Abstract
Fused in sarcoma (FUS) and TAR DNA-binding protein 43 (TDP-43) are RNA-binding proteins pathogenetically linked to amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD), but it is not known if they regulate the same transcripts. We addressed this question using crosslinking and immunoprecipitation (iCLIP) in mouse brain, which showed that FUS binds along the whole length of the nascent RNA with limited sequence specificity to GGU and related motifs. A saw-tooth binding pattern in long genes demonstrated that FUS remains bound to pre-mRNAs until splicing is completed. Analysis of FUS−/− brain demonstrated a role for FUS in alternative splicing, with increased crosslinking of FUS in introns around the repressed exons. We did not observe a significant overlap in the RNA binding sites or the exons regulated by FUS and TDP-43. Nevertheless, we found that both proteins regulate genes that function in neuronal development.
Fused in sarcoma (FUS, also referred to as Translocated in liposarcoma (TLS)), is a member of the FET family of RNA-binding proteins (RBP) that contain multiple domains with a potential for RNA binding, including an RRM domain, zinc-finger domain, and three RGG boxes1. Cytoplasmic inclusions containing FUS are the pathological hallmark of a subset of patients with amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD)2,3,4,5,6. ALS-associated mutations in FUS gene are most often located in the nuclear localization sequence, which inhibits the import of FUS protein into the nucleus and promotes the formation of cytoplasmic aggregates within affected neurons2,7. In cell culture, FUS was found to co-localize with TAR DNA-binding protein 43 (TDP-43), another RNA processing protein with mutations, and common pathologic inclusions in ALS and FTLD8,9. Biochemical studies confirmed that a fraction of TDP-43 is in complex with FUS10,11 and both proteins are part of the large Drosha complex that is involved in miRNA biogenesis12. However, the two proteins do not co-localize within the pathologic inclusions4, and studies in yeast show that TDP-43 and FUS do not influence the aggregation of each other13. It is therefore unclear if the two proteins cooperate in recognizing their RNA targets, and employ related mechanisms to regulate gene expression in the brain.
Here we performed individual-nucleotide resolution crosslinking and immunoprecipitation (iCLIP) with FUS, TDP-43 and U2 small nuclear RNA auxiliary factor 2 (U2AF65). We found that in contrast to the highly clustered binding of TDP-43 and U2AF65, FUS binding is distributed across the whole length of pre-mRNAs. All three proteins had increased RNA binding towards the 5′ end of introns, indicating that they are recruited to the nascent RNA soon after its transcription. Whereas binding of TDP-43 and U2AF65 is strongly determined by the RNA sequence, FUS has a very limited sequence preference for G-rich sequences. In agreement with the different sequence specificity of FUS and TDP-43, we did not find a significant overlap between their binding sites. Nevertheless, we found that both proteins regulate genes involved in neuronal development.
Results
FUS binds along the whole length of nascent RNA soon after its transcription
In order to compare the RNA binding of FUS and TDP-43, we determined their transcriptome-wide binding maps in E18 mouse brain using iCLIP14 (Supplementary Fig. S1a). As control, we performed iCLIP with a general splicing factor U2 small nuclear RNA auxiliary factor 2 (U2AF65) as well as without any antibody to test for non-specific binding. We identified 3.5, 4.4 and 4.0 million unique cDNAs with FUS, TDP-43 and U2AF65 iCLIP, respectively, but only 6,000 with the no-antibody control (Tables S1, S2). Since we obtained a similar number of iCLIP cDNAs with the three proteins, which all primarily bind to introns (Fig. 1a), we comparatively evaluated the binding of the three proteins in this study. A past study found that FUS binds to non-coding RNAs produced from the region 5′ to the CCND1 gene to repress transcription of this gene15. We detected binding of FUS to antisense RNAs in this region, but similar binding was also seen for TDP-43 or U2AF65 (data not shown). Interestingly, all three proteins had increased binding to antisense RNAs upstream of transcription start sites of protein-coding genes, and the enrichment of FUS was no greater than TDP-43 or U2AF65 (Fig. 1b).
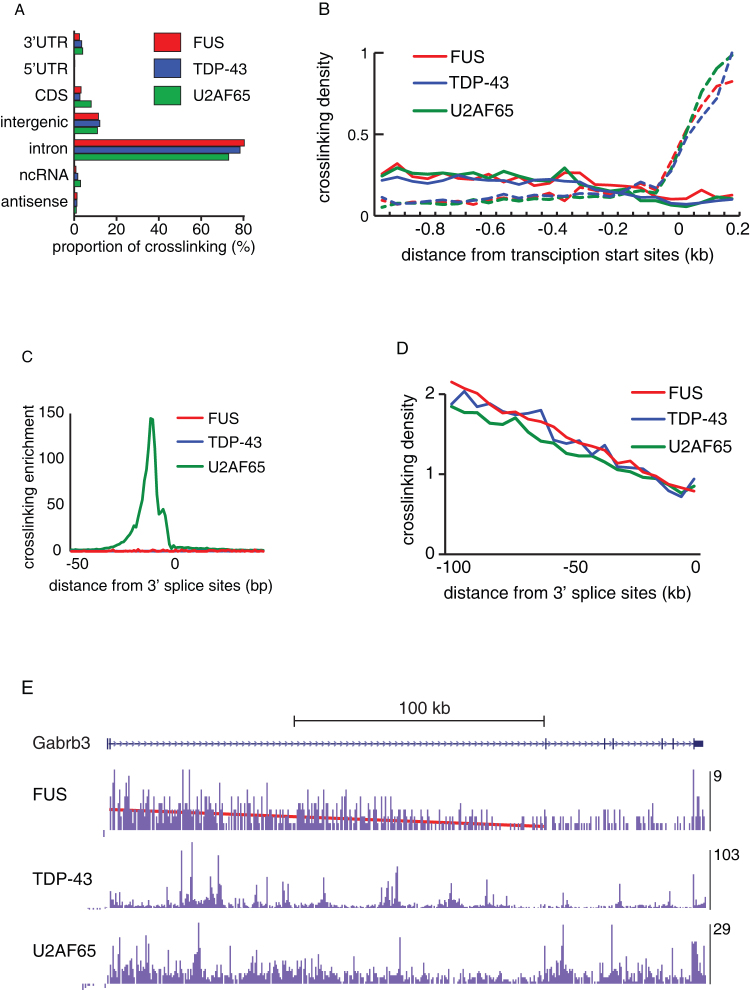
Figure 1. FUS binds along the whole length of nascent RNAs.
