Abstract
Understanding the basis of a successful clinical response after treatment with therapeutic cancer vaccines is essential for the development of more efficacious therapy. After vaccination with the single telomerase (hTERT) 16-mer peptide, GV1001, some patients experienced clinical responses and long-term survival. This study reports in-depth immunological analysis of the T-cell response against telomerase (hTERT) in clinically responding patients compared with clinical non-responders following vaccination with the single hTERT 16-mer peptide, GV1001. Extensive characterization of CD4+ T-cell clones specific for GV1001 generated from a lung cancer patient in complete remission after vaccination demonstrated a very broad immune response to this single peptide vaccine with differences in fine specificity, HLA restriction, affinity and function. Some CD4+ T-cell clones were cytotoxic against peptide-loaded target cells and also recognized processed recombinant hTERT protein. Furthermore, T-cell responses against several unrelated hTERT epitopes, some of which are novel, were detected, indicating extensive epitope spreading which was confirmed in other clinical responders. In contrast, patients responding immunologically, but not clinically, after vaccination did not display this intramolecular epitope spreading. Multifunctional CD4+ T-cell clones specific for novel hTERT epitopes were generated and shown to recognize a melanoma cell line. Pentamer analysis of T cells in peripheral blood also demonstrated the presence of an important CD8+ T-cell response recognizing an HLA-B7 epitope embedded in GV1001 not previously described. These results indicate that the highly diverse hTERT-specific T-cell response, integrating both T helper and CTL responses, is essential for tumor regression and the generation of long-term T-cell memory.
Keywords: T helper cells, T-cell epitopes, epitope spreading, hTERT, non-small cell lung cancer, peptide vaccination
Introduction
The reverse transcriptase subunit of telomerase (hTERT), is an attractive, almost universal antigen target in cancer being overexpressed in the majority of human tumors and absent in most normal adult tissues.1-4 In addition, the telomerase enzyme is expressed in cancer stem cells and targeting this antigen could therefore be an important tool to eliminate these cells which are not easily killed by conventional therapy.5-8 There has been conflicting evidence of efficacy of vaccination with the short peptide hTERT 540–548,9-12 however, vaccination with hTERT mRNA and long peptides have shown promise.13-19
GV1001 is a 16-mer peptide vaccine corresponding to amino acids 611–626 of hTERT. This is a highly promiscuous peptide that binds to a wide variety of HLA-class II molecules encoded by multiple alleles from the HLA-DR,-DP and –DQ loci, thereby eliciting T helper (Th) responses in up to 80% of vaccinated patients.18 GV1001 also harbours putative HLA-class I epitopes. Vaccination with GV1001 may therefore potentially elicit combined CD4+ and CD8+ T-cell responses. Such T-cell responses are considered to be crucial for tumor eradication and for generating long-term memory.
Phase I/II clinical trials in patients with pancreatic adenocarcinomas, non-small cell lung cancer (NSCLC) and malignant melanoma have demonstrated that vaccination is safe and induces immune responses in 50–80% of vaccinated patients. These trials demonstrated a clear correlation between GV1001-specific immune responses and survival. Long-term survivors harboured durable GV1001-specific T-cell responses with polyfunctional cytokine patterns.20 In a Phase I/II clinical trial of GV1001 vaccination in patients with advanced NSCLC, GV1001-specific T-cell responses were demonstrated in 13/24 evaluable patients (56%). During the > 9-y follow up, two of these patients developed complete remissions (CR) and continue to receive booster injections and demonstrate stable immune responses against GV1001.
Patients with CR and long-term immunological memory thus represent a unique opportunity to gain insight into the mechanisms behind tumor regressions observed after vaccination and the immunological consequence of vaccination. To this end, we studied in detail the immune response against hTERT in one of the two NSCLC patients with CR where multiple samples of peripheral blood mononuclear cells (PBMC) were still available for analysis. We first characterized the CD4+ T-cell response against the single Th vaccination peptide (GV1001), with respect to HLA-restriction, fine specificity, phenotype and function. We also investigated if cytotoxic T lymphocyte (CTL) epitopes embedded within GV1001 could be recognized by CD8+ T cells from the patient. Vaccination-induced tumor-specific CD4+ T cells homing to tumor can deliver inflammatory cytokines such as IFN-γ to the local mircroenvironment and thereby enhance cross-presentation by antigen-presenting cells (APCs) present.21,22 Peptide-based vaccination studies, and to a greater extent those using Th peptides, have shown that epitope spreading frequently occurs in patients with tumor regressions.23 We therefore studied T-cell responses against a library of overlapping hTERT peptides to look for reactivities against hTERT epitopes other than the vaccination peptide. The results were confirmed in three additional long-term surviving GV1001-vaccinated cancer patients.
The data reported here suggest that vaccination with hTERT Th peptides can generate strong, diverse T-cell responses integrating both the CD8+ and CD4+ T-cell compartments required for tumor eradication, resulting in intermolecular epitope spreading. Broad hTERT reactivity is associated with increased survival.
Results
Minimal hTERT peptide sequences recognized and T-cell affinity
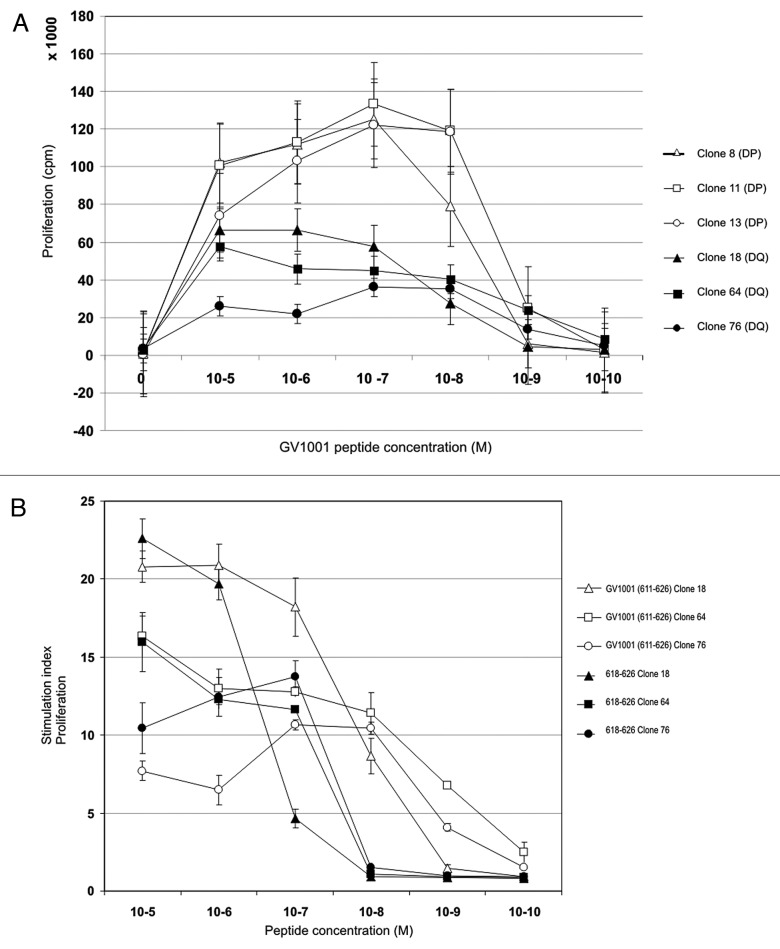
The 16-mer vaccination peptide GV1001 (611–626) is highly promiscuous and can be processed to fit into the peptide binding groove of different HLA-class I/II molecules. A broad immune response against GV1001 was previously demonstrated in this patient.14 GV1001-specific T cell clones derived from the patient recognized fragments of this peptide presented by HLA-DP*0401/02 and –DQ*04. CD4+ T-cell clones were also capable of killing autologous target cells pulsed with a 9-mer peptide derived from the C-terminal part of GV1001.14The minimal peptide sequences recognized by and the HLA restriction of these T-cell clones are shown in Fig. S1. The HLA-DP*0401/02 restricted T-cell clones recognize the 10-mer minimal sequence ALLTSRLRFI (615–624) shown in green, whereas the HLA-DQ*04 restricted T cell clones recognize the 9-mer overlapping minimal fragment (shown in blue), TSRLRFIPK (618–626). Clearly, the various T-cell clones and HLA class II molecules prefer different, but partly overlapping, fragments of GV1001.
