Abstract
Protein crystallization in cells has been observed several times in nature. However, owing to their small size these crystals have not yet been used for X-ray crystallographic analysis. We prepared nano-sized in vivo–grown crystals of Trypanosoma brucei enzymes and applied the emerging method of free-electron laser-based serial femtosecond crystallography to record interpretable diffraction data. This combined approach will open new opportunities in structural systems biology.
Protein crystallization occurs as a native process in vivo. Prominent examples of this biological self-assembly include storage proteins in seeds, enzymes within peroxisomes and insulin within secretory granules1. Cells seem to control interactions of these proteins through changes in the ionic environment, proteolysis of precursor proteins or specific binding partners. Although these structures regulate cellular functions, in vivo crystallization has been perceived as atypical behavior and has therefore been neglected in comparison to the considerable efforts devoted to understanding and optimizing in vitro protein crystallization for X-ray structure determination.
Heterologously expressed proteins are also able to form crystals within cells. In the baculovirus-Sf9 expression system, virions are coated with a crystalline polyhedrin matrix2, representing a functional biological crystallization system. Site-specific transposition of an expression cassette into a baculovirus shuttle vector replaces the polyhedrin gene with a gene of interest. The permanent activation of the polyhedrin gene promoter ensures high local protein concentrations, obviously one prerequisite for crystal formation in vivo. This system has been used in insect cells to successfully crystallize polyhedrin3 and chimeric proteins consisting of a polyhedrin part attached to the protein of interest4, as well as two polyhedrin-free subunits of calcineurin5. However, no structural data from in vivo–grown, polyhedrin-free crystals have been obtained so far, a gap largely attributed to the small size of the crystals, limited by the maximum diameter of the living cell.
In this study we present an approach for structural biology that combines the recently established method of serial femtosecond X-ray crystallography (SFX)6 and the use of in vivo–grown crystals to record diffraction data. The SFX method uses coherent X-ray pulses produced by an X-ray free-electron laser (FEL) that are over a billion times more brilliant than third-generation synchrotron sources and aims to overcome resolution limits imposed by radiation damage at conventional sources7. Using the ‘diffraction before destruction’ principle, diffraction patterns are collected with single, ultrafast pulses that essentially terminate before the onset of significant structural changes occurs, and the X-ray pulse finally vaporizes the sample6. The single-pulse diffraction pattern has been predicted to represent the undamaged crystal injected across the beam in a liquid microjet, depending on the pulse intensity and duration8. As a first proof of principle, a recent study has recorded tens of thousands of snapshots from individual crystals of photosystem I (PSI) at room temperature (17–20 °C), ranging in size from 200 nm to 2 µm (ref. 6). The successful structural investigation of this protein complex up to a resolution of 8.5 Å demonstrates the viability of using nanocrystals and the SFX method for structure determination.
We observed in vivo crystallization of polyhedrin-free, glycosylated cathepsin from Trypanosoma brucei (TbCatB) within Sf9 insect cells transfected with bmon14272 bacmid (Invitrogen) created by on site–specific transposition with pFastBac1 expression plasmid (Invitrogen) containing the Autographa californica multiple nuclear polyhedrosis virus polyhedrin (PH) promoter. Knockdown of TbCatB has been shown to be lethal for the parasite, which causes human African trypanosomiasis, one of the most important neglected diseases, affecting over 60 million people in central Africa9. Efficient and cost-effective drugs are not yet available, but cysteine proteases such as TbCatB have been identified as potential drug targets in protozoan parasites10.
