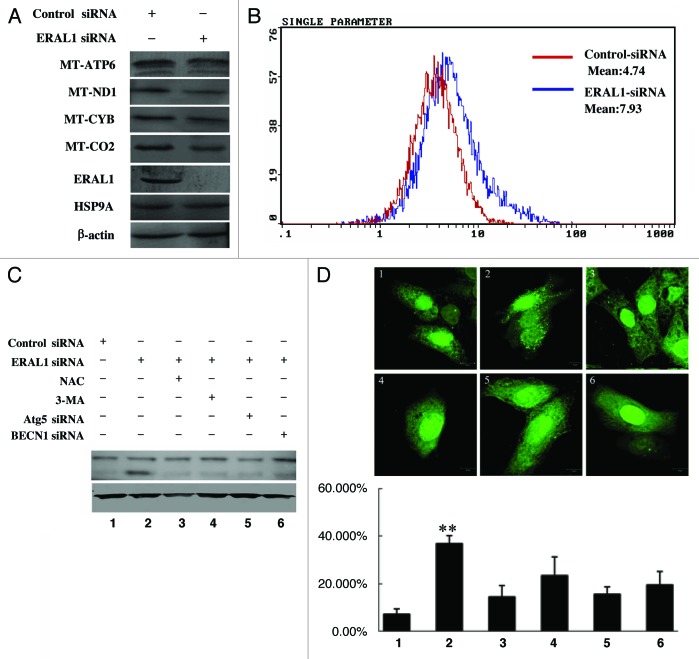
Recently, ERAL1 has been reported to be associated with the mitochondrial ribosome, and knockdown of ERAL1 would have an impact on mitochondrial protein translation.16,17 In the present study, HeLa cells were transfected with ERAL1 siRNA to downregulate ERAL1 expression. ERAL1 knockdown decreased the protein levels of MT-ATP6, MT-ND1, MT-CYB and MT-CO2 (Fig. 2A), which are integral parts of four mitochondrial respiratory chain complexes (I, III, IV and V), suggesting ERAL1 downregulation affects the function of the mitochondrial electron transfer chain (mETC) by suppressing protein synthesis in mitochondria. Reactive oxygen species (ROS), the byproduct of mETC, participate in many important intracellular processes, including autophagy induction.5,18,19 The ROS levels in ERAL1-knockdown cells were determined with the redox-responsive fluorescent dye H2-DCFDA. A significant increase in the fluorescence over the control was detected at 72 h post-ERAL1 siRNA transfection (Fig. 2B), indicating that ERAL1 downregulation induces ROS generation.
Figure 2. The autophagy induction by ERAL1 knockdown is dependent on ROS elevation. (A) Dysfunction of mitochondrial protein synthesis in HeLa cells with ERAL1 knockdown. HeLa cells were transfected with control siRNA or ERAL1 siRNA, respectively. At 72 h post-siRNA transfection, the cells were subjected to western blotting to detect the levels of indicated mitochondrial proteins. (B) ROS levels were elevated in HeLa cells with ERAL1 knockdown. HeLa cells were transfected with control siRNA (red) or ERAL1 siRNA (blue). At 72 h post-siRNA transfection, cells were incubated with H2-DCFDA (100 μM) for 30 min and then subjected to flow cytometric analysis for quantitative estimation of ROS levels. (C and D) Autophagy was induced by ERAL1 knockdown in a ROS-dependent manner. The LC3-I to LC3-II conversion in HeLa cells (C) and GFP-LC3 puncta formation in HeLa cells transfected with GFP-LC3 plasmid (D) were detected after the cells were treated for 72 h with the siRNA(s) and inhibitor as indicated. (1) Control siRNA; (2) ERAL1 siRNA; (3) ERAL1 siRNA and NAC; (4) ERAL1 siRNA and 3-MA; (5) ERAL1 siRNA and ATG5 siRNA; (6) ERAL1 siRNA and BECN1 siRNA. The percentage of GFP-LC3 puncta-positive cells was quantified as described under Materials and Methods. Representative data were from three independent experiments. The p value derived from Student’s t-test is **p < 0.001.
Tracking the conversion of LC3-I (18 kDa) to LC3-II (16 kDa) is indicative of autophagy activity. The LC3-I to LC3-II conversion was detected in ERAL1 siRNA-treated HeLa cells, which was blocked by the autophagy inhibitor 3-MA, as well as by the downregulation of ATG5 and BECN1, essential proteins needed to initiate autophagy (Fig. 2C; Fig. S1). To confirm the autophagy induction by ERAL1 knockdown, HeLa cells transfected with the GFP-LC3 plasmid were treated with control siRNA or ERAL1 siRNA respectively, and then subjected to confocal observation to check the GFP-LC3 puncta formation. The percentage of GFP-LC3 puncta-positive cells was significantly higher in the ERAL1 siRNA-treated cells than that in the control siRNA-treated cells. The GFP-LC3 puncta formation resulting from ERAL1 knockdown was attenuated by 3-MA treatment as well as by the downregulation of ATG5 or BECN1 (Fig. 2D). Furthermore, the ERAL1 knockdown-induced LC3-I to LC3-II conversion and GFP-LC3 puncta formation were suppressed by the ROS scavenger NAC (Fig. 2C and D), indicating that ERAL1 knockdown induced autophagy through elevating ROS levels.
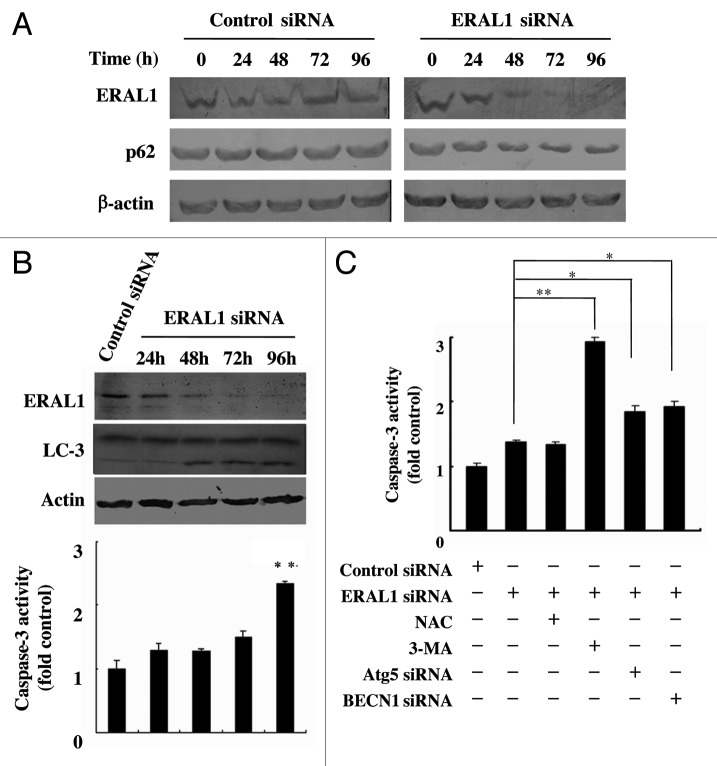
As a protein localized to the autophagosome via LC3 interaction and constantly degraded by the autophagy–lysosome system, SQSTM1/p62 is widely used as a marker for autophagic flux.20,21 It was found that SQSTM1 protein levels were downregulated in ERAL1 siRNA-treated HeLa cells in a time-dependent manner (Fig. 3A). Both SQSTM1 downregulation and LC3-I to LC3-II conversion were detected in HeLa cells at 48 h post-ERAL1 siRNA transfection (Fig. 3A and B), indicating that functional autophagy is induced by ERAL1 knockdown. Although it has been reported that CASP3/caspase-3 is activated in the ERAL1-knockdown cells,17 the CASP3 activity was not increased until 96 h post-ERAL1 siRNA transfection (Fig. 3B). Inhibition of autophagy by 3-MA, ATG5 siRNA or BECN1 siRNA led to a significant increase in CASP3 activity at 72 h post-ERAL1 siRNA transfection (Fig. 3C). These data suggest that autophagy was induced ahead of apoptosis and performed as a suppressor of apoptosis in the ERAL1-knockdown cells.
