Abstract
Tobacco smoke-induced accelerated cell senescence has been implicated in the pathogenesis of chronic obstructive pulmonary disease (COPD). Cell senescence is accompanied by the accumulation of damaged cellular components suggesting that in COPD, inhibition of autophagy may contribute to cell senescence. Here we look at whether autophagy contributes to cigarette smoke extract (CSE) - induced cell senescence of primary human bronchial epithelial cells (HBEC), and further evaluate p62 and ubiquitinated protein levels in lung homogenates from COPD patients. We demonstrate that CSE transiently induces activation of autophagy in HBEC, followed by accelerated cell senescence and concomitant accumulation of p62 and ubiquitinated proteins. Autophagy inhibition further enhanced accumulations of p62 and ubiquitinated proteins, resulting in increased senescence and senescence-associated secretory phenotype (SASP) with interleukin (IL)-8 secretion. Conversely, autophagy activation by Torin1, a mammalian target of rapamycin (mTOR inhibitor), suppressed accumulations of p62 and ubiquitinated proteins and inhibits cell senescence. Despite increased baseline activity, autophagy induction in response to CSE was significantly decreased in HBEC from COPD patients. Increased accumulations of p62 and ubiquitinated proteins were detected in lung homogenates from COPD patients. Insufficient autophagic clearance of damaged proteins, including ubiquitinated proteins, is involved in accelerated cell senescence in COPD, suggesting a novel protective role for autophagy in the tobacco smoke-induced senescence-associated lung disease, COPD.
Keywords: autophagy, COPD, p62, senescence, ubiquitin
Introduction
Chronic obstructive pulmonary disease (COPD) is caused by the noxious effects of tobacco smoke, which leads to airway epithelial cell injury and the induction of phenotypic changes such as squamous metaplasia and cellular senescence, which are assumed to be part of the adaptive response to toxic components such as reactive oxygen species (ROS).1 Recently, accelerated cell senescence has been demonstrated in COPD airway and lung and is now widely implicated in pathogenesis of COPD in terms of not only impaired cell repopulation following apoptotic cell loss but also aberrant cytokine secretions of senescence-associated secretory phenotype (SASP).2,3 SASP exert deleterious effects on the tissue microenvironment of neighboring cells, resulting in cancer development.4,5 Indeed, epidemiological studies indicate that COPD is one of the most important risk factors for lung cancer after smoking exposure, with airway inflammation as a mechanism for lung cancer recently proposed.6,7
Tobacco smoke induces both cell division-dependent replicative senescence, (linked to telomere shortening), as well as stress-induced premature senescence.8 The molecular mechanisms for regulation of cell senescence are complex and incompletely understood, but are generally thought to result from increased oxidative stress. One of the typical manifestations of cell senescence is accumulation of damaged proteins and organelles, occasionally associated with ubiquitinated aggregations.9 It has been widely proposed that the functional insufficiency of cellular cleaning and housekeeping mechanisms for protein and organelle degradation, including proteasome and autophagy pathways, play a pivotal role in the accumulations of deleterious cellular components and regulation of cell senescence.9,10
Proteasomes, with a narrow range of catalytic pore size, restricts the clearance of ubiquitin-conjugated soluble proteins, whereas autophagy is responsible for the degradation of aggregate-prone proteins and damaged organelles, suggesting the predominant contribution of autophagy to the regulation of cell senescence.11 Autophagy, a lysosomal degradation pathway, occurs not only continuously at basal levels for homeostatic turnover of cytoplasmic components but is also induced in response to a variety of stress stimuli including oxidative stress.12 Recent studies revealed interactions between the proteasome and autophagy pathways during degradation of ubiquitinated proteins by autophagy.13 p62/SQSTM1 has been shown to be an adaptor protein capable of binding both polyubiquitinated substrates and Atg8/LC3, an indispensable component for autophagosome formation, and hence is thought to play a key regulatory role in the delivery of ubiquitinated proteins for selective autophagic degradation. Therefore, accumulation of p62 and ubiquitinated proteins is proposed to be a representative indicator of insufficient autophagic clearance.
Collectively, it is reasonable to hypothesize that insufficiency of autophagic degradation of damaged proteins and organelles may be involved in the acceleration of cell senescence caused by tobacco smoke exposure. Recent studies have demonstrated that increased activation of autophagy occurs in lungs of patients with COPD, and concomitant induction of apoptosis was implicated in a part of the pathogenic sequence for COPD development.14 However, the implication of tobacco smoke exposure-induced autophagic regulation in clearance of damaged proteins for the regulation of human bronchial epithelial cell (HBEC) senescence has not been elucidated. In this context, we examined the regulatory role of autophagy in cigarette smoke extract (CSE) -induced cell senescence of primary HBEC.
Results
CSE transiently induces autophagy activation in human bronchial epithelial cells
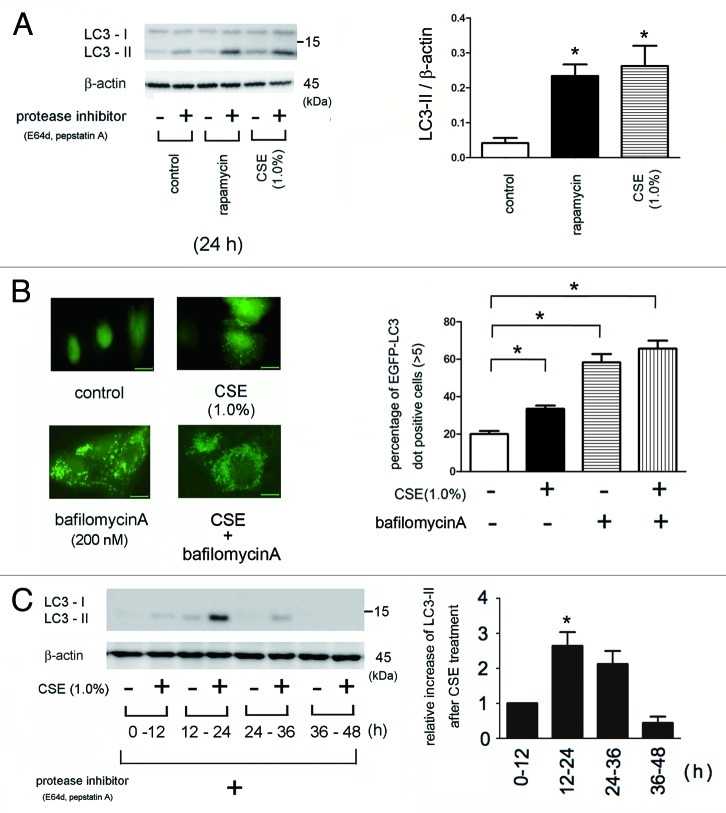
Western blotting for the conversion of LC3 from LC3-I (free form) to LC3-II (phosphatidylethanolamine-conjugated form) and detection of EGFP-LC3 dot formation by fluorescence microscopy were performed to evaluate the activation of autophagy.15 CSE significantly activated autophagy in BEAS-2B cells, consistent with previous reports (Fig. 1A,B).14 Treatment with rapamycin alone, an mTOR inhibitor, increased autophagy activation to a similar level as that of CSE (Fig. 1A), as previously described.16 CSE and bafilomycin A, an autophagy inhibitor via lysosomal proton pump inhibition, clearly induced EGFP-LC3 dot formation. CSE also induced autophagy in HBEC, which peaked 12 – 24 h after treatment and was barely detected after 36h (Fig. 1C), indicating that CSE-induced autophagy was transient.