(a) The proportion of cDNAs (out of all cDNAs that mapped to the mouse genome) from the FUS, TDP-43 and U2AF65 iCLIP experiments that mapped to different RNA regions. (UTR: untranslated region, ORF: open reading frame, ncRNA: non-coding RNA). (b) The map of FUS, TDP-43 and U2AF65 iCLIP crosslink sites at the 5′ regions of all protein-coding genes annotated by ENSEMBL 59, from 1 kb upstream of transcription start to 200 nts downstream. The data of each protein was first normalised to the average crosslinking on the sense strand within this region, and then to the crosslinking of U2AF65 within the interval 150-200 nts downstream of the transcription start site. Crosslinking distribution was then evaluated in 50 nt intervals. The solid lines show binding to antisense, and the dashed lines of the same colour show binding to the sense RNAs, relative to the orientation of the downstream gene. (c) The map of clustered crosslink sites (flank = 15) of FUS, TDP-43 and U2AF65 iCLIP identified at intron-exon junctions of all exons annotated by ENSEMBL 59. Crosslinking enrichment was determined by comparing to the average crosslinking of the same protein in the intronic region of 300–100 nts upstream of exons. (d) The average gene-normalized iCLIP cDNA density in 5 kb intervals of introns longer than 100 kb, at different distances relative to 3′ splice sites. (e) Gene-normalized cDNA density in 0.5 kb intervals within the Gabrb3 gene. The cDNA density in each interval was normalized to the average cDNA density within the whole gene. The maximum normalized cDNA density is shown at the right of each track.
We applied a peak-finding algorithm to identify clusters of crosslink nucleotides with significant enrichment of crosslink events relative to the local environment14. Using a flank size of 15 nucleotides on either side of crosslink sites identified only 1.7% of intronic FUS crosslink events, in agreement with the previous finding that FUS does not bind narrowly defined RNA sites16. Moreover, we saw no enrichment of FUS or TDP-43 crosslink clusters at 3′ splice sites, which contrasts a 150-fold of enrichment of intronic U2AF65 crosslink clusters (Fig. 1c). We also evaluated binding further from 3′ splice sites, which showed that the average crosslinking density of all three proteins was highest at the 5′ end of long introns, and decreased towards the 3′ end of introns, in a pattern similar to the results obtained by total RNA sequencing from the human brain17 (Fig. 1d). FUS binds along the whole length of pre-mRNAs, therefore this pattern is visible when analysing FUS crosslinking in individual genes (Fig. 1e). Taken together, iCLIP data indicate that the intronic binding of all three proteins corresponds to the abundance of the nascent RNA, which is higher at the 5′ end of long introns17. Thus, binding to promoter-associated antisense RNAs and the increased binding at the 5′ end of long introns demonstrate that FUS, TDP-43 and U2AF65 interact with the nascent RNA soon after its transcription.
FUS binds RNA with limited sequence specificity
A recent study used PAR-CLIP to find that FUS preferentially crosslinks to uridines in the loop region of stem-loop RNA structures16. However, crosslinking methods can introduce RNA sequence and structure biases, therefore it is important to compare data of the multiple proteins18. We compared the probability of single-strandedness around the crosslink sites of FUS, TDP-43 and U2AF65, which showed that all three proteins had an increased probability of single-strandedness directly at the crosslink sites compared to surrounding region (Fig. 2a). Since this increase is common to the three proteins, it indicates a general preference of protein crosslinking to single-stranded RNA. The probability of single-strandedness was greatest for U2AF65 (where it continued around the crosslink sites) and lowest for TDP-43, which most likely reflects the preference of U2AF65 for single-stranded pyrimidine tracts, and the capacity of TDP-43 to bind double-stranded nucleic acids19. In contrast to the past study, we did not observe a significant decrease in single-strandedness around the FUS crosslink sites. Taken together, since the preferential crosslinking of FUS to single-stranded RNA could be of technical nature and we find little evidence for decreased single-strandedness in the surrounding RNA, iCLIP does not conclusively support the preferential binding of FUS to stem-loop RNA structures.
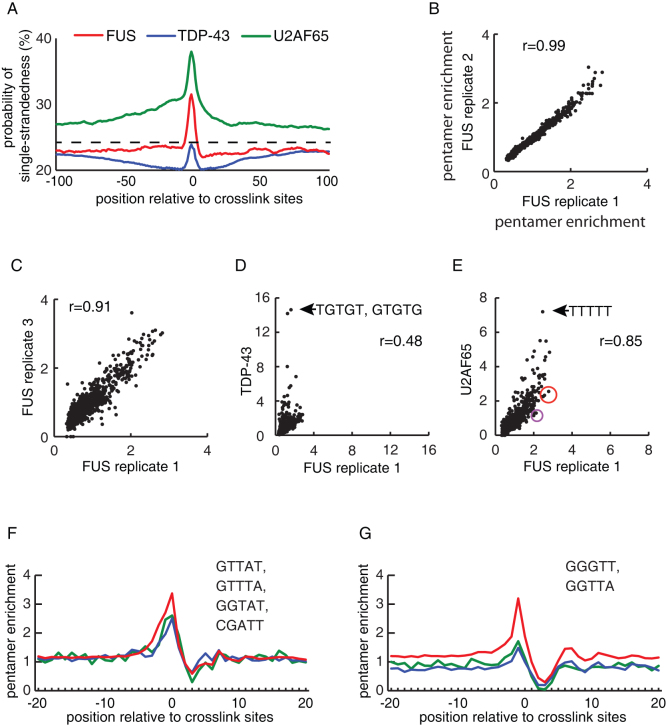
Figure 2. Analysis of RNA sequence and structure specificity of FUS.
(a) Analysis of the probability of single-strandedness at positions −100 to +100 around the FUS, TDP-43 and U2AF65 crosslink sites. We calculated the probability of tetramers being located in single-stranded regions in 70-nucleotide sliding windows using RNAplfold, as described previously50,51. (b–e) Comparison of pentamer enrichment at crosslink sites between replicate FUS experiments, or between FUS, TDP-43 and U2AF65 iCLIP. Pearson correlation coefficient (r) is shown. Pentamers centered at positions −4 to 0 relative to the crosslink site were evaluated and enrichment is determined by comparison to randomised positions in the same genomic regions. The sequences of the most enriched pentamers at TDP-43 (d) and U2AF65 (e) crosslink sites are shown, and two groups of pentamers circled in (e) are evaluated further in panels 2f and 2g. (f) Position of the average enrichment of the four listed pentamers (circled in red in Fig. 2e). The middle position of the pentamers is evaluated relative to the position of cross-link sites, and enrichment is determined by comparison to randomised positions in the same genomic regions. (g) Position of the average enrichment of the two listed pentamers (circled in purple in Fig. 2e). The middle position of the pentamers is evaluated relative to the position of cross-link sites, and enrichment is determined by comparison to randomised positions in the same genomic regions.