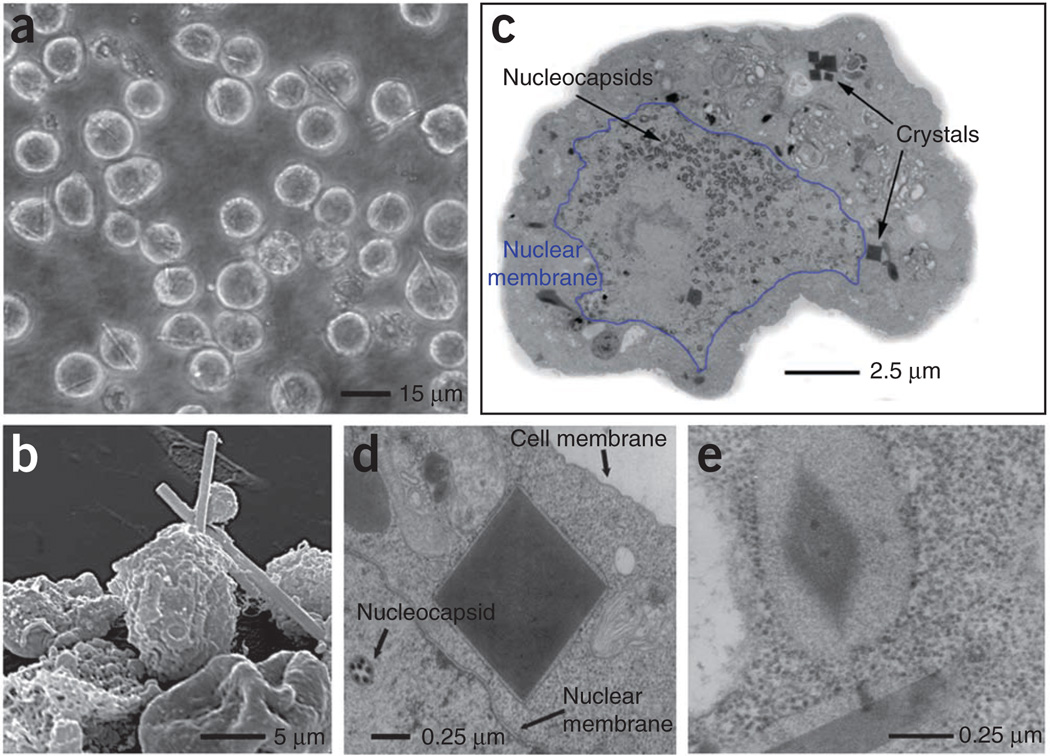
Approximately 70 h after infection, the formation of needle-shaped microstructures was visible by light microscopy in Sf9 cells infected with recombinant baculovirus containing the gene encoding the pre-pro form of TbCatB (including the TbCatB signal sequence, the pro-peptide and the mature enzyme sequence of TbCatB; Fig. 1a). Electron microscopy (EM) revealed a damaged cell surface and sharp, needle-like crystals 10–15 µm in length and 0.5–1 µm in width spiking out of the cells (Fig. 1b). Capsids were visible within the nucleus, and crystals appeared as defined dark areas with sharp square edges within the cytoplasm (Fig. 1c). Membranes decorated with ribosomes surrounding the crystals indicate an origin of crystal formation within the endoplasmic reticulum (Fig. 1d). An ordered lattice structure observed at higher magnification identified these particles as protein crystals (Fig. 1e).
Figure 1.
Light microscopic and EM analysis of Sf9 insect cells with embedded in vivo crystals. (a) Light micrograph of Sf9 cells infected with TbCatB virus 90 h after infection. (b) Transmission EM (TEM) micrograph of an embedded and sectioned infected Sf9 cell with crystals cut perpendicular to the long axis of the needle. Nuclear membrane is outlined in blue. (c) Scanning EM micrograph of a group of Sf9 cells infected with TbCatB virus 80 h after infection. (d) TEM micrograph of a sectioned sample, showing a crystal cut perpendicular to the long axis of the needle with surrounding membrane between nuclear and cell membrane. (e) TEM micrograph showing the lattice structure of a crystal and a longitudinal section of a second crystal (both crystals are surrounded by membrane).
Usually, protein accumulation is inhibited by the ‘unfolded protein response’ (UPR), in which atypical or misfolded proteins inhibit further protein biosynthesis11. This transcriptional regulation fails in the case of the polyhedrin promoter, leading to an enormous increase of TbCatB concentrations that finally provokes crystallization. This interpretation is consistent with the observation that changing the signal sequence from the trypanosomal to the insect cell signal peptide led to secretion of soluble TbCatB without crystal formation (data not shown). In contrast to polyhedrin3, all crystals appeared without embedded virions. During the progress of infection, the number of crystals continuously increased, until more than 70% of the cells contained one or more crystals. If released during infection-mediated lysis, crystals either remained attached to cell remnants or floated freely within the medium.