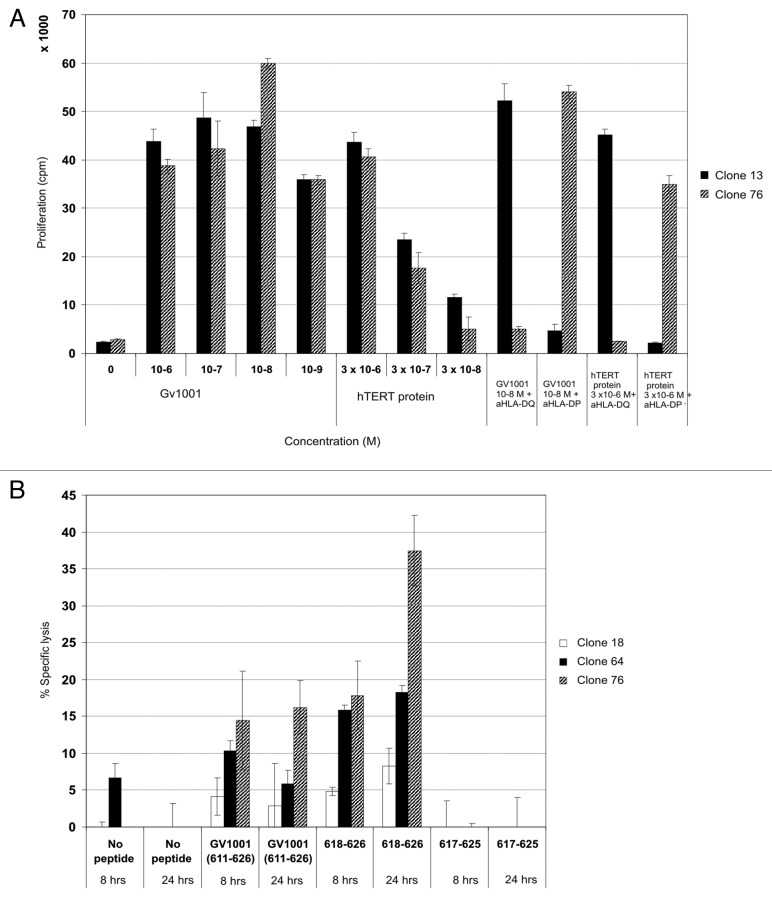
Since recognition of different fragments may influence the affinity of the interactions, we performed dose-response experiments with GV1001 using three different HLA-DP restricted and three different HLA –DQ restricted CD4+ T cell clones (Fig. 1A). Interestingly, two distinct response patterns emerged; the HLA-DP restricted clones were highly proliferative and demonstrated a bell-shaped response curve with optimal responses at 10−7 M of GV1001. The HLA-DQ restricted clones were generally less proliferative and did not demonstrate typical bell-shaped curves, but responded in a dose-related fashion over the whole concentration range of peptide used. The differences in response pattern make it difficult to evaluate the affinity of the clones. It is feasible that the negative effect of high peptide doses on the HLA-DP restricted clones may reflect a higher affinity and a propensity to enter into anergy at higher doses. The HLA-DQ restricted T-cell clones recognizing the peptide 618–626 at concentrations down to 10−7 and 10−8 M also demonstrate cytotoxicity14,20. In Figure 1B, the difference in proliferation of HLA-DQ restricted T-cell clones against peptides GV1001 and 618–626 (9-mer) is shown. The increased proliferation in response to GV1001 compared with 618–626 could either be due to the shorter peptide being more sensitive to proteolysis than GV1001 or reflect differences between direct binding and processing leading to different efficacy of presentation. The T-cell clones were also shown to recognize APCs loaded with recombinant hTERT protein even at nanomolar concentrations (Fig. 2A). Unfortunately, due to very high spontaneous 51Cr-release of EBV-LCLs loaded with hTERT protein we were unable to demonstrate lysis of these APCs.
Figure 1. Vaccine-specific HLA-DQ4 and –DP4 restricted CD4+ T-cell clones display high affinity for GV1001 peptide but also recognize a nested 9-mer epitope. The relative peptide affinity of GV1001-specific T cell clones was tested in proliferation assays against titrated peptide concentrations from 10−5-10−10 M (A). Open labels show HLA-DP4 restricted T cell clones and closed labels show HLA-DQ4 restricted T cell clones. (B) shows the relative affinity of HLA-DQ4 restricted T-cell clones for GV1001 compared with nested 9-mer 618–626 tested in proliferation assays. Open labels show T-cell clones tested against GV1001 peptide and closed labels show T cell clones tested against peptide 618–626.
Figure 2. Cytotoxic CD4+ CTL clones specific for GV1001 lyse peptide-loaded target cells most efficiently with nested 9-mer epitope. Both HLA-DP and –DQ restricted T-cell clones recognize processed hTERT protein. GV1001-specific T-cell clones were also tested for proliferation against EBV-LCL loaded with recombinant hTERT protein at low concentrations (A). One representative HLA-DP restricted clone is shown (black bars) and one HLA-DQ restricted clone (hatched bars). (B) shows 8- and 24-h 51Cr-release assays for HLA-DQ restricted CTL clones tested against autologous EBV-LCL loaded with peptides GV1001 (611–626) and nested epitope 618–626. Peptide 617–625 was used as a negative control. Effector: Target ratio was 100:1. Black bars represent clone 18, dotted bars clone 64, and hatched bars clone 76.
Cytotoxic CD4+ T-cell clones
Paradoxically, when tested in 51Cr-release assays the HLA-DQ restricted T-cell clones are more cytotoxic in response to the 9-mer peptide, 618–626, than to GV1001 indicating that peptide presentation in this case is more efficient with the shorter peptide (Fig. 2B). Target cells loaded with control peptide 617–625 were not recognized. Different T-cell clones were cytotoxic to varying degrees with clone 76 inducing the highest lysis. The effector-to-target (E:T) ratio was high (100:1), and lysis was found to be dose-dependent when the ratio was titrated for clone 76 (data not shown).
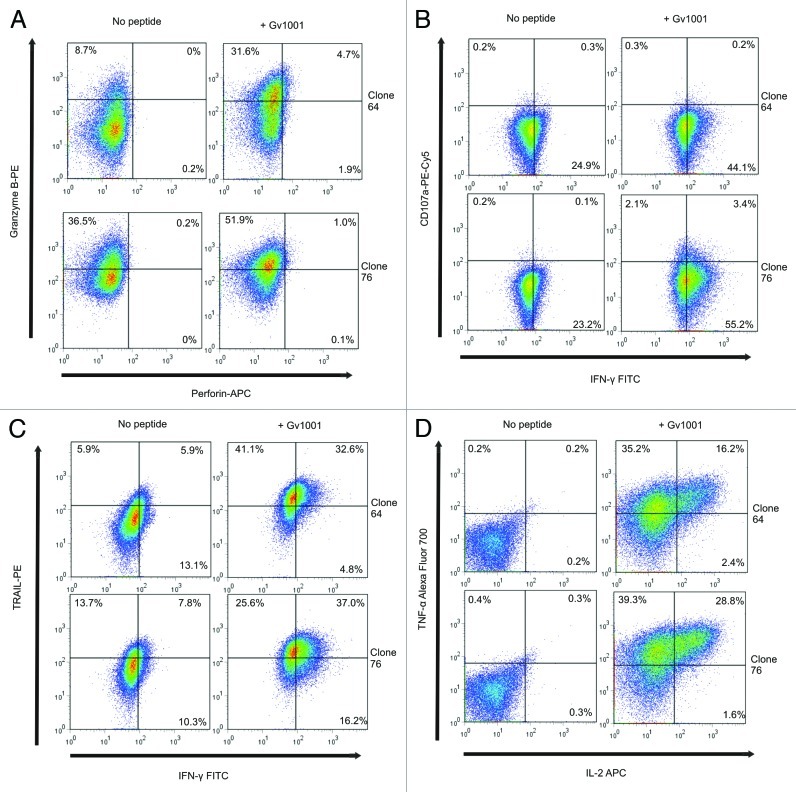
Clones 64 and 76 were further tested to determine what their killing mechanisms could be (Fig. 3). After overnight peptide stimulation, the clones were shown to express granzyme B but not perforin (Fig. 3A). This agrees with data from other groups showing that cytotoxic CD4+ T-cell clones can be perforin-independent.24 Furthermore, the degranulation marker CD107a expression was analyzed in the two T-cell clones. Clone 64 was CD107a negative and only around 5% of clone 76 expressed CD107a (Fig. 3B). The loss of granzyme B and expression of the degranulation marker may not be concomitant events as recently shown for activated CD4+ CD25+ T cells selectively killing B lymphocytes.25 Moreover, the clones stained very positively for TRAIL with more than 60% increase in expression in clone 64 in the presence of peptide (Fig. 3C). Clone 76 showed high baseline TRAIL expression with a 30% upregulation upon incubation with peptide. This clone also produced high levels of cytokines in the presence of peptide with around 70% of the cells producing TNF-α and 30% of these also producing IL-2 (Fig. 3D). In addition, around 55% of these cells produced IFN-γ (Fig. 3B and C), meaning that the T-cell clone is multifunctional. Cytokine levels were somewhat lower for clone 64, with around 50% of cells producing TNF-α, 16% of these also producing IL-2 and close to 40% of the cells producing IFN-γ. Both clones 64 and 76 stained negatively for both Fas and FasL (data not shown), agreeing with data demonstrating the importance of this pathway in murine CD4+ CTLs, whereas human CD4+ T cells preferentially use the same pathways as CD8+ T cells.26 Taken together, these data suggest that the cytotoxicity of these CD4+ CTL clones is mediated by granzyme B, TRAIL and cytotoxic cytokines TNF-α and IFN-γ.
Figure 3. CD4+ CTL clones express granzyme B, TRAIL, TNF-α and low levels of CD107a, but not perforin. Expression of Granzyme B, perforin, TRAIL, CD107a, IFN-γ, TNF-α and IL-2 was measured by flow cytometry after overnight incubation of T cell clones with peptide GV1001- loaded autologous EBV-LCL. Effector:target ratio was 1:3. Top panels in A-D show T-cell clone 64 and bottom panels show T-cell clone 76. Left panels show responses against non-peptide loaded controls and right panels show GV1001 peptide-loaded target cells.
Pentamer-positive T cells
Despite the demonstration of a diverse response against the vaccination peptide it was likely that T cells of other specificities were involved in the anti-tumor response. We therefore studied T-cell reactivity against additional hTERT epitopes.