To study the sequence-specificity of FUS, we evaluated the occurrence of pentamers overlapping with crosslink sites. We observed a high correlation in pentamer occurrence between the replicate FUS experiments (r>0.91, Pearson correlation coefficient, Fig. 2b, c), a low correlation with TDP-43 (r = 0.48, Fig 2d) and a surprisingly high correlation between FUS and U2AF65 (0.85, Fig. 2e). Uridine-rich pentamers had the higest occurrence at FUS and U2AF65 crosslink sites (Fig. 2e). Whereas, the pentamers consisting purely of pyrimidines were more enriched at U2AF65 crosslink sites, the pentamers containing two (circled in red in Fig. 2e) or three (circled in purple in Fig. 2e) purines were more enriched at FUS crosslink sites. Enrichment of the first pentamer group was seen for all three proteins (Fig. 2f), therefore we cannot exclude the possibility that it represents a sequence bias of crosslinking. However, pentamers containing GG or GGG upstream of the uridines were enriched at FUS, but not TDP-43 and U2AF65 crosslink sites (Fig. 2g). Most of these pentamers contained a GGU motif, indicating that this motif increases the affinity of FUS for RNA. Nevertheless, the 2-fold enrichment of these pentamers in the area overlapping the FUS crosslink sites (Fig. 2e) contrasts the 15-fold enrichment of GU-rich pentamers at the TDP-43 crosslink sites (Fig. 2d), indicating that FUS binds RNA with limited sequence specificity.
Since we did not find any overlap between sequence specificity of FUS and TDP-43, we instead evaluated if the two proteins overlapped in binding to longer RNA regions. For this purpose, we identified 0.5 kb regions within genes longer than 50 kb that had at least 3-fold enriched normalized crosslink density compared to the rest of the gene. We then compared the overlap between these regions of the three proteins. This showed that regions with enriched FUS binding overlapped with a higher proportion of U2AF65 than TDP-43 binding regions (Supplementary Fig. S1b). Taken together, our analyses of iCLIP data indicate that FUS and TDP-43 generally bind to distinct sites on pre-mRNAs.
FUS binds to long RNA regions around the repressed exons
FUS is essential for the viability of inbred mice, as FUS null mice (FUS−/−) show marked chromosomal instability and immune defects and die within 16 hours of birth20. Outbred FUS−/− mice show male sterility and enhanced sensitivity to radiation21. We isolated RNA from the brain tissue of day 18 embryo FUS−/− (KO, n = 3) and wildtype (WT, n = 3) littermates from an inbred strain20. We analyzed alternative splicing using Affymetrix high-resolution splice-junction microarrays. Since past studies have suggested that FUS plays a role in transcriptional regulation22, we first analysed transcript-level changes using ASPIRE 3 software14. Microarray probes detected signal of 25,539 genes, but only ten of these (including the FUS gene) showed at least a two-fold change in transcript levels in the FUS−/− brains that was significant (p-value<0.05, Student's t test, one-tailed, unequal variance, Table S3). Due to the limited extent of transcript-level changes, we proceeded to evaluate changes in alternative splicing.
We identified splicing changes in 68 alternative cassette exons, and 48 other types of splicing changes when using ASPIRE software with a threshold of |ΔIrank|≥1. Notably, 69% (47/68) of the changed cassette exons increased their inclusion in the FUS−/− brain. Moreover, when using a higher threshold (|ΔIrank|≥1.5), 77% (6/26) of the cassette exons increased their inclusion. This indicates that FUS primarily represses exon inclusion in WT brain. To validate the splicing changes in FUS−/− brain, we assessed 21 of the cassette exons using RT-PCR and capillary electrophoresis. The expected alternative isoforms were detected for 17 of these exons, and 14 of these showed a significant change that agreed with change detected by microarray, resulting in an 82% (14/17) validation rate (Supplementary Fig. S2 and Table S4). In addition, we validated splicing changes in six variable-length exons, a retained intron and a terminal exon within the Ewing sarcoma breakpoint region 1 (Ewsr1) gene, which encodes a FET-family protein homologous to FUS (Supplementary Fig. S2 and Table S4).
To explore the possibility that some splicing changes were an indirect effect of proximity to post-natal death, we compared our data with changes in the brain of Nova1/Nova2−/− mice, which also die within 16 hours of birth23. Nova1 and Nova2 are neuron-specific proteins containing three KH-family RNA-binding domains, which regulate brain-specific splicing of genes with synapse-related functions24. RNA from E18 Nova1/Nova2−/− mouse brains was analysed using the same type of splice junction microarray as our study25, and we detected 304 splicing changes using ASPIRE 3 analysis (|ΔIrank|≥1). Of these, only five changes were also present in the FUS−/− mice, and two of these changes were in the opposite direction (data not shown). This indicates that the splicing changes in these mouse models most likely reflect a specific role of these proteins in splicing regulation, rather than non-specific ante-mortem changes.
To study if the exons regulated by FUS have an increased incidence of FUS crosslinking, we first identified FUS crosslink clusters using the flank side of 200 nucleotides and FDR < 5% on the combined replicate experiments, which identified 49879 FUS crosslink clusters. We also identified TDP-43 and U2AF65 crosslink clusters in the same way, to compare their distribution around the exons regulated by FUS. Five out of the 20 exons with lower ( , dIrank<−1.5) inclusion in WT than FUS−/− brain overlapped with a FUS crosslink cluster, which was a significant increase compared to control exons (p-value = 0.0003, Fisher's exact test, Table 1, Figs. 3, 4). In contrast, TDP-43 crosslink clusters were most often positioned far from the exons regulated by FUS, and were not enriched at the repressed exons (Fig. 4, Table 1). Interestingly, the number of FUS and U2AF65 crosslink sites around the repressed exons was highly correlated (r = 0.94, Pearson correlation coefficient): all of the FUS-regulated cassette exons that contained a proximal FUS crosslink cluster also contained a proximal U2AF65 crosslink cluster (Table 2), whereas 67% (26/39) of the remaining repressed exons lacked a U2AF65 crosslink cluster. However, U2AF65 crosslink clusters were most often positioned at the 3′ splice site, whereas the FUS crosslink clusters covered several hundred nucleotide long regions (Figs. 3, 4). The correlation between the crosslink density of U2AF65 and FUS, in spite of their binding to different RNA regions, might result from the variable abundance of the intronic RNA that flanks the repressed exons: introns that are slowly spliced are available for protein binding for longer, resulting into increased crosslink density of proteins.
, dIrank<−1.5) inclusion in WT than FUS−/− brain overlapped with a FUS crosslink cluster, which was a significant increase compared to control exons (p-value = 0.0003, Fisher's exact test, Table 1, Figs. 3, 4). In contrast, TDP-43 crosslink clusters were most often positioned far from the exons regulated by FUS, and were not enriched at the repressed exons (Fig. 4, Table 1). Interestingly, the number of FUS and U2AF65 crosslink sites around the repressed exons was highly correlated (r = 0.94, Pearson correlation coefficient): all of the FUS-regulated cassette exons that contained a proximal FUS crosslink cluster also contained a proximal U2AF65 crosslink cluster (Table 2), whereas 67% (26/39) of the remaining repressed exons lacked a U2AF65 crosslink cluster. However, U2AF65 crosslink clusters were most often positioned at the 3′ splice site, whereas the FUS crosslink clusters covered several hundred nucleotide long regions (Figs. 3, 4). The correlation between the crosslink density of U2AF65 and FUS, in spite of their binding to different RNA regions, might result from the variable abundance of the intronic RNA that flanks the repressed exons: introns that are slowly spliced are available for protein binding for longer, resulting into increased crosslink density of proteins.
