Abstract
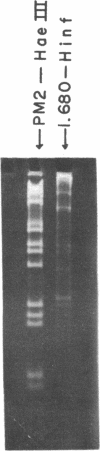
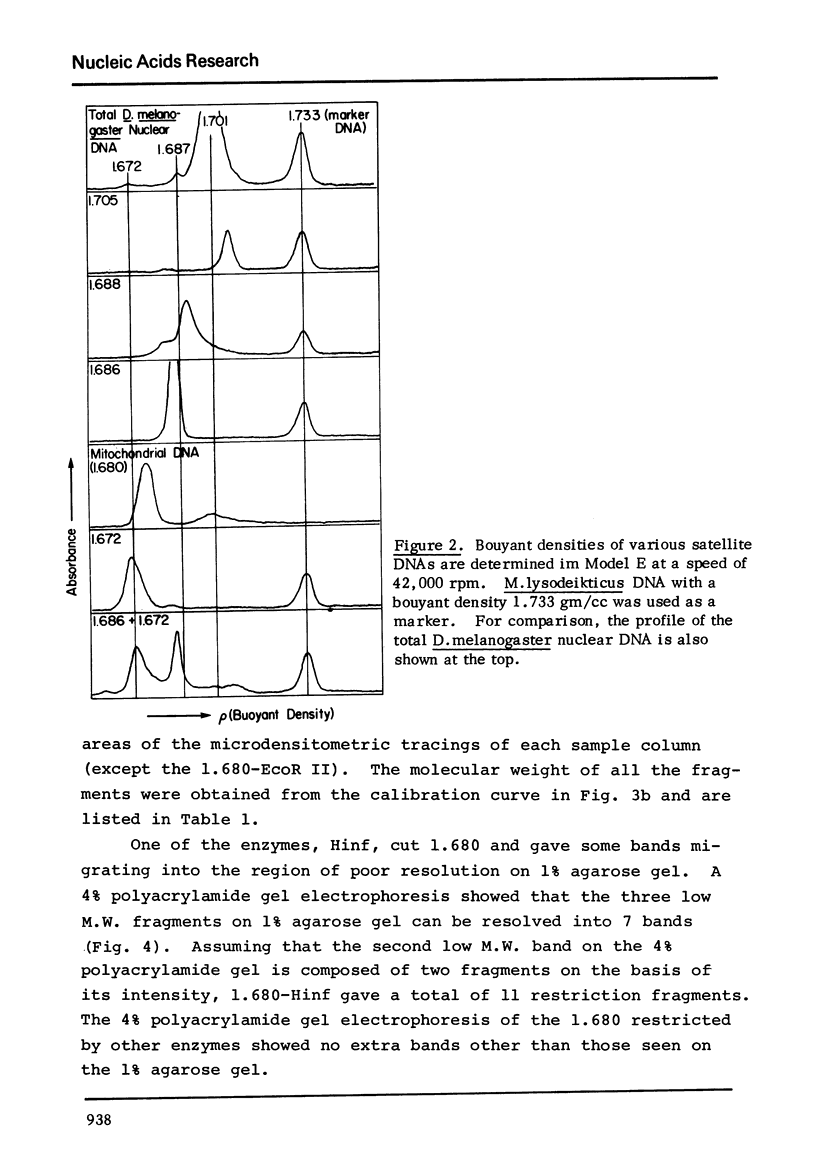
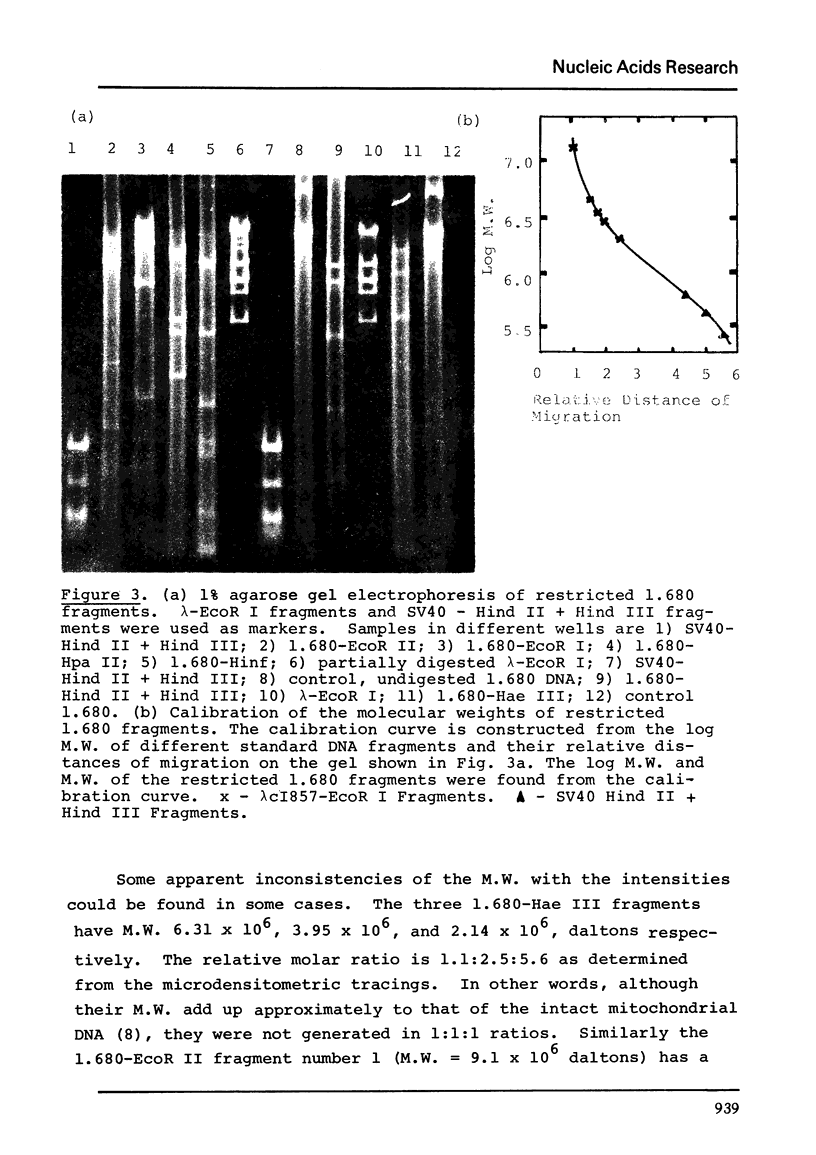
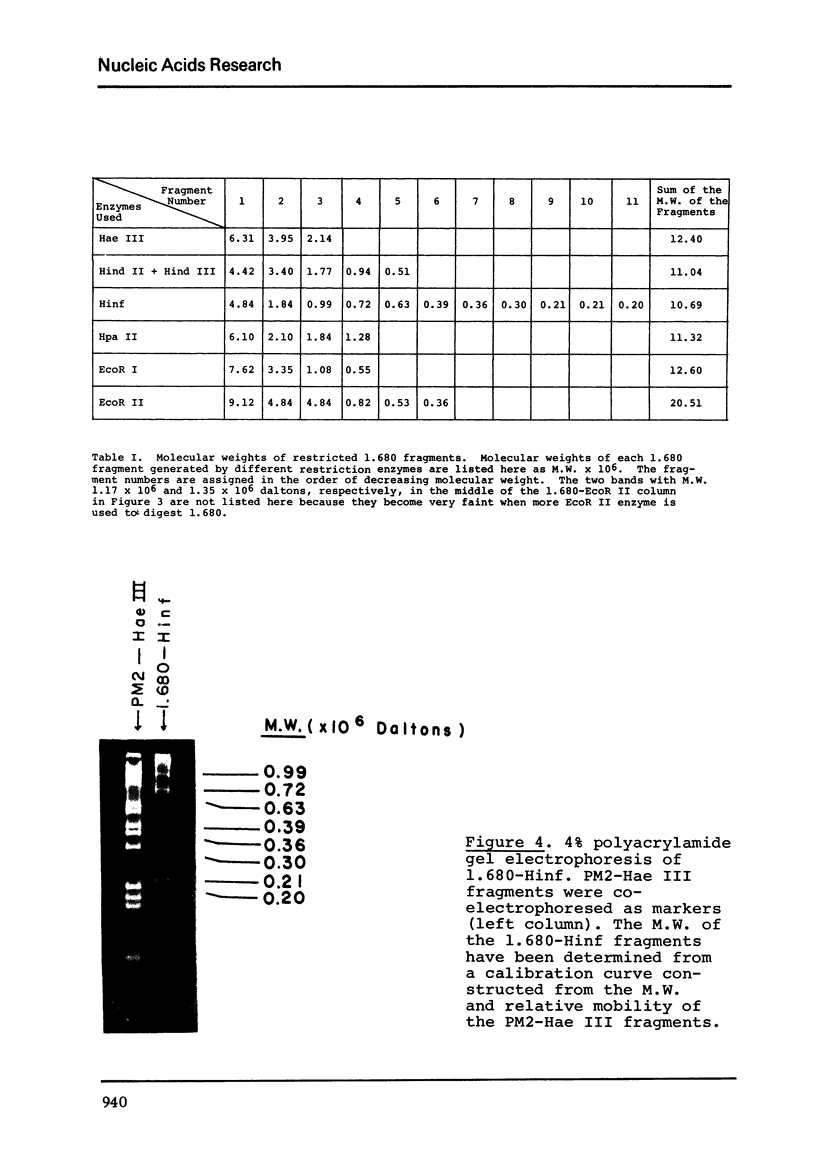
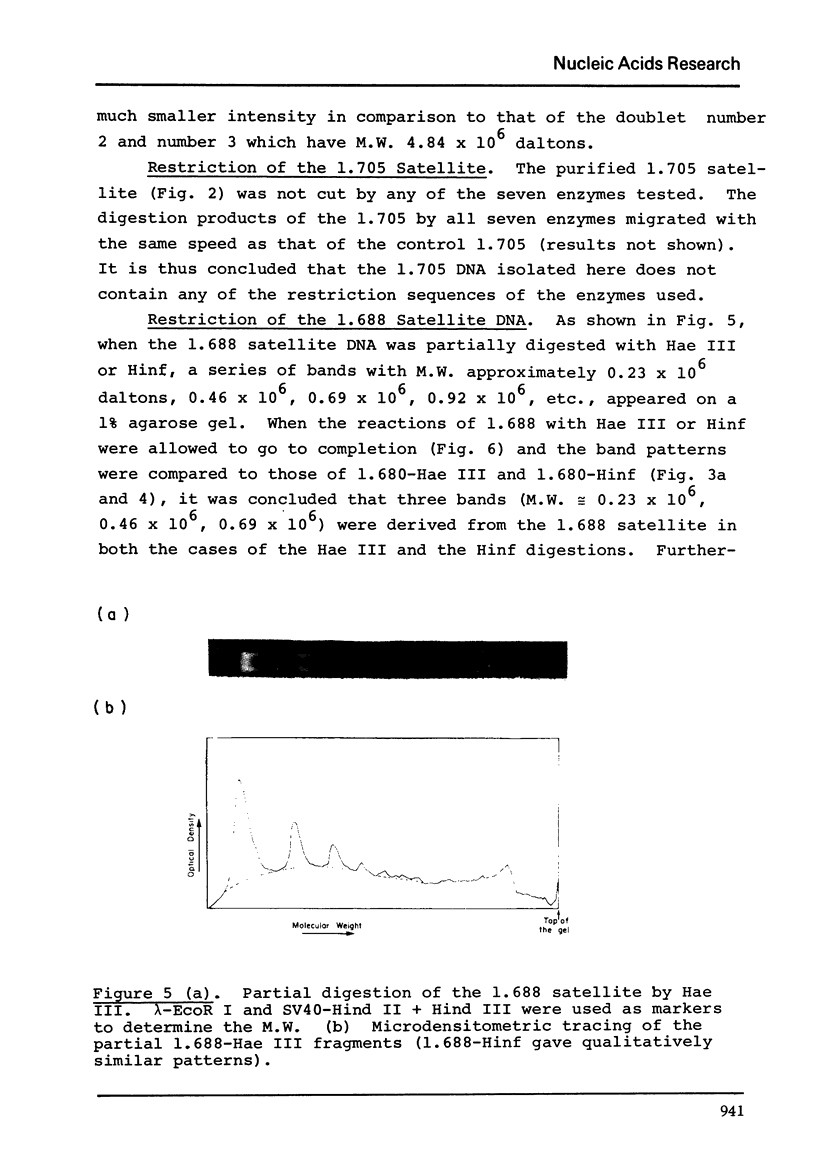
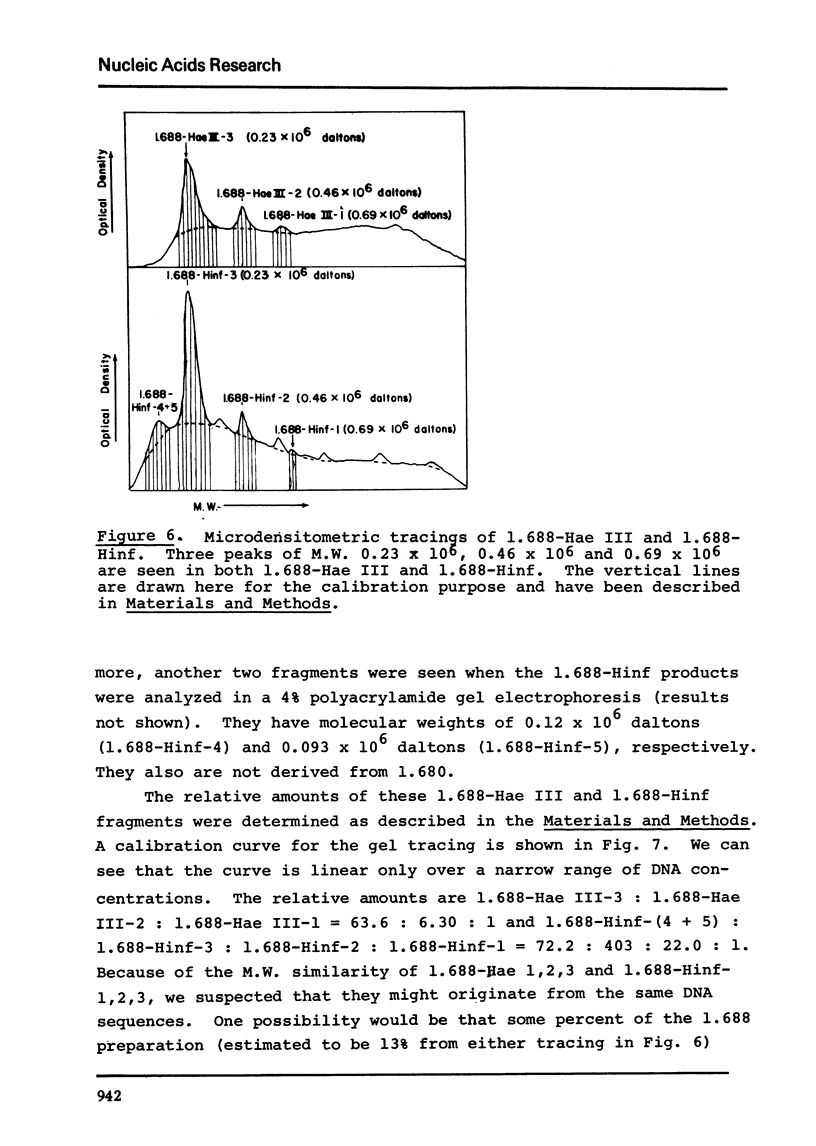
The five satellite DNAs of Drosophila melanogaster have been isolated by the combined use of different equilibrium density gradients and hydrolyzed by seven different restriction enzymes; Hae III, Hind II + Hind III, Hinf, Hpa II, EcoR I and EcoR II. The 1.705 satellite is not hydrolyzed by any of the enzymes tested. Hae III is the only restriction enzyme that cuts the 1.672 and 1.686 satellites. The cleavage products from either of these reactions has a heterogeneous size distribution. Part of the 1.688 satellite is cut by Hae III and by Hinf into three discrete fragments with M.W. that are multiples of 2.3 X 10(5) daltons (approximately 350 base pairs). In addition, two minor bands are detected in the 1.688-Hinf products. The mole ratios of the trimer, dimer and monomer are: 1:6.30 : 63.6 for 1.688-Hae III and 1 : 22.0 : 403 for 1.688-Hinf. Circular mitochondrial DNA (rho = 1.680) is cut into discrete fragments by all of the enzymes tested and molecular weights of these fragments have been determined.