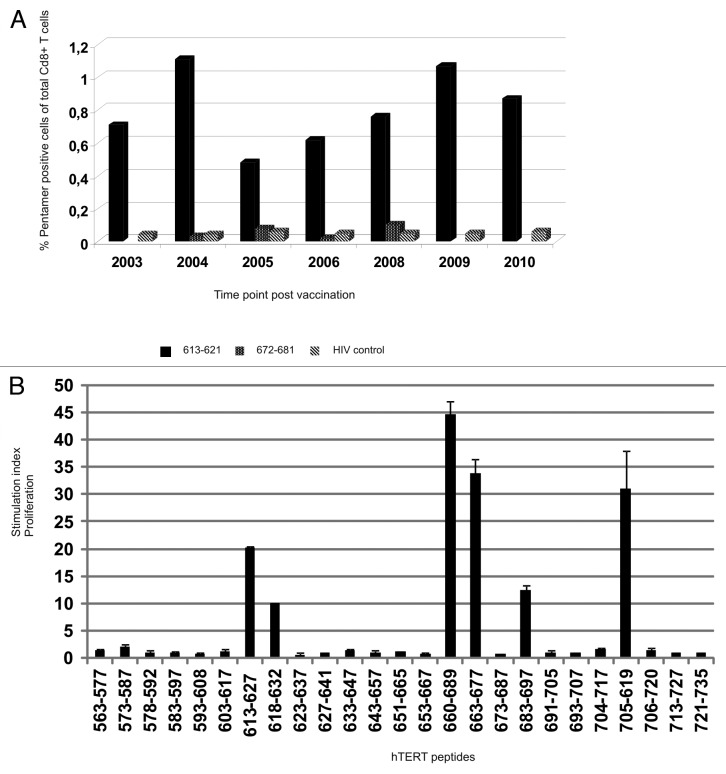
To look for possible involvement of hTERT-specific CD8+ CTLs we performed hTERT pentamer analysis of PBMCs from various time points according to availability. Vaccination started in 2002 and post-vaccination samples from years between 2003 and 2010 were tested. As the patient was HLA-A*0201 negative, we tested the presence of T cells recognizing hTERT peptide 613–621, a nested epitope of GV1001, presented on HLA-B*0702. PBMCs were thawed and stained directly without stimulation. As a negative control HIV-peptide pentamer was used for each time point. The seven time points tested from 2003 to 2010 demonstrated the presence of a stable pentamer-positive population of 0.5–1.1% of CD8+ T cells (Fig. 4A and see Fig. S2 for plots). We also detected a small population of 0.10% of CD8+ T cells specific for another HLA-B*0702 hTERT peptide, 672–681. This population could not be expanded and functionally tested, but the peptide is a nested epitope of a 30-mer peptide (660–689) inducing strong T-cell responses (Fig. 4B). Lacking a pre-vaccination sample for comparison, we cannot say if the CD8+ T-cell population recognizing this novel, CTL epitope within the vaccination peptide, is vaccine-induced, but its presence remains stable from 2003, 1 y after the onset of vaccination, until last measured in 2010.
Figure 4.T-cell reactivity against multiple unrelated hTERT Th and CTL epitopes. (A) shows pentamer analysis of hTERT-specific HLA-B*0702 restricted CD8+ T cells post vaccination. PBMCs were stained ex vivo with two pentamers with novel hTERT peptides 613–621 and 672–681 which were nested epitopes of GV1001 and 660–689, respectively. Vaccination started in 2002 and post vaccination samples from the years indicated were tested. Black bars represent T cells specific for peptide 613–621-specific, dotted bars peptide 672–681 and hatched bars the HIV control peptide. (B) shows a summary of all post-vaccination-T cell responses detected against several unrelated 15- and 30-mer hTERT peptides. PBMCs from different post-vaccination sample time points were pre-stimulated with 25 overlapping hTERT peptides and tested for proliferation against single peptides.
Broad reactivity against hTERT
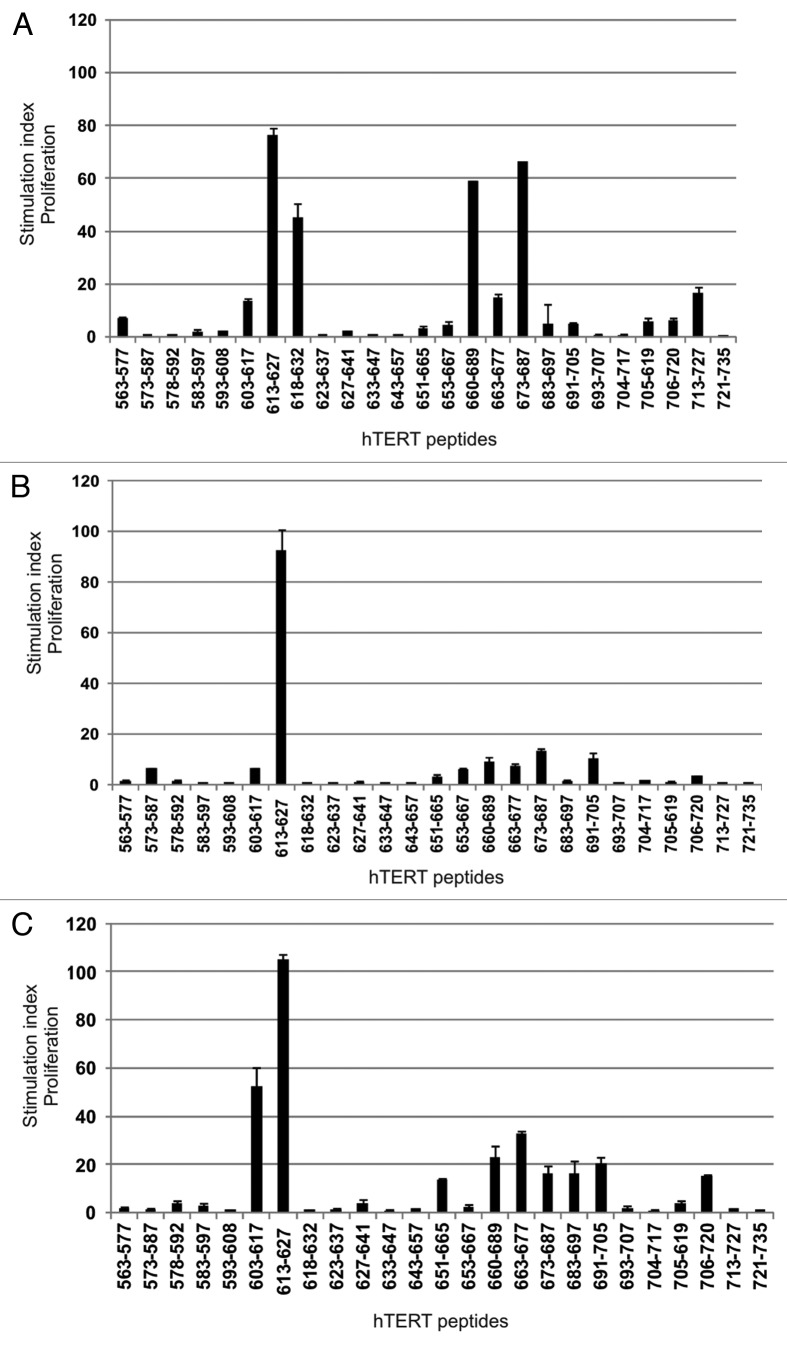
We looked at potential epitope spreading within hTERT using a set of overlapping peptides covering part of the active site of hTERT (amino acids 563–735). The peptides tested were previously shown to be recognized by T cells from patients vaccinated with dendritic cells (DCs) loaded with hTERT mRNA.27 PBMCs from different time points were stimulated with 24 15-mer hTERT peptides from the overlapping peptide library and one 30-mer hTERT peptide before proliferative T-cell responses were measured against each of these. Figure 4B shows a summary of the strongest responses detected against each peptide.
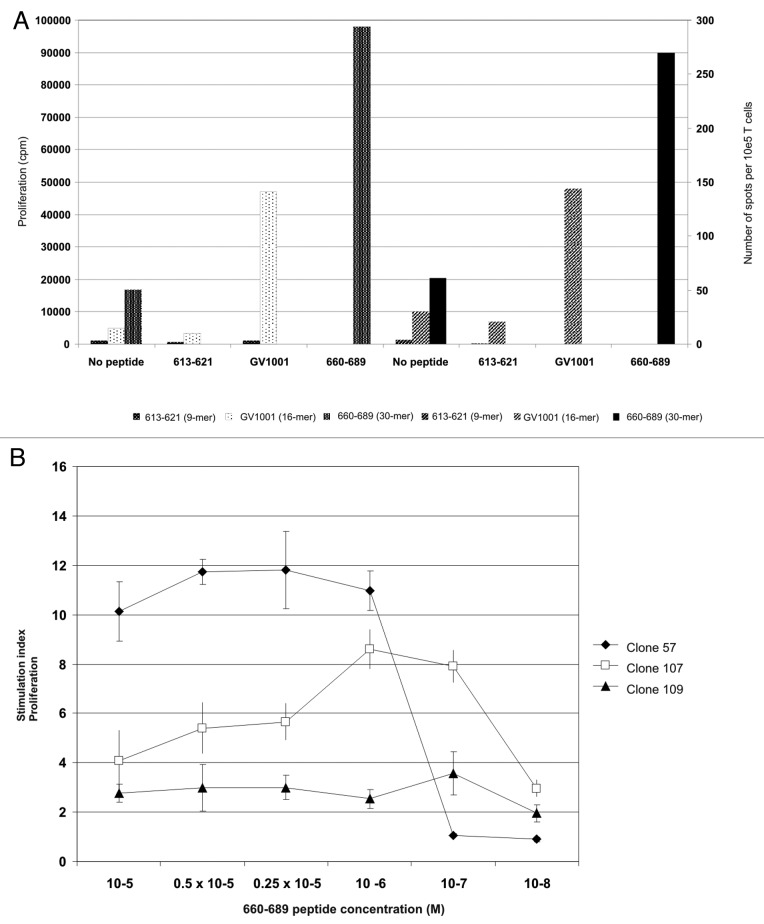
We detected proliferative T-cell responses against six 15-mer hTERT peptides from the overlapping peptide library, including a GV100-overlapping peptide, 613–627, and a 30-mer hTERT peptide 660–689. The 30-mer peptide and its fragment 663–677 induced the strongest proliferation, even surpassing that of GV1001, which was used for vaccination. This was also the case for peptide 705–719, a T-cell epitope previously identified by others.28 In addition, responses against peptides 573–587, 618–632, and 683–697 were detected. Peptide 618–632 is also partially overlapping with GV1001 and includes the entire sequence of CTL epitope 618–626. These are peptides recognized by T-cells in samples from several different patients (unpublished data and ref. 27) and shown to be promiscuous with respect to HLA class II molecules as well as being processed into shorter peptides acting as CTL epitopes. The strong proliferative response against vaccination peptide GV1001 and 30-mer peptide 660–689 also correlated with IFN-γ secretion measured by ELISPOT assays testing the same T cells in parallel (Fig. 5A). A frequency of 0.27% of T cells produced IFN-γ against peptide 660–689 vs. 0.14% against GV1001. Both cytokine secretion and proliferation were stronger in response to the 660–689 peptide than to GV1001. The 9-mer peptide, 613–621, embedded in GV1001, was unable to induce such responses. Despite the lack of pre-vaccination samples to prove this is vaccine-induced, it is likely that this response has further developed as a result of the strong and long lasting vaccination response against GV1001.