Table 1. The number of FUS-regulated exons with proximal crosslink clusters of FUS, TDP-43 or U2AF65. The exons with lower ( , dIrank<−1.5) or higher (
, dIrank<−1.5) or higher ( , dIrank>1.5) inclusion in WT than FUS−/− brain are evaluated, and compared to control exons (all alternative exons detected by microarray). The last column shows the total number of these exons, and the first three columns show the number of these exons that contain FUS, TDP-43 or U2AF65 crosslink cluster within 250nt from the middle of the exon. Crosslink clusters were determined using flank = 200 and FDR<0.05. The star indicates that the proportion of exons repressed by FUS with a FUS crosslink cluster (5/20) is significantly increased compared to control exons with a FUS crosslink cluster (177/5779) (p-value = 0.0003, Fisher's exact test).
, dIrank>1.5) inclusion in WT than FUS−/− brain are evaluated, and compared to control exons (all alternative exons detected by microarray). The last column shows the total number of these exons, and the first three columns show the number of these exons that contain FUS, TDP-43 or U2AF65 crosslink cluster within 250nt from the middle of the exon. Crosslink clusters were determined using flank = 200 and FDR<0.05. The star indicates that the proportion of exons repressed by FUS with a FUS crosslink cluster (5/20) is significantly increased compared to control exons with a FUS crosslink cluster (177/5779) (p-value = 0.0003, Fisher's exact test).
| FUS | TDP-43 | U2 | total | |
|---|---|---|---|---|
 |
5* | 2 | 8 | 20 |
 |
0 | 1 | 2 | 6 |
| control | 177 | 273 | 1098 | 5779 |
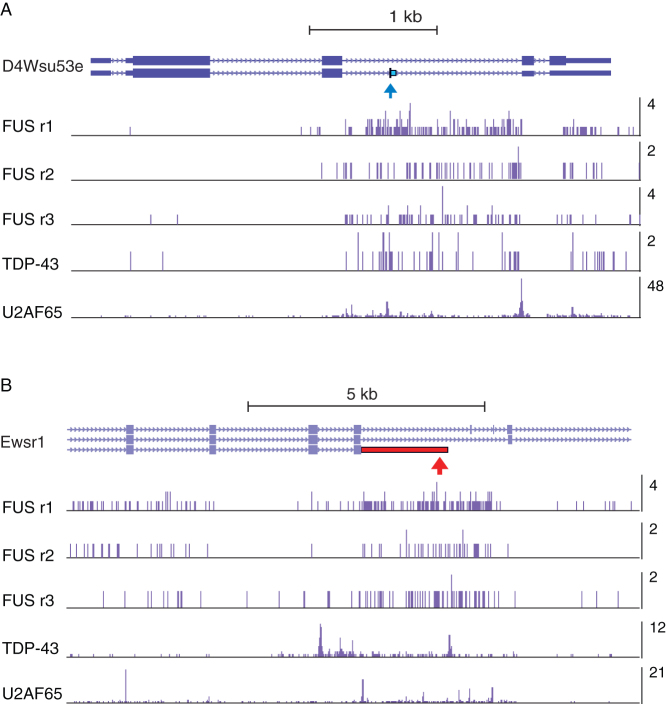
Figure 3. FUS, TDP-43 and U2AF65 crosslinking around two exons regulated by FUS.
(a) Crosslinking density within the D4Wsu53e gene. The exon repressed by FUS is marked by a blue arrow. The bar graph shows the cDNA count at individual crosslink sites in replicate FUS iCLIP experiments, and the grouped TDP-43 and U2AF65 iCLIP experiments. (b) Crosslinking density within the Ewsr1 gene. The exon enhanced by FUS is marked by a red arrow. The bar graph shows the cDNA count at individual crosslink sites in replicate FUS iCLIP experiments, and the grouped TDP-43 and U2AF65 iCLIP experiments.
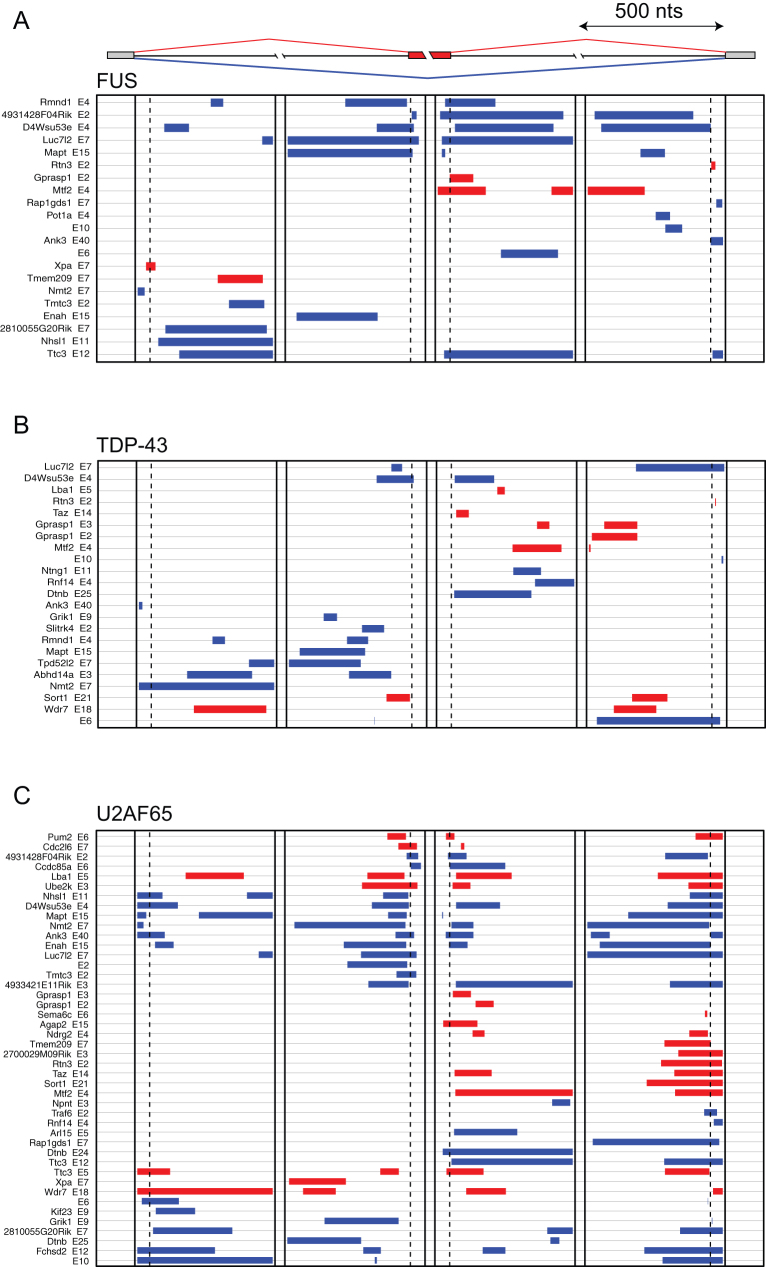
Figure 4. Analysis of iCLIP crosslink clusters around the exons regulated by FUS.