Figure 1. Autophagy activation in response to CSE exposure. (A) western blotting (WB) using anti-LC3 and anti-β-actin in control treated (lane 1, 2), rapamycin treated (1 μM) (lane 3, 4), CSE treated (lane 5 to 6) in the presence or absence of protease inhibitors (E64d 10 μg/ml and pepstatin A10 μg/ml) in Beas2B cells. Protein samples were collected after 24 h treatment with indicated concentrations of CSE. Shown is a representative experiment of three similar results. The right panel is the densitometric analysis of WB from three independent experiments. LC3-II / β-actin in the presence of protease inhibitors was subtracted by LC3-II / β-actin without protease inhibitors. *p < 0.05. (B) Fluorescence microscopic detection of pEGFP-LC3 dot formation in BEAS-2B cells: BEAS-2B cells with stable expression of pEGFP-LC3 were treated with bafilomycin A (200 nM), CSE (1.0%), or both bafilomycin A and CSE for 24h. Photomicrographs are taken at the same magnification. (Original magnification, 1000X) Bar = 10μm. The right panel is the percentage of positive cells with more than five dot formations and data were collected from three independent experiments. *p < 0.05. (C) WB using anti-LC3 and anti-β-actin for indicated time points of control or CSE (1.0%) treatment in the presence of protease inhibitors (E64d and pepstatin A) in HBEC from non-COPD patients. CSE treatment was started from 0h to sample collection and protease inhibitors were added during indicated time period. Shown is a representative experiment of three similar results. The lower panel is the relative increase of LC3-II after CSE exposure taken from densitometric analysis of WB. *p < 0.05.
Autophagy activation regulates CSE-induced cell senescence in human bronchial epithelial cells
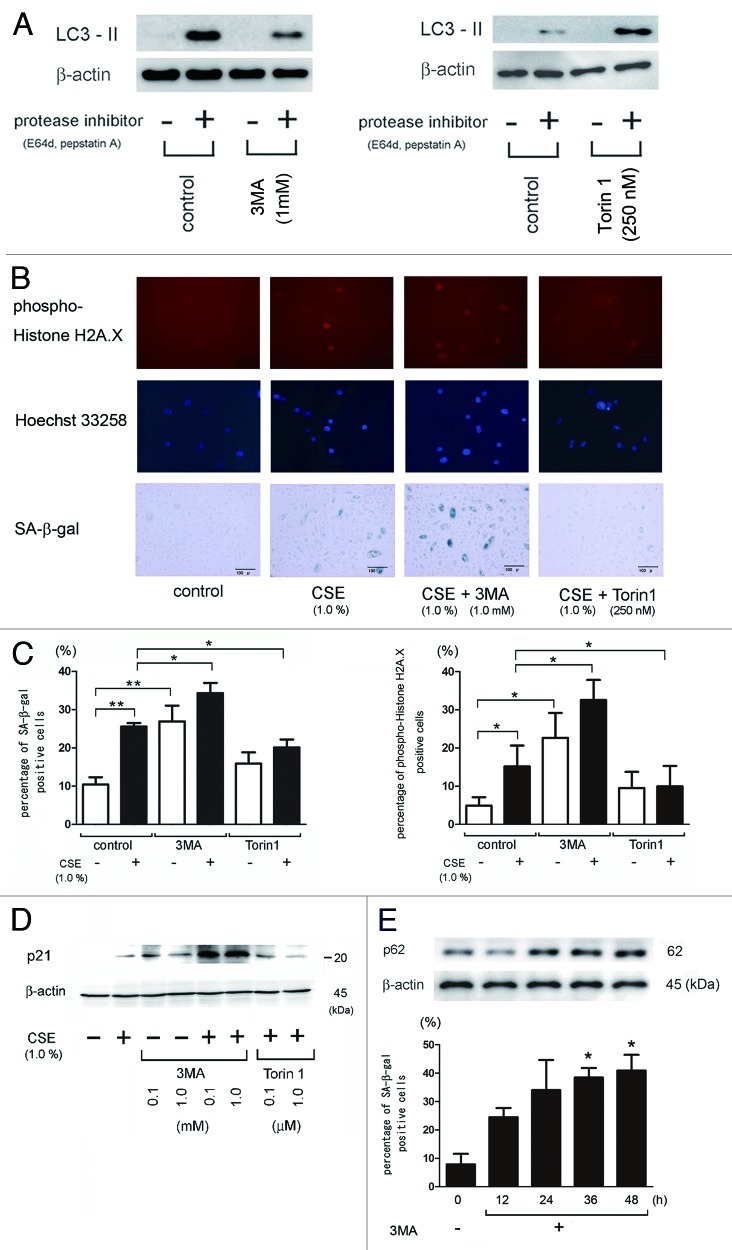
To elucidate the regulatory role of autophagy in CSE-induced HBEC senescence, we used 3MA and Torin1 as an inhibitor and an inducer of autophagy, respectively. 3MA inhibited autophagy flux and Torin1, a selective and potent small molecule inhibitor of mTOR, effectively induced autophagy (Fig. 2A).17 CSE (1.0%) significantly increased the percentage of senescent cells as measured by both an increase in phospho-Histone H2A.X (Ser139) staining of DNA damage and by SA-β-Gal staining, which were enhanced in the presence of 3MA, an autophagic sequestration blocker (Fig. 2B and C). Western blotting for p21/waf-1, a senescence-associated cyclin-dependent kinase inhibitor, also demonstrated enhanced expression with CSE treatment, particularly in the presence of 3MA (Fig. 2D). In contrast, Torin1 suppressed CSE-induced HBEC senescence since it significantly reduced phospho-Histone H2A.X (Ser139) staining, SA-β-Gal staining, and p21/waf-1 expression (Fig. Two B, C and D). Interestingly, 3MA alone induced HBEC senescence to the similar extent as CSE exposure (Fig. 2C, D). To further clarify the direct link between functional impairment of autophagy and HBEC senescence, a time course of 3MA effect on HBEC senescence was evaluated without CSE exposure. 3MA significantly increased the percentage of SA-β-Gal positive staining in a time dependent manner and 3MA-mediated autophagy inhibition was confirmed by accumulations of p62 (Fig. 2E).
Figure 2. Involvement of autophagy in regulation of CSE-induced HBEC senescence. (A) The left panel is western blotting (WB) using anti-LC3 and anti-β-actin in control treated (lane 1, 2) and 3MA treated (1 mM) (lane 3, 4) and in the presence or absence of protease inhibitors (E64d 10 μg/ml and pepstatin A10 μg/ml) in HBEC. The right panel is western blotting (WB) using anti-LC3 and anti-β-actin in control treated (lane 1, 2) and Torin1 (250 nM) treated (lane 3, 4) in the presence or absence of protease inhibitors (E64d 10 μg/ml and pepstatin A10 μg/ml) in HBEC. Protein samples were collected after a 24 h treatment. Shown is a representative experiment of three similar results. (B) Photographs of immunofluorescent staining of phospho-Histone H2A.X (Ser139) (upper panels), Hoechst 33258 staining (middle panels), and senescence associated β-galactosidase (SA-β-gal) staining (lower panels) of control or CSE (1.0% for 48 h) treated HBEC in the presence or absence of 3MA (1.0 mM) and Torin1 (250 nM). Hoechst 33258 staining corresponds to phospho-Histone H2A.X staining. Bar = 50μm in IF and 100μm in SA-β-gal staining. (C) Shown in left panel is the percentage (± SEM) of SA-β-gal positive cells from three independent experiments. Open bar is no treatment and filled bar is CSE (1.0% for 48 h) treated in HBEC. *p < 0.05. **p < 0.001. Shown in right panel is the percentage (± SEM) of phospho-Histone H2A.X (Ser139) positive cells from three independent experiments. Open bar is no treatment and filled bar is CSE (1.0% for 48 h) treated in HBEC. *p < 0.05. (D) western blot (WB) using anti-p21 or β-actin in control treated (lane 1), CSE treated (lane 2), control treated in the presence of 3MA (1.0mM) (lane 3,4), CSE treated in the presence of 3MA (1mM) (lane 5,6), and CSE treated in the presence of torin1(lane 7,8) in HBEC. Protein samples were collected after 48 h treatment with CSE. Shown is a representative experiment of three similar results. (E) WB using anti-p62 or anti-β-actin of cell lysates from HBEC for indicated time points of control or 3MA (1mM) treatment (upper panels). SA-β-gal staining of indicated time points after 3MA (1mM) treatment in HBEC. Shown in lower panel is the percentage (± SEM) of SA-β-gal positive cells from three independent experiments. *p < 0.05.