To isolate in vivo crystals, we lysed cells and subjected them to differential centrifugation, resulting in ~5 × 105 purified crystals from 106 cells obtained after 8 d in suspension culture. These crystals were stable in deionized water, high- or low-salt solutions and alkaline buffers but became soluble in buffers at pH values below 4. Analysis of the solubilized protein confirmed that the major constituent of the crystals was glycosylated TbCatB comprising the C-terminal residues 63 to 93 of the propeptide and the mature enzyme sequence (residues 94 to 336; Supplementary Note and Supplementary Fig. 1).
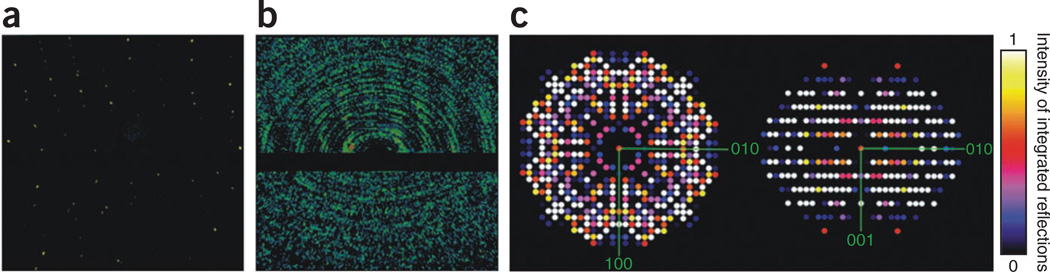
Synchrotron radiation–based experiments showed that the isolated TbCatB in vivo–grown crystals were too small to generate suitable X-ray diffraction at beamline X13 of DORIS III (HASYLAB/DESY, Hamburg, Germany), probably owing to technical limitations of the beamline associated with the crystal size rather than to the diffraction quality of the crystal. However, strong diffraction was observed at the Linac Coherent Light Source (LCLS, Menlo Park, California, USA). Purified in vivo crystals were crushed by vigorous stirring with glass beads to increase the particle density and injected across the FEL beam in a vacuum in a stream of water using a liquid jet (Supplementary Fig. 2). Single-pulse diffraction patterns were recorded at up to 7.5-Å resolution, corresponding to the technical resolution limit determined by the photon energy of the available X-ray pulses (2.0 keV, λ = 6.2 Å) and the detector geometry. During 23.1 min, 83,224 frames were obtained. After background subtraction, 988 of the frames contained distinct diffraction signals of three or more Bragg spots, each corresponding to a ‘snapshot’ diffraction pattern from a different randomly oriented crystal (Fig. 2a). Quality measures indicate that the FEL dataset conforms to diffraction data from a macromolecular crystal (Supplementary Fig. 3 and Supplementary Table 1). In the limited time available for data collection, it was not possible to obtain a complete dataset, as indicated by the granularity in the summed powder diffraction rings (Fig. 2b) and by the statistics of the dataset (Supplementary Table 1). As the redundancy was far too low for structure-factor extraction by Monte Carlo integration12, an overall Rsplit value (T.A.W. et al., unpublished data) was not calculated. Detailed comparison to corresponding values of a similar number of diffraction patterns recorded from photosystem I nanocrystals using SFX6 showed that the data quality improves with the number of patterns, confirming that additional diffraction data will result in a complete TbCatB dataset. However, 879 (89%) of the 988 recorded diffraction patterns were indexed without unit cell constraints. The raw lattice parameters were a = 122.9 Å, b = 123.6 Å, c = 53.4 Å, α = 90.3°, β = 90.2° and γ = 90.3°; therefore, the unit cell was assigned to be tetragonal, with a = b = 123.3 Å and c = 53.4 Å. Multiple measurements of individual Bragg reflections were averaged, and intensities from symmetry-equivalent reflections were merged according to the point group 4/mmm; a pseudo-precession pattern of the [001] zone is shown in Figure 2c. Our results reveal that structural information can be obtained from in vivo grown crystals of a glycosylated protein, providing a second independent proof of the SFX method.
Figure 2.
Serial femtosecond crystallography of in vivo TbCatB crystals. (a) Diffraction pattern of a TbCatB in vivo crystal recorded from a single shot of 70 fs FEL X-rays. (b) Sum of 988 single-shot FEL diffraction patterns from TbCatB crystals in different orientations. The lower panel of the detector was shifted to achieve higher resolution (Online Methods). At the edge of the detector, a maximum resolution of 7.5 Å was obtained. (c) Precession-style image of the [001] zone for TbCatB, obtained by merging SFX data from 328 in vivo crystal patterns indexed with unit cell constraints. Intensities of integrated reflections were normalized to values between 0 and 1 on a linear scale.