The map of crosslink clusters at positions within 500 nt of alternative exons and flanking exons. Crosslink clusters were determined using flank = 200 and FDR<0.05. The exons are grouped by sequential analysis of crosslink cluster positions in three regions: group 1 is identified by clusters within the exon; group 2 by clusters downstream of the exon; and group 3 by clusters upstream of the exon. The cassette exons with ΔIrank > 1 (enhanced exons, red clusters) or ΔIrank < −1 (repressed exons, blue clusters) in analysis of FUS−/− brain RNA that contain at least one crosslink cluster in these regions are shown. The positions of FUS (a), TDP-43 (b) and U2AF65 (c) crosslink clusters are shown, and the gene symbol and exon number (according to genomic annotation) is shown on the left. Only the FUS-regulated exons that contain crosslink clusters for the corresponding RBP in the evaluated regions are shown.
Table 2. The list of FUS-regulated exons with proximal crosslink clusters of FUS. The genomic positions of the exons are shown, as well as the number of FUS, TDP-43 or U2AF65 crosslink sites in crosslink clusters located within 250nt from the middle of the exon. Crosslink clusters were determined using flank = 200 and FDR<0.05.
| cross-linking sites | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| chr | start | end | strand | Gene | exon type | effect | FUS | TDP-43 | U2AF65 |
| chr4 | 134481443 | 134481943 | + | D4Wsu53e | Cassette |  |
89 | 13 | 98 |
| chr16 | 94614470 | 94614970 | + | Ttc3 | Cassette |  |
24 | 0 | 19 |
| chr6 | 38515406 | 38515906 | + | Luc7l2 | Cassette |  |
19 | 2 | 22 |
| chr10 | 5917827 | 5918327 | + | Rmnd1 | Cassette |  |
17 | 2 | 3 |
| chr8 | 107812815 | 107813315 | − | 4931428F04Rik | Cassette |  |
9 | 0 | 9 |
| chr11 | 104179222 | 104179722 | + | Mapt | Cassette |  |
7 | 2 | 9 |
| chr5 | 108497932 | 108498432 | + | Mtf2 | Cassette |  |
5 | 0 | 7 |
| chr1 | 183841540 | 183842040 | − | Enah | Cassette |  |
4 | 0 | 21 |
| chrX | 132277732 | 132278232 | + | Gprasp1 | Cassette |  |
2 | 0 | 7 |
| chr15 | 3941706 | 3942206 | − | AW549877 | Bleeding |  |
9 | 5 | 18 |
| chrX | 132376016 | 132376516 | + | Gprasp1 | Bleeding |  |
3 | 0 | 0 |
| chr11 | 4979238 | 4979738 | − | Ewsr1 | Intron |  |
60 | 3 | 31 |
| chr1 | 58477677 | 58478177 | − | Clk1 | Intron |  |
9 | 0 | 40 |
| chr9 | 103258106 | 103258606 | − | Cdv3 | 3′ variable |  |
3 | 0 | 3 |
| chr1 | 164603984 | 164604484 | − | Bat2d | 5′ variable |  |
8 | 0 | 12 |
| chr12 | 71367229 | 71367729 | − | Trim9 | Terminal |  |
1 | 0 | 8 |
| chr18 | 38456210 | 38456710 | + | Rnf14 | Promoter |  |
2 | 0 | 0 |
FUS regulates genes involved in neuronal development
To understand the biological relevance of splicing regulation by FUS, we performed GO term analysis of genes containing FUS-regulated exons, and compared these with all alternative exons that were detected by the microarray. This identified a significant enrichment of genes associated with positive regulation of cell adhesion, negative regulation of apoptosis, neuronal development and axonogenesis (Table S5). Since the GO annotation is incomplete, we referred to the relevant publications to confirm well-defined neuronal functions of 19 proteins encoded by the FUS-regulated transcripts (Table 3). 10 of these regulate axonogenesis, neurite outgrowth or axon guidance, and 10 are associated with neurologic disorders. We don't find a significant overlap between exons regulated by FUS identified in this study and those regulated by TDP-43 in human neuroblastoma cells26,27. Nevertheless, we find that both proteins primarily regulate genes involved in neuronal development.
Table 3. Neurologic disease and neuronal functions of proteins encoded by the alternative mRNA isoforms regulated by FUS.
| Gene symbol | Gene name | Neurologic disease* | Neuronal function | Pubmed PMID |
|---|---|---|---|---|
| Ablim1 | Actin binding LIM protein 1 | Axon guidance | 11163266 | |
| Adnp | Activity-dependent neuroprotector homeobox | Neurogenesis | 17363064 | |
| 17222401 | ||||
| Ank3 | Ankyrin 3 node of Ranvier (ankyrin G), | BPD | Membrane-cytoskeleton linker | 18711365 |
| Apc | Adenomatous polyposis col | Axonogenesis, neuronal differentiation | 16185824, | |
| 21915313 | ||||
| Cdh2 | Cadherin 2, type 1, N-cadherin (neuronal) | Synapse morphology, axon outgrowth | 21452216 | |
| 22019151 | ||||
| Enah | Enabled homolg | Axon guidance | 15371503 | |
| Ewsr1 | Ewing sarcoma breakpoint region 1 | FTD | RNA regulation, transcription | 21856723 |
| Grik1 | glutamate receptor, ionotropic, kainate 1 | EPI, BPD,SCH | Signalling | 19941835 |
| 17698324 | ||||
| Mapt | Microtubule associated protein tau | FTD, AD, CBD, PSP, PD, PiD | Microtubule stability | 18688091 |
| Ndrg2 | NDRG family member 2 | FTD, AD | Neurite outgrowth | 20886841 |
| 15207261 | ||||
| Ndrg2 | NDRG family member 2 | FTD, AD | Neurite outgrowth | 20886841 |
| 16033798 | ||||
| Ntng1 | Netrin G1 | RS | Axon guidance | 14595443 |
| 16502428 | ||||
| Picalm | Phosphatidylinositol binding clathrin assembly protein | AD | Endocytosis, dendritic growth | 20080314 |
| Rasgrf1 | Ras protein-specific guanine nucleotide-releasing factor 1 | Synaptic transmission, long-term memory | 9384379 | |
| Sema6c | Semaphorin 6c | Axonogenesis | 12110693 | |
| Slitrk4 | SLIT and NTRK-like family, member 4 | Suppression of neurite outgrowth | 14550773 | |
| Sort1 | Sortilin 1 | FTD | Binds neuropeptides | 21092851 |
| 21087763 | ||||
| Tsc1 | Tuberous sclerosis 1 | TSC | Axonogenesis | 18794342 |
| Ttc3 | Tetratricopeptide repeat domain 3 | DS | Neuronal development | 20059950 |
| 17488780 |
*AD = Alzheimer's disease; BPD = bipolar disorder; CBD = Cortico-basal degeneration; DS = Down syndrome; EPI = epilepsy; FTD = frontotemporal dementia; PiD = Pick disease; PD = Parkinson's disease; PSP = progressive supranuclear palsy; RS = Rett syndrome; SCH = schizophrenia; TSC = Tuberous sclerosis complex.