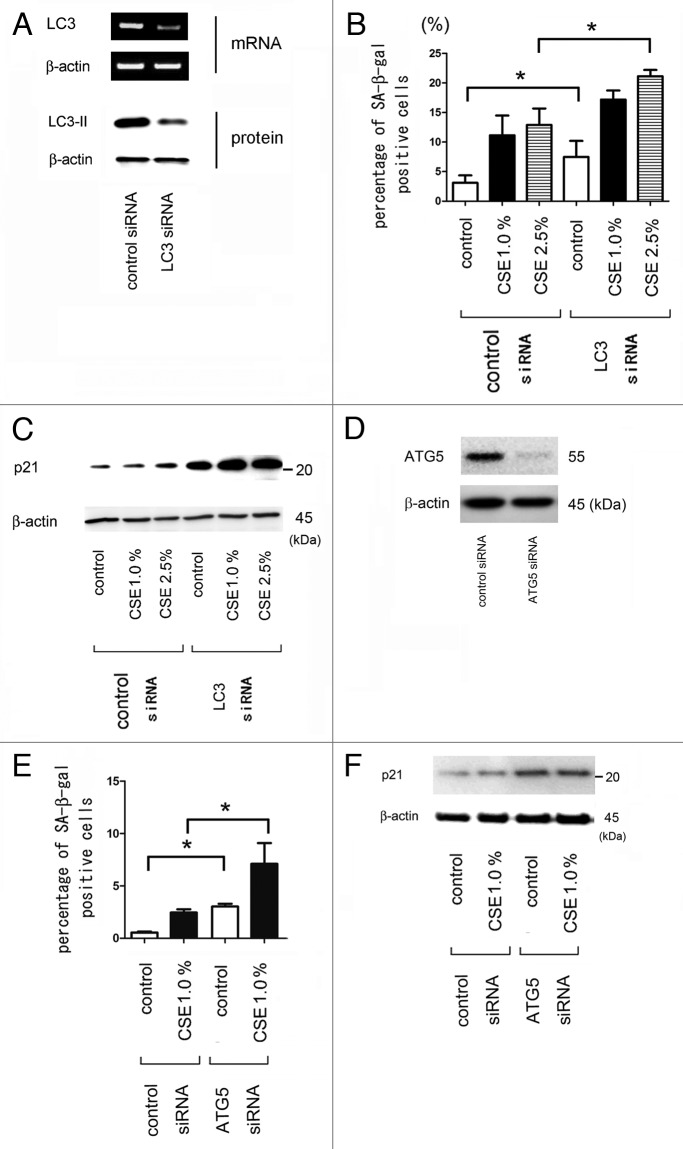
Next, HBEC were transfected with siRNA to LC3B, which inhibits autophagosome formation. RT-PCR and western blotting of HBEC showed that LC3B siRNA efficiently reduced both mRNA and protein levels of LC3B (Fig. 3A). Knockdown of LC3 was further confirmed by the lack of dot formation of EGFP-LC3 in stably transfected BEAS-2B cells (data not shown). Knockdown of LC3B resulted in an increased percentage of senescent cells in response to CSE treatment in HBEC as determined by an increase in SA-β-Gal staining (Fig. 3B) and p21/waf-1 expression (Fig. 3C). LC3B knockdown alone also enhanced cell senescence in the absence of CSE treatment (Fig. 3B and C). To further confirm the involvement of autophagy, HBEC were also transfected with siRNA to ATG5, which is an essential component for autophagy. In concordance with the data of LC3 siRNA, ATG5 knockdown significantly enhanced CSE-induced HBEC senescence by means of SA-β-Gal staining (Fig. 3E) and p21/waf-1 expression (Fig. 3F), suggesting that autophagy activation negatively regulates CSE-induced HBEC senescence.
Figure 3. Effect of LC3 and ATG5 knock down on CSE-induced cell senescence in HBEC. (A) RT-PCR (upper panel), using primers to LC3B and β-actin, was performed from total RNA harvested from control siRNA (lane 1) and LC3B siRNA (lane 2) transfected HBEC after 48 h incubation. WB (lower panel) using LC3 and anti- β-actin of cell lysates from control siRNA (lane 1) and LC3 siRNA (lane 2) transfected HBEC after 48 h in the presence of protease inhibitors (E64d 10 μg/ml and pepstatin A10 μg/ml). (B) Senescence associated β-galactosidase (SA-β-gal) staining of control or LC3B siRNA transfected HBEC. Shown in panel is the percentage (± SEM) of SA-β-gal positive cells from three independent experiments. Open bar is no treatment and filled bar is CSE (1.0% for 48 h) treated and horizontal crosshatched bar is CSE (2.5% for 48 h) treated. *p < 0.05. (C) western blot (WB) using anti-p21 or β-actin in control or LC3B siRNA transfected HBEC. HBEC were treated with control (lane 1, 4), CSE (1.0%) (lane 2, 5), and CSE (2.5%) (lane 3, 6). Protein samples were collected after 48 h treatment with CSE. Shown is a representative experiment of 3 showing similar results. (D) western blot (WB) using ATG5 and anti- β-actin of cell lysates from control siRNA (lane 1) and ATG5 siRNA (lane 2) transfected HBEC after 48 h. (E) Senescence associated β-galactosidase (SA-β-gal) staining of control or ATG5 siRNA transfected HBEC. Shown in panel is the percentage (± SEM) of SA-β-gal positive cells from three independent experiments. Open bar is no treatment and filled bar is CSE (1.0% for 48 h) treated. *p < 0.05. (F) western blot (WB) using anti-p21 or β-actin in control or ATG5 siRNA transfected HBEC. HBEC were treated with control (lanes 1 and 4), and CSE (1.0%) (lanes 2 and 4). Protein samples were collected after 48 h treatment with CSE. Shown is a representative experiment of three similar results.