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blumenfled M., Forrest H. S. Is Drosophila dAT on the Y chromosome? Proc Natl Acad Sci U S A. 1971 Dec;68(12):3145–3149. doi: 10.1073/pnas.68.12.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M. R. Bovine satellite I DNA consists of repetitive units 1,400 base pairs in length. Nature. 1974 Sep 27;251(5473):288–292. doi: 10.1038/251288a0. [DOI] [PubMed] [Google Scholar]
- Botchan M., Kram R., Schmid C. W., Hearst J. E. Isolation and chromosomal localization of highly repeated DNA sequences in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971 Jun;68(6):1125–1129. doi: 10.1073/pnas.68.6.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W. Restriction and modification of DNA: enzymes and substrates. Introductory remarks. Fed Proc. 1974 May;33(5):1125–1127. [PubMed] [Google Scholar]
- Brown W. M., Vinograd J. Restriction endonuclease cleavage maps of animal mitochondrial DNAs. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4617–4621. doi: 10.1073/pnas.71.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna K. J., Sack G. H., Jr, Nathans D. Studies of simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol. 1973 Aug 5;78(2):363–376. doi: 10.1016/0022-2836(73)90122-8. [DOI] [PubMed] [Google Scholar]
- Endow S. A., Polan M. L., Gall J. G. Satellite DNA sequences of Drosophila melanogaster. J Mol Biol. 1975 Aug 25;96(4):665–692. doi: 10.1016/0022-2836(75)90145-x. [DOI] [PubMed] [Google Scholar]
- Gall J. G., Atherton D. D. Satellite DNA sequences in Drosophila virilis. J Mol Biol. 1974 Jan 5;85(4):633–664. doi: 10.1016/0022-2836(74)90321-0. [DOI] [PubMed] [Google Scholar]
- Gall J. G., Cohen E. H., Polan M. L. Reptitive DNA sequences in drosophila. Chromosoma. 1971;33(3):319–344. doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- Hearst J. E., Hanocq F., Kram R. The molecular organization of the very rapidly renaturing DNA sequences in Drosophila melanogaster: further evidence for a class of non-satellite simple sequence DNA. Biochimie. 1974;56(6-7):955–965. doi: 10.1016/s0300-9084(74)80517-1. [DOI] [PubMed] [Google Scholar]
- Hörz W., Hess I., Zachau H. G. Highly regular arrangement of a restriction-nuclease-sensitive site in rodent satellite DNAs. Eur J Biochem. 1974 Jun 15;45(2):501–512. doi: 10.1111/j.1432-1033.1974.tb03575.x. [DOI] [PubMed] [Google Scholar]
- Laird C. D., Chool W. Y., Cohen E. H., Dickson E., Hutchinson N., Turner S. H. Organization and transcription of DNA in chromosomes and mitochondria of Drosophila. Cold Spring Harb Symp Quant Biol. 1974;38:311–327. doi: 10.1101/sqb.1974.038.01.035. [DOI] [PubMed] [Google Scholar]
- Loening U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem J. 1967 Jan;102(1):251–257. doi: 10.1042/bj1020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteuil S., Hamer D. H., Thomas C. A., Jr Regular arrangement of restriction sites in Drosophila DNA. Cell. 1975 Aug;5(4):413–422. doi: 10.1016/0092-8674(75)90060-4. [DOI] [PubMed] [Google Scholar]
- Peacock W. J., Brutlag D., Goldring E., Appels R., Hinton C. W., Lindsley D. L. The organization of highly repeated DNA sequences in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:405–416. doi: 10.1101/sqb.1974.038.01.043. [DOI] [PubMed] [Google Scholar]
- Rae P. M. Chromosomal distribution of rapidly reannealing DNA in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1970 Oct;67(2):1018–1025. doi: 10.1073/pnas.67.2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasch E. M., Barr H. J., Rasch R. W. The DNA content of sperm of Drosophila melanogaster. Chromosoma. 1971;33(1):1–18. doi: 10.1007/BF00326379. [DOI] [PubMed] [Google Scholar]
- Sederoff R., Lowenstein L. Polypyrimidine segments in Drosophila melanogaster DNA: II. Chromosome location and nucleotide sequence. Cell. 1975 Jun;5(2):183–194. doi: 10.1016/0092-8674(75)90026-4. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Corden J. L., Sahasrabuddhe C. G., Van Holde K. E. Chromatographic separation of chromatin subunits. Biochem Biophys Res Commun. 1974 Dec 23;61(4):1193–1198. doi: 10.1016/s0006-291x(74)80410-9. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Long range periodicities in mouse satellite DNA. J Mol Biol. 1975 May 5;94(1):51–69. doi: 10.1016/0022-2836(75)90404-0. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Walker P. M. "Repetitive" DNA in higher organisms. Prog Biophys Mol Biol. 1971;23:145–190. doi: 10.1016/0079-6107(71)90019-8. [DOI] [PubMed] [Google Scholar]
- Zeiger R. S., Salomon R., Peacock A. C. Isolation of mouse satellite deoxyribonucleic acid by composite polyacrylamide gel electrophoresis. Biochemistry. 1971 Nov;10(23):4219–4223. doi: 10.1021/bi00799a010. [DOI] [PubMed] [Google Scholar]