Figure 5. Peptide-specific T-cell proliferation correlates with IFN-γ secretion and give rise to high affinity CD4+ T-cell clones. (A) shows hTERT-specific T-cell proliferation and IFN-γ production in response to peptide-loaded autologous PBMC. The x-axis shows peptide, the Y axis proliferation measured in cpm and the z-axis shows number of spots detected per 105 T cells in an IFN-γ ELISPOT on the same cells. White dotted bars represent proliferation of 613–621-stimulated T cells, black dotted bars the proliferation of GV1001-stimulated T cells, and small-dotted bars 660–689-stimulated T cells, respectively. Large hatched bars represent the IFN-γ secretion from 613–621-stimulated T cells, small hatched bars the GV1001-stimulated T cells and black bars the 660–689-stimulated T cells, respectively. (B) shows proliferation of three representative CD4+ T-cell clones against autologus EBV-LCL loaded with titrated concentrations of peptide 660–689. Closed squares show T cell clone 57, open squares show T cell clone 107 and closed triangles show T cell clone 109.
CD4+ T-cell clones recognizing a new hTERT epitope
As the 30-mer peptide 660–689 contained several nested Th and CTL epitopes, we further investigated responses against this peptide. T cells were cloned by limiting dilution after one peptide pre-stimulation and 119/152 (78%) of the clones generated were peptide-specific, indicating that these cells must have been present at a high frequency in the patient. The T-cell clones were phenotyped by flow cytometry and shown to be CD4+ CD8- (data not shown). All T-cell clones tested against a panel of EBV-LCL homozygous for HLA-alleles in common with the patient were found to be HLA-DRB1*0801 restricted (Fig. S3). The T-cell epitope recognized has previously been shown to be naturally processed as clones specific for the same peptide sequence derived from another patient were found to recognize target cells loaded with hTERT protein in an HLA-DR*0801 restricted manner (data not shown). Additionally, vaccination with DCs loaded with hTERT mRNA also induce responses against this epitope.27 T-cell clones were further tested against titrated concentrations of peptide 660–689 and data from three representative clones are shown (Fig. 5B). The more proliferative clone 57, displayed intermediate antigen affinity and responded to peptide concentrations down to 10−6 M, but the response dropped drastically at 10-fold lower concentration. Clones 107 and 109 were of high affinity and displayed low but stable peptide-specific proliferation even at peptide concentrations down to 10−7 and 10−8 M, respectively.
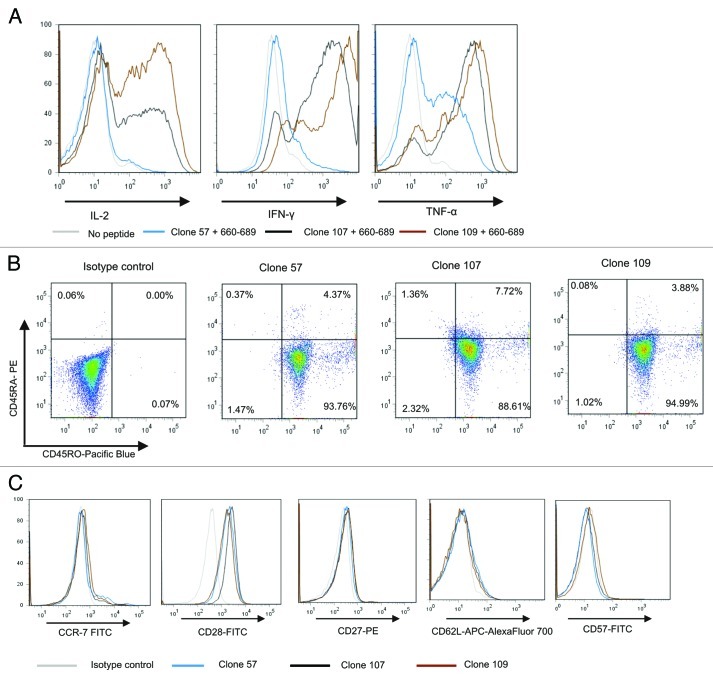
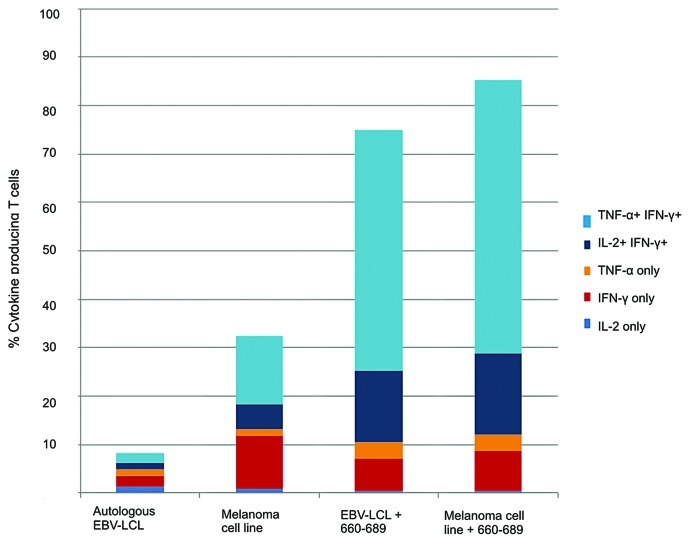
The T-cell clones displayed an inflammatory cytokine profile upon activation as seen in Figure 6A. Clone 57 secreted mainly TNF-α, low levels of IFN-γ and no IL-2, whereas the higher affinity clones 107 and 109 secreted high levels of all three cytokines upon peptide stimulation. When analyzed by flow cytometry, the three clones displayed the same phenotypes, being CD45RO+, CD45RA-, CD28+, CCR7low, CD27low, CD62L-, CD57- (Fig. 6B and C). This is consistent with an early effector memory (EM) phenotype analogous to that previously described for CD8+ T cells by several groups29-31 and more recently for CD4+ T cells.32 The high-affinity clone 109 which produced high levels of cytokines upon peptide recognition was tested against a HLA-DR*0801 positive melanoma cell line, ESTDAB-100. The melanoma cell line was recognized resulting in the same multifunctional cytokine production pattern as demonstrated upon peptide recognition (Fig. 7 and Fig. S4). These results confirm that the hTERT epitope is naturally processed from endogenous hTERT protein in this melanoma cell line.
Figure 6. Multifunctional HLA-DR*08 restricted CD4+ T cell clones specific for novel hTERT epitope display an inflammatory cytokine profile and early effector memory phenotype. (A) shows intracellular cytokine staining of three representative HLA-DR*08 restricted CD4+ T cell clones upon overnight stimulation with hTERT 30-mer peptide 719–20-loaded autologus EBV-LCL. Clones were tested for production of cytokines IL-2, IFN-γ and TNF-α. Non-peptide loaded EBV-LCLs were used as negative control. (B) and (C) show the T-cell phenotype. T cells were tested for CD45RA and CD45RO expression (B) and CCR7, CD28, CD27, CD62L, CD57 (C). Grey lines represent no peptide (A) or isotype controls (C), blue lines clone 57 (A and C), black lines clone 107 (A and C) and brown lines clone 109 (A and C).
Figure 7. HLA-DR*0801 restricted CD4+ T cell clone specific for novel hTERT epitope recognizes melanoma cell line. Intracellular cytokine staining of the hTERT-specific HLA-DR*08 restricted CD4+ T cell clone 109 was performed after overnight stimulation with autologus EBV-LCL or the HLA-DR*0801 melanoma cell line, ESTDAB-100. As positive controls, the EBV-LCL and ESTDAB-100 were both loaded with hTERT 30-mer peptide 660–689.
We wanted to investigate whether the widespread hTERT reactivity seen in this long-term surviving lung cancer patient was also present in other clinical responders who had undergone vaccination with the GV1001 peptide. Frozen blood samples from several post-vaccination time points from three different patients were available for study. PBMCs were prestimulated with and tested against the same overlapping hTERT peptide library as the lung cancer patient described above (Fig. 8). The results in this figure confirm the observation of widespread reactivity seen in the lung cancer patient described in Figure 4B. This indicates that widespread reactivity against the hTERT protein may be generally associated with long-term survival in GV1001-vaccinated patients. These results further extend the spectrum of hTERT epitopes recognized since between 10–17 peptides (Fig. 8, A-C) induced T-cell proliferation in samples from these three patients. The sequence of peptides 603–617, 613–627, and 618–632 overlap with the GV1001 peptide sequence (611–626). The reactivity against these peptides reflects cross-reactivity of T-cells raised against GV1001. The extensive overlap between the epitopes recognized, indicate that some of these epitopes may be more immunogenic than others. The results in Figure 8 represent the highest SI observed for each peptide in any of the blood samples analyzed. We observed considerable variation in SI between sample time points (data not shown), indicating a dynamic process of T-cell clonal expansion and contraction during the time frame represented by the sample time points.
Figure 8. T-cell reactivity against multiple unrelated hTERT Th and CTL epitopes in several long-term surviving GV1001-vaccinated patients. PBMCs from different post-vaccination sample time points were pre-stimulated with 25 overlapping hTERT peptides and tested for proliferation against single peptides. The graphs show a summary of post-vaccination-T cell responses detected against several unrelated 15- and 30-mer hTERT peptides. Two melanoma patients (A and B) and one colon cancer patient (C) were tested.
For comparison, we tested available post-vaccination blood samples from other GV1001-vaccinated cancer patients who had a documented immunological response to GV100118,33 without any evident clinical benefit from the response. Ten patients, six with melanoma and four with lung cancer, were tested against GV1001 (611–626) and five of the other hTERT peptides frequently recognized in the long-term survivors tested (Fig. 9). The T-cell responses against these peptides are clearly different from those observed in the long-term survivors with only sporadic responses against peptides other than GV1001 (Fig. 9; Fig. S5).
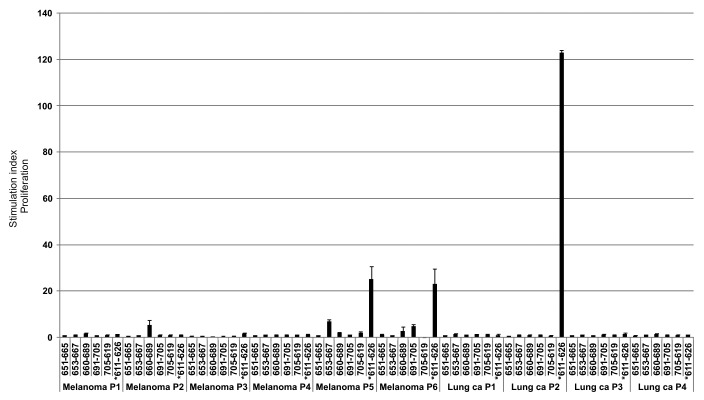
Figure 9. T-cell reactivity against hTERT in short-term surviving GV1001-vaccinated patients is less broad. PBMCs from post-vaccination sample time points were pre-stimulated with 6 hTERT peptides from our test panel frequently inducing responses in long-term survivors. T-cells were then tested for proliferation against single peptides. The graph shows a summary of post-vaccination-T cell responses detected against the different hTERT peptides. Six melanoma patients and four lung cancer patients were tested.* indicates the GV1001 peptide sequence (611–626).
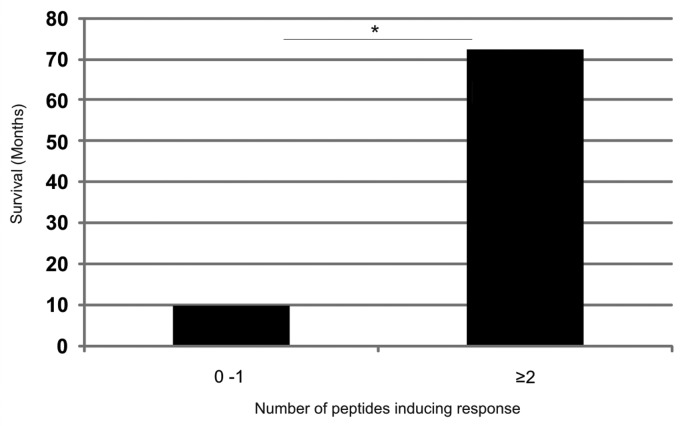
When the patient survival was analyzed according to how many peptides each patient recognized, the patient were split into two groups; patients with responses against 0–1 peptides in addition to GV1001 and patients with responses against ≥ 2 peptides (Fig. 10). The first group had a median survival of 10 mo compared with the second group which displayed a median survival of 72.5 mo. This difference was statistically significant (p = 0.005). As we observe T-cell responses against the test panel of hTERT peptides in individuals patients who both have advanced cancer and have been vaccinated with GV1001, we wanted to investigate whether we could detect similar responses in healthy donors who neither had cancer, nor had been vaccinated with GV1001. No such responses were observed in blood samples from six healthy donors (Fig. S6). Taken together, these results demonstrate that T-cell responses against our overlapping hTERT peptide library are associated with long-term survival in cancer patients following GV1001 vaccination.
Figure 10. Increased survival correlates with the recognition of an increased number of additional hTERT peptides in patients responding immunologically against the GV1001vaccine. Five of the most frequently recognized hTERT peptides from our overlapping peptide library (653–667, 651–665, 660–689, 691–705, 705–719) were tested for their capacity to stimulate T cells in ten melanoma and lung cancer patients who had responded immunologically against the GV1001 vaccine but displayed below median survival. The T-cell responses were compared with those of four clinical responders/long-term survivors who also responded immunologically against the GV1001 vaccine. Patients were grouped according to whether they recognized 0–1 peptides (n = 10) or ≥ 2 peptides (n = 4) in addition to GV1001 with a median survival of 10 mo and 72.5 mo, respectively. The Mann-Whitney U test was used for testing the correlation between survival and T-cell responses against an increased number of peptides.*p = 0.005
Discussion
Initial studies demonstrated a very broad T-cell response against the vaccination peptide, GV1001, in a lung cancer patient after CR. The CD4+ T-cell clones generated displayed very diverse specificity and function. First, we showed that CD4+ T-cell clones specific for the vaccination peptide had different fine specificity recognizing different minimal peptide sequences and were restricted to different HLA molecules. The T-cell clones described have very different response patterns with HLA-DP restricted clones being unresponsive to high concentrations of peptide whereas HLA-DQ restricted T-cell clones responded over the whole range of concentrations in a dose-dependent manner. This indicates that the HLA-DQ restricted, cytotoxic T-cell clones may have lower peptide affinity. The T-cell clones were also shown to recognize low concentrations of processed recombinant hTERT protein.
Interestingly, two of the HLA-DQ restricted CD4+ T cell clones recognizing both GV1001 and the shorter nested CTL epitope were shown to be cytotoxic. Our results suggest that the cytotoxicity of these CD4+ T-cell clones is mediated by granzyme B, TRAIL and the cytotoxic cytokines TNF-α and IFN-γ, whereas it is independent of perforin and the Fas/FasL pathway. Granzyme B-dependent and perforin-independent killing by CD4+ T cells has also been demonstrated by Gondek et al.24 as a mechanism of regulatory T cells (T reg) to suppress effector T cells. This mechanism could also mediate cytotoxicity in Th1- like CD4+ T cells, but has not previously been demonstrated in human cells. Previous studies have established constitutive TRAIL expression in human CD4+ T cells, further upregulated upon stimulation through CD3, as an important pathway of T cell mediated cytotoxicity.34 Both T-cell clones tested express TRAIL, but clone 76 demonstrates high basal level of TRAIL. TRAIL expression is further increased upon incubation with peptide indicating that the upregulation is antigen-dependent.
Pentamer analysis of PBMCs from different time points after vaccination revealed the presence of a stable population of circulating hTERT-specific HLA-B7 restricted CTLs in this patient. This hTERT 613–621 CTL epitope has not previously been described, but is predicted by the SYFPEITHI algorithm. The frequency of CTLs specific for this epitope remained stable over a period of 7 y, indicating that these CTLs have been induced and maintained by vaccination with the long GV1001 peptide. This population may have contributed to the clinical success of vaccination in this case. This may have important clinical implications since HLA-B7 is present in approximately 20% of Caucasians. In addition, a second minor CTL population specific for another novel CTL eptiope, 672–681, was detected at some time points. Interestingly, this latter epitope also presented by HLA-B7, is embedded in the 30-mer peptide 660–689 inducing the strongest T-cell response and containing two distinct Th epitopes.
As the broad and durable T-cell response against GV1001 concomitant with tumor regression may have prepared the ground for epitope spreading through the release of endogenous hTERT under inflammatory conditions, we also studied T-cell responses against an overlapping hTERT peptide library. Others have demonstrated intramolecular eptiope spreading in 75- 84% of patients vaccinated with Th epitopes from the Her2/neu protein35,36 and more recently also intermolecular epitope spreading when vaccination was combined with monoclonal antibody against the same protein.37 We detected peptide-specific T-cell proliferation against several novel hTERT Th peptides unrelated to GV1001, indicating that hTERT is largely immunogenic and that extensive epitope spreading has taken place. Several of these epitopes were independently verified, since they were recognized by T cells from a cancer patient having undergone vaccination with hTERT transfected DCs.27
T-cell clones generated against the 30-mer peptide 660–689 were specific for the 663–677 fragment and restricted to HLA-DR*0801. The 660–689 peptide was also recognized in a HLA-DR*0101 and –DR*0401 restricted fashion by T-cell lines from a patient vaccinated with hTERT transfected DCs and by HLA-DQ*0604 and HLA-DR*1401 restricted T-cell clones from one of the melanoma patients and the colon cancer patient described here (unpublished results), demonstrating promiscuous binding to HLA class II molecules encoded by several alleles. Three clones with different peptide affinity were chosen for further study and shown to display an inflammatory cytokine profile upon peptide stimulation. They all displayed an early effector memory phenotype (CD45RO+,CCR7-, CD28+) indicating that these cells are not exhausted like the cytotoxic CD4+ T cells described in patients with chronic viral infections.38 Importantly, one of these CD4+ T-cell clones with high peptide affinity also recognized a HLA-DR*0801 positive melanoma cell line. This direct tumor cell recognition by hTERT-specific CD4+ T cells could have important implications for the in vivo anti-tumor response in melanoma patients as malignant melanomas are often positive for HLA class II molecules.39
The strong T-cell responses against multiple hTERT epitopes 5–6 y after tumor eradication suggest that they may have been important in tumor rejection, but we were unable to formally demonstrate that these T cells have expanded as result of vaccination as there was no pre-vaccination sample left from the patient. Recently, we demonstrated long-term immunological memory in pancreas cancer patients seven years after vaccination with long synthetic peptides representing point mutations in p21 Ras.40 Results from Williams and colleagues in a transgenic mouse model, suggest that memory CD4+ T cells with high functional avidity survive preferentially and show virtually no reduction in numbers during the memory maintenance phase.41
The breadth and magnitude of the CD4+ T cell responses presented here after vaccination with a single 16-mer peptide are associated with complete tumor remission of a stage IV non-small cell lung cancer. We have only studied the reactivity against a few hTERT epitopes located within the sequence of the active site of the enzyme, but this does not exclude that T cells of other specificities could be involved in tumor eradication.
Available blood samples from other long-term survivors vaccinated with GV1001 enabled us to confirm and extend the findings of the existence of widespread hTERT reactivity from the first lung cancer patient analyzed. Responses against the hTERT peptides in the test panel were detected using the same methods as for immunomonitoring of GV1001 vaccine responses. The data from four long-term surviving patients clearly show that the amino acid sequence covered by our overlapping peptide library contains a surprisingly high number of immunogenic epitopes. Since pre vaccine samples were not available for analysis, we are unable to determine if the observed immune responses against GV1001 unrelated epitopes were present before vaccination or were generated as a result of the vaccination, i.e., by “epitope spreading.” Analysis of responses in a blood sample taken in week 2 post start of vaccination from the melanoma patient shown in Figure 8A, does however indicate that responses against two of the peptides were already present at that time, 3 weeks before the first immune response against GV1001 was detectable.18 This analysis thus reveals a complex picture of the immune responses against hTERT in this patient, where some epitopes have spontaneously elicited an immune response prior to vaccination, while other responses may have been induced by the vaccination. In a novel sensitive bioluminescence mouse model, it was recently demonstrated that during elimination of large established tumors, tumor-specific T cells show an oscillating pattern of expansion and contraction often followed by a rebound, until full tumor rejection is obtained.42 This melanoma patient had a progressive widespread metastatic melanoma with very high tumor load at the onset of vaccination and chemotherapy. The tumor slowly regressed over a period of > 61 mo during which the blood samples used for analysis in this study were taken. This clinical situation thus resembles the experimental model described by Charo et al.42 Taking into account the uncontrolled and presumably suboptimal way the immune responses to the novel set of endogenous hTERT peptides have been induced, and the fact that the patient had concomitant bulky tumor masses, it is not surprising that the analysis of the immune responses revealed a highly dynamic situation similar to that described in animal models.
The main focus in the field of cancer immunotherapy has been on the stimulation of CTL responses and much less has therefore been known about the role of CD4+ T lymphocytes in anti-tumor responses. Recent mouse studies of adoptive CD4+ T-cell transfer have confirmed that they are capable of tumor eradication independently of CD8+ T cells. Xie et al. demonstrated how naïve tumor-specific transgenic CD4+ T cells could differentiate into both Th1 and CTLs in vivo after adoptive transfer into tumor-bearing lymphopenic hosts and cause tumor regression.43 In addition, other mouse studies have established the importance of antigen-specific CD4+ T cells for migration of CD8+ CTLs into tumor or infected tissue through the secretion of IFN-γ and induction of local chemokine secretion in infected tissue.44,45 The recent accumulated evidence of the important and very diverse function of CD4+ T cells in anti-tumor responses should contribute to improved effort in documenting these responses in the clinic. This knowledge will be crucial for the efficient manipulation of CD4+ and not only CD8+ T cells to generate more successful immunotherapy. A number of the novel epitopes described here are candidates for second generation hTERT peptide vaccines and are soon entering planned clinical trials. Active vaccination with a combination of several long peptides containing multiple epitopes is expected to induce a broader immune response and subsequently an improved clinical response.
Materials and Methods
Patient characteristics
A 54-y old male patient diagnosed with inoperable NSCLC/lung adenocarcinoma stage IIIA included in a phase I/II telomerase peptide vaccination trial as patient number 710, CTN-2000 was tested as a clinical responder in this study.14,33 CT (CT) scans demonstrated complete response with the regression of a 32 x 32 mm pulmonary tumor and mediastinal lymph node metastasis. Regular follow-up CT scans showed that the patient is tumor-free 10 y after vaccination start and booster vaccinations every 6 mo are continued.
Malignant melanoma patients included in this study had advanced stage IV melanoma (M1B or M1C) and had received concomitant treatment with temozolomide and hTERT peptide GV1001 (Trial identification number: NCT01247623). Temozolomide was administered 200 mg/m2 orally for 5 d every fourth week, and GV1001 as eight injections over 11 weeks. Immune response was evaluated by delayed type hypersensitivity, T-cell proliferation, and cytokine assays. Immunological responders continued monthly vaccination. Samples from six immunological vaccine responders (P3, P5, P6, P7, P8, P9), but clinical non-responders, were tested as well as two patients responding both immunologically and clinically, P19 and P22.18
Four stage III lung cancer patients treated with the GV1001 vaccine in combination with chemoradiotherapy in the CTN-2006 study (Trial identification number: NCT00509457) were also tested in the current study.33 The patients tested (P104–106, P108) all responded immunologically against the vaccine, but had below median survival and were included in the clinical non-responder group in this study.
A colon cancer patient vaccinated with GV1001 on a compassionate use basis and reported in a previous study of T-cell cytokine profiles as patient H.20 The patient first underwent surgery to remove primary tumor (adenocarcinoma, Duke C), but relapsed with liver metastases (Duke D) after 1 y. He was operated by hemihepatectomi before he started vaccination with GV1001. The patient still receives booster vaccinations once a year and has no evidence of tumor after 92 mo of vaccination.
The clinical trials were approved by the Norwegian Medicines Agency, the Committee for Medical Research Ethics, Region South and the Hospital Review Board and performed in compliance with the World Medical Association Declaration of Helsinki. Informed consent was obtained from the patients.
Peripheral blood was collected from anonymous, healthy donors at The Oslo Blood Bank, Oslo University Hospital, Norway, after informed consent.
Vaccine
The patients were vaccinated with telomerase peptide GV1001provided by GemVax AS. GV1001 is corresponding to the 16-amino acid residue 611–626 fragment (EARPALLTSRLRFIPK) of the hTERT protein (hTERT sequence, GenBank accession number: AB085628). The vaccine was administered by intra-dermal injection and 30 μg of Leucomax, granulohcyte-macrophage colony-stimulating factor (GM-CSF), (Schering-Plough) was injected at the vaccination site 5–15 min before each vaccine as adjuvant. Further details of the treatment have been previously described.14,18,33,20
Analysis of T-cell responses against hTERT
PBMCs collected during and after booster vaccinations were available for analysis. The PBMCs had been isolated and frozen as previously described.14 In brief, 50 ml of Acid Citrate Dextrose-blood was collected and PBMCs were isolated 0.5–3 h later by centrifugation over Lymphoprep (Axis-Shield). The PBMCs were frozen in RPMI-1640 (PAA Laboratories) with 10% human serum albumin (HSA, (Baxter)) and 10% dimethyl sulfoxide (DMSO, (Sigma-Aldrich)) for storage in liquid nitrogen. Viability of cells upon thawing was found to be between 94–97% as assessed by the trypan blue exclusion test.
Thawed PBMCs were stimulated once in vitro with peptide at 2 x 106 cells/ml in CellGro DC medium (CellGenix). 20U/ml IL-2 (Chiron) was added on day 3 cultured and this protocol was later modified to also include the addition of 5 ng/ml IL-7 (R&D Systems Inc.) on day 3. PBMCs from various time points were stimulated with an overlapping hTERT 15-mer peptide library or a 30-mer hTERT peptide covering the hTERT amino acid sequence 563–735 (GenBank accession number: AB085628) and then tested in proliferation assays (3H-Thymidine). For proliferation assays, T cells were seeded in triplicates in 96-well U-bottomed microtiter plates, generally at 5 104 cells per well (c/w). The same number of irradiated (30 Gy), autologous PBMCs was added for use as APCs. Proliferation was measured at day 3 after labeling with 3.7 104 Bq 3H-Thymidine (Laborel, Oslo, Norway) overnight before harvesting. T-cell proliferation assays were performed in triplicates.
All peptides were purchased from ProImmune Ltd. Recombinant hTERT protein was produced as previously described.14 Stimulatory Index (SI) was defined as proliferation with peptide divided by proliferation without peptide and standard deviations were calculated. SI ≥ 2 was considered a positive response.
51Cr-relase assays
51Cr-release cytotoxicity assay was performed as previously described.14 Autologous EBV-transformed B lymphoblastoid cell lines (EBV-LCL) target cells were pulsed with 10 µM peptide for about 0.5–1.0 h at 37°C. Effector T-cell clones were added at 2 x 105 (E:T ratio 100:1) and the plate was left at 37°C. Supernatants were harvested after 8–24 h and radioactivity was measured in a Packard Topcount microplate scintillation counter (Packard Instrument Company).
Generation of T cell clones against hTERT peptides GV1001 (611–626) and 660–689
T cells were cloned by limiting dilution as previously described46 for the generation of GV1001-specific clones. Cytokine additions were modified to 50 U/ml IL-2 (Chiron) and 5 ng/ml IL-7 (R&D Systems Inc.) for the generation of peptide 660–689 specific clones in order to favor the growth of CD8+ T cells. T-cell clones were expanded using irradiated PBMCs as feeder cells, PHA (Oxoid Ltd) and IL-2, then screened for peptide specificity in proliferation assays. GV1001-specific T-cell clones were also screened against a set of truncated peptides T2 and T8-T14 (for sequences, see Fig. S2) for identification of the minimal sequences required for peptide recognition. Patient lymphocytes were serotyped for HLA class I and genotyped for HLA class II by the National tissue typing laboratory and found to be HLA-A1 and –B7, -DRB1*08, -DQB1*04 and -DPB1*0401/0402.
IFN-γ ELISPOT
The IFN-γ ELISPOT assays were performed essentially as previously described.47 Monoclonal antibody against human IFN-γ (Mabtech) was diluted with PBS to a final concentration of 5 µg/ml. 96-well MultiScreen-HA plates (Millipore) were coated with antibody by adding 75 µl/well of the stock solution and incubated overnight at 4°C. The following day, plates were stored at room temperature for 1 h before washing wells six times with PBS 200 µl/well to remove excess antibody. To block unspecific binding, plates were incubated for 1–2 h at 37°C with 100 µl per well of CellGro DC medium plus 10% human serum (HS; Baxter) Thawed and washed autologous PBMCs were enumerated and added to the pre-coated wells at 5 x 105 cells/well. The responder T cells were harvested, washed, enumerated and transferred in CellGro DC medium (CellGenix) in triplicates to the wells containing autologous PBMCs at 1 x 105cells per well. Negative controls with T cells only and PBMCs only and positive controls with T cells + PBMC + Staphylococcus enterotoxin C3 (SEC3; Toxin Technologies) were included. After overnight incubation at 37°C with 5% CO2 in a humidified incubator, the plates were washed six times with PBS. Between the second and third wash, the plates were incubated for 10 min at room temperature. To each well, 75 µl of a stock solution of 1 µg/ml of biotinylated antibody against human IFN-γ (Mabtech) was added and plates were incubated for 2 h at room temperature. Following six repeated washings, plates were incubated for 1 h with 75 µl per well of streptavidin-ALP (Mabtech) from a stock solution (diluted 1:1000 in PBS plus 1% HSA). To remove excess antibody, the wells were again washed six times with PBS. Then, after adding 75 µl of substrate BCIP/NBT (Sigma-Aldrich) to each well, plates were incubated for 5–20 min. When spots appeared, water was added to stop the reaction. Spots were enumerated using an automated analyzer, CTL IMMUNOSPOT S5 VERSA-02–9030 (Cellular Technology Ltd).
Flow cytometry
Pentamer staining was performed on previously frozen patient PBMCs. Phycoerythrin (PE)-conjugated pentamers were manufactured by ProImmune. Pentamer with HIV peptide TPGPGVRYPL-B*0702 was used as a negative control. PBMCs were screened for reactivity against B*0702 pentamers with hTERT peptides 613–621 and 672–681. The cells were incubated with the protein kinase inhibitor, dasatinib (LC Labotatories), for 30 min at 37°C, then washed once before staining as previously described.48 Pentamer staining was performed as previously reported27 before staining with anti-CD4-Fluorescein isothiocyanate (FITC), anti-CD19-FITC, anti-CD8-Allophycocyanin (ACP) and anti-CD3-Pacific Blue (PB) (all from eBioscience). Cells were fixed in either BD Fix/Perm reagent or 1% paraformaldehyde (PFA). Newly thawed and rested T-cell clones were phenotyped. CD45RA-RPE (DAKO), CD45RO-PB (BioLegend) CCR7-FITC (eBioscience), CD28-FITC (eBioscience), CD27-PE (eBioscience), CD62L-APC-Alexa Fluor 750 (AF750) (eBioscience), CD57-FITC (eBioscience). Corresponding isotype controls were in each case purchased from the same company. For intracellular staining, T cells were stimulated overnight with autologous EBV-LCL loaded with peptide or melanoma cell line ESTDAB-100 which was a kind gift from Prof. Graham Pawelec, University of Tübingen, Germany. The T cell-to-target ratio was 1:3 in the presence of Brefeldin A at 10 μg/ml and BD Golgistop at a 1/1000 dilution. CD107a-PE-Cy5 (BD Bioscience) was added for the last hour of the incubation. Cells were stained for CD3 (eBioscience), CD4, CD8, TRAIL-PE (eBioscience), Perforin-APC, Granzyme B-PE (eBioscience), IFN-γ-FITC eBioscience, IL-2-APC (eBioscience) and TNF-α-AlexaFuor700 (BioLegend) using the BD Cytofix/Cytoperm kit according to the manufacturer's instructions. All antibodies and reagents for intracellular cytokine staining were purchased from BD Bioscience, except where noted. Fifty to one hundred thousand CD8+ T cells were acquired per sample for the detection of pentamer positive populations using a BD LSR II flow cytometer and the data were analyzed using FlowJo software (Treestar Inc.).
Statistics
The non-parametric Mann-Whitney U test was used to analyze both patient survival according to the number of hTERT peptides inducing T-cell responses and the differences in the mean magnitude of T-cell responses (SI) in short-term survivors vs. long-term survivors. At a p-value < 0.05, the differences were considered statistically significant.
Supplementary Material
Disclosure of Potential Conflicts of Interest
Gustav Gaudernack is a member of the advisory board for Kael-GemVax (patent holders for GV1001).
Acknowledgments
We are grateful to Paal F. Brunsvig, and Steinar Aamdal and Bjarte Solheim for clinical follow-up of the patients and Hiroko Solvang for statistical advice. We thank Dr Graham Pawelec for providing the melanoma cell line, ESTDAB-100 and Kael-GemVax for providing the GV1001 peptide. This work was supported by a grant from the Regional Health Authority of South Eastern Norway (Helse Sør-Øst RHF).
Glossary
Abbreviations:
- APC
antigen presenting cell
- CR
complete remission
- CTL
cytotoxic T lymphocyte
- DC
dendritic cell
- DMSO
dimethyl sulfoxide
- EBV-LCL
Epstein-Barr virus-transformed lymphoblastoid cell lines
- HLA
Human leukocyte antigen
- HSA
Human serum albumin
- IFN-γ
interferon-γ
- IL-2
interleukin-2
- hTERT
human reverse transcriptase
- NSCLC
non-small cell lung cancer
- PBMC
peripheral blood mononuclear cell
- SI
stimulation index
- Th
T helper
- TNF-α
tumour necrosis factor-α
- TRAIL
TNF-related apoptosis-inducing ligand
- Treg
T regulatory
Footnotes
Previously published online: www.landesbioscience.com/journals/oncoimmunology/article/20426
References
- 1.Harley CB, Kim NW, Prowse KR, Weinrich SL, Hirsch KS, West MD, et al. Telomerase, cell immortality, and cancer. Cold Spring Harb Symp Quant Biol. 1994;59:307–15. doi: 10.1101/SQB.1994.059.01.035. [DOI] [PubMed] [Google Scholar]
- 2.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–5. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 3.Shay JW. Telomerase in human development and cancer. J Cell Physiol. 1997;173:266–70. doi: 10.1002/(SICI)1097-4652(199711)173:2<266::AID-JCP33>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 4.Counter CM, Meyerson M, Eaton EN, Ellisen LW, Caddle SD, Haber DA, et al. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene. 1998;16:1217–22. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- 5.Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–33. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 6.Karimi-Busheri F, Rasouli-Nia A, Mackey JR, Weinfeld M. Senescence evasion by MCF-7 human breast tumor-initiating cells. Breast Cancer Res. 2010;12:R31. doi: 10.1186/bcr2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marian CO, Cho SK, McEllin BM, Maher EA, Hatanpaa KJ, Madden CJ, et al. The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clin Cancer Res. 2010;16:154–63. doi: 10.1158/1078-0432.CCR-09-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marian CO, Wright WE, Shay JW. The effects of telomerase inhibition on prostate tumor-initiating cells. Int J Cancer. 2010;127:321–31. doi: 10.1002/ijc.25043. [DOI] [PubMed] [Google Scholar]
- 9.Aloysius MM, Mc Kechnie AJ, Robins RA, Verma C, Eremin JM, Farzaneh F, et al. Generation in vivo of peptide-specific cytotoxic T cells and presence of regulatory T cells during vaccination with hTERT (class I and II) peptide-pulsed DCs. J Transl Med. 2009;7:18. doi: 10.1186/1479-5876-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen DY, Vance BA, Thompson LB, Domchek SM, Vonderheide RH. Differential lysis of tumors by polyclonal T cell lines and T cell clones specific for hTERT. Cancer Biol Ther. 2007;6:1991–6. doi: 10.4161/cbt.6.12.5078. [DOI] [PubMed] [Google Scholar]
- 11.Vonderheide RH, Domchek SM, Schultze JL, George DJ, Hoar KM, Chen DY, et al. Vaccination of cancer patients against telomerase induces functional antitumor CD8+ T lymphocytes. Clin Cancer Res. 2004;10:828–39. doi: 10.1158/1078-0432.CCR-0620-3. [DOI] [PubMed] [Google Scholar]
- 12.Wenandy L, Sørensen RB, Sengeløv L, Svane IM. thor Straten P, Andersen MH. The immunogenicity of the hTERT540-548 peptide in cancer. Clin Cancer Res. 2008;14:4–7. doi: 10.1158/1078-0432.CCR-07-4590. [DOI] [PubMed] [Google Scholar]
- 13.Bernhardt SL, Gjertsen MK, Trachsel S, Møller M, Eriksen JA, Meo M, et al. Telomerase peptide vaccination of patients with non-resectable pancreatic cancer: A dose escalating phase I/II study. Br J Cancer. 2006;95:1474–82. doi: 10.1038/sj.bjc.6603437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunsvig PF, Aamdal S, Gjertsen MK, Kvalheim G, Markowski-Grimsrud CJ, Sve I, et al. Telomerase peptide vaccination: a phase I/II study in patients with non-small cell lung cancer. Cancer Immunol Immunother. 2006;55:1553–64. doi: 10.1007/s00262-006-0145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domchek SM, Recio A, Mick R, Clark CE, Carpenter EL, Fox KR, et al. Telomerase-specific T-cell immunity in breast cancer: effect of vaccination on tumor immunosurveillance. Cancer Res. 2007;67:10546–55. doi: 10.1158/0008-5472.CAN-07-2765. [DOI] [PubMed] [Google Scholar]
- 16.Su Z, Dannull J, Yang BK, Dahm P, Coleman D, Yancey D, et al. Telomerase mRNA-transfected dendritic cells stimulate antigen-specific CD8+ and CD4+ T cell responses in patients with metastatic prostate cancer. J Immunol. 2005;174:3798–807. doi: 10.4049/jimmunol.174.6.3798. [DOI] [PubMed] [Google Scholar]
- 17.Parkhurst MR, Riley JP, Igarashi T, Li Y, Robbins PF, Rosenberg SA. Immunization of patients with the hTERT:540-548 peptide induces peptide-reactive T lymphocytes that do not recognize tumors endogenously expressing telomerase. Clin Cancer Res. 2004;10:4688–98. doi: 10.1158/1078-0432.CCR-04-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyte JA, Gaudernack G, Dueland S, Trachsel S, Julsrud L, Aamdal S. Telomerase peptide vaccination combined with temozolomide: a clinical trial in stage IV melanoma patients. Clin Cancer Res. 2011;17:4568–80. doi: 10.1158/1078-0432.CCR-11-0184. [DOI] [PubMed] [Google Scholar]
- 19.Hunger RE, Kernland Lang K, Markowski CJ, Trachsel S, Møller M, Eriksen JA, et al. Vaccination of patients with cutaneous melanoma with telomerase-specific peptides. Cancer Immunol Immunother. 2011;60:1553–64. doi: 10.1007/s00262-011-1061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kyte JA, Trachsel S, Risberg B. thor Straten P, Lislerud K, Gaudernack G. Unconventional cytokine profiles and development of T cell memory in long-term survivors after cancer vaccination. Cancer Immunol Immunother. 2009;58:1609–26. doi: 10.1007/s00262-009-0670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanderlugt CL, Miller SD. Epitope spreading in immune-mediated diseases: implications for immunotherapy. Nat Rev Immunol. 2002;2:85–95. doi: 10.1038/nri724. [DOI] [PubMed] [Google Scholar]
- 22.Le Bon A, Etchart N, Rossmann C, Ashton M, Hou S, Gewert D, et al. Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat Immunol. 2003;4:1009–15. doi: 10.1038/ni978. [DOI] [PubMed] [Google Scholar]
- 23.Ribas A, Timmerman JM, Butterfield LH, Economou JS. Determinant spreading and tumor responses after peptide-based cancer immunotherapy. Trends Immunol. 2003;24:58–61. doi: 10.1016/S1471-4906(02)00029-7. [DOI] [PubMed] [Google Scholar]
- 24.Gondek DC, Lu LF, Quezada SA, Sakaguchi S, Noelle RJ. Cutting edge: contact-mediated suppression by CD4+CD25+ regulatory cells involves a granzyme B-dependent, perforin-independent mechanism. J Immunol. 2005;174:1783–6. doi: 10.4049/jimmunol.174.4.1783. [DOI] [PubMed] [Google Scholar]
- 25.Zhao DM, Thornton AM, DiPaolo RJ, Shevach EM. Activated CD4+CD25+ T cells selectively kill B lymphocytes. Blood. 2006;107:3925–32. doi: 10.1182/blood-2005-11-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanai F, Ishii E, Kojima K, Hasegawa A, Azuma T, Hirose S, et al. Essential roles of perforin in antigen-specific cytotoxicity mediated by human CD4+ T lymphocytes: analysis using the combination of hereditary perforin-deficient effector cells and Fas-deficient target cells. J Immunol. 2003;170:2205–13. doi: 10.4049/jimmunol.170.4.2205. [DOI] [PubMed] [Google Scholar]
- 27.Suso EM, Dueland S, Rasmussen AM, Vetrhus T, Aamdal S, Kvalheim G, et al. hTERT mRNA dendritic cell vaccination: complete response in a pancreatic cancer patient associated with response against several hTERT epitopes. Cancer Immunol Immunother. 2011;60:809–18. doi: 10.1007/s00262-011-0991-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroers R, Shen L, Rollins L, Rooney CM, Slawin K, Sonderstrup G, et al. Human telomerase reverse transcriptase-specific T-helper responses induced by promiscuous major histocompatibility complex class II-restricted epitopes. Clin Cancer Res. 2003;9:4743–55. [PubMed] [Google Scholar]
- 29.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 30.Romero P, Zippelius A, Kurth I, Pittet MJ, Touvrey C, Iancu EM, et al. Four functionally distinct populations of human effector-memory CD8+ T lymphocytes. J Immunol. 2007;178:4112–9. doi: 10.4049/jimmunol.178.7.4112. [DOI] [PubMed] [Google Scholar]
- 31.Ben-Smith A, Gorak-Stolinska P, Floyd S, Weir RE, Lalor MK, Mvula H, et al. Differences between naive and memory T cell phenotype in Malawian and UK adolescents: a role for Cytomegalovirus? BMC Infect Dis. 2008;8:139–48. doi: 10.1186/1471-2334-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrington LE, Janowski KM, Oliver JR, Zajac AJ, Weaver CT. Memory CD4 T cells emerge from effector T-cell progenitors. Nature. 2008;452:356–60. doi: 10.1038/nature06672. [DOI] [PubMed] [Google Scholar]
- 33.Brunsvig PF, Kyte JA, Kersten C, Sundstrøm S, Møller M, Nyakas M, et al. Telomerase peptide vaccination in NSCLC: a phase II trial in stage III patients vaccinated after chemoradiotherapy and an 8-year update on a phase I/II trial. Clin Cancer Res. 2011;17:6847–57. doi: 10.1158/1078-0432.CCR-11-1385. [DOI] [PubMed] [Google Scholar]
- 34.Kayagaki N, Yamaguchi N, Nakayama M, Kawasaki A, Akiba H, Okumura K, et al. Involvement of TNF-related apoptosis-inducing ligand in human CD4+ T cell-mediated cytotoxicity. J Immunol. 1999;162:2639–47. [PubMed] [Google Scholar]
- 35.Disis ML, Grabstein KH, Sleath PR, Cheever MA. Generation of immunity to the HER-2/neu oncogenic protein in patients with breast and ovarian cancer using a peptide-based vaccine. Clin Cancer Res. 1999;5:1289–97. [PubMed] [Google Scholar]
- 36.Disis ML, Gooley TA, Rinn K, Davis D, Piepkorn M, Cheever MA, et al. Generation of T-cell immunity to the HER-2/neu protein after active immunization with HER-2/neu peptide-based vaccines. J Clin Oncol. 2002;20:2624–32. doi: 10.1200/JCO.2002.06.171. [DOI] [PubMed] [Google Scholar]
- 37.Disis ML, Wallace DR, Gooley TA, Dang Y, Slota M, Lu H, et al. Concurrent trastuzumab and HER2/neu-specific vaccination in patients with metastatic breast cancer. J Clin Oncol. 2009;27:4685–92. doi: 10.1200/JCO.2008.20.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Appay V, Zaunders JJ, Papagno L, Sutton J, Jaramillo A, Waters A, et al. Characterization of CD4(+) CTLs ex vivo. J Immunol. 2002;168:5954–8. doi: 10.4049/jimmunol.168.11.5954. [DOI] [PubMed] [Google Scholar]
- 39.Deffrennes V, Vedrenne J, Stolzenberg MC, Piskurich J, Barbieri G, Ting JP, et al. Constitutive expression of MHC class II genes in melanoma cell lines results from the transcription of class II transactivator abnormally initiated from its B cell-specific promoter. J Immunol. 2001;167:98–106. doi: 10.4049/jimmunol.167.1.98. [DOI] [PubMed] [Google Scholar]
- 40.Wedén S, Klemp M, Gladhaug IP, Møller M, Eriksen JA, Gaudernack G, et al. Long-term follow-up of patients with resected pancreatic cancer following vaccination against mutant K-ras. Int J Cancer. 2011;128:1120–8. doi: 10.1002/ijc.25449. [DOI] [PubMed] [Google Scholar]
- 41.Williams MA, Ravkov EV, Bevan MJ. Rapid culling of the CD4+ T cell repertoire in the transition from effector to memory. Immunity. 2008;28:533–45. doi: 10.1016/j.immuni.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Charo J, Perez C, Buschow C, Jukica A, Czeh M, Blankenstein T. Visualizing the dynamic of adoptively transferred T cells during the rejection of large established tumors. Eur J Immunol. 2011;41:3187–97. doi: 10.1002/eji.201141452. [DOI] [PubMed] [Google Scholar]
- 43.Xie Y, Akpinarli A, Maris C, Hipkiss EL, Lane M, Kwon EK, et al. Naive tumor-specific CD4(+) T cells differentiated in vivo eradicate established melanoma. J Exp Med. 2010;207:651–67. doi: 10.1084/jem.20091921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong SB, Bos R, Sherman LA. Tumor-specific CD4+ T cells render the tumor environment permissive for infiltration by low-avidity CD8+ T cells. J Immunol. 2008;180:3122–31. doi: 10.4049/jimmunol.180.5.3122. [DOI] [PubMed] [Google Scholar]
- 45.Nakanishi Y, Lu B, Gerard C, Iwasaki A. CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature. 2009;462:510–3. doi: 10.1038/nature08511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kyte JA. Cancer vaccination with telomerase peptide GV1001. Expert Opin Investig Drugs. 2009;18:687–94. doi: 10.1517/13543780902897631. [DOI] [PubMed] [Google Scholar]
- 47.Gjertsen MK, Saeterdal I, Saebøe-Larssen S, Gaudernack G. HLA-A3 restricted mutant ras specific cytotoxic T-lymphocytes induced by vaccination with T-helper epitopes. J Mol Med (Berl) 2003;81:43–50. doi: 10.1007/s00109-002-0390-y. [DOI] [PubMed] [Google Scholar]
- 48.Kyte JA, Kvalheim G, Aamdal S, Saebøe-Larssen S, Gaudernack G. Preclinical full-scale evaluation of dendritic cells transfected with autologous tumor-mRNA for melanoma vaccination. Cancer Gene Ther. 2005;12:579–91. doi: 10.1038/sj.cgt.7700837. [DOI] [PubMed] [Google Scholar]
- 49.Lissina A, Ladell K, Skowera A, Clement M, Edwards E, Seggewiss R, et al. Protein kinase inhibitors substantially improve the physical detection of T-cells with peptide-MHC tetramers. J Immunol Methods. 2009;340:11–24. doi: 10.1016/j.jim.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.