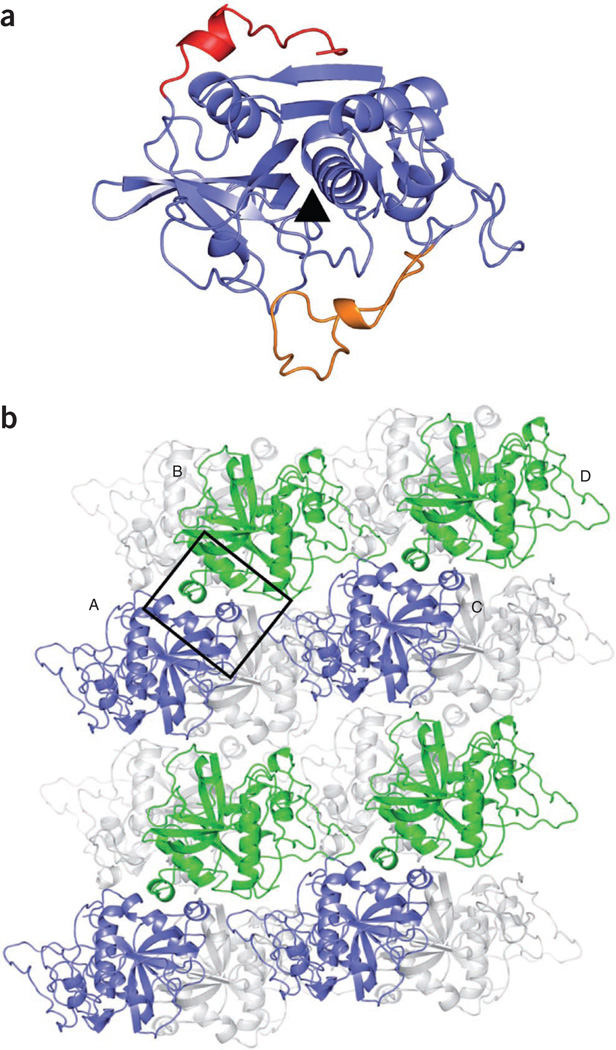
In addition, PNGase F–treated TbCatB from isolated and solubilized in vivo crystals was recrystallized in vitro using the vapor diffusion technique. X-ray diffraction data were collected at the Swiss Light Source (Villigen, Switzerland). The structure was determined by molecular replacement using the bovine CatB structure (PDB 1QDQ) as a model and refined to 2.55-Å resolution (Supplementary Table 2). The final structure of TbCatB (PDB 3MOR) shows the characteristic papain–cathepsin B fold, including 16 of the pro-peptide residues (Ser78 to Pro93; Fig. 3a), and is highly similar to a structure of nonglycosylated recombinant TbCatB that has been independently reported13 (PDB 3HHI). Here we refer to our own structural data, as we prepared our crystals from protein obtained from the in vivo–grown crystals.
Figure 3.
Structure of solubilized and recrystallized TbCatB solved by conventional X-ray crystallography. (a) Ribbon tracing of TbCatB, showing the pro-peptide in red and the occluding loop in orange. The black triangle indicates the position of the catalytic cleft. (b) Cartoon representation of the crystal packing. The crystals contain two molecules in their asymmetric unit and four molecules in the unit cell. This diagram of crystal contacts shows four molecules labeled A–D. In the foreground, every second TbCatB molecule is shown in blue or green, respectively. The major contact area is boxed. Gray ribbons represent additional molecules in the crystal packing.
Despite deglycosylation treatment, our TbCatB structure still contains a glycan side chain at Asn216, clearly identified in the electron density. This suggests that, although deglycosylation was effective as judged from SDS-PAGE results (Supplementary Fig. 1c), it removed only the glycan at Asn76. As the essential crystal contacts primarily involve hydrophobic patches within the persisting pro-peptide of TbCatB (Fig. 3b), this pro-peptide might also influence crystal formation in vivo (Supplementary Note). Moreover, the structure shows distinct differences from human CatB (PDB 1GMY), which are particularly important to consider for rational drug discovery investigations (Supplementary Note).