Figure 3. Autophagy suppresses the apoptosis induction in ERAL1 siRNA-treated HeLa cells. (A) SQSTM1/p62 protein levels were decreased in ERAL1 siRNA-treated HeLa cells. HeLa cells were transfected with control siRNA or ERAL1 siRNA. The levels of SQSTM1/p62 in HeLa cells were detected by western blot at the indicated time points after siRNA transfection. (B) Autophagy was induced ahead of apoptosis. The LC3-I to LC3-II conversion (top panel) and CASP3/Caspase-3 activity (bottom panel) in HeLa cells were detected at the indicated time points after ERAL1 siRNA transfection. (C) Inhibition of autophagy enhanced the CASP3 activation. HeLa cells were treated for 72 h with the siRNA(s) and inhibitor as indicated. CASP3 activities in cell lysates were determined as described under Materials and Methods. * and **, different from ERAL1 siRNA alone (column 2). The p values derived from Student’s t-test are *p < 0.01 and **p < 0.001.
ERAL1 knockdown induces autophagy through the ROS-mediated activation of the TP53-DRAM1 pathway
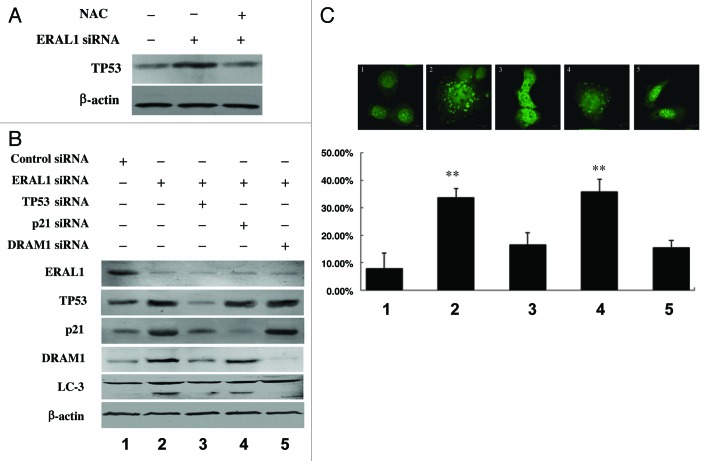
It has been reported that ROS can act as an upstream signal to trigger the TP53 signaling pathway, and nuclear TP53 activates autophagy by inducing its downstream genes, including DRAM1 (damage-regulated autophagy modulator 1).22-29 It was found that TP53 protein levels were upregulated in ERAL1 siRNA-treated HeLa cells, whereas the TP53 upregulation was blocked by NAC, a specific scavenger of ROS, suggesting that ROS are involved in the upregulation of TP53 by ERAL1 knockdown (Fig. 4A). When TP53 expression was suppressed by its specific siRNA, LC3-I to LC3-II conversion, as well as GFP-LC3 puncta formation, in ERAL1 siRNA-treated HeLa cells was attenuated (Fig. 4B and C). The expression levels of both DRAM1 and CDKN1A/p21, the downstream target genes of TP53, were upregulated in HeLa cells treated with ERAL1 siRNA. The ERAL1 knockdown-induced LC3-I to LC3-II conversion and GFP-LC3 puncta formation in HeLa cells were significantly blocked by DRAM1 siRNA, but not by CDKN1A/p21 siRNA (Fig. 4B and C). These data indicate that the ROS-mediated activation of TP53-DRAM1 pathway is required for the autophagy induction by ERAL1 knockdown.
Figure 4. Activation of the TP53-DRAM1 pathway is required for the autophagy induction by ERAL1 knockdown. (A) TP53 expression was upregulated by ERAL1 knockdown in a ROS-dependent manner. HeLa cells were transfected with ERAL1 siRNA in the presence or absence of NAC. At 72 h post-siRNA transfection, cells were subjected to western blotting to detect the TP53 protein levels. (B and C) The LC3-I to LC3-II conversion in HeLa cells (B) and GFP-LC3 puncta formation in HeLa cells transfected with a GFP-LC3 plasmid (C) were detected after the cells were treated for 72 h with the siRNA(s) as indicated. (1) control siRNA; (2) ERAL1 siRNA; (3) ERAL1 siRNA and TP53 siRNA; (4) ERAL1 siRNA and CDKN1A/p21 siRNA; (5) ERAL1 siRNA and DRAM1 siRNA. The percentage of GFP-LC3 puncta-positive cells was quantified as described under Materials and Methods. Representative data were from three independent experiments. The p value derived from Student’s t-test is **p < 0.001.
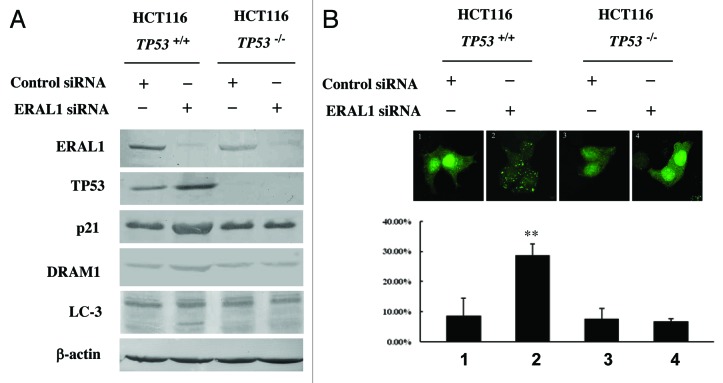
TP53 is essential for the autophagy induction by ERAL1 knockdown
HCT-116 TP53+/+ and HCT-116 TP53−/− cells were used to further confirm the role of TP53 in the autophagy induction by ERAL1 knockdown. Except for the deletion of the TP53 gene, HCT-116 TP53−/− cells have the same genomic background as that of HCT-116 TP53+/+ cells. ERAL1 siRNA treatment induced LC3-I to LC3-II conversion and GFP-LC3 puncta formation in HCT-116 TP53+/+ cells, but not in HCT-116 TP53−/− cells (Fig. 5A and B). With the knockdown of ERAL1, both TP53 and DRAM1 expression were elevated in HCT-116 TP53+/+ cells, while DRAM1 expression was not upregulated in HCT-116 TP53−/− cells with TP53 deletion (Fig. 5A and B). These data suggest that TP53 play an essential role in the autophagy induction by ERAL1 knockdown.
Figure 5. TP53 is essential for the autophagy induction by ERAL1 knockdown. (A) HCT-116 TP53+/+ and HCT-116 TP53−/− cells were transfected with ERAL1 siRNA. At 72 h post-siRNA transfection, cells were subjected to western blotting to detect the levels of the indicated proteins. (B) HCT-116 TP53+/+ and HCT-116 TP53−/− cells were transfected with GFP-LC3 plasmid and then treated with control siRNA or ERAL1 siRNA, respectively. At 72 h post-siRNA transfection, GFP-LC3 puncta formation in the cells was detected by confocal microscopy. The percentage of GFP-LC3 puncta-positive cells was quantified as described under Materials and Methods. Representative data were from three independent experiments. The p value derived from Student’s t-test is **p < 0.001.