Discussion
In this study we aimed to understand how FUS interacts with nascent RNA to regulate gene expression in the brain. We studied the RNA binding of FUS using iCLIP and determined its role in regulating alternative splicing in the mouse brain, thereby providing insights both into mechanisms of FUS-dependent splicing regulation, as well as its function in neuronal biology. FUS crosslinking is enriched at GGU motifs, but this enrichment is only two-fold, which might explain why FUS does not bind clearly defined RNA sites; instead, its binding is enriched over long intronic regions. Analysis of FUS−/− brain identified a role of FUS in regulating splicing of specific alternative exons, and showed that the repressed exons were often flanked by long FUS crosslink clusters. Interestingly, the intensity of FUS crosslinking around the regulated exons was closely correlated with the intensity of U2AF65 crosslinking at the 3′ splice site.
Our data does not indicate a strong preference of FUS for stem-loop RNA structures that were observed in a past study16. We found that uridine-rich pentamers were enriched at crosslink sites of FUS, TDP-43 and U2AF65, but we recently demonstrated that UV-C crosslinking has a slight preference for uridines18, therefore the uridine enrichment at FUS crosslink sites could partly reflect a sequence bias of UV-C crosslinking. However, pentamers containing a GGU motif were specifically enriched at the FUS crosslink sites, indicating that GGU motif confers specificity to RNA recognition by FUS. This agrees with in vitro selection and NMR studies, which showed that the zinc finger-like domain in FUS recognises GGUG motif28,29. Moreover, GGT motif is also the critical part of sequences that were recently demonstrated to be important for interaction between FUS and single-stranded DNA30, therefore it appears likely that FUS recognises similar sequence motifs in RNA and single-stranded DNA.
The sequence specificity of FUS is very limited compared to U2AF65 and TDP-43, therefore FUS has a more widespread crosslinking in pre-mRNAs. This results in a saw-tooth pattern of crosslinking density in long genes that reflects the increased abundance of nascent RNA towards the 5′ end of long introns17. Interestingly, when summarizing iCLIP data over multiple introns, the saw-tooth binding pattern in long genes becomes apparent also in U2AF65 and TDP-43 iCLIP. U2AF65 is a splicing factor that recruits U2 snRNP to the nascent transcripts, therefore its increased binding to long introns could lead to recognition of cryptic splicing elements. Active repression of cryptic splicing elements might therefore be particularly important for long genes. Therefore, long genes could be more sensitive to perturbations in RBPs that affect binding of U2AF65 to pre-mRNA; this could potentially explain the greater sensitivity of long genes to TDP-43 knockdown31.
We did not identify an increased association between FUS and promoter-associated RNAs compared to TDP-43 and U2AF65. However, this does not exclude the possibility that FUS plays a specific role by linking promoter-associated RNAs to chromatin-associated proteins, such as the CREB-binding protein (CBP)15. Recently, it was shown that incompletely spliced transcripts are retained on the chromatin until fully spliced32. Moreover, it was shown that knockdown of FUS leads to accumulation of incompletely spliced transcripts33. We find that introns flanking the exons repressed by FUS have an increased crosslink density of FUS (and partly also U2AF65), indicating that these introns have a slow splicing kinetics and are therefore bound to FUS for a longer time than the rest of the pre-mRNA. Taken together, our data are consistent with a model where FUS regulates alternative splicing by binding introns that are slowly spliced, possibly by maintaining chromatin retention of these introns.
We find that FUS and TDP-43 generally bind distinct RNA sites and regulate distinct alternative exons. This agrees with their differential mRNA binding that was recently documented in motoneuron-like NSC-34 cells34. Nevertheless, both proteins regulate splicing of genes enriched in functions related to neuronal development and neurodegenerative diseases26. For instance, FUS regulates two exons in enabled homolog (ENAH), an actin regulatory protein involved in cell motility, axon guidance, neural tube closure, and cell-cell adhesion35,36,37,38,39. Both exons are also associated with metastatic progression40,41. FUS also promotes inclusion of exon 3 in activity-dependent neuroprotective protein (ADNP) that contains the start codon. The protein-coding transcript is the main isoform in the wild-type brain, but represents only 23% of the transcripts made in the FUS−/− brain. ADNP−/− mice die at embryonic day 942, and a neuroprotective peptide NAP (Davunetide), derived from ADNP, is being used in pilot and clinical trials for treatment of neurodegenerative disorders43. FUS promotes inclusion of exon 21 of Sortilin 1 (Sort1), which is, interestingly, silenced by TDP-4331,44. Sort1 is a type 1 receptor, which has been functionally associated with progranulin and FTLD, and exon 21 was reported to regulate trafficking of Sort145. In agreement with the FUS knockdown study in hippocampal neurons46, we also identified a modest splicing change in exon 10 of microtubule associated protein Tau (Mapt), which codes for the second microtubule binding domain47. Finally, we found that FUS promotes the use of an alternative terminal exon in EWSR1, indicating cross-regulation among the FET protein family members. In conclusion, our study places FUS alongside TDP-43 in regulating splicing of transcripts associated with neuronal development and neurodegenerative diseases.
Methods
iCLIP
We used the iCLIP method as described previously48, with the following modifications. Following dissociation and UV-crosslinking of embryonic day 18 (E18) mouse brain, iCLIP immunoprecipitation was performed with protein A Dynabeads (Invitrogen) conjugated to rabbit anti-FUS (Novus Biologicals, NB100-565) or rabbit anti-TDP-43 (Proteintech, 10782-2-AP) or without conjugation of any antibody, or protein G Dynabeads (Invitrogen) conjugated to mouse anti-U2AF65 (U4758, Sigma). The size of the protein-RNA complexes present in the high RNase condition corresponded to a single protein molecule bound to the RNA, and a shift upwards on the gel was seen under low RNase conditions, corresponding to the proteins that were crosslinked to longer RNA molecules (Supplementary Fig. S1a). The region corresponding to 80–130 kDa, 50–100 kDa and 70–120 kDa complexes was excised from the membrane to isolate the RNA bound to FUS, TDP-43 and U2AF65, respectively.