The advantages of in vivo crystallization are (i) production of post-translationally modified proteins of interest, (ii) easy isolation by spinning down the crystals after cell lysis, (iii) the possibility of preliminarily analyzing crystals by EM and (iv) a narrow crystal size distribution that is ideal for SFX applications. As the use of non-insect cell signal peptides is explored further, it seems likely that more proteins will form in vivo crystals within insect cells. Recently, we obtained similar results with an inosine monophosphate dehydrogenase from trypanosomes (TbIMPDH). Under comparable experimental conditions to those reported for TbCatB, TbIMPDH in vivo crystals appeared after 5 to 6 d. Besides TbCatB and calcineurin, this is a third example of in vivo–grown crystals from a heterologously expressed protein in Sf9 insect cells. TbIMPDH crystals were isolated and subjected to a brief SFX diffraction test at LCLS. The ability of these in vivo crystals to diffract up to a resolution of more than 8 Å (data not shown) strongly supports a more general applicability of the approach described in this study.
Although progress has been made in using microfocus beamlines that allow data collection from crystals only 1 µm in size, radiation damage is an inherent problem of X-ray crystallography14. Typically, large and well-ordered crystals of 20–500 µm are required for conventional X-ray crystallography, often a serious challenge to grow15. Therefore, the unique combination of in vivo crystallization and serial femtosecond crystallography offers notable new possibilities for proteins that do not form crystals suitable for conventional X-ray diffraction in vitro, extending the available methods of structural biology particularly when, in due time, shorter wavelengths of the FEL will provide higher-resolution data.
ONLINE METHODS
Cloning
The gene coding for the pre-pro-form of TbCatB (GeneDB Tb927.6.560) was amplified by PCR using primers 5′-TAGGATCCATGCATCTCATGCGTGCCT-3′ (sense) and 5′-TAACTCGAGCTACGCCGTGTTGGGTG-3′ (antisense), and AccuPrime Taq DNA polymerase (Invitrogen) according to the manufacturer’s instructions. After subcloning (TOPO-TA cloning kit, Invitrogen) into XL1-Blue–competent Escherichia coli cells (Stratagene), plasmid DNA purification (QIAprep spin miniprep kit, Qiagen) and digestion with BamHI and XhoI, the extracted gel fragment (QIAquick gel extraction kit, Qiagen) was cloned into pFastBac1 expression plasmid (Invitrogen) and transformed into DH10Bac-competent E. coli cells (Invitrogen) according to the manufacturer’s instructions. The TbCatB gene, containing its own signal peptide sequence, replaced the polyhedrin gene, including the polyhedrin nuclear import signal.
Recombinant Bacmid DNA was purified according to the Bacmid isolation protocol (Invitrogen) using the QIAprep spin miniprep kit (Qiagen) and subsequently used for PCR analysis of the cloned sequence. Correctness of the PCR products was verified by sequencing. Bacmid DNA was then used for lipofection with Sf9 insect cells grown in serum-free medium at 27 °C to generate recombinant virus stock according to the Bac-to-Bac manual (Invitrogen). This stock was used to generate a high-titer virus stock for further infections (titer: 1 × 108 plaque-forming units (p.f.u.) ml−1).
Expression of TbCatB
Recombinant virus stock was used to infect a 70%-confluent monolayer culture of Sf9 insect cells (Invitrogen), grown in serum-free medium at 27 °C with a multiplicity of infection of 0.1 p.f.u. per cell. After 72–96 h (upon visual inspection of the amount of crystals within cells by light microscopy), we harvested the cells by rinsing and scraping them from the surface of the cell flask and then centrifuging the cell suspension at 1,000g for 5 min. For greater amounts of protein material, suspension cultures with agitation at 110 r.p.m. at 27 °C were infected and harvested as described above.
Electron microscopy
For TEM, infected insect cells of a 75-mm2 confluent monolayer culture were fixed using 2% (vol/vol) glutaraldehyde in 0.2 M sodium cacodylate buffer containing 0.12 M sucrose for 1 h at 41 °C. After washing four times (10 min each) and storing overnight in sodium cacodylate buffer, cells were postfixed in 1.5% (wt/vol) osmium tetroxide and stained in 0.5% (wt/vol) uranyl acetate. Dehydration in ethanol, clearing in propylene oxide, embedding in Agar 100 resin and sectioning were performed according to standard procedures. Sections were stained in 5% (wt/vol) uranyl acetate and 0.4% (wt/vol) lead citrate. For scanning electron microscopic analysis, the same fixation and staining protocol was applied up to 70% (vol/vol) ethanol. Cells were then placed on polylysine-covered coverslips and dehydrated with 96% and 100% ethanol for 8 h each. Critical-point drying and gold-palladium sputter staining were performed using standard protocols. TEM was performed using a Zeiss EM10 device; for scanning electron microscopy, a Cambridge Stereo Scan 250 Mk2 device was used.
Isolation and solubilization of crystals
Harvested cells were washed twice in PBS and lysed in RIPA lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% (vol/vol) Nonidet P-40, 0.25% (wt/vol) sodium deoxycholate, 0.1% (wt/vol) SDS, 0.01% (wt/vol) sodium azide, pH 7.4), intensely vortexed and incubated on ice for 15 min. To reduce viscosity, we added 25 units ml−1 Benzonase (Novagen) before incubating the mixture for 30 min at 37 °C or overnight at 4 °C. The sample was then washed several times with PBS. The pellet was resuspended in solubilization buffer (50 mM sodium acetate, pH 3.5), incubated for 15 min on ice and centrifuged at 16,000g for 15 min. The supernatant was concentrated using 10-kDa NMWL-Centricon devices, thereby changing the buffer to 50 mM sodium acetate, 4 mM DTT, 0.5 mM EDTA (pH 5.5) or another appropriate buffer, depending on the application.
Serial femtosecond crystallography
TbCatB in vivo crystals were analyzed by the SFX method6 at LCLS in the SLAC National Accelerator Laboratory (Menlo Park, California, USA)16. The X-ray FEL generated intense quasi-monochromatic X-ray pulses of 67-fs duration with 6.7 × 1012 photons per pulse and a wavelength of 6.2 Å (2 keV). Focused onto the crystal, a single pulse reaches an intensity of ~0.5 × 1018 W cm−2 and a dose to the sample of about 1 to 3 GGy, equivalent to 100 times the Garman safe limit7, depending on the number of TbCatB monomers within the unit cell. The detailed electron and photon beam parameters are summarized in Supplementary Table 3. To ensure that there was always, on average, one crystal passing through the 100-fl X-ray interaction volume of the liquid jet, corresponding to the theoretically required optimal volume/hit rate ratio, we had to increase the crystal number density before measurement. Therefore, ~5 × 107 TbCatB crystals isolated from a single 100-ml culture of 2 × 106 virus-infected insect cells per ml were crushed by vigorous vortexing with glass beads for 30 min at room temperature, concentrated by centrifugation for 30 s at 14,500g and filtered through a 2-µm filter (A-430, Upchurch Scientific). Thus, ~109 crystals in 1 ml ddH2O were obtained.
The experiment was performed at the Atomic, Molecular and Optical Science beamline17 using the CFEL-ASG multipurpose (CAMP) instrument18. A liquid microjet19 focused by a coaxial flow of gas to a diameter of about 4 µm at a flow rate of 15 µl min−1 was used to introduce crystals stored in a sample loop6 into the FEL interaction region. The interaction region of the X-rays and the crystals was located in the continuous liquid column, before the Rayleigh break-up of the jet into drops. Testing of TbCatB microcrystal delivery through the gas-focused jet at SLAC in the PULSE Institute enabled selection of the optimum jet nozzle before installation on the CAMP apparatus. In 23.1 min, 83,224 frames were recorded with a pair of movable, high-frame-rate, low-noise pn junction charge-coupled device (pnCCD) detector modules18 (each panel consisted of 512 pixels × 1,024 pixels, each 75 µm2 × 75 µm2 in area) operating at the 60-Hz repetition rate of the FEL pulses. The upper panel of the detector was located 65 mm from the interaction point; the lower panel was mounted to z = 68 mm to record scattering from 3.51° to 49.02° in the vertical plane. The 988 identified single-crystal diffraction patterns were processed to subtract background noise and then indexed by an autoindexing procedure based on DirAx20 to determine the unit cell parameters and therefore the symmetry of the lattice. Details of data processing are given in ref. 6.
Activity assay
The cathepsin B activity kit (BioVision) was used to determine the enzyme activity of solubilized TbCatB. This assay uses the preferred cathepsin B substrate sequence RR labeled with amino-4-trifluoromethyl coumarin. Fluorescence emission at 495 nm was measured using a PerkinElmer LS 55 fluorescence spectrometer with an excitation wavelength of 400 nm. The chemical identity of the included CatB inhibitor was not released by the manufacturer.