Pharmacological inhibition of mitochondrial protein synthesis induces autophagy through the ROS-TP53-DRAM1 pathway
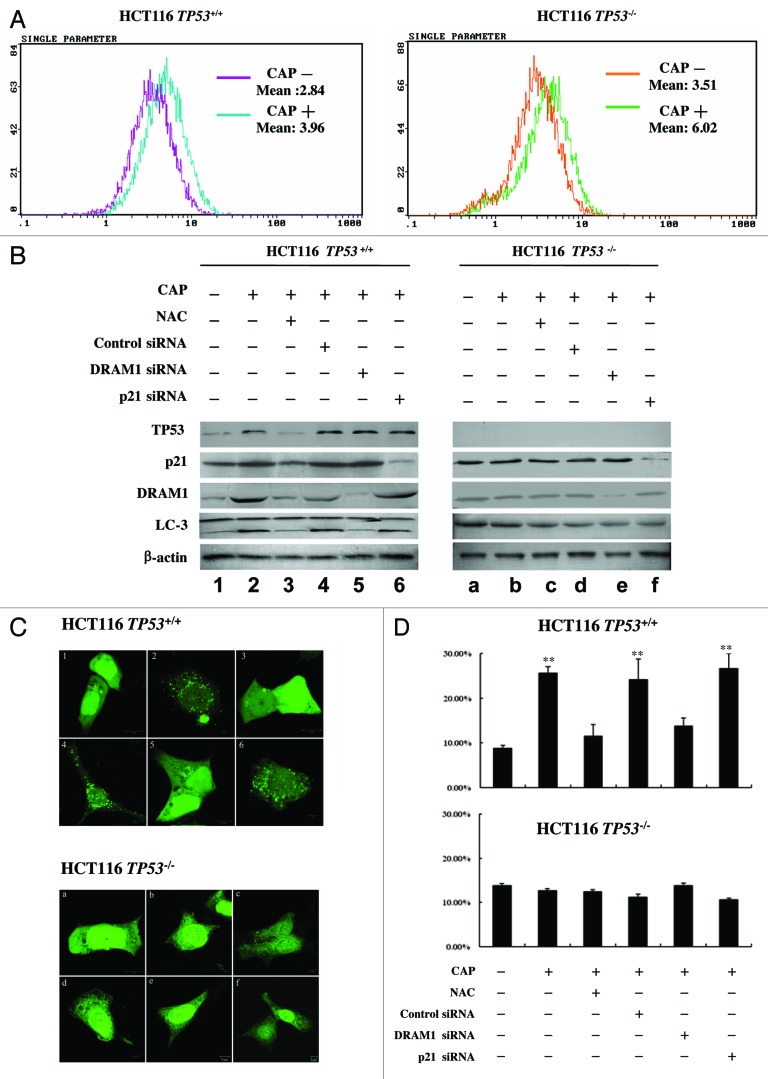
Chloramphenicol is a specific mitoribosome inhibitor that blocks mitochondrial protein synthesis.30-32 We wondered if CAP could induce autophagy as ERAL1 knockdown did. The HCT-116 TP53+/+ and HCT-116 TP53−/− cells were treated with CAP at a concentration that had no toxicity on cell viability (50 μg/ml). After CAP treatment for 48 h, the cells were incubated with H2-DCFDA for 30 min following by flow cytometry to determine ROS levels. As shown in Figure 6A, CAP treatment elevated ROS levels in both HCT-116 TP53+/+ and HCT-116 TP53−/− cells. The expression levels of TP53 and its downstream genes, CDKN1A/p21 and DRAM1, were upregulated in the CAP-treated HCT-116 TP53+/+ cells, whereas expression levels of CDKN1A/p21 and DRAM1 were not upregulated in the CAP-treated HCT-116 TP53−/− cells (Fig. 6B, lines 2 and b). The TP53, CDKN1A/p21 and DRAM1 upregulation in CAP-treated HCT-116 TP53+/+ cells was suppressed by NAC (Fig. 6B, line 3), indicating that ROS function as upstream effectors for the activation of TP53 signaling. Meanwhile, CAP induced LC3-I to LC3-II conversion in HCT-116 TP53+/+ cells but not in HCT-116 TP53−/− cells (Fig. 6B, lines 2 and b). The CAP-induced LC3-I to LC3-II conversion in HCT-116 TP53+/+ cells could be blocked by NAC treatment or DRAM1 knockdown, but not by CDKN1A/p21 knockdown (Fig. 6B, lines 3–6). On the contrary, LC3 remained inactive in HCT-116 TP53−/− cells after CAP treatment (Fig. 6B, lines b-f). HCT-116 TP53+/+ and HCT-116 TP53−/− cells were transfected with the GFP-LC3 plasmid and then subjected to CAP treatment. After CAP treatment for 48 h, GFP-LC3 puncta formation was significantly detected in HCT-116 TP53+/+ cells, but not in HCT-116 TP53−/− cells. NAC and DRAM1 siRNA, but not CDKN1A/p21 siRNA, significantly inhibited the GFP-LC3 puncta formation in CAP-treated HCT-116 TP53+/+ cells (Fig. 6C and D). In addition, SQSTM1 protein levels were decreased in HCT-116 TP53+/+ cells post CAP treatment in a time-dependent manner (Fig. S2). These data indicate that the dysfunction of mitochondrial protein synthesis by CAP induced autophagy through ROS elevation and ROS-mediated activation of the TP53-DRAM1 pathway.
Figure 6. CAP induces autophagy through the ROS-TP53-DRAM1 pathway. (A) The ROS elevation by CAP treatment. HCT-116 TP53+/+ and HCT-116 TP53−/− cells were treated with or without CAP (50 μg/ml) for 48 h and then incubated with H2-DCFDA (100 μM) for 30 min. Cells were subjected to flow cytometric analysis for quantitative estimation of ROS levels. (B) The induction of LC3-I to LC3-II conversion by CAP treatment. HCT-116 TP53+/+ and HCT-116 TP53−/− cells were treated with CAP (50 μg/ml) in the present of NAC or after the transfection of control siRNA, DRAM1 siRNA or CDKN1A/p21 siRNA as indicated. After 48 h treatment with CAP, cells were lysed and subjected to western blotting to detect the levels of indicated proteins. (C and D) The induction of GFP-LC3 puncta formation by CAP treatment. HCT-116 TP53+/+ and HCT-116 TP53−/− cells were transfected with the GFP-LC3 plasmid and then treated with CAP (50 μg/ml) in the present of NAC or after the transfection of siRNA as indicated in (B). After 48 h treatment with CAP, cells were fixed and GFP-LC3 puncta signals were detected by confocal microscopy (C). The percentage of GFP-LC3 puncta-positive cells was quantified as described under Materials and Methods (D). Representative data were from three independent experiments. The p value derived from Student’s t-test is **p < 0.001.
TP53 is activated by ROS at both transcriptional and post-translational levels
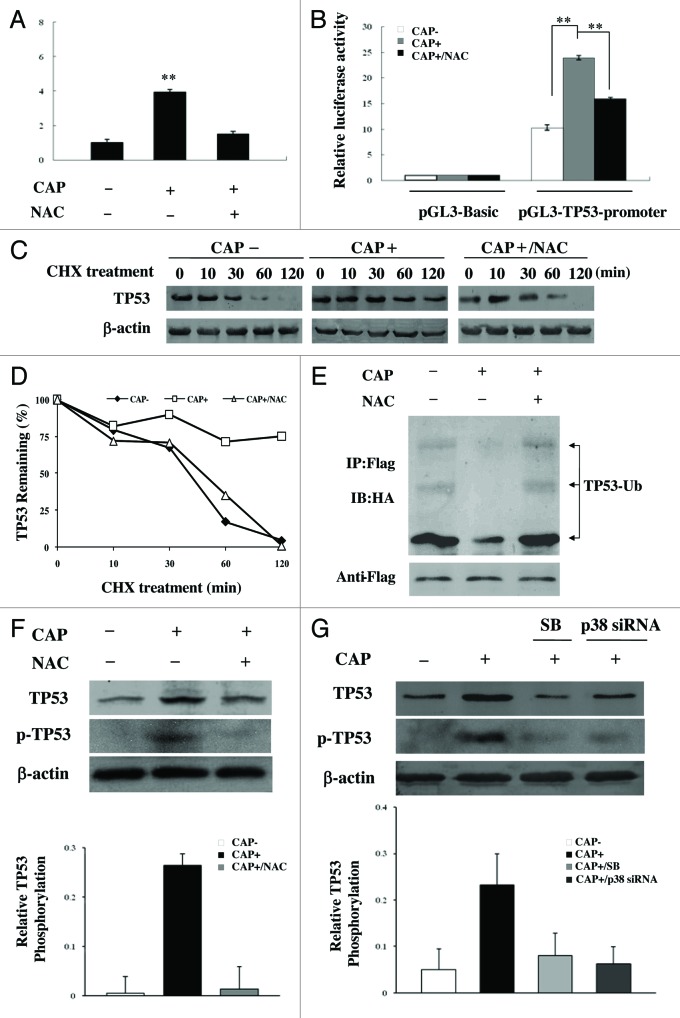
The expression of TP53, a critical guardian of genomic stability and cell growth, is tightly controlled at the transcriptional and post-translational levels. TP53 mRNA levels were increased in CAP-treated HCT-116 TP53+/+ cells, whereas the elevation of TP53 mRNA levels was inhibited by the ROS scavenger NAC (Fig. 7A). With luciferase as reporter gene under the TP53 promoter, we found that the activity of the TP53 promoter was increased under CAP treatment, which was blocked in the presence of NAC (Fig. 7B). TP53 mRNA levels as well as TP53 promoter activity were also upregulated in the ERAL1 siRNA-treated HeLa cells in a ROS-dependent manner (Fig. S3). These data suggest that, under mitochondrial protein synthesis inhibition, ROS induced TP53 expression at the transcription level through enhancing TP53 promoter activity.
Figure 7.TP53 is activated by ROS at both transcriptional and post-translational levels in CAP-treated cells. (A) TP53 mRNA levels in CAP-treated cells. HCT-116 TP53+/+ cells were treated with CAP (50 μg/ml) for 48 h in the presence or absence of NAC. TP53 mRNA levels were analyzed by Real-Time PCR. (B) TP53 promoter activity in CAP-treated cells. HeLa cells were transfected with pGL3-Basic vector or pGL3-TP53-promoter together with a β-gal expressing plasmid, and then treated with CAP (50 μg/ml) for 48 h in the presence or absence of NAC. Luciferase activity was measured and normalized to transfection efficiency with β-galactosidase activity as internal control. (C) and (D) TP53 stability in CAP-treated cells. HCT-116 TP53+/+ cells were treated with CAP for 48 h in the absence or presence of NAC, and then incubated with CHX (20 μM). The TP53 protein levels were detected by western blotting at indicated time points after the addition of CHX (C). The densitometry analysis was performed to quantify the TP53 downregulation following CHX treatment (normalized to β-actin) (D). (E) TP53 ubiquitination in CAP-treated cells. HCT-116 TP53+/+ cells were cotransfected with HA-Ubiquitin and TP53-Flag plasmid, and then treated with CAP for 48 h in the absence or presence of NAC. Cells were treated with MG132 for 2 h, then lysed and subjected to immunoprecipitation with anti-Flag agarose beads, followed by western blotting analysis with anti-HA antibody (top panel). The same membrane was stripped and reprobed with anti-Flag antibody (bottom panel); (F) TP53 phosphorylation in CAP-treated cells. HCT-116 TP53+/+ cells were treated with CAP (50 μg/ml) for 48 h in the absence or presence of NAC and then subjected to western blotting to detect the levels of TP53 and p-TP53 (Ser15) respectively. Densitometric measurements represent relative TP53 phosphorylation as normalized against total TP53 levels (bottom panel). (G) The role of MAPK14/p38 in TP53 phosphorylation in CAP-treated cells. HCT-116 TP53+/+ cells were treated with CAP (50 μg/ml) in the presence of the MAPK14 inhibitor SB202190 (SB) or after the transfection of MAPK14-specific siRNA. After 48 h treatment with CAP, cells were lysed and subjected to western blotting to detect the levels of TP53 and p-TP53 (Ser15) respectively. Densitometric measurements represent relative TP53 phosphorylation as normalized against total TP53 levels (bottom panel). Representative data were from three independent experiments.
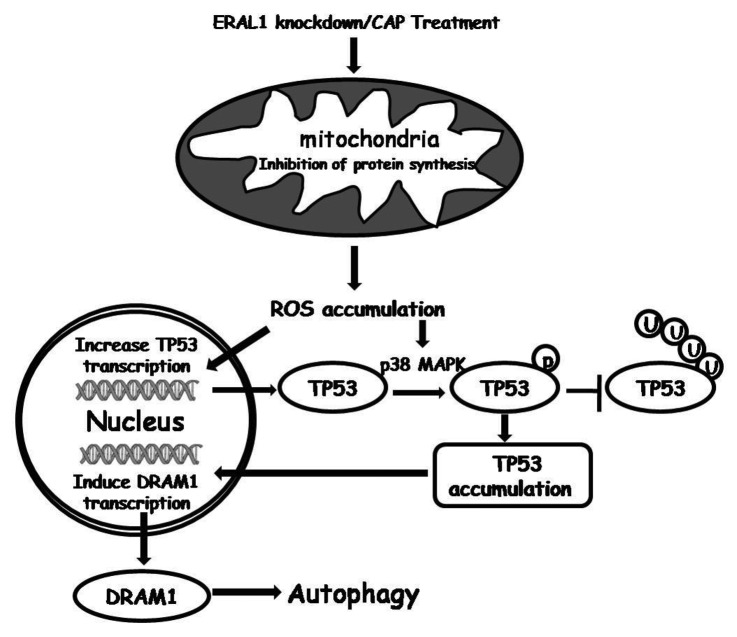
The stability and activity of TP53 are regulated by its post-translational ubiquitination and ubiquitin-dependent degradation through the 26S proteasome.33-35 As TP53 activation is required for the autophagy induction as described above, the life span of TP53 was tested when mitochondrial protein synthesis was inhibited. HCT-116 TP53+/+ cells were treated with or without CAP, and then incubated with CHX (20 μM). The protein levels of TP53 were detected by western blotting at different time points after CHX treatment. It was found that the half-life of TP53 was significantly increased in CAP-treated cells, while the increase of TP53 stability was suppressed in the presence of NAC (Fig. 7C and D), suggesting TP53 is stabilized due to the ROS accumulation resulting from mitochondrial protein synthesis inhibition. Furthermore, it was found that the ubiquitination of TP53 was decreased in CAP-treated HCT-116 TP53+/+ cells, which was restored in the presence of NAC (Fig. 7E). It has been reported that ROS are implicated in the phosphorylation of TP53 via protein kinases, including MAPK14/p38, to stabilize the TP53 protein by interfering with TP53 ubiqitination.34-36 We found that CAP induced phosphorylation of TP53 at Ser15, one of the TP53 phosphorylation sites by MAPK14/p38. The TP53 phosphorylation, as well as its accumulation, in CAP-treated cells was suppressed by NAC, MAPK14-specific siRNA and the MAPK14 inhibitor SB202190 (Fig. 7F and G), suggesting that the ROS-MAPK14 pathway is involved in TP53 activation. In ERAL1 siRNA-treated HeLa cells, the stability of TP53 was increased by ROS through the same mechanism, the induction of MAPK14-mediated TP53 phosphorylation and the suppression of TP53 ubiquitination (Fig. S4). These data indicate that ROS not only enhance TP53 expression at the transcriptional level but also increase TP53 stability at the post-translational level, leading to the activation of TP53 and its downstream gene DRAM1 to induce autophagy in response to the inhibition of protein synthesis in mitochondria (Fig. 8).
Figure 8. Autophagy induction pathway under the inhibition of mitochondrial protein synthesis.
Discussion
The mitochondria genome encodes 13 proteins essential for the mitochondrial respiration chain.37 Translation of the mitochondrial DNA (mtDNA) protein-encoding genes is performed in the mitochondrial matrix by a specific protein-synthesis system, which is composed of tRNAs and rRNAs synthesized from the corresponding mitochondrial genes and a number of proteins encoded by nuclear DNA, including mitoribosome proteins, aminoacyl-tRNA synthetases, the translation initiation, elongation and termination factors, and a large number of unidentified factors such as mitoribosome assembly factors.37,38 ERA is a conserved protein family with a N-terminal GTPase domain and a C-terminal hnRNPK homology (KH) domain. Bacterial ERA is required for the maturation of the 16S rRNA and the assembly of the 30S ribosomal subunit.10,11 ERAL1, the human ERA homolog, has been identified as a novel nuclear-encoded mitoribosome assembly factor associated with mitochondrial 12S rRNA, and plays an important role in the formation of functional 28S mitoribosomal small subunit.16,17 It has been reported that ERAL1 knockdown induces CASP3 activation in HeLa cells.17 In the present study, we confirmed that knockdown of ERAL1 inhibited protein synthesis in mitochondria, resulting in dysfunction of the mitochondrial respiration chain and leading to apoptotic cell death when galactose was supplied as sugar substrate, which needs to be metabolized via mitochondrial oxidation. However, there was no significant difference in apoptotic rate between the HeLa-shERAL1 and HeLa-shNC cells cultured in a normal glucose medium. Instead, autophagosome formation and LC3-I to LC3-II conversion, the hallmarks of autophagy, were detected in HeLa-shERAL1 cells but not in HeLa-shNC cells. Furthermore, LC3-I to LC3-II conversion, GFP-LC3 puncta formation and SQSTM1 degradation were detected in the ERAL1 siRNA-treated HeLa cells, indicating that functional autophagy is induced by ERAL1 knockdown. There was no significant change in intracellular ATP levels while autophagy was induced in the ERAL1-knockdown HeLa cells cultured in a normal glucose medium (Fig. S5), which may due to the compensation of ATP production by glycolysis, suggesting that the autophagy induction by ERAL1 downregulation did not result from the alteration in ATP levels. Both LC3-I to LC3-II conversion and SQSTM1 degradation were induced ahead of the increase of CASP3 activity, and inhibition of autophagy could promote the induction of CASP3 activity in the ERAL1 siRNA-treated HeLa cells. Therefore, we propose that the autophagy is induced by ERAL1 knockdown and performs as a suppressor of apoptosis, although ERAL1 knockdown may be able to induce apoptosis eventually due to the dysfunction of the mitochondrial respiration chain.
TP53 is a tumor suppressor gene involved in various cellular processes. Growing evidence indicates that TP53 plays an important role in autophagy regulation.22-26DRAM1 is a TP53 target gene involved in autophagy. The autophagy mediated by TP53 can be markedly inhibited by DRAM1 knockdown.27-29 The expression levels of TP53 and its target genes, CDKN1A/p21 and DRAM1, were elevated in ERAL1 siRNA-treated HeLa cells, while the autophagy induced by ERAL1 knockdown was suppressed by downregulation of TP53 and DRAM1, but not CDKN1A/p21. Furthermore, ERAL1 knockdown induced LC3-I to LC3-II conversion and GFP-LC3 puncta formation in HCT-116 TP53+/+ but not in HCT-116 TP53−/− cells, which have the same genomic background as that of HCT-116 TP53+/+ cells except the deletion of the TP53 gene. Chloramphenicol, as a specific inhibitor of mitochondrial ribosomes, is used to study the dysfunction of mitochondrial protein synthesis. It is interesting that, similar to ERAL1 knockdown, CAP treatment induced LC3-I to LC3-II conversion and GFP-LC3 puncta formation in HCT-116 TP53+/+ but not in HCT-116 TP53−/− cells. The CAP-induced autophagy in HCT-116 TP53+/+ cells was also suppressed by the downregulation of DRAM1. These data indicate that mitochondrial protein synthesis inhibition induced autophagy through the activation of the TP53-DRAM1 pathway.
ROS levels were elevated by both ERAL1 knockdown and CAP treatment. The autophagy induced by either ERAL1 knockdown or CAP treatment was suppressed by NAC, a specific scavenger of ROS, suggesting that ROS elevation functions as an oxidative stress to trigger the activation of autophagy. ROS are implicated in autophagy regulation through distinct mechanisms, depending on cell types and stimulation conditions. Nutrient starvation leads to mitochondrial ROS production to promote autophagosome formation by regulating the protease activity of ATG4.39 In malignant glioma, ROS disrupt mitochondrial membrane potential and induce autophagy through inhibiting AKT1-MTOR signaling.40 In the present study, we found that, in either ERAL1 siRNA or CAP-treated cells, ROS promoted TP53 transcription by enhancing TP53 promoter activity and increased TP53 stability by suppressing TP53 ubiquitination (Fig. 7; Figs. S3 and S4). ROS stimulated the MAPK14-mediated TP53 phosporylation at serine 15, a key target during the TP53 activation process. It has been reported that TP53-dependent transactivation is activated by the serine 15 phosphorylation, which decreases binding of TP53 to its negative regulator MDM2 and increases binding to the CITED/p300 coactivator protein.41,42 Therefore, in response to mitochondrial protein synthesis inhibition, ROS elevation acts as an upstream signal of TP53 activation to induce autophagy through the TP53-DRAM1 pathway.
In the present paper, we have demonstrated that the dysfunction of mitochondrial protein synthesis caused by CAP treatment or the knockdown of ERAL1, a novel mitoribosome assembly factor, induces autophagy through the ROS-TP53-DRAM1 pathway (Fig. 8). Meanwhile, we do not rule out other mechanisms contributing to the autophagy induction, as it has been reported that CAP can induce expression of ATG12, which plays an essential role during the activation of mammalian autophagy.43 ROS resulting from mitochondrial protein synthesis inhibition may oxidize proteins and damage organelles, while the autophagy induced by ROS may function as a cell-survival mechanism, which is able to degrade oxidized proteins, eliminate the ROS-producing mitochondria, and remove the damaged organelles (e.g., mitochondria and ER). Although swollen mitochondria were observed, mitophagy was rarely detected by electron microscopy in HeLa-shERAL1 cells (Fig. S6). By western blotting analysis, we found that HeLa cells are free of PARK2/PARKIN (data not shown), which can be selectively recruited to impaired mitochondria to promote their autophagy (mitophagy).44,45 The role of mitochondrial protein synthesis inhibition in mitophagy, especially PARK2-dependent mitophagy, remains to be studied in the future. As autophagy is induced by ROS through the TP53-DRAM1 pathway, mitochondrial protein synthesis inhibition may have different cellular effects dependent on the activation of TP53-DRAM1 signaling. It was observed that, when treated with CAP (50 μg/ml), the HCT-116 TP53−/− cells, which are resistant to autophagy induction due to TP53 deletion, were more susceptible to apoptosis than HCT-116 TP53+/+ cells (Fig. S7). However, when treated with a high concentration of CAP (200 μg/ml or more), both HCT-116 TP53+/+ and HCT-116 TP53−/− cells were more likely to undergo apoptosis due to the severe cellular toxicity (data not shown). These data suggest that the autophagy induction serves as a protective cellular mechanism under mitochondrial protein synthesis inhibition, while the apoptotic cell death will occur when the stress reaches a level beyond control. Dysfunction of mitochondrial protein synthesis caused by mutations either at the MRPs, tRNAs or translation factors has been involved in various human disorders, including maternally inherited nonsyndromic sensorineural deafness, aminoglycoside-induced deafness, and neonatal encephalopathy.37,38,46 mtDNA deletion, which also induces dysfunction of mitochondrial protein synthesis, is involved in widespread multisystemic disorders.47 Our findings in the present study have shed light on the mechanism by which mitochondrial protein synthesis inhibition induces autophagy. The physiological and pathological roles of the autophagy induction by mitochondrial protein synthesis disorders remain to be further studied.
Material and Methods
Cell culture
HeLa cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 mg/L streptomycin in an environment of 5% CO2 at 37°C. HCT116 TP53+/+ and HCT116 TP53−/− cells were grown in McCoy's 5A medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 mg/L streptomycin in an environment of 5% CO2 at 37°C.
Reagents and Antibodies
N-acetyl cysteine (NAC) (A9165), H2-DCFDA (D6883), 3-methyladenine (3-MA) (M9281) and cycloheximide (CHX) (C1988) were from Sigma. Galactose (0637) was from Amresco. Chloramphenicol (CAP) (220551) was from Calbiochem. The specific MAPK14/p38 inhibitor SB202190 (559388) was from Merck. Lipofectamine 2000 (11668-019) was from Invitrogen. The CASP3 colorimetric kit (BF3100) was from R&D System. Flag-tagged protein purification kit (IPK002) was from Macgene. Antibodies used in this study include: Anti-ERAL1 (18881) was from IBL; anti-TP53 (sc-6243), β-actin (sc-81178), MT-CYB (sc-9509), HSPA9 (sc-33575) and MT-CO2 (sc-65239) were purchased from Santa Cruz; anti-DRAM1 (ab68987), MT-ATP6 (ab101908) and MT-ND1 (ab74257) were from Abcam; anti-LC3 (L8918), Flag-Tag (F1804) and HA-Tag (H3663) were from Sigma; anti-CDKN1A/p21 (K0081-3) was from MBL; anti-phospho-TP53 (Ser15) (9286) and anti-SQSTM1/p62 (5114) were from Cell Signaling. ATP-Lite Assay Kit (TOO7) was from Vigorous Bitotechnology.
RNA interference
ERAL1 was downregulated with either short hairpin RNA (shRNA) or short interfering RNA (siRNA). The ERAL1 shRNA and siRNA have the same targeting sequence 5′-GGU GCC CAA AGA AUC UUA UGU-3′. The ERAL1 shRNA and control scrambled shRNA plasmids were constructed by GenePharma. The plasmids were transfected into HeLa cells with LipofectamineTM 2000 (Invitrogen). The transfected cells were treated with G418-sulfate (600 μg/mL) (Merck) 48 h post-transfection. G418-sulfate-resistant clones were selected to establish the stable cell line with ERAL1 downregulation. The sequences of siRNAs used in this study were as follows: si-Negative control (NC): UUC UUC GAA CGU GUC ACG U; si-ERAL1: GGU GCC CAA AGA AUC UUA UGU; si-DRAM1: CCA CAG AAA UCA AUG GUG A; si-ATG5: GGC UUA UCC AAU UGG CCU ACU GUU; si-BECN1: CAG UUU GGC ACA AUC AAU A; si-TP53: CUA CUU CCU GAA AAC AAC G; si-CDKN1A/p21: GGA GUC AGA CAU UUU AAG; si-MAPK14/p38 was purchased from Santa Cruz (sc-29433).
Plasmids
The cDNA encoding wild-type ERAL1 was cloned into the pCMV-tag4 vector, creating the pCMV-tag4-ERAL1-Flag to express ERAL1 fused with Flag tag at the C terminus. The Mut-ERAL1 plasmid was generated from pCMV-tag4-ERAL1-Flag with the shRNA-targeting sequence G GTGCCCAAAGAATCT TAT GT mutated to G GTACCTAAGGAGTCA TAT GT. Wild type TP53 cDNA was inserted into the pCMV-Tag4 vector, creating the plasmid named TP53-Flag to express TP53 fused with the Flag tag at the C terminus. The GFP-LC3 plasmid was a kind gift from Dr. Tamotsu Yoshimori (National Institute for Basic Biology, Japan), the pGL3-TP53-promoter was from Dr. Daniel S. Peeper (The Netherlands Cancer Institute) and the HA-Ubiquitin plasmid was from Dr. Xiaobo Qiu (Beijing Normal University).
Electron microscopy examination
Cells were collected and fixed with 2% paraformaldehyde and 0.1% glutaraldehyde in 0.1 M sodium cacodylate for 2 h, post-fixed with 1% OsO4 for 1.5 h, washed and finally stained for 1 h in 3% aqueous uranyl acetate. The samples were then washed again, dehydrated with graded alcohol and embedded in Epon-Araldite resin (Canemco, 034). Ultrathin sections were cut on a Reichert ultramicrotome, counterstained with 0.3% lead citrate and examined on a HC JEM-1230 electron microscope.
Western blotting
Cells were lysed in ice-cold RIPA lysis buffer (150 mM NaCl, 1% NP-40, 50 mM Tris–HCl, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 mM Deoxycholic acid and 1mM EDTA) and clarified at 12,000 × g for 10 min. Protein samples were subjected to SDS-PAGE. After electrophoresis, proteins were transferred to a PVDF transfer membrane (Amersham Biosciences, RPN303F). Blots were then incubated with primary antibodies using the manufacturer’s protocol followed by appropriate horseradish peroxidase-conjugated secondary antibody. Immunostained proteins were then visualized by 3, 3′-Diaminobenzidine tetrahydrochloride (Golden Bridge, ZLI-9033) or on X-ray film using the ECL detection system.
Staining of autophagosomes with GFP-LC3
Cells transfected with plasmid expressing GFP-LC3 (2 μg) were transfected with ERAL1 siRNA for 72 h or treated with CAP (50 μg/ml) for 48 h, respectively. The fluorescence of GFP-LC3 was observed and the number of GFP-LC3-labeled vacuoles (autophagosomes) was counted using a Laser Scaning Confocal Microscope (LSM 510 Meta, Zeiss). More than 200 transfected cells were counted and only cells with at least 10 spots were counted as positive. The reported results were from at least 3 independent experiments.
Analysis of ROS production
Cells were transfected with ERAL1 siRNA for 72 h or treated with CAP (50 μg/ml) for 48 h. The cells were washed with PBS and then incubated in the dark with an oxidation-sensitive fluorescent probe dye, 2′7′-dichlorodihydrofluorescein diacetate (H2-DCFDA) (100 μM) for 30 min at 37°C. H2-DCFDA can be oxidized by cellular peroxides to the fluorescent compound 2,7-dichlorofluorescein (DCF). The fluorescence intensity of DCF was measured by flow cytometry with an excitation wavelength of 480 nm and an emission wavelength of 525 nm.
CASP3 activity assay
The activity of CASP3 was evaluated using a Caspase-3 Colorimetric Assay kit (BF3100) (R&D Systems). In brief, cells (2 × 106) were collected by centrifugation, rinsed with PBS, and then added to 50 μl of ice-cold cell lysis buffer (provided in the kit). After incubation on ice for 10 min, the samples were centrifuged at 10,000 × g for 2 min at 4°C. Then 50 μl of supernatant from each sample was transferred to a 96-well, flat-bottom microtiter plate (BD). Subsequently, 50 μl of 2 × Reaction Buffer (with freshly prepared DTT) and 5 μl of Caspase-3 Colorimetric Substrate (DEVD-pNA) were added. Samples were incubated at 37°C for 2 h before the absorbance was read on a TECAN Safire2 plate reader (Tecan Austria GmbH) at 405 nm, with the sample without DEVD-pNA as blank control.
Apoptosis assay
The ERAL1 siRNA or CAP-treated cells were fixed overnight in 70% ethanol. The cells were washed twice with PBS and then incubated in PBS containing RNase A (10 μg/ml) for 30 min at 37°C. The cells were then stained with propidium iodide (0.5 mg/ml) for 30 min at 4°C. The percentage of apoptotic cells were identified by the sub-G1 DNA content measured with flow cytometry.
Promoter activity analysis
HeLa cells (2 × 105 cells) were seeded in 24-well plates and grown for 24 h. The pGL3-basic vector (0.2 μg) or pGL3-TP53-promoter plasmid (0.2 μg) was transfected into cells together with the β-gal expressing plasmid (0.1 μg) using Lipofectamine 2000 (Invitrogen, 11668-019). Cells were treated with CAP in the presence or absence of NAC at 24 h post-plasmid transfection. After 48 h treatment with CAP, cells were washed twice with PBS, and lysed with 100 μl of passive lysis buffer (Promega, E1941). Luciferase activity was measured and normalized for efficiency of transfection by using the ratio of luciferase to β-gal activity. For each transfected cell line, the results were compared with the mean of pGL3-basic vector control levels and expressed as fold activity relative to pGL3-basic.
Analysis of TP53 ubiquitination
HCT-116 TP53+/+ cells were cotransfected with HA-Ubiquitin and TP53-Flag plasmid, and then treated with CAP (50 μg/ml). After 48 h treatment with CAP, cells were treated with 10 μM MG132 for 2 h and then lysed with RIPA buffer. The lysates were incubated with anti-Flag agarose beads (Macgene, China, IPK002) at 4°C overnight. Beads-bound proteins were then eluted in SDS sample buffer and subjected to western blotting analysis with anti-HA antibody to detect ubiquitinated proteins. The same membrane was stripped and reprobed with anti-Flag antibody to determine the TP53-Flag levels.
Statistical analyses
Values in the graph are shown as the mean ± SD of at least three experiments. Analysis of variance was used to assess the statistical significance of the differences, with a p value of < 0.01 being considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported in part by the National Nature Science Foundation of China (Nos. 31070714 and 30770030), the Fundamental Research Funds for the Central Universities (105566GK) and the Beijing NOVA Program (No. 2005B47). We thank Dr. Tamotsu Yoshimori (National Institute for Basic Biology) for providing the GFP-LC3 expression plasmid, Dr. Daniel S. Peeper (The Netherlands Cancer Institute) for providing the pGL3-TP53 promoter plasmid, Dr. Xiaobo Qiu (Beijing Normal University) for providing the HA-Ubiquitin expression plasmid, and Dr. Yusheng Cong (Beijing Normal University) for providing HCT-116 TP53+/+ and HCT-116 TP53−/− cells.
Glossary
Abbreviations:
- ERA
Escherichia coli Ras-like protein
- KH domain
hnRNPK homology domain
- mtDNA
mitochondrial DNA
- TP53
tumor protein 53
- MRPs
mitochondrial ribosomal proteins
- mETC
mitochondrial electron transfer chain
- ROS
reactive oxygen species
- H2-DCFDA
2′7′-dichlorodihydrofluorescein diacetate
- DRAM1
damage-regulated autophagy modulator 1
- CAP
chloramphenicol
- NAC
N-acetyl cysteine
- 3-MA
3-methyladenine
- CHX
cycloheximide
- MAPK
mitogen-activated protein kinase
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/20250
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/20250
References
- 1.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherra SJ, 3rd, Dagda RK, Chu CT. Review: autophagy and neurodegeneration: survival at a cost? Neuropathol Appl Neurobiol. 2010;36:125–32. doi: 10.1111/j.1365-2990.2010.01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez-Enriquez S, He L, Lemasters JJ. Role of mitochondrial permeability transition pores in mitochondrial autophagy. Int J Biochem Cell Biol. 2004;36:2463–72. doi: 10.1016/j.biocel.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17:422–7. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Kenmochi N, Suzuki T, Uechi T, Magoori M, Kuniba M, Higa S, et al. The human mitochondrial ribosomal protein genes: mapping of 54 genes to the chromosomes and implications for human disorders. Genomics. 2001;77:65–70. doi: 10.1006/geno.2001.6622. [DOI] [PubMed] [Google Scholar]
- 7.Pel HJ, Grivell LA. Protein synthesis in mitochondria. Mol Biol Rep. 1994;19:183–94. doi: 10.1007/BF00986960. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien TW. Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene. 2002;286:73–9. doi: 10.1016/s0378-1119(01)00808-3. [DOI] [PubMed] [Google Scholar]
- 9.Chen SM, Takiff HE, Barber AM, Dubois GC, Bardwell JC, Court DL. Expression and characterization of RNase III and Era proteins. Products of the rnc operon of Escherichia coli. J Biol Chem. 1990;265:2888–95. [PubMed] [Google Scholar]
- 10.Tu C, Zhou X, Tarasov SG, Tropea JE, Austin BP, Waugh DS, et al. The Era GTPase recognizes the GAUCACCUCC sequence and binds helix 45 near the 3′ end of 16S rRNA. Proc Natl Acad Sci U S A. 2011;108:10156–61. doi: 10.1073/pnas.1017679108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma MR, Barat C, Wilson DN, Booth TM, Kawazoe M, Hori-Takemoto C, et al. Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol Cell. 2005;18:319–29. doi: 10.1016/j.molcel.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 12.Britton RA, Chen SM, Wallis D, Koeuth T, Powell BS, Shaffer LG, et al. Isolation and preliminary characterization of the human and mouse homologues of the bacterial cell cycle gene era. Genomics. 2000;67:78–82. doi: 10.1006/geno.2000.6243. [DOI] [PubMed] [Google Scholar]
- 13.Ingram GC, Simon R, Carpenter R, Coen ES. The Antirrhinum ERG gene encodes a protein related to bacterial small GTPases and is required for embryonic viability. Curr Biol. 1998;8:1079–82. doi: 10.1016/s0960-9822(98)70445-2. [DOI] [PubMed] [Google Scholar]
- 14.Akiyama T, Gohda J, Shibata S, Nomura Y, Azuma S, Ohmori Y, et al. Mammalian homologue of E. coli Ras-like GTPase (ERA) is a possible apoptosis regulator with RNA binding activity. Genes Cells. 2001;6:987–1001. doi: 10.1046/j.1365-2443.2001.00480.x. [DOI] [PubMed] [Google Scholar]
- 15.Gohda J, Nomura Y, Suzuki H, Arai H, Akiyama T, Inoue J. Elimination of the vertebrate Escherichia coli Ras-like protein homologue leads to cell cycle arrest at G1 phase and apoptosis. Oncogene. 2003;22:1340–8. doi: 10.1038/sj.onc.1206287. [DOI] [PubMed] [Google Scholar]
- 16.Dennerlein S, Rozanska A, Wydro M, Chrzanowska-Lightowlers ZM, Lightowlers RN. Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem J. 2010;430:551–8. doi: 10.1042/BJ20100757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchiumi T, Ohgaki K, Yagi M, Aoki Y, Sakai A, Matsumoto S, et al. ERAL1 is associated with mitochondrial ribosome and elimination of ERAL1 leads to mitochondrial dysfunction and growth retardation. Nucleic Acids Res. 2010;38:5554–68. doi: 10.1093/nar/gkq305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brady NR, Hamacher-Brady A, Westerhoff HV, Gottlieb RA. A wave of reactive oxygen species (ROS)-induced ROS release in a sea of excitable mitochondria. Antioxid Redox Signal. 2006;8:1651–65. doi: 10.1089/ars.2006.8.1651. [DOI] [PubMed] [Google Scholar]
- 19.Azad MB, Chen Y, Gibson SB. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxid Redox Signal. 2009;11:777–90. doi: 10.1089/ars.2008.2270. [DOI] [PubMed] [Google Scholar]
- 20.Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010;584:1374–8. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 21.Ichimura Y, Komatsu M. Selective degradation of p62 by autophagy. Semin Immunopathol. 2010;32:431–6. doi: 10.1007/s00281-010-0220-1. [DOI] [PubMed] [Google Scholar]
- 22.Balaburski GM, Hontz RD, Murphy ME. p53 and ARF: unexpected players in autophagy. Trends Cell Biol. 2010;20:363–9. doi: 10.1016/j.tcb.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin S. p53, Autophagy and tumor suppression. Autophagy. 2005;1:171–3. doi: 10.4161/auto.1.3.2051. [DOI] [PubMed] [Google Scholar]
- 24.Liu B, Chen Y, St Clair DK. ROS and p53: a versatile partnership. Free Radic Biol Med. 2008;44:1529–35. doi: 10.1016/j.freeradbiomed.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G. Autophagy regulation by p53. Curr Opin Cell Biol. 2010;22:181–5. doi: 10.1016/j.ceb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Tasdemir E, Chiara Maiuri M, Morselli E, Criollo A, D’Amelio M, Djavaheri-Mergny M, et al. A dual role of p53 in the control of autophagy. Autophagy. 2008;4:810–4. doi: 10.4161/auto.6486. [DOI] [PubMed] [Google Scholar]
- 27.Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–34. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 28.Criollo A, Dessen P, Kroemer G. DRAM: a phylogenetically ancient regulator of autophagy. Cell Cycle. 2009;8:2319–20. doi: 10.4161/cc.8.15.9153. [DOI] [PubMed] [Google Scholar]
- 29.Kerley-Hamilton JS, Pike AM, Hutchinson JA, Freemantle SJ, Spinella MJ. The direct p53 target gene, FLJ11259/DRAM, is a member of a novel family of transmembrane proteins. Biochim Biophys Acta. 2007;1769:209–19. doi: 10.1016/j.bbaexp.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karbowski M, Kurono C, Wozniak M, Ostrowski M, Teranishi M, Soji T, et al. Cycloheximide and 4-OH-TEMPO suppress chloramphenicol-induced apoptosis in RL-34 cells via the suppression of the formation of megamitochondria. Biochim Biophys Acta. 1999;1449:25–40. doi: 10.1016/s0167-4889(98)00167-0. [DOI] [PubMed] [Google Scholar]
- 31.Leiter LM, Thatte HS, Okafor C, Marks PW, Golan DE, Bridges KR. Chloramphenicol-induced mitochondrial dysfunction is associated with decreased transferrin receptor expression and ferritin synthesis in K562 cells and is unrelated to IRE-IRP interactions. J Cell Physiol. 1999;180:334–44. doi: 10.1002/(SICI)1097-4652(199909)180:3<334::AID-JCP4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 32.Li CH, Tzeng SL, Cheng YW, Kang JJ. Chloramphenicol-induced mitochondrial stress increases p21 expression and prevents cell apoptosis through a p21-dependent pathway. J Biol Chem. 2005;280:26193–9. doi: 10.1074/jbc.M501371200. [DOI] [PubMed] [Google Scholar]
- 33.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 35.Brooks CL, Gu W. New insights into p53 activation. Cell Res. 2010;20:614–21. doi: 10.1038/cr.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim SJ, Hwang SG, Shin DY, Kang SS, Chun JS. p38 kinase regulates nitric oxide-induced apoptosis of articular chondrocytes by accumulating p53 via NFkappa B-dependent transcription and stabilization by serine 15 phosphorylation. J Biol Chem. 2002;277:33501–8. doi: 10.1074/jbc.M202862200. [DOI] [PubMed] [Google Scholar]
- 37.Pérez-Martínez X, Funes S, Camacho-Villasana Y, Marjavaara S, Tavares-Carreón F, Shingú-Vázquez M. Protein synthesis and assembly in mitochondrial disorders. Curr Top Med Chem. 2008;8:1335–50. doi: 10.2174/156802608786141124. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs HT. Disorders of mitochondrial protein synthesis. Hum Mol Genet. 2003;12(Spec No 2):R293–301. doi: 10.1093/hmg/ddg285. [DOI] [PubMed] [Google Scholar]
- 39.Scherz-Shouval R, Shvets E, Elazar Z. Oxidation as a post-translational modification that regulates autophagy. Autophagy. 2007;3:371–3. doi: 10.4161/auto.4214. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Kong X, Kang J, Su J, Li Y, Zhong J, et al. Oxidative stress induces parallel autophagy and mitochondria dysfunction in human glioma U251 cells. Toxicol Sci. 2009;110:376–88. doi: 10.1093/toxsci/kfp101. [DOI] [PubMed] [Google Scholar]
- 41.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–34. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 42.Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN. Phosphorylation of p53 serine 15 increases interaction with CBP. J Biol Chem. 1998;273:33048–53. doi: 10.1074/jbc.273.49.33048. [DOI] [PubMed] [Google Scholar]
- 43.Prigione A, Cortopassi G. Mitochondrial DNA deletions and chloramphenicol treatment stimulate the autophagic transcript ATG12. Autophagy. 2007;3:377–80. doi: 10.4161/auto.4239. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka A. Parkin-mediated selective mitochondrial autophagy, mitophagy: Parkin purges damaged organelles from the vital mitochondrial network. FEBS Lett. 2010;584:1386–92. doi: 10.1016/j.febslet.2010.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller C, Saada A, Shaul N, Shabtai N, Ben-Shalom E, Shaag A, et al. Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann Neurol. 2004;56:734–8. doi: 10.1002/ana.20282. [DOI] [PubMed] [Google Scholar]
- 47.Suomalainen A, Isohanni P. Mitochondrial DNA depletion syndromes--many genes, common mechanisms. Neuromuscul Disord. 2010;20:429–37. doi: 10.1016/j.nmd.2010.03.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.