High-throughput sequencing was done either using 50 cycles on Illumina GAII or 70 cycles on Illumina MiSeq and the barcode sequences corresponding to the individual experiment were as described (Table S1). The randomers were registered and the barcodes were removed before mapping the sequences to the genome sequence (mm9/NCBI37) allowing two mismatches using Bowtie version 0.12.7 (command line: -v 2 -m 1 -a --best --strata). The nucleotide preceding the iCLIP cDNAs mapped by Bowtie was used to define the crosslink sites identified by truncated cDNAs. The method for the randomer evaluation, annotation of genomic segments and identification of significantly clustered crosslinking events was performed with FDR<0.05 and a maximum spacing of 15 or 200 nt (as indicated in the text), as described earlier14,49, except that the Ensembl 59 gene annotation was used and that the positions of crosslink sites were randomised within the whole gene, rather than within individual introns. Unless information on replicates is specifically shown, the replicate iCLIP experiments for the same protein were grouped before performing the analyses.
Analysis, normalisation, and identification of regions with enriched crosslinking density
To identify regions with increased crosslink density within pre-mRNAs, we studied genes longer than 50 kb that were highly transcribed in the brain. These genes were selected based on five-fold higher density of iCLIP cDNAs compared to the average iCLIP cDNA density in all ENSEMBL genes. The genes were fragmented into 0.5 kb regions. To normalise the saw-tooth effect of intronic read density (see Fig. 1e), we first analysed the read density in total RNAseq data from fetal human brain15 in all introns longer than 100 kb. The read density in these introns increased by a factor of 0.0077 per 1 kb distance from the 3′ splice site. Thus, the read density increases by a factor of (1 + 0.00385 * intron_1len,kb) as the distance from the 3′ end of intron increases. The read density in genes after normalising the saw-tooth effect is therefore:
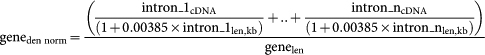 |
geneden norm: cDNA density in each gene, normalized by the relative increase in cDNA density per intron increases by a factor of (1 + 0.00385 * intron_1len,kb)
genelen: gene length
intron_1: the first intron in the gene, including also the preceding exon
intron_n: the last intron in the gene, including also the preceding exon
intron_1len, kb: length of the intron_1 in kilobases
intron_1cDNA: sum of cDNA counts within the intron_1
For Fig. 1d and 1e, we used the gene-normalized read density to determine the relative read density at each intronic region:
regionden, gene norm: cDNA density in each region, normalized by the cDNA density of the whole gene
regioncDNA: sum of cDNA counts within a region
regionlen: region length
To study crosslinking density independently of the saw-tooth effect, we performed regional normalization of iCLIP cDNA density using the following formula:
regionden, region norm: cDNA density in each region, normalized by cDNA density of the whole gene, as well by the increase in intronic cDNA density at a distance from the 3′ splice site
regiondist, kb: distance of the 3′ end of region from the 3′ splice site in kilobases
We used the regionally normalized data in genes longer than 50 kb to identify 0.5 kb regions of enriched crosslink density. These were identified by finding the 0.5 kb intervals where the regionally normalized crosslinking density was higher by at least a factor of three compared to the average crosslinking density in the whole gene.
RNA sequence and structure analysis
We first identified a set of pentamers that were most enriched in the region of −30 to +30 nucleotides around the crosslink sites in the iCLIP data of a specific RBP compared to the other two RBPs. In this way we avoided identifying motifs that would result from sequence biases associated with UV-C crosslinking or other aspects of iCLIP method. We then evaluated the positional enrichment of these sets of pentamers around the crosslink sites. This was determined by comparing the occurrence at each pentamer around the true crosslink site with the occurrence at randomized crosslink positions. The positions of crosslink sites were randomised within the same regions of the gene (i.e., within the same intron, CDS or UTR). To determine the single-strandedness probability around crosslink sites, we calculated unpaired probability of all tetramers in the regions using RNAplfold program with default parameters, as described previously50,51.
Splice-junction microarray
RNA was isolated from E18 whole brain of three FUS−/− and three wild-type littermates and the cDNA samples were prepared using the GeneChip WT cDNA Synthesis and Amplification Kit (Affymetrix 900673), followed by GeneChip Hybridization, Wash, and Stain Kit (Affymetrix 900720) using Affymetrix GeneChip Fluidics Station 450. Mouse high-resolution AltSplice splice-junction microarrays (Affymetrix) were then scanned on Affymetrix GeneChip Scanner 3000 7G. Data was analyzed with version 3 of ASPIRE (Analysis of SPlicing Isoform Reciprocity), which determined ΔIrank value, which was used to rank the splicing changes based on their significance14. Changes with |ΔIrank|≥1 were further validated, and were used for iCLIP binding and GO term analysis. GO term analysis was performed as described previously26.
RT-PCR
Total RNA was extracted using the miRNeasy Kit (Qiagen) and 200 ng of total RNA was used for reverse transcription using Superscript III (Invitrogen) according to the manufacturer's instructions. For analysis of splicing, PCR was performed using Immomix (Bioline) using primers listed in Table S3. The PCR products were visualized using QIAxcel capillary electrophoresis system (Qiagen). To calculate exon inclusion/exclusion, the value of each peak was first divided by the size of the amplicon (nucleotide number) it represented to even out the changes brought about by stronger staining of longer amplicons. The percentage of the peak representing exon inclusion was obtained from the ratio of size modified values for the exon inclusion peak divided by the sum of size modified values for peaks representing exon inclusion and skipping. Splicing change was calculated by subtracting the exon inclusion in the knockdown cells from the inclusion in wild-type cells (thus, a positive ΔI represents exons enhanced by TDP-43, and negative those silenced by TDP-43).
Gene ontology (GO) analysis
GO term enrichment analysis was performed on exons with |ΔIrank|≥1 in annotated genes (108 exons), and all exons in annotated genes that had sufficient signal on the microarray were used as controls (15975). To make GO annotation exon-centric, we multiplied each gene annotation record to include all exons on the splicing microarray for the gene, and the p-value was calculated as described previously52.
Author Contributions
B.R., L.E.E., M.M. and J.T. carried out iCLIP, microarray and PCR experiments, R.F. prepared the mouse brain samples, G.R., Y.S., N.H. and G.K.B. performed the computational analyses, B.R., L.E.E. and J.U. prepared the manuscript and L.W.T., C.E.S., T.C., T.T., B.Z., L.W.S. and J.U. supervised the project.