Deglycosylation
Solubilized protein in 10 mM HEPES and 20 mM NaCl (pH 7) was deglycosylated overnight at 25 °C by addition of 1 unit µg−1 N-glycosidase F (PNGase F, New England Biolabs). Sample was then concentrated using 10 kDa NMWL-Centricon devices to a concentration of 2 mg ml−1 in 10 mM HEPES and 20 mM NaCl, pH 7.
Recrystallization and structure determination
We grew all crystals with the sitting-drop vapor diffusion technique by mixing TbCatB (2 mg ml−1 in 10 mM HEPES and 20 mM NaCl, pH 7) and precipitant solution (22–30% polyethylene glycol 3000 (wt/vol) and 200–400 mM (NH4)2HPO4) in a 1:1 ratio. Crystals were flash-frozen for X-ray data collection at 100 K using 20% (vol/vol) glycerol as cryoprotectant.
Diffraction data were collected at beamline X06DA at the Swiss Light Source (Villigen, Switzerland) using a data collection wavelength of 1.000 Å and a MarCCD detector (Mar Research), and processed with XDS21. Initial phases were obtained by molecular replacement with PHASER22 using the bovine CatB structure23 as a search model (PDB 1QDQ). The structure was refined using rigid-body refinement and simulated annealing with PHENIX24. Subsequent refinement was carried out by alternating rounds of model-building with Coot25 and restrained refinement using two-fold noncrystallographic symmetry restraints with Refmac26. The final model had a low free R-factor of 24.3% and good stereochemistry (Supplementary Table 2). The structure reported here was aligned with the TbCatB structure determined in ref. 13 (PDB 3HHI) using the secondary structure–matching superposition protocol of the program Coot25. The buried surface area was calculated with AREAIMOL27. Figure 3a,b was prepared with PyMOL (DeLano Scientific)28.
Supplementary Material
Acknowledgments
FEL experiments were carried out at LCLS in June 2010 (TbCatB) and in August 2011 (TbIMPDH), a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The X-ray diffraction experiments on recrystallized TbCatB crystals were carried out at beamline X06DA of the Swiss Light Source (Villigen, Switzerland). This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft (DFG), from the Swedish Research Council, from the Knut och Alice Wallenbergs Stiftelse, from the European Research Council, as well as by US National Science Foundation award MCB-1021557. R.K. received a fellowship from the Landesgraduiertenförderung Baden-Württemberg. L.R., D. Rehders and C. Betzel thank the German Federal Ministry for Education and Research for funding (grants 01KX0806 and 01KX0807). Support from the Hamburg Ministry of Science and Research and Joachim Herz Stiftung as part of the Hamburg Initiative for Excellence in Research and the Hamburg School for Structure and Dynamics in infection, and from the DFG Cluster of Excellence “Inflammation at Interfaces” (EXC 306) is gratefully acknowledged.
Footnotes
Accession codes. Protein Data Bank: 3MOR (coordinates and structure factors for in vitro–recrystallized TbCatB).
Note: Supplementary information is available on the Nature Methods website.
AUTHOR CONTRIBUTIONS
R.K., K.C. and L.R. contributed equally to this work. R.K. performed the in vivo crystallization experiments under the supervision of M.D.; K.C. prepared samples for synchrotron X-ray crystallography and collected and analyzed data under the supervision of T.S.; H.N.C. and J.C.H.S. conceived the SFX experiment, which was designed with P.F., A.B., R.A.K., J.S., D.P.D., U.W., R.B.D., M.J.B., I.S., H.F. and J.H.; FEL samples were prepared by L.R., D. Rehders and C. Betzel; SFX experiments were carried out by L.R., K.N., H.N.C., D.P.D., F.S., M.L., T.A.W., A.A., M.J.B., C.Y.H., R.G.S., U.W., A.B., R.A.K., R.B.D., N.C., R.L.S., L.L., J.D., M.S.H., C. Bostedt, J.D.B., S. Boutet and G.J.W.; beamline setup was done by C. Bostedt, J.D.B., S. Boutet, G.J.W. and M.M. The delivery system was developed and operated by R.B.D., D.P.D., U.W., J.C.H.S., P.F., L.L. and R.L.S.; S.W.E., B.E., L.F., H.G., A.H., R.H., G.H., H.H., P.H., N.K., C.R., D. Rolles, B.R., A.R., H.S., L.S., J.U., C.G.W. and G.W. operated the CAMP instrument and the pn junction charge-coupled devices and developed the software for pnCCD readout. Diffraction instrumentation was developed and calibrated by H.N.C., A.B., A.A., J.S., D.P.D., U.W., R.B.D., M.J.B., L.G., J.H., M.M.S., N.T., J.A., S.S., S. Bajt, M.B. and J.C.H.S. Data were analyzed by T.A.W., K.N., F.S., A.B., R.A.K., A.A., F.R.N.C.M., A.V.M., L.L., N.C., L.F., N.K., G.W., P.H., C.C., I.S., T.E., J.H., S.K., X.W., H.N.C. and J.C.H.S. The manuscript was prepared by L.R., M.D., C. Betzel and T.S. with discussion and improvements from all authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Doye JPK, Poon WCK. Curr. Opin. Colloid Interface Sci. 2006;11:40–46. [Google Scholar]
- 2.Rohrmann GF. J. Gen. Virol. 1986;67:1499–1513. doi: 10.1099/0022-1317-67-8-1499. [DOI] [PubMed] [Google Scholar]
- 3.Coulibaly F, et al. Nature. 2007;446:97–101. doi: 10.1038/nature05628. [DOI] [PubMed] [Google Scholar]
- 4.Ijiri H, et al. Biomaterials. 2009;30:4297–4308. doi: 10.1016/j.biomaterials.2009.04.046. [DOI] [PubMed] [Google Scholar]
- 5.Fan GY, et al. Microsc. Res. Tech. 1996;34:77–86. doi: 10.1002/(SICI)1097-0029(19960501)34:1<77::AID-JEMT11>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 6.Chapman HN, et al. Nature. 2011;470:73–77. doi: 10.1038/nature09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owen RL, Rudino-Pinera E, Garman EF. Proc. Natl. Acad. Sci. USA. 2006;103:4912–4917. doi: 10.1073/pnas.0600973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman HN, et al. Nat. Phys. 2006;2:839–843. [Google Scholar]
- 9.Mackey ZB, O’Brien TC, Greenbaum DC, Blank RB, McKerrow JH. J. Biol. Chem. 2004;279:48426–48433. doi: 10.1074/jbc.M402470200. [DOI] [PubMed] [Google Scholar]
- 10.Bryant C, et al. Bioorg. Med. Chem. Lett. 2009;19:6218–6221. doi: 10.1016/j.bmcl.2009.08.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitamura M. Int. Rev. Immunol. 2011;30:4–15. doi: 10.3109/08830185.2010.522281. [DOI] [PubMed] [Google Scholar]
- 12.Kirian RA, et al. Opt. Express. 2010;18:5713–5723. doi: 10.1364/OE.18.005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerr ID, et al. PLoS Negl. Trop. Dis. 2010;4:e701. doi: 10.1371/journal.pntd.0000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cusack S, et al. Nat. Struct. Biol. 1998;5(suppl.):634, 637. doi: 10.1038/1325. [DOI] [PubMed] [Google Scholar]
- 15.Mueller M, Jenni S, Ban N. Curr. Opin. Struct. Biol. 2007;17:572–579. doi: 10.1016/j.sbi.2007.09.004. [DOI] [PubMed] [Google Scholar]
References
- 16.Emma R, et al. Nat. Photonics. 2010;4:641–647. [Google Scholar]
- 17.Bozek JD. Eur. Phys. J. Spec. Top. 2009;169:129–132. [Google Scholar]
- 18.Strüder L, et al. Nucl. Instrum. Methods Phys. Res. A. 2010;614:483–496. [Google Scholar]
- 19.DePonte DP, et al. J. Phys. D Appl. Phys. 2008;41 195505. [Google Scholar]
- 20.Duisenberg AJM. J. Appl. Cryst. 1992;25:92–96. [Google Scholar]
- 21.Kabsch W. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 22.Read RJ. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto A, et al. J. Biochem. 2000;127:635–643. doi: 10.1093/oxfordjournals.jbchem.a022651. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, et al. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 27.Collaborative Computational Project, Number 4. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. [Google Scholar]
- 28.DeLano WE. The PyMOL Molecular Graphics System. San Carlos, California: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.