Autophagy regulates accumulations of ubiquitinated protein and p62 in response to CSE exposure
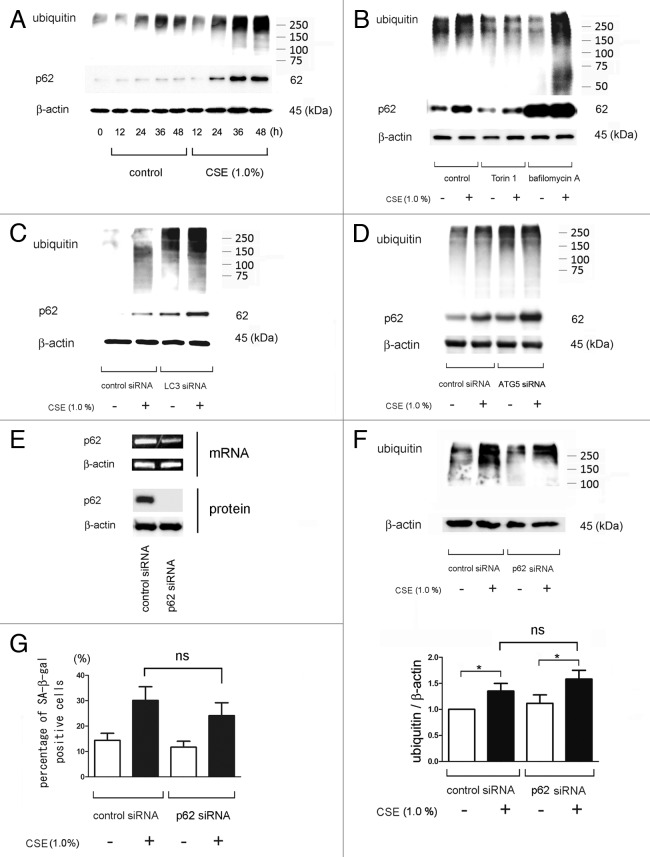
We next examined the changes of expression levels of ubiquitinated proteins and p62 as indicator for autophagic clearance during CSE treatment. CSE (1.0%) induced accumulation of both ubiquitinated proteins and p62 in a time dependent manner, especially after 24 h incubation, which corresponds to a decline of autophagy activity (Fig. 1C and 4A). Degradation of ubiquitinated proteins and p62 was inhibited by bafilomycin A, LC3B and ATG5 knock down (Fig. 4B-D). Conversely, Torin1 clearly reduced the accumulations of ubiquitinated proteins and p62 in response to CSE exposure (Fig. 4B).
Figure 4. Accumulations of ubiquitinated protein and p62 in response to CSE exposure in HBEC. (A) WB using anti-ubiquitin, anti-p62, or anti- β-actin of cell lysates from HBEC for indicated time points of control or CSE (1.0%) treatment. Shown is a representative experiment of 3 showing similar results. (B) WB using anti-ubiquitin, anti-p62, or anti- β-actin. HBEC were exposed to CSE (1.0% for 48 h) in the absence (lane 1, 2) or presence of torin1 (250 nM) (lane 3, 4) and bafilomycin A (200 nM) (lane 5, 6). (C) WB using anti-ubiquitin, anti-p62, or anti- β-actin of cell lysates from control siRNA or LC3B siRNA transfected HBEC. CSE (1.0% for 48 h) treatment was started 24 h post-siRNA transfection. (D) WB using anti-ubiquitin, anti-p62, or anti- β-actin of cell lysates from control siRNA or ATG5 siRNA transfected HBEC. CSE (1.0% for 48 h) treatment was started 24 h post-siRNA transfection. (E) RT-PCR (upper panel), using primers to p62 and β-actin, was performed from total RNA harvested from control siRNA (lane 1) and p62 siRNA (lane 2) transfected HBEC after 48 h incubation. WB (lower panel) using anti-p62 and anti-β-actin of cell lysates from control siRNA (lane 1) and LC3 siRNA (lane 2) transfected HBEC after 48 h incubation. Shown is a representative experiment of 3 showing similar results. (F) WB using anti-ubiquitin and anti- β-actin of cell lysates from control siRNA or p62 siRNA transfected HBEC. CSE (1.0% for 48 h) treatment was started 24 h post-siRNA transfection. In the lower panel is the average (± SE) taken from densitometric analysis of WB of four independent experiments shown as relative expression of ubiquitin compared with β-actin. Open bar is no treatment and filled bar is CSE (1% for 48 h). (G) Shown in panel is the percentage (± SEM) of SA-β-gal positive cells from three independent experiments. Open bar is no treatment and filled bar is CSE (1.0% for 48 h) treated HBEC. ns = not significant
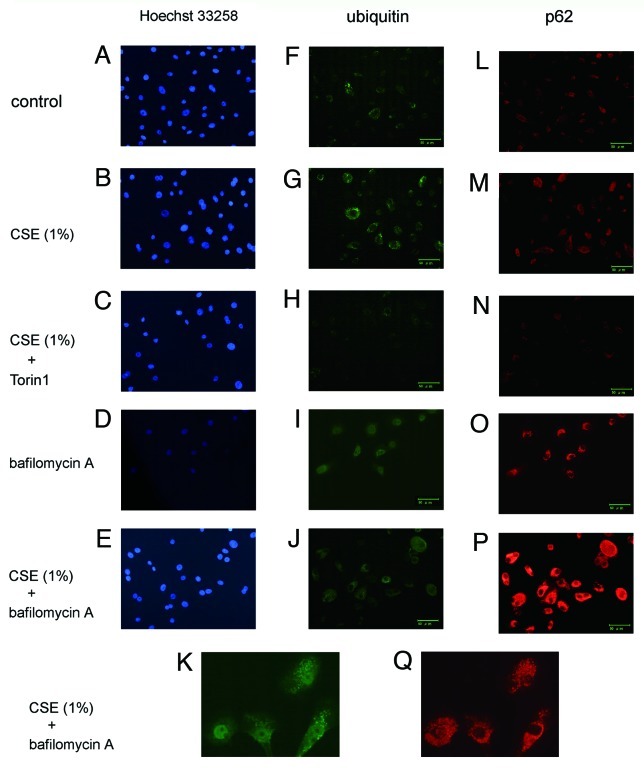
To elucidate the involvement of p62 in ubiquitinated protein accumulation in the progression of cellular senescence, we employed p62 siRNA. HBECs were transfected with siRNA targeting p62, and RT-PCR and WB analysis 48 h after transfection showed that p62 siRNA very efficiently reduced the amount of p62 mRNA and protein (Fig. 4E). Knock-down of p62 did not effect CSE-induced accumulation of ubiquitinated proteins and cell senescence (Fig. 4F, G). Interestingly, p62 knockdown led to a slight but insignificant suppression of senescence both in the presence and absence of CSE (Fig. 4G). Immunofluorescence staining demonstrated that enhanced expression of cytoplasmic ubiquitin, p62, and protein aggregations were induced in response to CSE exposure (Fig. 5G and M) and a marked enhancement of ubiquitin and p62 positive aggregations was observed in the presence of bafilomycin A (Fig. 5I-K and O-Q). Torin1 clearly suppressed accumulations of p62 and ubiquitinated protein (Fig. 5H, N). The changes of cytoplasmic accumulation in response to CSE and autophagy regulation occurred similarly in both ubiquitin and p62. Taken together, these data support an important regulatory role for autophagy in the clearance of ubiquitinated aggregates via a p62-mediated pathway.
Figure 5. Immunofluorescence staining of ubiquitin and p62 in HBEC. Photomicrographs of nuclear staining with Hoechst 33258 (panels A to E), immunofluorescence staining with anti-ubiquitin (panels F to K), and anti-p62 (panels L to Q). HBEC were control treated (A, F, L) or CSE (1.0%) treated (B, C, E, G, H, J, K, M, N, P, and Q), in the presence of Torin1 (250 nM)(C, H, N), or bafilomycin A (200 nM) (D, E, I to K, O to Q). All photomicrographs are taken at the same magnification. Original magnification is × 200. Bar = 50 μm. K and Q are high magnification view.
Autophagy regulates the senescence-associated secretory phenotype of IL-8 expression in human bronchial epithelial cells
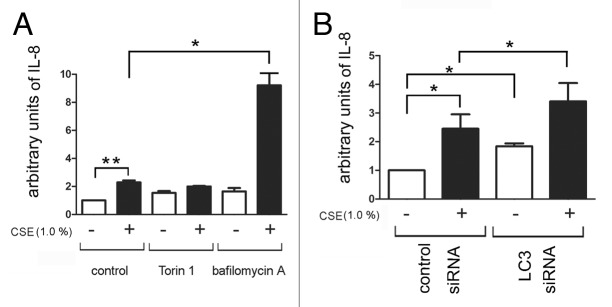
We evaluated IL-8 secretion in response to CSE exposure, since increased IL-8 secretion is known to be associated with cell senescence.5 CSE significantly induced the secretion of IL-8, and IL-8 secretion was enhanced in the presence of bafilomycin A (Fig. 6A). Autophagic regulation of IL-8 secretion was further demonstrated by siRNA-mediated knock down of LC3B, which showed clearly enhanced IL-8 secretion in response to CSE exposure (Fig. 6B).
Figure 6. IL-8 secretion by autophagy inhibition in CSE-induced senescent HBEC. For preparation of the conditioned medium, HBEC were treated with CSE (1.0%) in the absence or presence of Torin1 (250 nM) and bafilomycin A (200 nM) for 24h, washed three times with PBS, then incubated in fresh BEGM for 48h. (A) ELISA showing IL-8 in conditioned media with or without CSE-treatment in the absence (lane 1, 2), or presence of Torin1 (lane 3, 4) and bafilomycin A (lane 5, 6). Shown is the arbitrary units ± SEM *p < 0.05. **p < 0.001. (B) ELISA showing IL-8 in conditioned media from control siRNA (lane 1, 2) or LC3B siRNA (lane 3, 4) transfected HBEC. CSE (1.0% for 48 h) treatment was started 24 h post-siRNA transfection. Shown is the arbitrary units ± SEM *p < 0.05.
Impaired Autophagy induction in response to CSE exposure in HBEC from COPD patients
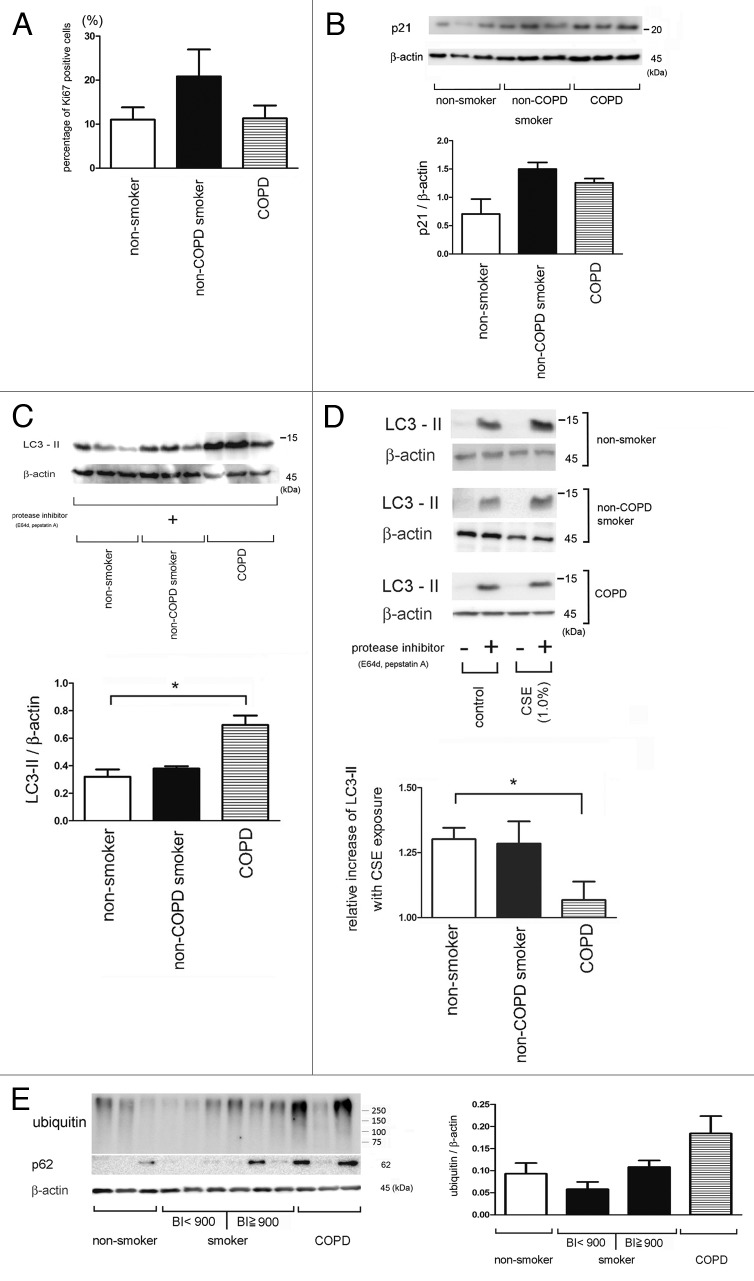
We have compared baseline activity and induction of autophagy in response to CSE exposure in HBEC. Our results suggest that autophagy may play a role in COPD pathogenesis. Initially, we compared cell proliferation, senescence, and oxidative stress, respectively, in HBEC (Table 1). Cell proliferation tended to be increased in HBEC from non-COPD smokers compared with HBEC from non-smokers and COPD patients (Fig. 7A). In contrast, carbonylated proteins were increased in HBEC from COPD patients compared with HBEC from non-COPD patients, reflecting increased oxidative stress (published data).24 However, there were no significant differences in p21 expression levels (Fig. 7B). Baseline autophagy activity (without CSE treatment) in HBEC from COPD patients was significantly increased compared with those of HBEC from non-smokers and non-COPD smokers (Fig. 7C). In contrast, autophagy induction in response to CSE exposure was significantly attenuated in HBEC from COPD patients compared with that in HBEC from non-smokers and non-COPD smokers (Fig. 7D). To further elucidate the functional status of autophagy in lung tissue, we evaluated the expression levels of ubiquitinated proteins and p62 in lung homogenates from non-smokers, non-COPD smokers, and COPD patients. Relatively high accumulations of ubiquitinated proteins and p62 were detected in lung homogenates from COPD patients, compared with those detected in lung homogenates from heavy smokers (Brinkman index ≥ 900) and non-smokers/light smokers (Brinkman index < 900) (Fig. 7E and Table 2). Importantly, no significant age-related differences were observed between each group (Table 1 and 2). Taken together, these data suggest that insufficient autophagy activation in response to cigarette smoke exposure may be responsible for accumulation of ubiquitinated proteins and p62 in lung homogenates from COPD patients.
Table 1. Patient characteristics (for HBEC).
| Non-smoker (n = 5) |
Non-COPD smoker (n = 5) |
COPD (n = 5) |
p value | |
|---|---|---|---|---|
| Age, years |
63.8 ± 14.2 |
67.2 ± 8.2 |
66.0 ± 4.8 |
NS |
| Male, % of group |
20 |
80 |
100 |
NA |
| Brinkman Index |
0 |
518 ± 374.4 |
1010 ± 391.2 |
NA |
| FEV1/FVC | 73.8 ± 3.9 | 79.1 ± 1.3 | 60.2 ± 12.7 | NA |
Abbreviations: COPD, chronic obstructive pulmonary disease; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; NA, not assessed; NS, not statistically significant. Values are mean ± SD.
Figure 7. Autophagy activity in HBEC and lung homogenates from COPD patients. (A) Ki67 staining. Shown in panel is the average percentage (± SEM) of Ki67 positive cells in HBEC isolated from non-smoker (n = 5), smoker without COPD (n = 5), and COPD (n = 5). (B) WB using anti-p21 and anti-β-actin of cell lysates from HBEC isolated from nonsmokers (lane 1, 2, 3), non-COPD smokers (lane 4, 5, 6), and COPD patients (lane 7, 8, 9). The lower panel is the average (± SEM) taken from densitometric analysis of WB. Presented cases are non-smoker (n = 5), smoker without COPD (n = 5), and COPD (n = 5). (C) WB using anti-LC3 and anti-β-actin without CSE treatment in the presence of protease inhibitors (E64d and pepstatin A) in HBEC from nonsmokers (lane 1, 2, 3), non-COPD smokers (lane 4, 5, 6), and COPD patients (lane 7, 8, 9). The lower panel is the average (± SEM) taken from densitometric analysis of WB. Presented cases are non-smoker (n = 5), smoker without COPD (n = 5), and COPD (n = 5). (D) WB using anti-LC3 and anti-β-actin of control or CSE (1.0%) treatment in the presence or absence of protease inhibitors (E64d and pepstatin A) in HBEC (left panel). The lower panel is the average (± SEM) of relative increase in LC3-II normalized to β-actin following CSE exposure compared with control treated HBEC in the presence of protease inhibitors, which are taken from densitometric analysis of WB. Presented cases are non-smoker (n = 5), smoker without COPD (n = 5), and COPD (n = 5). (E) WB using anti-ubiquitin, anti-p62, or anti- β-actin of lung homogenates from non-smoker (n = 3), smoker without COPD (Brinkman Index < 900)(n = 3), smoker with COPD (Brinkman Index ≧900)(n = 3), and COPD (n = 3). The right panel is the average (± SEM) taken from densitometric analysis of WB using anti-ubiquitin.
Table 2. Patient characteristics (for lung homogenates).
| Non-smoke (n = 3) |
Non-COPD smoker |
COPD (n = 3) |
p value | ||
|---|---|---|---|---|---|
| BI<900 (n = 3) |
BI≧900 (n = 3) |
||||
| Age, years |
60.3±18 |
74.0±1.73 |
62.3±12.7 |
66.3±7.6 |
NS |
| Male, % of group |
0 |
100 |
100 |
100 |
NA |
| Brinkman Index |
0 |
506.7±100.7 |
983.3±76.4 |
1,030±246.4 |
NA |
| FEV1/FVC | 75.1±5.1 | 75.1±2.8 | 79.3±0.8 | 62.1±5.7 | NA |
Abbreviations: BI, Brinkman index; COPD, chronic obstructive pulmonary disease; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; NA, not assessed; NS, not statistically significant. Values are mean ± SD.
Discussion
Although the molecular mechanisms for cellular senescence is incompletely understood, intracellular protein and organelle degradation pathways, in terms of quality control, have been widely implicated in the regulation of cell senescence. Accumulations of damaged cellular components are proposed to be involved in the decline of cellular function, resulting in cell senescence.9,18 We show autophagy inhibition with both 3MA treatment or LC3 knockdown induced cell senescence, suggesting pivotal involvement of autophagy in the regulation of cell senescence in HBEC. Interestingly, we elucidated that the CSE-mediated activation of autophagy was transient and that insufficient autophagic clearance, at least partly, accounted for an increase in the percentage of senescent HBEC by CSE treatment. We consider that our findings can be understood to be an extension of recent interesting findings of autophagy induction in response to CSE exposure in association with apoptosis and also speculate that the difference of CSE concentration was a critical determination for different cell fates between apoptosis and senescence.14,19 Our findings of simultaneous accumulations of p62 and ubiquitinated proteins after CSE treatment is of particular interest, because p62 accumulation has been recognized as a plausible hallmark of impairment or deficiency of autophagy, in the setting of selective degradation of ubiquitinated proteins.13
Ubiquitinated proteins are originally designated for proteolytic degradation in the proteasome system, a multicatalytic complex, which is major intracellular proteolytic machinery and is also responsible for a diverse array of cell functions and fates.10 It remains likewise unresolved whether proteasome activity simply declines with aging. However, accumulation of ubiquitin-conjugated proteins is an age-related feature of senescent cells, especially in pathological conditions such as neurodegenerative disorders.13 Recently, impairment of proteasome activity has been demonstrated in COPD lung, which was associated with pathological increase of endoplasmic reticulum (ER) stress and apoptosis.20 Our in vitro experiments demonstrated accumulations of ubiquitin-conjugated protein in response to CSE exposure, which were clearly enhanced by inhibition of autophagy and decreased by Torin1-mediated activation of autophagy. Simultaneous accumulations of ubiquitinated proteins and p62 support the notion that not only decreased proteasome activity but also impairment of autophagic degradation is responsible for the accumulations of ubiquitinated proteins. Knock-down of LC3 enhanced accumulations of both ubiquitinated proteins and p62 even without exposure to CSE (Fig. 6C), suggesting the pivotal role of LC3 for autophagic degradation of ubiquitinated proteins through p62 binding, which is consistent with prior reports.13 In contrast, p62 knockdown demonstrated no clear impact on baseline levels of ubiquitinated proteins, suggesting that p62 is mainly employed for the degradation of stress-induced ubiquitinated proteins and that different types of adaptor protein might also exist for autophagic degradation of ubiquitinated proteins in physiological conditions.
Increase in LC3-II / LC3-I has been reported in COPD lung tissues, which are recognized as markers for increased activation of autophagy.14 Accordingly, we have also performed anti-LC3 western blotting using lung homogenates. However, LC3-II expression in lung homogenates was not detected, regardless of diagnosis (data not shown). We speculate that LC3-II is rapidly processed in lung tissue in the absence of protease inhibitors in our system.16 Furthermore, LC3-II bands were barely detected in the absence of protease inhibitors in cultured HBEC. Previous studies demonstrated high levels of baseline LC3-II conversion in COPD lung, which was due to a decrease in the processing of autophagosomes in alveolar macrophages from smokers.21 However, our observations of elevated LC3-II conversion in HBEC in the absence of CSE exposure were detected only in the presence of in vitro protease inhibitors, demonstrating an increase of baseline autophagy flux in HBEC from COPD patients, suggesting the potential for increased autophagy in COPD lung in our system. Practically, detection of LC3-II by western blotting in the presence of protease inhibitors (E64d and pepstatin A) to prevent further degradation is a standard method to precisely evaluate the activation of autophagy flux, especially in in vitro culture models.16 It is attractive to superimpose the oxidant/antioxidant imbalance theory in COPD pathogenesis on the mechanisms for increased baseline autophagy flux.22 We postulate that HBEC from COPD patients are already stressed by both extrinsic and intrinsic ROS by several mechanisms, including insufficient amounts of anti-oxidant proteins and excessive oxidant release from damaged mitochondria,23 even after smoking cessation, resulting in increase of baseline autophagy. In support of this notion, carbonylated proteins, reflecting oxidative stress, were apparently increased in HBEC from COPD patients compared with those from non-COPD patients.24
In spite of increased baseline activity, the accumulations of ubiquitinated proteins and p62 in COPD lung homogenates may be explained by insufficient autophagic clearance of smoking-induced damaged cellular compartments in COPD lungs (Fig. 7C). It is likely that the attenuation of autophagy flux in response to CSE exposure may reflect an insufficient reserve of autophagy activation in HBEC from COPD patients, resulting in accumulations of ubiquitinated-protein and p62. Accumulations of ubiquitinated protein and p62 were also demonstrated in response to CSE in alveolar macrophages from non-smokers and also in alveolar macrophages from smokers without CSE exposure, indicating that HBEC and alveolar macrophages may, at least partly, share similar mechanisms of impairment for induction of autophagic degradation, which is potentially involved in the pathogenic sequence of COPD.21 Interestingly, a recent paper demonstrated that ROS-mediated SUMOylation of transglutaminase was responsible for autophagy inhibition through downregulation of beclin1 in airway epithelial cells, however whether this is a cystic fibrosis-specific mechanism remains to be elucidated.25
Accelerated cellular senescence has been widely implicated in the pathogenesis of COPD in terms of both impaired cell repopulation and aberrant cytokine secretions of SASP.2-4 Autophagy inhibition with bafilomycin A and LC3B knockdown with CSE treatment clearly enhanced IL-8 secretion in conditioned medium, which appeared to be corresponding to the accelerated cellular senescence accompanied by SASP. Conversely, autophagy has also been demonstrated to be activated during senescence progression and aggressively contributes to the development of cellular senescence with SASP of cytokine secretions including IL-6 and IL-8 in the setting of oncogene-induced senescence (OIS) in human fibroblasts.5 Autophagy-mediated protein turnover has been postulated to be necessary for active protein synthesis of these cytokines, which are required for the execution and maintenance of OIS.26 In our system, autophagy-mediated protein turnover seems to be unimportant for senescence progression and IL-8 secretion, suggesting the nature of autophagy in cell senescence regulation is cell-type or inducer-specific. Interestingly, p62 has also been demonstrated to be responsible for the upregulation of inflammatory cytokines via activation of NF-κB,27 and NF-κB is also involved in the regulation of cell senescence.28 Although p62 knock down delineated the tendency of senescence suppression (Fig. 4E), further studies are required to address the more direct involvement of p62 in the regulation of cell senescence and SASP.
In summary, we demonstrated the involvement of autophagy in the regulation of CSE-induced HBEC senescence and SASP. The accumulation of both ubiquitinated proteins and p62 suggest that induction of autophagic degradation of damaged proteins in response to CSE exposure might be a crucial determinant for cell senescence, and insufficiency of autophagy is potentially involved in COPD pathogenesis. We understand the potential limitations of using large airway bronchial epithelial cells as an experimental model to elucidate the pathogenesis of small airway disease and emphysema, therefore more relevant cell culturing models using small airway epithelial cells or alveolar epithelial cells are needed in future studies to further confirm the physiological relevance of our results. However, our finding of an inhibitory effect of Torin 1 on CSE-induced accumulations of ubiquitinated protein and cellular senescence suggests that optimal levels of autophagy induction is a potentially effective medical intervention to reduce the accumulation of damaged proteins and organelles, resulting in amelioration of the tobacco smoking related senescence associated lung disease, COPD.
Material and Methods
Cell culture, antibodies, and reagents
Normal and COPD airways were collected from 1st through 4th order bronchi from pneumonectomy and lobectomy specimens from resections performed for primary lung cancer. Informed consent was obtained from all surgical participants as part of an approved ongoing research protocol by the ethical committee of Jikei University School of Medicine. HBEC were isolated with protease treatment and freshly isolated HBEC were plated onto rat-tail collagen type I-coated (10 μg/ml) dishes, incubated overnight, and then the medium was changed to bronchial epithelial growth medium (BEGM, Clonetics). Cultures were characterized immunohistochemically using anti-cytokeratin antibodies (Lu-5, BioCare Medical) and anti-vimentin (Sigma-Aldrich), as previously described.29 HBEC showed > 95% positive staining with anti-cytokeratin and < 5% positive staining with the anti-vimentin antibody (data not shown). HBEC were serially passaged and used for experiments until passage 3. The majority of experiments for autophagy induction and evaluation of cell senescence were performed with HBEC from non-COPD patients. Bronchial epithelial cell line BEAS-2B cells were cultured in RPMI1640 with 10% FCS and penicillin-streptomycin. Antibodies used were rabbit anti-LC3 (Novus), rabbit anti-ATG5 (Novus), mouse anti-p21 (Cell signaling Technology), rabbit anti-phospho-histone H2A.X (Ser139) (Cell signaling Technology), mouse anti-poly and mono ubiquitin (Enzo Life Sciences), rabbit anti-p62 (MBL, Nagoya, Japan), and mouse anti-β-actin (Santa Cruz). Pepstatin A (Peptide institute), E64d (Peptide institute), Bafilomycin A (R&D Systems), 3-methyladenine (3-MA), an autophagic sequestration blocker, (Affinity BioReagents), rapamycin (Sigma Aldrich), and rat-tail type-I collagen (Sigma-Aldrich) were purchased. Torin 1, a selective and potent small molecule inhibitor of mTOR as inducers of autophagy, was kindly provided by Drs Gray and Sabatini (Whitehead Institute).
Preparation of cigarette smoke extract
Cigarette smoke extract (CSE) was prepared as previously described with minor modifications.21 Forty milliliters of cigarette smoke was drawn into the syringe and slowly bubbled into sterile serum-free cell culture media in a 15-ml BD falcon tube. One cigarette was used for the preparation of 10 mL of solution. CSE solution was filtered (0.22 μm) to remove insoluble particles and was designated as a 100% CSE solution.
SA-β-Gal staining
Senescence associated β-galactosidase (SA-β-Gal) staining was performed using HBEC (2 x 105) grown on 12-well culture plates according to the manufacturer’s instructions (β-galactosidase staining kit, BioVision Research Products).
Western blotting
HBEC grown on 12-well culture plates were treated with CSE for the indicated time points and lysed in RIPA buffer (Thermo Fisher Scientific) with protease inhibitor cocktail (Roche Diagnostics) and 1mM sodium orthovanadate (Wako). HBEC were also lysed in Laemmli sample buffer for LC3 detection. Lung homogenates were prepared using a Bioruptor UCD250 (Cosmo Bio) and sonication of lung tissue was performed according to the manufacturer’s instructions. Western blotting was performed as previously described with minor modification.1 After transfer to PVDF membrane (Immobilon-P), blotting with specific primary antibodies were performed overnight at 4þC. Proteins were detected by HRP-conjugated secondary antibody (Bethyl Laboratories) followed by chemiluminescence detection (ECL; GE Healthcare) with a LAS-4000 UV mini system (Fujifilm). The conversion of LC3 from LC3-I (free form) to LC3-II (phosphatidylethanolamine-conjugated form) represents a key step in autophagosome formation15 and detection of LC3-II by western blotting in the presence of protease inhibitors (E64d and pepstatin A) to prevent further degradation is a standard method to evaluate the activation of autophagy.
Plasmids, siRNA, and transfection
The LC3 cDNA was the kind gift of Dr. Mizushima (Tokyo Medical and Dental University) and Dr. Yoshimori (Osaka University), and was cloned into pEGFP-C1 vector (Clontech).15 pEGFP-LC3 plasmid was transfected into Beas-2B cells using Lipofectamine 2000 (Invitrogen) and stably expressing clones were selected by culturing with G418 (1.0 mg/ml) containing medium. The p62, LC3B, ATG5, p21, and negative control siRNAs were purchased (Applied Biosystems) and transfections of HBEC were performed with the Amaxa Nucleofector (Lonza), using matched optimized transfection kits for airway epithelial cells.
RNA isolation, polymerase chain reaction
RNA isolation, reverse transcription and polymerase chain reaction were performed as previously described. The primers used were LC3 sense primer, 5′- GATGTCCGACTTATTCGAGAGC -3′, LC3 antisense primer, 5′-TTGAGCTGTAAGCGCCTTCTA -3′; p62 sense primer, 5′- CCAGTGACGAGGAATTGACAA -3′, p62 antisense primer, 5′- CATCGCAGATCACATTGGGG -3′; β−actin sense primer, 5′-TGACGGGGTCACCCACACTGTGCC-3′, β-actin antisense primer, 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3′. These primer sets yielded PCR products of 167 bp, 156 bp, and 662 bp for LC3, p62, and β-actin respectively. Aliquots of the PCR products were subjected to agarose gel electrophoresis in TRIS-acetate-EDTA buffer and visualized by ethidium bromide staining. Primer sequences for LC3 and p62 were from Primer Bank (http://pga.mgh.harvard.edu/primerbank).
IL-8 enzyme-linked immunosorbent assay
HBEC were incubated with 1.0% CSE for 24 h in the presence or absence of Torin1 and bafilomycin A, and washed three times with PBS. To collect the conditioned medium, HBEC were incubated in serum-free DMEM for 48h in the presence or absence of Torin1 and bafilomycin A respectively. CSE (1.0% for 48 h) treatment was started 24 h post-siRNA transfection. Culture medium was collected from similarly treated cells. IL-8 was measured in conditioned media with an IL-8 Quantikine ELISA kit (R and D Systems).
Statistics
Data are shown as the average (± SEM) taken from at least three independent experiments. Student’s t-test was used for comparison of two data sets, analysis of variance for multiple data sets. Tukey’s or Dunn’s test were used for parametric and nonparametric data, respectively. Significance was defined as p < 0.05. Statistical software used was Prism v.5 (GraphPad Software, Inc.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Acknowledgments
We wish to thank Stephanie Cambier of UCSF for technical support, Dr. Mizushima (Tokyo medical and dental University, Tokyo, Japan) and Dr. Yoshimori (Osaka University, Osaka, Japan) for providing LC3 cDNA, and Drs. Gray and Sabatini (Whitehead Institute, MA) for providing Torin1. This work was supported by grants from the Jikei University Research fund to JA and KK, Uehara memorial Foundation to JA, The Research Funding for Longevity Sciences (22–11) from NCGG to KN and a Grant-In-Aid for Scientific Research from the Ministry of Education to JA, HH, TN, KN, and KK.
Footnotes
These authors contributed equally to this work
Previously published online: www.landesbioscience.com/journals/oncoimmunology/article/20297
References
- 1.Araya J, Cambier S, Markovics JA, Wolters P, Jablons D, Hill A, et al. Squamous metaplasia amplifies pathologic epithelial-mesenchymal interactions in COPD patients. J Clin Invest. 2007;117:3551–62. doi: 10.1172/JCI32526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsuji T, Aoshiba K, Nagai A. Cigarette smoke induces senescence in alveolar epithelial cells. Am J Respir Cell Mol Biol. 2004;31:643–9. doi: 10.1165/rcmb.2003-0290OC. [DOI] [PubMed] [Google Scholar]
- 3.Tsuji T, Aoshiba K, Nagai A. Alveolar cell senescence in patients with pulmonary emphysema. Am J Respir Crit Care Med. 2006;174:886–93. doi: 10.1164/rccm.200509-1374OC. [DOI] [PubMed] [Google Scholar]
- 4.Coppé JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adcock IM, Caramori G, Barnes PJ. Chronic obstructive pulmonary disease and lung cancer: new molecular insights. Respiration. 2011;81:265–84. doi: 10.1159/000324601. [DOI] [PubMed] [Google Scholar]
- 7.Brenner DR, McLaughlin JR, Hung RJ. Previous lung diseases and lung cancer risk: a systematic review and meta-analysis. PLoS One. 2011;6:e17479. doi: 10.1371/journal.pone.0017479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito K, Barnes PJ. COPD as a disease of accelerated lung aging. Chest. 2009;135:173–80. doi: 10.1378/chest.08-1419. [DOI] [PubMed] [Google Scholar]
- 9.Rajawat YS, Hilioti Z, Bossis I. Aging: central role for autophagy and the lysosomal degradative system. Ageing Res Rev. 2009;8:199–213. doi: 10.1016/j.arr.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Jung T, Catalgol B, Grune T. The proteasomal system. Mol Aspects Med. 2009;30:191–296. doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Rubinsztein DC, Mariño G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–95. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 12.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 14.Chen ZH, Kim HP, Sciurba FC, Lee SJ, Feghali-Bostwick C, Stolz DB, et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS One. 2008;3:e3316. doi: 10.1371/journal.pone.0003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–32. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salminen A, Kaarniranta K. Regulation of the aging process by autophagy. Trends Mol Med. 2009;15:217–24. doi: 10.1016/j.molmed.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Hwang JW, Chung S, Sundar IK, Yao H, Arunachalam G, McBurney MW, et al. Cigarette smoke-induced autophagy is regulated by SIRT1-PARP-1-dependent mechanism: implication in pathogenesis of COPD. Arch Biochem Biophys. 2010;500:203–9. doi: 10.1016/j.abb.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malhotra D, Thimmulappa R, Vij N, Navas-Acien A, Sussan T, Merali S, et al. Heightened endoplasmic reticulum stress in the lungs of patients with chronic obstructive pulmonary disease: the role of Nrf2-regulated proteasomal activity. Am J Respir Crit Care Med. 2009;180:1196–207. doi: 10.1164/rccm.200903-0324OC. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Monick MM, Powers LS, Walters K, Lovan N, Zhang M, Gerke A, et al. Identification of an autophagy defect in smokers’ alveolar macrophages. J Immunol. 2010;185:5425–35. doi: 10.4049/jimmunol.1001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cantin AM. Cellular response to cigarette smoke and oxidants: adapting to survive. Proc Am Thorac Soc. 2010;7:368–75. doi: 10.1513/pats.201001-014AW. [DOI] [PubMed] [Google Scholar]
- 23.Mammucari C, Rizzuto R. Signaling pathways in mitochondrial dysfunction and aging. Mech Ageing Dev. 2010;131:536–43. doi: 10.1016/j.mad.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara H, Araya J, Takasaka N, Fujii S, Kojima J, Yumino Y, et al. Involvement of creatine kinase B in cigarette smoke-induced bronchial epithelial cell senescence. Am J Respir Cell Mol Biol. 2012;46:306–12. doi: 10.1165/rcmb.2011-0214OC. [DOI] [PubMed] [Google Scholar]
- 25.Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina D, Settembre C, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol. 2010;12:863–75. doi: 10.1038/ncb2090. [DOI] [PubMed] [Google Scholar]
- 26.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–31. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 27.Lee HM, Shin DM, Yuk JM, Shi G, Choi DK, Lee SH, et al. Autophagy negatively regulates keratinocyte inflammatory responses via scaffolding protein p62/SQSTM1. J Immunol. 2011;186:1248–58. doi: 10.4049/jimmunol.1001954. [DOI] [PubMed] [Google Scholar]
- 28.Kriete A, Mayo KL. Atypical pathways of NF-kappaB activation and aging. Exp Gerontol. 2009;44:250–5. doi: 10.1016/j.exger.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Araya J, Cambier S, Morris A, Finkbeiner W, Nishimura SL. Integrin-mediated transforming growth factor-beta activation regulates homeostasis of the pulmonary epithelial-mesenchymal trophic unit. Am J Pathol. 2006;169:405–15. doi: 10.2353/ajpath.2006.060049. [DOI] [PMC free article] [PubMed] [Google Scholar]