Supplementary Material
Supplementary figures and tables
Acknowledgments
We thank Melis Kayikci for analysing the microarray data, and Chris Sibley, Kathi Zarnack and Julian König for advice and comments on the manuscript. This work was supported by the European Research Council (206726-CLIP), the Medical Research Council [grant number U105185858], Slovenian Research Agency (P2-0209, J2-2197, L2-1112, Z7-3665, J3-4026), Wellcome Trust and MRC Strategic Grant Award (089701/Z/09/Z), Motor Neuron Disease Association, Heaton-Ellis Trust, Psychiatry Research Trust, Grant-in-Aid for Scientific Research from MEXT and CREST, JST and the Nakajima Foundation scholarship to YS and Otto Likar scholarship to MM.
Data access: The mouse brain FUS, TDP-43 and U2AF65 iCLIP data are available from arrayexpress with accession number: E-MTAB-1223 and the FUS WT and KO mouse brain AltSplice microarray data are available from arrayexpress with accession number: E-MTAB-1229.
References
- Nguyen C. D. et al. Characterization of a family of RanBP2-type zinc fingers that can recognize single-stranded RNA. J Mol Biol 407, 273–283 (2011). [DOI] [PubMed] [Google Scholar]
- Vance C. et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie I. R., Rademakers R. & Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol 9, 995–1007 (2010). [DOI] [PubMed] [Google Scholar]
- Urwin H. et al. FUS pathology defines the majority of tau- and TDP-43-negative frontotemporal lobar degeneration. Acta Neuropathol 120, 33–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelaar H. et al. Frequency of ubiquitin and FUS-positive, TDP-43-negative frontotemporal lobar degeneration. J Neurol 257, 747–753 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M. et al. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 132, 2922–2931 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D. et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. Embo J 29, 2841–2857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J. et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M. et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006). [DOI] [PubMed] [Google Scholar]
- Ling S. C. et al. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci U S A 107, 13318–13323 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Shanware N. P., Bowler M. J. & Tibbetts R. S. Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J Biol Chem 285, 34097–34105 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory R. I. et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 (2004). [DOI] [PubMed] [Google Scholar]
- Sun Z. et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol 9, e1000614 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J. et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17, 909–915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454, 126–130 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoell J. I. et al. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol 18, 1428–1431 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameur A. et al. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol 18, 1435–1440 (2011). [DOI] [PubMed] [Google Scholar]
- Sugimoto Y. et al. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biology Aug 3; 13(8), R67–.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo P. H., Doudeva L. G., Wang Y. T., Shen C. K. & Yuan H. S. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res 37, 1799–1808 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks G. G. et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet 24, 175–179 (2000). [DOI] [PubMed] [Google Scholar]
- Kuroda M. et al. Male sterility and enhanced radiation sensitivity in TLS(-/-) mice. Embo J 19, 453–462 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law W. J., Cann K. L. & Hicks G. G. TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Brief Funct Genomic Proteomic 5, 8–14 (2006). [DOI] [PubMed] [Google Scholar]
- Ruggiu M. et al. Rescuing Z+ agrin splicing in Nova null mice restores synapse formation and unmasks a physiologic defect in motor neuron firing. Proc Natl Acad Sci U S A 106, 3513–3518 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J. et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet 37, 844–852 (2005). [DOI] [PubMed] [Google Scholar]
- Licatalosi D. D. et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey J. R. et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14, 452–458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten J. T. & Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet 27, 89–97 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerga A. et al. Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol Chem 276, 6807–6816 (2001). [DOI] [PubMed] [Google Scholar]
- Iko Y. et al. Domain architectures and characterization of an RNA-binding protein, TLS. J Biol Chem 279, 44834–44840 (2004). [DOI] [PubMed] [Google Scholar]
- Tan A. Y., Riley T. R., Coady T., Bussemaker H. J. & Manley J. L. TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc Natl Acad Sci U S A 109, 6030–6035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M. et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14, 459–468 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, Dev M. et al. Transcript Dynamics of Proinflammatory Genes Revealed by Sequence Analysis of Subcellular RNA Fractions. Cell 150, 279–290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichmann D. S. & Harland R. M. fus/TLS orchestrates splicing of developmental regulators during gastrulation. Genes Dev 26, 1351–1363 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C. et al. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J Biol Chem 287, 15635–15647 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. A. et al. Ena/VASP proteins can regulate distinct modes of actin organization at cadherin-adhesive contacts. Mol Biol Cell 17, 1085–1095 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V., Bauer C., Yin M. & Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell 100, 209–219 (2000). [DOI] [PubMed] [Google Scholar]
- Lanier L. M. et al. Mena is required for neurulation and commissure formation. Neuron 22, 313–325 (1999). [DOI] [PubMed] [Google Scholar]
- Gertler F. B., Niebuhr K., Reinhard M., Wehland J. & Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell 87, 227–239 (1996). [DOI] [PubMed] [Google Scholar]
- Gertler F. & Condeelis J. Metastasis: tumor cells becoming MENAcing. Trends Cell Biol 21, 81–90 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski A. V. et al. Ena/VASP Is Required for neuritogenesis in the developing cortex. Neuron 56, 441–455 (2007). [DOI] [PubMed] [Google Scholar]
- Di Modugno F. et al. Molecular cloning of hMena (ENAH) and its splice variant hMena+11a: epidermal growth factor increases their expression and stimulates hMena+11a phosphorylation in breast cancer cell lines. Cancer Res 67, 2657–2665 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhasov A. et al. Activity-dependent neuroprotective protein: a novel gene essential for brain formation. Brain Res Dev Brain Res 144, 83–90 (2003). [DOI] [PubMed] [Google Scholar]
- Gozes I. NAP (davunetide) provides functional and structural neuroprotection. Curr Pharm Des 17, 1040–1044 (2011). [DOI] [PubMed] [Google Scholar]
- Hu F. et al. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron 68, 654–667 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermey G. et al. Characterization of sorCS1, an alternatively spliced receptor with completely different cytoplasmic domains that mediate different trafficking in cells. J Biol Chem 278, 7390–7396 (2003). [DOI] [PubMed] [Google Scholar]
- Orozco D. et al. Loss of fused in sarcoma (FUS) promotes pathological Tau splicing. EMBO reports (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo J. M., Noble W. & Martin T. R. RNA and protein-dependent mechanisms in tauopathies: consequences for therapeutic strategies. Cell Mol Life Sci 64, 1701–1714 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J. et al. iCLIP--transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J Vis Exp (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol 8, e1000530 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. et al. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science 329, 439–443 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart S. H., Hofacker I. L. & Stadler P. F. Local RNA base pairing probabilities in large sequences. Bioinformatics 22, 614–615 (2006). [DOI] [PubMed] [Google Scholar]
- Tollervey J. R. et al. Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res 21, 1572–1582 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables