Abstract
Summary: The TAR RNA binding protein (TRBP) has emerged as a key player in many cellular processes. First identified as a cellular protein that facilitates the replication of human immunodeficiency virus, TRBP has since been shown to inhibit the activation of protein kinase R (PKR), a protein involved in innate immune responses and the cellular response to stress. It also binds to the PKR activator PACT and regulates its function. TRBP also contributes to RNA interference as an integral part of the minimal RNA-induced silencing complex with Dicer and Argonaute proteins. Due to its multiple functions in the cell, TRBP is involved in oncogenesis when its sequence is mutated or its expression is deregulated. The depletion or overexpression of TRBP results in malignancy, suggesting that the balance of TRBP expression is key to normal cellular function. These studies show that TRBP is multifunctional and mediates cross talk between different pathways. Its activities at the molecular level impact the cellular function from normal development to cancer and the response to infections.
INTRODUCTION
Despite the large amount of information obtained from the Human Genome Project, which was completed in 2003, it has become evident that the sequence of our genes is not sufficient to predict the complexity of our cells. Some groups have since turned to the transcriptome, because the expressed genetic material is important, while others have focused on the proteome. More recently, the interactome has been defined as connections between various molecules in the cell with individual molecules acting sequentially or simultaneously in different pathways. With every level of cell biology that we scrutinize, it becomes increasingly apparent that a good understanding of cellular processes requires us to take a step back and look at the outcome of molecular interactions, rather than dissecting a single molecule.
Double-stranded RNA (dsRNA) binding proteins (dsRBPs) play central roles in many cellular processes by bridging RNAs and proteins to form ribonucleoprotein (RNP) complexes (5, 26, 107). Their common feature is the presence of one or several dsRNA binding domains (dsRBDs), which mediate their interaction with dsRNA (6, 21, 37, 75, 135). Most cellular processes require multiple dsRBPs, and individual dsRBPs are involved in multiple pathways. dsRBPs are involved in nearly all aspects of the cellular life cycle, from transcription and translation to development and immunity (56, 96, 118, 129). One dsRBP that has received much attention in the past few years because of its role in RNA interference (RNAi) is the trans-activation response (TAR) RNA binding protein (TRBP).
TRBP AS A dsRNA BINDING PROTEIN
Identification
The cDNAs coding for TRBP1 and TRBP2 were cloned based on a protein-RNA binding screen using the highly structured human immunodeficiency virus type 1 (HIV-1) TAR RNA as a probe (52–54). The two proteins are identical except that TRBP2 has 21 additional amino acids at the N-terminal end due to the translation of an alternative first exon (4, 53) (Fig. 1A). TRBP was one of the first proteins involved in the identification of dsRBDs (51, 135). It has two dsRBDs and a C-terminal region called Medipal [Merlin, Dicer, protein kinase R [PKR] activator [PACT] liaison], which mediates protein-protein interactions (31, 40, 51, 84). Analyses of the natural RNA structures bound by TRBP indicated that the protein preferentially binds to double-stranded G-C-rich sequences (51, 89). In the context of synthetic small dsRNAs, TRBP favors asymmetric perfectly matched sequences independently of the sequence length (59, 78, 113).
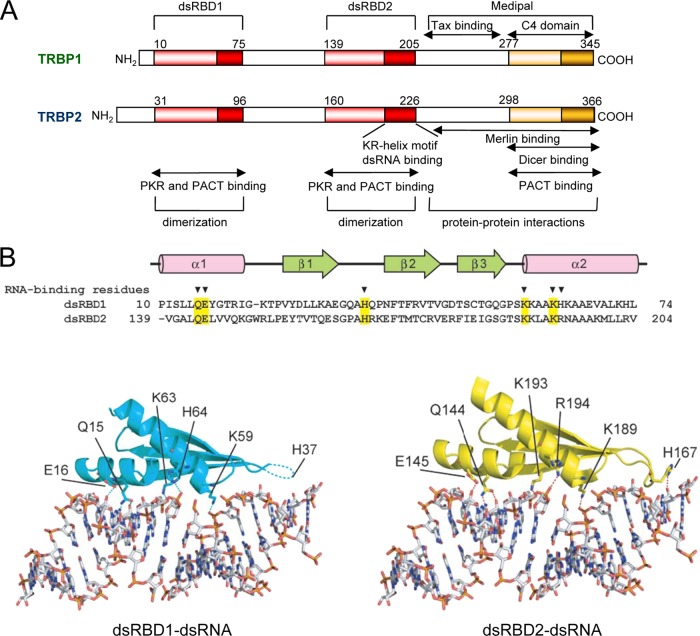
Fig 1.
Structural organization of TRBP. (A) Structural organization of TRBP1 and TRBP2. dsRBDs mediate RNA binding and are represented in red. Dark red represents the RNA binding motif, which is fully functional only in dsRBD2 and has been named the KR-helix motif. The Medipal domain is involved in protein-protein interactions. The orange domain (also called C4) has a weak homology with dsRBDs and does not bind RNA but mediates Dicer and PACT interactions. Sites of homo- and heterodimerizations are indicated. The dark red and dark orange domains are basic domains with an α-helix structure. (B) Comparison of the dsRNA binding surfaces of the dsRBDs of TRBPs. (Top) Sequence of dsRBD1 and dsRBD2 of TRBP (amino acid numbers refer to TRBP1). The dsRNA binding residues are indicated with arrowheads. The conserved dsRNA binding residues are shaded in yellow. (Bottom left) Docking model of the TRBP dsRBD1 with dsRNA. The residues with side chains that interact directly with dsRNA are shown in stick representations. The hydrogen bonds are shown by dashed lines. (Bottom right) Structure of the dsRBD2-dsRNA complex. (Adapted from reference 146 with permission of Wiley-Blackwell [copyright 2010 The Protein Society].)
Structural Domains
TRBP has three structural domains: dsRBD1 is located between amino acids (aa) 31 and 96 in TRBP2, and dsRBD2 is located between aa 160 and 226 (Fig. 1A). The third domain, named the C4 domain (30), is sometimes called half dsRBD or dsRBD type B (aa 298 to 366), because it has some structural homology with dsRBDs in its C-terminal basic portion. However, in TRBP and other dsRBPs, this domain does not bind RNA (66, 68, 135, 145, 146). In contrast, it mediates protein-protein interactions, and the larger region (aa 237 to 366) encompassing C4 is now referred to as the Medipal region (84). For TRBP-PACT and TRBP-Dicer interactions, the C4 domain is sufficient for binding (30, 84, 135), but a larger region has been defined for the TRBP-Merlin interaction (87). Future refined mapping will determine if the Merlin-interacting region can also be reduced to the C4 domain. In TRBP, dsRBD2 has a stronger dsRNA binding activity than dsRBD1. This difference has been attributed to a 15-aa peptidic sequence within dsRBD2 called the KR-helix motif, which can bind dsRNA by itself (31, 40, 51, 146). The entire protein or the two dsRBDs expressed in tandem have a much higher affinity for dsRNA than either dsRBD alone, indicating that the two domains cooperate for dsRNA binding (31, 51, 146).
Crystal Structure
The crystal structure of dsRBD1 and the solution structure of dsRBD2 have revealed that dsRBD1 and dsRBD2 of TRBP exhibit an α-β-β-β-α fold, which is common to dsRBDs (146) (Fig. 1B). In the context of the KR-helix peptide binding to TAR RNA, each K and R residue is required for RNA interactions (31, 40). For general dsRNA binding by dsRBD1 of TRBP1, the important residues have been mapped to Q15, E16, H37, K59, K63, and H64. The corresponding residues in dsRBD2 (Q144, E145, H167, K189, K193, and R194) are involved in the binding of that domain to dsRNA (146). Mutations of H37, K59, and K63 in dsRBD1 or H167, K189, and K193 in dsRBD2 prevented dsRNA binding (146). In TRBP2, the position numbers are increased by 21 aa. In addition, the thermal stability of dsRBD2 is higher than that of dsRBD1 due to a W residue at position 152 in TRBP1 and at position 173 in TRBP2 (146).
PHYLOGENY OF THE TRBP PROTEIN AND GENE
Evolution and Phylogeny of the TRBP Protein
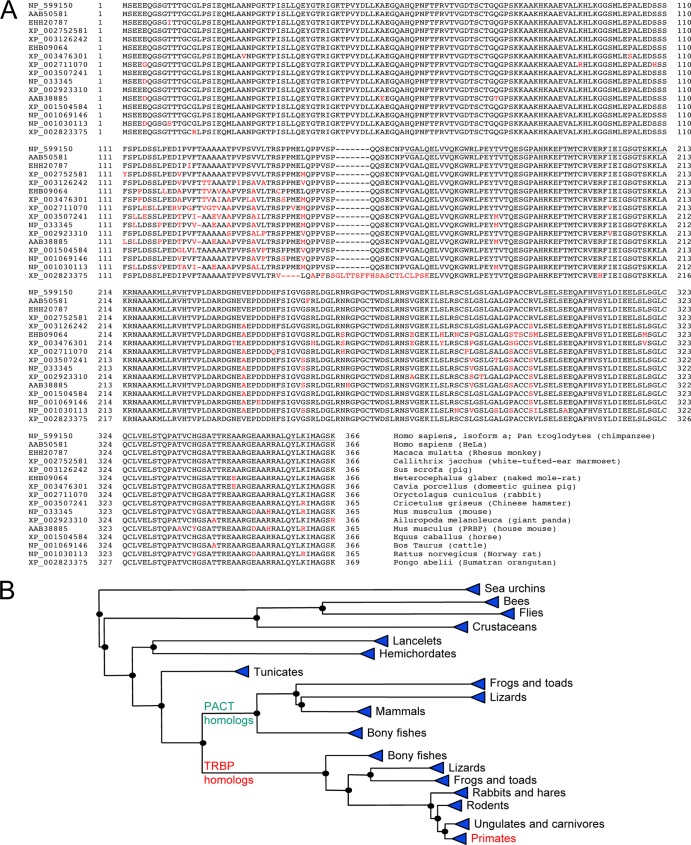
Following the identification of human TRBP, the cDNA coding for protamine-1 (Prm1) RNA binding protein (PRBP), the murine homolog of TRBP, was cloned from a mouse testis library, and the protein was identified as a key player in translational control during spermatogenesis (16, 89). TRBP2 and PRBP are highly homologous, having 93 to 94% identity and 94 to 96% homology, depending on the subspecies sequenced. With advances in the sequencing of many other organisms, TRBP has been found in several mammalian species with a minimum of 92% identity and no more than 11 gaps (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Fig. 2A). Homologs of TRBP are also found in fish, lizards, crustaceans, insects, and worms, and their functions have been well studied in Drosophila sp. (named Loquacious), in Caenorhabditis elegans (named RDE-4), and, recently, in shrimps (named Fc- and Mj-TRBP) (14, 44, 105, 113, 143, 144). The identification of a TRBP homolog in tunicates, the closest living invertebrate relative of vertebrates (32), and the presence of both TRBP and a related protein, PACT, in bony fishes suggests that the separation between TRBP and PACT occurred between invertebrates and vertebrates (Fig. 2B). Within vertebrates, birds lack a TRBP homolog but conserve RNAi activity (15, 98), suggesting that the function has been replaced by related proteins.
Fig 2.
Evolution and phylogeny of TRBP. (A) Homologies between mammalian TRBP2 proteins. Alignments were performed by using BLASTP (http://www.ncbi.nlm.nih.gov/blast/) and COBALT multiple alignments (110). Different amino acids compared to human TRBP2 are shown in red. GenBank accession numbers are indicated on the left. The name of the species used is shown on the right. The human clone in HeLa cells is the first identified protein (52). Mouse PRBP is the first identified murine protein (89). Underlined are dsRBD1 (aa 31 to 96), dsRBD2 (aa 160 to 226), and the C4 domain of Medipal (aa 298 to 366), with amino acid numbers referring to TRBP2. (B) Phylogenetic tree of TRBP2 obtained from COBALT multiple alignments. The phylogenetic tree was created by using the fast minimum-evolution method (33), showing evolutionary distance according to data reported previously by Grishin (61). The branches on the right of the PACT homologs represent proteins that have more homology with PACT than with TRBP2. The branches on the right of the TRBP homologs are more closely related to TRBP2. Proteins between sea urchins and tunicates are related to both PACT and TRBP2.
tarbp2 Gene Characterization
TRBPs and PRBP are encoded by the tarbp2 gene located on human chromosome 12 and mouse chromosome 15, and one or two pseudogenes, respectively, are present in each species (38, 82, 147). For the human gene, two adjacent promoters initiate the transcription of alternative first exons for TRBP1 and TRBP2 mRNAs. The TRBP1 promoter is upregulated by NF-Y transcription factors, which influences the expression of both isoforms, as observed by the low expression levels of the proteins in astrocytic cells (3, 4). The knockout of the tarbp2 gene in mice results in a growth defect, and males are sterile, displaying severe oligospermia. In addition, tarbp2−/− mice die at weaning, indicating an important role during development (148). In humans, TRBP RNA expression levels are high in the testis and low in the brain (3, 87, 132). This expression profile suggests a particular role for TRBP in undifferentiated cells and cancer cells, as certain testis-associated genes are upregulated in malignant cells (18).
REGULATION OF VIRUSES
Activity in HIV-1 Replication
The first identified function of TRBP was to stimulate the expression of the HIV-1 promoter in human cells, an activity partially linked to the presence of the TAR RNA structure (52). Although HIV-1 is produced at very low levels in murine cells and in human astrocytes, exogenous TRBP enhances both expression from the HIV-1 promoter and viral production in these cells (7, 38, 109). This increased HIV-1 expression level is ascribed to the inhibition of PKR (8, 25, 29) and to a direct activity of TRBP that relieves the translational block caused by the TAR RNA structure (2, 36). The transfection of small interfering RNAs (siRNAs) against TRBP decreases the level of HIV-1 production in HeLa cells, and the transduction of short hairpin RNAs (shRNAs) against TRBP leads to the long-term inhibition of HIV-1 replication in human SupT1 lymphocytic cells, indicating that the protein contributes to viral expression and replication (24, 39), but these findings have not yet been reproduced in primary cells. This property seems to be mediated mainly by a decreased TRBP-PKR interaction and a consequent enhancement of PKR activation, which prevents viral translation, but could also be due to TRBP's interactions with other molecules (24, 109, 127).
Activity in Expression of Other Viruses
TRBP also enhances expression from the early promoter of the simian virus 40 (SV40) and the Visna virus promoter but not the immediate-early promoter from simian cytomegalovirus (52). In the case of the human T-cell leukemia virus type I (HTLV-I) promoter, TRBP increases the basal expression level of the promoter but inhibits Tax-mediated trans-activation by direct TRBP-Tax interactions (35, 52). It is currently not clear if the observed effect of TRBP on these promoters occurs directly on transcription or is a translational effect, but the inhibition of Tax has been linked to a nuclear localization signal and Tax binding domain in TRBP located between aa 211 and 269 in TRBP1 (35).
REGULATION OF THE INTERFERON RESPONSE
Binding to PKR
PKR is central in the host innate immune response as an interferon (IFN)-inducible dsRBP that has a serine/threonine kinase activity (48, 102, 125). PKR is activated after binding to low levels of dsRNA through its two dsRBDs and by dimerization. Once active, PKR phosphorylates its substrate, the alpha subunit of eukaryotic translation initiation factor 2 (eIF2α). eIF2α phosphorylation dramatically alters the efficiency and rate of translational initiation and is a critical component of antiviral and cell growth pathways (46, 48). In addition to its role in translational regulation in response to dsRNA, several other activators, such as growth factors, cytokines, proinflammatory stimuli, and oxidative stress, also activate PKR. PKR is therefore implicated in the integration and transmission of signals to the translational machinery as well as to factors such as STAT, IFN regulatory factor 1, p53, Jun N-terminal protein kinase (JNK), p38, ATF-3, and NF-κB (48, 121, 125). PKR overexpression decreases—whereas its inhibition increases—the expression levels of several viruses (34, 72, 106, 109, 117). The cellular function of PKR on cell growth appears to be more complicated, because its upregulation or overexpression can inhibit the growth of some tumor cells, whereas an upregulation of PKR has been identified in some malignancies (13, 104, 141). Whether the overall level of PKR is increased or not, its activation is highly regulated by viral and cellular proteins and RNAs (1, 26, 47–49, 56, 117).
The inhibition of PKR activation by TRBP was first identified by its ability to restore the growth of a vaccinia virus lacking the E3L protein and was ascribed to dsRNA sequestration by TRBP (27, 112). Further studies using immunoprecipitation, in vitro interactions, and yeast and mammalian two-hybrid assays showed that the two proteins bind directly through their dsRBDs, leading to the inhibition of PKR function (8, 12, 29, 64). Therefore, it is likely that TRBP inhibits PKR function both by dsRNA sequestration and by direct protein-protein interactions.
Consequences of PKR Inhibition by TRBP
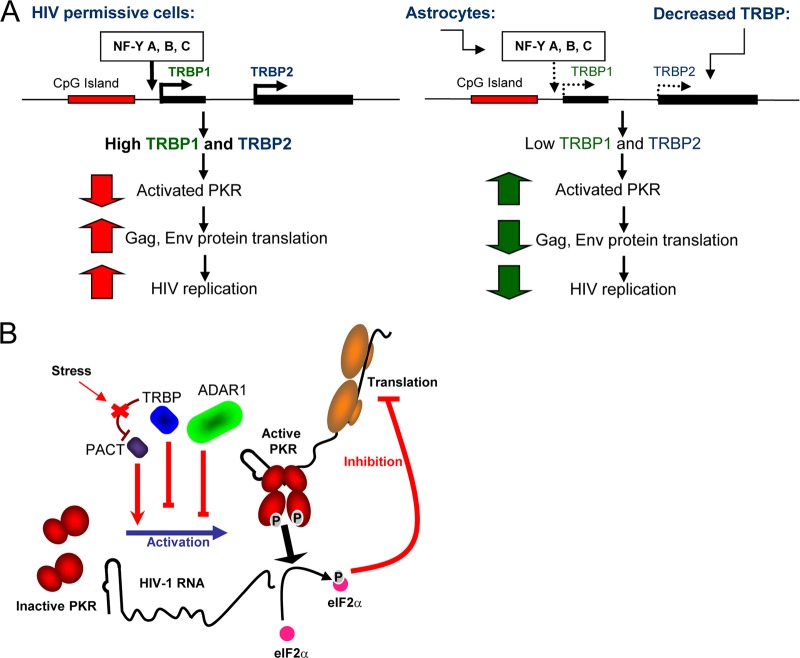
A comparison of levels of HIV-1 production in cells producing high or low levels of TRBP demonstrates the consequence of the TRBP inhibition of PKR in vivo. HeLa, HEK293T, or lymphocytic T-cell lines efficiently replicate the virus. Astrocytes are brain cells that can be infected with HIV-1 but produce very small amounts of virus (58). Studies using astrocytoma cells showed that there is a major block in the HIV-1 translation of structural proteins due to a heightened PKR activation in these cells after the transfection of an HIV-1 molecular clone (57, 109). This high level of PKR activation correlates with a weak expression of TRBP in these cells compared to that in HeLa cells or lymphocytes and the consequent absence of a PKR-squelching mechanism. The transfection of TRBP is able to partially rescue HIV-1 expression in these cells (25, 109). The TRBP expression level in astrocytes is low because of the weak induction of its promoter. In turn, the poor stimulation of the TRBP1 promoter in astrocytes compared to that in lymphocytes is due to the reduced amount of the NF-Y transcription factor in these cells (3, 4, 109) (Fig. 3A). This model of the consequences of low TRBP expression levels on viral replication in brain cells suggests additional roles for TRBP in the physiological and pathological pathways in which PKR is involved.
Fig 3.
Role of TRBP in regulating HIV-1 expression and replication through PKR regulation. (A) Control of HIV replication in astrocytes compared to HIV-permissive cells through TRBP expression. In HIV-permissive cells, the NF-Y transcription factors activate the TRBP1 promoter, which results in the production of TRBP1 and TRBP2. TRBP binds to PKR and inhibits its activation. This results in the translation of HIV-1 proteins and high-level HIV-1 replication. Astrocytes express low levels of NF-Y transcription factors, which results in low TRBP promoter expression and low TRBP protein production levels. Low TRBP protein expression levels can also be obtained by using siRNAs against TRBP mRNA. Low TRBP expression levels result in high levels of PKR activation, a block of HIV-1 protein production, and low levels of HIV-1 replication (3, 4, 24, 109, 127). (B) Control of PKR activation by TRBP, ADAR1, and PACT during HIV-1 expression. PKR is activated by HIV-1 TAR RNA. This activation is inhibited by TRBP and ADAR1 but enhanced by PACT. TRBP also inhibits PACT activity, which can be restored by oxidative stress (25, 26, 29, 30, 84). (Adapted from reference 25.)
DAMPENING OF THE STRESS RESPONSE
PACT and the Stress Response
PACT is a dsRBP that was cloned based on its interaction with PKR in a yeast two-hybrid screen of a human cDNA library using inactive PKR as bait (116). In contrast to the inhibition of PKR by TRBP, mammalian cells transfected with PACT show an enhanced phosphorylation of PKR and eIF2α, which inhibits translation. PACT expressed in bacteria, purified and devoid of any contaminating dsRNA, also activates PKR (116). In contrast to transfection assays that used a truncated PACT at its C-terminal end (68, 116, 119), wild-type (wt) PACT activates the translation of SV40 or HIV promoter-driven luciferase genes, which raises questions about the physiological conditions under which PACT activates PKR (28, 84, 91). Assays using wt and endogenous proteins demonstrated that PACT and PKR activator X (RAX), its murine homolog, are proapoptotic proteins (64, 68, 70, 115, 119) and stress-modulated physiological activators of PKR (28, 115). Stress signals lead to the phosphorylation of PACT on serines 18 and 287, and PACT and RAX need to be phosphorylated to mediate PKR activation (11, 120).
Modulation of the Stress Response by TRBP-PACT Interactions
TRBP and PACT are highly homologous and interact through three domains, the two dsRBDs and the C4 domain in Medipal (79, 84). The stress modulation of PACT activity is explained by its interaction with TRBP under nonstress conditions (28). While the native form of PACT does not constitutively activate PKR in most cells, a 13-aa C-terminally truncated mutant (called PACT305 or PACTΔ13) is able to do so (28, 119). Both forms increase levels of PKR phosphorylation in astrocytes, suggesting that the weak expression of TRBP in these cells plays a role in enhancing the PACT activation of PKR (28, 84). Indeed, PACTΔ13 is a mutant that weakly binds to TRBP and constitutively activates PKR in all cells. In the absence of stress, PKR activation by wt PACT occurs only when there are low levels or an absence of TRBP such as in astrocytes, in cells treated with siRNAs against TRBP, or in tarbp2−/− cells (28). PACT activity was also restored by a stress that phosphorylates serine 287 on PACT and disrupts the TRBP-PACT heterodimer, indicating that TRBP controls the PACT activation of PKR by a stress-reversible process (28, 133). Together with TRBP and ADAR1, PACT is involved in the overall regulation of HIV-1 gene expression by PKR during viral replication and may also be involved in other cellular processes (25, 26) (Fig. 3B). The modulation of the TRBP-PACT interaction by stress may also influence several other cellular processes in which both proteins are involved, including RNA interference (RNAi) and oncogenesis.
TRBP FUNCTION IN RNA INTERFERENCE
TRBP in RNA Interference
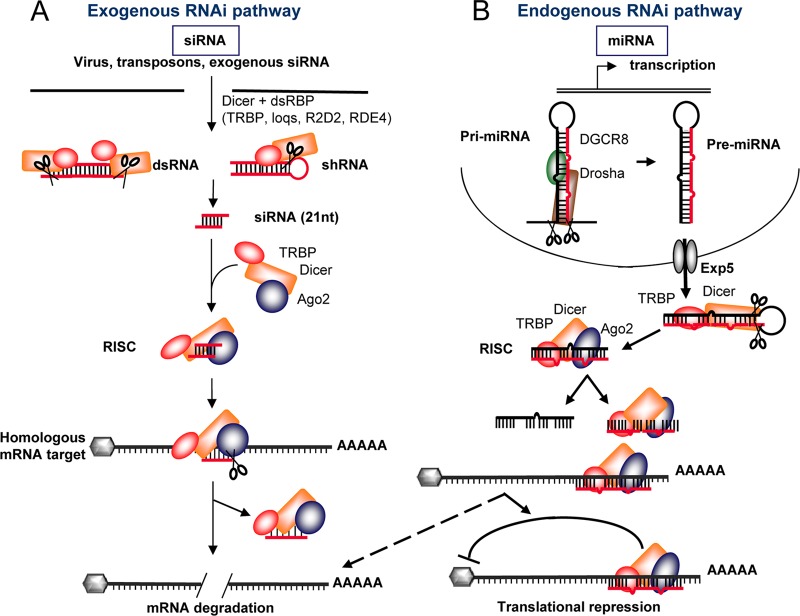
In addition to its key roles in HIV replication and the regulation of innate immune pathways, in 2005, TRBP was found to be associated with the RNAi machinery (22, 65, 122). RNAi has emerged as a crucial regulatory pathway in all eukaryotic cells and is continually being implicated in new mechanisms of cellular function, proliferation, differentiation, and defense (17, 41, 62, 74, 83, 93, 136). Since RNAi was first described in the nematode C. elegans (43), the key proteins of the RNA-induced silencing complex (RISC) have been identified as being Dicer, Argonaute (Ago), and a small dsRNA binding protein. These three proteins are highly conserved among eukaryotes and represent the minimal RISC-loading complex (RLC). In C. elegans and mammals, only one isoform of Dicer has been identified and is central to RNAi activity. In Drosophila melanogaster, two Dicer proteins have been identified, Dicer-1 and Dicer-2. While Dicer-1 is specialized for the processing of microRNAs (miRNAs), Dicer-2 is involved primarily in the formation of the RISC and in carrying out silencing functions (71). In contrast, two Ago isoforms exist in Drosophila sp., compared to four in mammals and C. elegans. While these proteins are highly similar, each isoform appears to have some specificity of function (67, 69). The dsRBPs associated with Ago and Dicer differ from one organism to another. In C. elegans, this protein is RDE-4; in Drosophila, it is either R2D2 or Loquacious (Loqs), and in humans, it is TRBP (22, 44, 65, 94, 138) (Fig. 4). Although these proteins undoubtedly form a complex with Ago2, as shown by immunoprecipitation (22, 65, 92), it is yet unclear whether TRBP directly binds Ago2 or whether Dicer forms a bridge between the two proteins, as no direct interaction has been shown so far.
Fig 4.
TRBP in the exogenous and endogenous RNAi pathways. (A) Exogenous pathway mediated by siRNAs and shRNAs. Perfectly matched dsRNAs are bound by Dicer and TRBP and cleaved by Dicer to form 21- to 23-nucleotide (nt) siRNAs. The complex then recruits Ago2 and forms the RISC. After strand separation, the complex hybridizes to the complementary sequence in an mRNA. Ago2 mediates the cleavage of the mRNA. (B) Endogenous pathway mediated by miRNAs. Primary mRNAs (pri-miRNAs) are synthesized in the nucleus. They are bound by Drosha and DGCR8. Drosha cleaves them to form the precursor miRNA (pre-miRNA), which is an imperfectly matched dsRNA. Pre-miRNAs are exported to the cytoplasm via exportin5 (Exp5), where they are bound by TRBP and Dicer. Dicer cleaves them to form miRNAs. The RISC is then formed with Ago2. After strand separation, one miRNA strand targets the corresponding mRNA. The translation of this mRNA is generally repressed by a process that involves the recruitment of additional factors and the formation of processing bodies, but in some cases, cleavage of the target mRNA may occur (dotted arrow).
Part of the RNA-Induced Silencing Complex
Further studies have demonstrated that in vitro, TRBP, Dicer, and Ago2 are able to spontaneously assemble, form the RLC, and carry out RISC activity by processing precursor miRNA (pre-miRNA) and cleaving target mRNAs (85, 97). Whereas two studies suggested that this complex remains intact with multiple rounds of cleavage (60, 90), others indicated that Ago2 is associated more with mature RNA sequences and tends to dissociate after miRNA loading, suggesting that TRBP and Dicer are essential for processing shRNAs/miRNAs but do not participate in target cleavage (63, 97, 99). While few in vivo studies have been carried out, results indicate that the presence of TRBP is crucial for RNAi activity. Reconstitution in the yeast Saccharomyces cerevisiae showed that Dicer, TRBP, and Ago2 are necessary and sufficient to recapitulate functional human RNAi (137). Cells derived from tarbp2−/− mice exhibit virtually no RNAi activity, but this activity is restored by exogenous TRBP (30). In addition, the C4 domain of TRBP, which is crucial for TRBP binding to Dicer, is essential for this restoration of RNAi activity (30). The cellular localizations of TRBP and Dicer indicate a colocalization in the cytoplasm but not in processing bodies, further supporting a major function in RLC formation and miRNA cleavage rather than target cleavage mediated by Ago2 (30, 111).
Role in miRNA Processing
Interestingly, the exact function of TRBP in RNAi remains unclear. While in C. elegans, RDE-4 is crucial for the sensing and cleavage of long dsRNA into siRNA, human TRBP appears to act independently of the length of the siRNA (113). TRBP binds to Dicer within the ATPase/helicase domain in a unique domain between the DExH/D and helicase subdomains (30). Interestingly, when this domain is inactivated in Dicer, the protein is unable to process thermodynamically unstable hairpins (134), whereas when it is deleted or bound to TRBP, Dicer exhibits an enhanced cleavage rate (95). Furthermore, Dicer-TRBP heterodimers are required to sense siRNA asymmetry (108). The two proteins act in concert, and while Dicer is able to preselect effective siRNAs (126), TRBP enhances Dicer-catalyzed miRNA-processing kinetics (20) and acts downstream of siRNA/miRNA production for the selection and incorporation of the guide sequence into the RLC (19, 78, 113). Knockdown studies of TRBP have demonstrated that TRBP contributes to the accumulation of siRNAs and miRNAs and also functions in sensing the target mRNA to effect silencing (19, 22, 65, 90).
Structure of the RISC-Loading Complex
Recently, several groups have examined the structural characteristics of the RLC and have modeled the three-dimensional aspects of the complex from electron microscopy analyses. Dicer, Ago2, and TRBP assemble in a 1:1:1 stoichiometric ratio (97), and the association of TRBP with Dicer yields an “L-shaped” complex (85). Another study identified Dicer alone with an L-shaped structure with a short base. This base represents the DExH/D helicase and ATPase domain of Dicer (142). In agreement with the finding that this domain of Dicer is involved in binding to the TRBP C4 domain (30), when the RLC does not contain TRBP, the Ago-Dicer interaction is weakened, suggesting that TRBP stabilizes the complex. In addition, that study suggested that TRBP contributes to RNAi in several respects. First, the binding of TRBP to Dicer may facilitate the retention of siRNA and may help to orient the binding of Ago2 toward the correct end of the siRNA. Second, TRBP could contribute to the selection of the appropriate strand for loading into the RISC. This hypothesis is further supported by the fact that TRBP recognizes asymmetrical dsRNA molecules better and specifically binds to the 3′ overhang (59).
Relationship between PACT and TRBP in RNAi
Finally, PACT, which has also been recovered in Dicer-containing complexes, contributes to the stabilization and activity of Dicer (79, 90). Although it remains unclear whether TRBP or dsRNA mediates the recruitment of PACT to Dicer, PACT enhances the activity of pre-miRNA processing and the efficiency of siRNA to some extent, and both TRBP and PACT stabilize Dicer (22, 81, 90, 101). In one study, the contribution of PACT was minor, whereas TRBP seemed to be crucial in those assays (90). In another study, PACT contributed to shRNA-mediated silencing but had no influence on siRNA-mediated silencing (79). In contrast, in tarbp2−/− cells, where PACT is present, shRNAs were inactive, suggesting that in these cells, PACT cannot replace the RNAi function of TRBP (30). Overall, PACT is found in the 500-kDa complex with TRBP, Dicer, and Ago2 and contributes to the processing by Dicer and the function of miRNAs and shRNAs, but TRBP seems to have the central role, as shown by in vitro reconstitution and the lack of functional rescue by PACT in tarbp2−/− cells (30, 97). More in vivo studies are required to determine if the association between TRBP and Ago2 is maintained during the target RNA cleavage or translation inhibition step in the miRNA process.
ROLE AND REGULATION OF TRBP IN CELL GROWTH AND CANCER
Overexpression and Malignancy
Several lines of evidence indicate that TRBP has a role in promoting cell growth during development and in cancer. Mice in which the tarbp2 gene has been homozygously disrupted have a growth defect, suggesting an important role during development (148). The overexpression of TRBP in murine cells induces a transformed phenotype, and the injection of these TRBP-overexpressing cells into nude mice induces tumor formation (8). This activity is attributed to PKR inhibition, since similar results were observed with a transdominant, catalytically inactive PKR mutant (80, 103). The growth-promoting function and oncogenicity of TRBP are regulated by a tumor suppressor called Merlin or Schwannomin, encoded by the neurofibromatosis type 2 (nf2) gene. Mutations in the nf2 gene result in brain tumors (123, 140). Multiple protein interactions could explain the role of Merlin as a tumor suppressor (130). Merlin and TRBP interact directly through aa 237 to 366 in TRBP2 and aa 288 to 595 in Merlin, located in their C-terminal ends. Merlin causes TRBP ubiquitination, thereby targeting the protein to the proteasome pathway for degradation (87, 88), and expression levels of TRBP are known to be low in brain tissue (3, 132). This, along with the fact that disruption of Merlin results in brain-specific tumors, suggests that the maintenance of low levels of TRBP is essential for normal brain function and may be mediated by Merlin's activity.
Disruption of the miRNA Pathway
Recent studies shine additional light on the mechanism by which the overexpression of TRBP, its translational modifications, or its mutations contribute to cell growth and cancer-related disorders. Growth induction by TRBP was shown to occur through the mitogen-activated protein (MAP) kinase (MAPK) pathway, which induces the phosphorylation of TRBP by the Erk kinase (114). The phosphorylation of TRBP on serine residues increases the stability of the TRBP-Dicer complex. As a result, certain miRNAs known to promote cell growth, miR-17, miR-20a, and miR-92a, are more expressed, whereas levels of let7 miRNA, which acts as a tumor suppressor, are downregulated. These experiments suggest that a more stable TRBP-Dicer complex promotes cell growth. However, another study found that both wt and phosphomimic TRBPs enhanced Dicer kinetics similarly, suggesting that an increased TRBP-Dicer stability cannot solely be the cause of the observed changes in miRNA expression and cell growth (20).
Truncated TRBP and miRNA Expression
In the colorectal cancer cell line Co115 and in the endometrial cancer cell line SKUT-1B, the deletion or addition, respectively, of one C in the tarbp2 gene creates a truncated TRBP protein (101). These cells have a severely impaired processing of primary miRNAs into precursors, which results in decreased expression levels of miRNAs with potential tumor suppressor capabilities, such as let7, miR205, miR26a, miR125a, and miR125b. Truncated TRBP is unable to maintain Dicer stability, whereas the wt protein does. The coexpression of the wild-type tarbp2 gene restores pre-miRNA levels in the cells and reduces the tumorigenicity of the corresponding cells. Although the frequency of these mutations in colorectal cancer has been questioned, their presence was confirmed by another study (50). Taken together, those studies suggest that both impaired TRBP and overexpressed, phosphorylated TRBP promote cell growth by opposite mechanisms, leading to either impaired miRNA processing or decreased miRNA synthesis. Interestingly, in either case, levels of let7 miRNA were significantly decreased. Whether the increased cell growth is due mainly to decreased let7 miRNA levels or whether other mechanisms are involved remains to be determined.
Additional correlations between TRBP expression and mutations have been observed. For example, increased expression levels of miRNAs, Dicer, TRBP, and Ago2 have been shown for prostate cancer cells compared to normal tissue (45). A mutation in the tarbp2 gene was also found in a gastric carcinoma, a cancer with microsatellite instability and a deregulation of miRNA expression (77). Further studies are needed to confirm the relevance and the role of these mutations in cancer development and to determine if TRBP deregulations contribute to additional cancers. The overexpression, low expression levels, truncations, or mutations of TRBP correlate with cell cycle dysfunction and cancer development, suggesting that an optimal threshold of TRBP seems to be required for appropriate cell growth (Fig. 5).
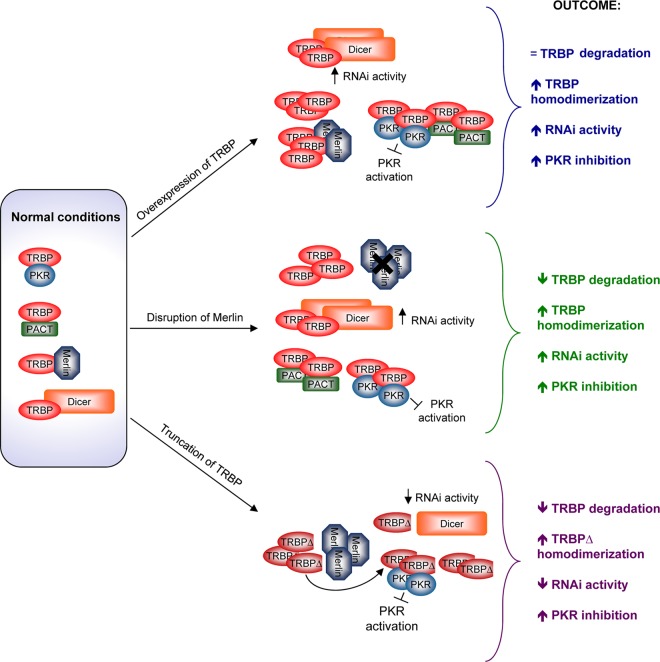
Fig 5.
TRBP involvement in malignant phenotypes. (Left) In cells, TRBP can be bound to Merlin, Dicer, PKR, and PACT. These interactions are reversible and regulated. (Top) When TRBP is overexpressed, it binds Dicer for increased RNAi activity, it fully represses PACT and PKR, and it is not fully inactivated by Merlin. This results in a transformed phenotype. (Middle) When the nf2 gene, coding for Merlin, is disrupted in brain cancers, TRBP becomes overexpressed by a lack of degradation, which results in increased RNAi and increased PKR inhibition. (Bottom) When TRBP is truncated in its C terminus, as seen in some cancer cells, it no longer binds to Dicer, which results in decreased RNAi. It still binds to PKR and inactivates it, but its affinity for PACT is decreased. It no longer interacts with Merlin, which cannot degrade it. As a consequence, TRBP overexpression, Merlin inactivation, or TRBP truncation results in malignant phenotypes by different mechanisms.
TRBP-MEDIATED CROSS TALK BETWEEN MULTIPLE PATHWAYS
We have only just started to unravel the mysteries of the cellular interactome. While most functions of TRBP-interacting factors identified in a previous proteomic study remain to be fully characterized (23), some ribonucleoprotein complexes formed around TRBP have revealed important interconnected biological processes. Indeed, by interacting with multiple cellular RNAs and proteins, TRBP bridges multiple pathways that can no longer be considered isolated entities (Table 1). The protein can be associated with different targets at the same time in the cell and mediates cross talk that may affect cell survival (Fig. 6).
Table 1.
Molecular partners and inhibitors of TRBP
| Molecular partner or inhibitor | Activity of TRBP | Reference(s) |
|---|---|---|
| Cellular partner | ||
| TRBP | Forms homodimers | 84 |
| PACT | Sequesters PACT and prevents PKR activation | 28, 79, 84, 133 |
| PKR | Inhibits PKR activation | 8, 12, 29, 64 |
| Dicer | Acts as a cofactor for RNAi | 22, 30, 65 |
| Merlin | Is a substrate for ubiquitination | 87, 88 |
| siRNA/shRNA/miRNA | Loads small RNAs into the RISC | 19, 20, 78, 108, 113 |
| Viral factor | ||
| HTLV-I Tax | Inhibits Tax-mediated transactivation of the LTRa | 35, 41 |
| Ebola virus VP35 | Is a target of VP35-mediated RNAi suppression | 42 |
| HIV-1 TAR | Alleviates translational block of mRNA and is a target of TAR-mediated RNAi suppression | 52–54 |
| vsiRNA/vmiRNA | Loads small RNAs into the RISC | 19, 20, 78, 108, 113 |
| Synthetic compound | ||
| Enoxacin | Is a substrate of enoxacin-mediated enhancement of miRNA activity | 100 |
| siRNA/shRNA targeting TRBP | Is a target of RNAi and results in decreased function | 22, 24, 39, 65 |
LTR, long terminal repeat.
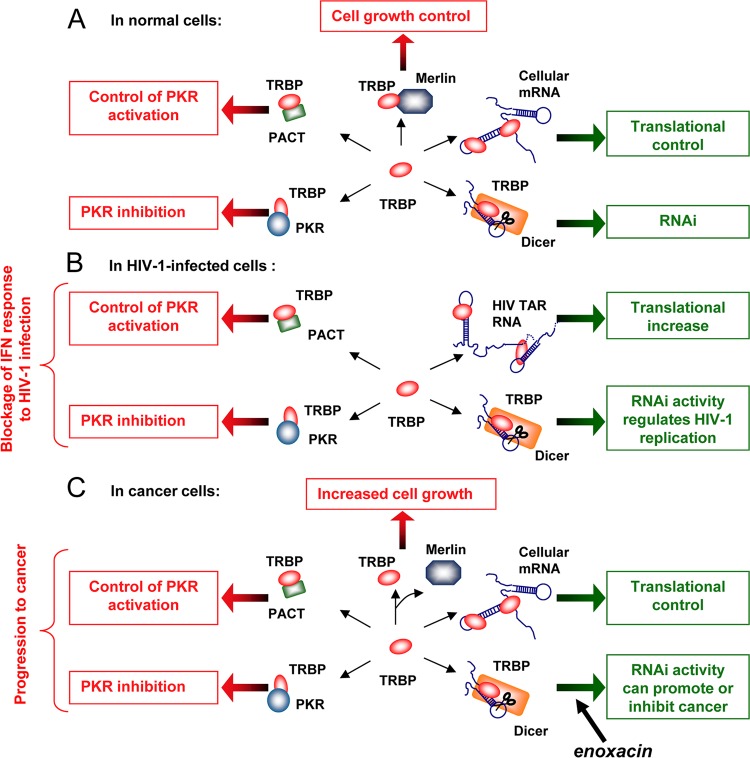
Fig 6.
Known and possible interactions and functions of TRBP under normal conditions, in HIV-infected cells, and in cancer cells. (A) Known and possible interactions and functions of TRBP under normal conditions. TRBP interacts with cellular RNAs and miRNAs as well as with Dicer, PKR, and PACT to regulate translation, RNAi, and PKR activation. Its binding to Merlin mediates its degradation and control of cell growth. (B) Known and possible interactions and functions of TRBP in HIV-1-infected cells. TRBP interacts with HIV-1 TAR RNA to increase the translation of HIV-1 mRNAs, while it still binds to cellular mRNAs and miRNAs. TRBP binds to Dicer, PKR, and PACT to regulate RNAi and PKR activation. Its interaction with Merlin has not been tested in infected cells. (C) Known and possible interactions and functions of TRBP in cancer cells. The disruption between TRBP and Merlin leads to an increased TRBP concentration and increased cell growth. The overexpression or truncation of TRBP results in increased binding to cellular mRNA and miRNA, which disturbs translational control. Increased or decreased binding to Dicer may disturb RNAi activity and lead to cancer. The small molecule enoxacin enhances miRNA processing by TRBP and inhibits cancer growth (100). Increased binding to PKR and PACT inhibits PKR activation.
From Viral RNA to PKR Inhibition
Although TRBP was originally isolated as a protein that binds a viral RNA, its primary role is not to interact with HIV-1, a virus which likely emerged in humans about a century ago (139). This interaction, which favors HIV-1 replication, quickly emerged as a coevolutionary mechanism in which the virus coopts a cellular function for its own benefit. This benefit was first identified as a mechanism which blocks part of the antiviral innate immune function by binding to PKR (2, 26) but also impacts the general regulation of PKR activation during development and cell growth (48).
Enhancing or Preventing Translation during Development
Murine PRBP/TRBP was first found to repress Prm1 RNA translation by binding to the structured 3′ untranslated region (3′UTR) (89). This activity is required for proper spermatogenesis during development. The disruption of the tarbp2 gene causes a growth defect and indicates a role for the protein in enhancing translation (148). At the time when those studies were conducted, TRBP was not known to be part of the RISC. A review of those results suggests that TRBP could enhance translation by inhibiting PKR, unfolding structured RNA, or activating the MAPK pathway, as shown in other studies (29, 36, 114). In a different spatiotemporal context, it could repress the expression of some transcripts when bound to their 3′UTRs as a RISC component (22, 30, 65, 99) or by enhancing the production of specific miRNAs (101, 114). The appropriate expression and the multifunctional activity of TRBP are likely required for normal developmental processes.
From miRNAs to Viral siRNAs
The RNAi pathway is implicated in all aspects of cellular function (83). This has made the role of TRBP at the center of miRNA biogenesis all the more important (22, 30, 65). The dual requirement of TRBP for both HIV-1 replication and RNAi function, together with a possible role of RNAi as an innate immune pathway in mammals (9, 86), does not appear to be compatible (55). In the case of HIV-1, it is clear that TRBP enhances viral expression and replication (7, 8, 24, 25, 29, 38, 39, 52, 109, 127), which suggests that in this context, TRBP may be more frequently associated with the PKR ribonucleoprotein complex and the TAR RNA (2, 26) than with the RISC. Alternatively, HIV-1 might use cellular miRNAs and the RISC to enhance its own replication, as does hepatitis C virus (24, 55, 73). The characterization of HIV-1 TAR RNA as an RNAi suppressor, which sequesters TRBP and prevents the virus from being cleaved by RNAi, increases the difficulty of reconciling the TRBP enhancement of HIV-1 replication and the role of RNAi as a defense mechanism against the virus (10). In contrast, Ebola virus limits RNAi efficiency, and TRBP is a direct target of the viral VP35 protein (42). Indeed, VP35 acts as an RNAi suppressor by directly or indirectly binding to TRBP, suggesting that this interaction prevents RISC activity. Several mammalian viruses, mostly herpesviruses, express viral siRNAs (vsiRNAs), which control their own expression. In the case of herpesviruses, these vsiRNAs regulate the viral life cycle by allowing a switch between lytic and latent cycles, but the role of TRBP in this switch is not known (62). The effect of the overexpression or inhibition of TRBP has not been studied for other viral infections besides HIV. These assays would give insight into the role of TRBP-mediated cross talk between different pathways during various viral infections. It would also help to resolve the issue of the controversial debate on innate immunity mediated by RNAi in response to viral infections in mammals (62, 128).
Cooperation or Competition between TRBP and PACT?
TRBP and PACT have evolved from a common ancestor in invertebrates and have separated in vertebrates (Fig. 2B). They have the same protein organization and are highly homologous. Because of this similarity, they have been proposed to be interchangeable in development and RNAi but to have opposite functions in regard to PKR activation (64, 79, 90). However, their functional similarities or differences are not easy to evaluate considering that the two proteins bind to each other and regulate each other's functions (28, 79, 84, 133). In the context of PKR regulation, it now seems clear that the two proteins have opposite functions due to their C4 domains, but PACT can exert its PKR-activating function only in the absence of TRBP or under stress conditions, which dissociates the interaction between the two proteins (28, 133). Within the RISC, whether PACT can replace TRBP as a partner for Dicer and reconstitute the RLC is still under debate. Some groups, but not all, have found PACT to be associated with the RISC (79, 90). Furthermore, it has not been determined whether PACT interacts with Dicer directly or through TRBP or dsRNA, and so far, no in vitro reconstitution has been shown solely with PACT, Dicer, and Ago2. The idea of interchangeability between TRBP and PACT arose from the fact that both tarbp2−/− and prkra−/− (PACT knockout) mice are viable, with defects in spermatogenesis and ear development, respectively (124, 148). Considering that TRBP is involved in so many different processes, it seems surprising that TRBP knockout mice could survive, if only until weaning. In addition, TRBP knockout cells derived from these mice, which exhibit no RNAi activity (30), survive extremely well in cell cultures and become immortalized in the absence of any transforming agent (our unpublished observations). However, the fact that tarbp2−/− mice die at weaning demonstrates that the proposed compensatory effect of PACT is only partial during development and not sufficient to allow survival. These remaining questions indicate that more in vitro and in vivo studies need to be done to determine if PACT is an essential protein, a TRBP substitute, or a contributing protein that cooperates or competes with TRBP within the RISC and during development.
Combining miRNA Disruption and PKR Inhibition in Cancer
The association of either the overexpression, modification, or mutation of TRBP with several types of cancer raises important questions about TRBP-mediated oncogenesis. The inhibition of PKR by TRBP contributes to tumorigenesis in mice (8), and phosphorylated TRBP contributes to accelerated cell growth following the activation of the MAPK pathway (114). In addition, mutations of TRBP that inhibit its binding to Dicer and Merlin lead to a disruption of the miRNA pathway (30, 87, 101). These mechanisms are not exclusive and likely contribute to cell growth control during normal development and to oncogenesis when they are disturbed or uncontrolled. Furthermore, the involvement of TRBP and Ago1 in the transcriptional gene silencing of the RASSF1A promoter, which is epigenetically silenced in cancer, may lead to the discovery of additional roles of TRBP in cancer cells (76). More studies of these mechanisms could show a more important role of TRBP in a larger variety of cancers. The potential of small molecules to inhibit cancer cell growth by binding TRBP and restoring a normal miRNA pathway might also have an impact on PKR activation in these cells (100, 131).
CONCLUSIONS AND PERSPECTIVES
The dsRBP TRBP has emerged as an essential protein. It is critical for the survival of the entire organism and is heavily involved in spermatogenesis and cell growth. More specifically, it has been shown to be a crucial regulator of many cellular functions: TRBP is involved in the regulation of the IFN pathway, is essential for HIV replication, has a function in the stress response by sequestering PACT, is an essential component of the RNAi machinery, and contributes to cell growth and cancer. In most of these pathways, TRBP is a hub of interactions connecting various pathways in cell growth and in cellular responses to infection. Some of these interactions have been well characterized, but many more have been recently identified and need to be functionally characterized (23). The comparison between the proteins and RNAs that interact with TRBP in normal and pathological contexts will give further insight into the various cellular functions of this multifunctional protein.
The plurifunctionality of TRBP may have important implications for therapies targeting TRBP. On the one hand, the knocking down of TRBP may be extremely beneficial in situations of infections with oncogenic viruses, as TRBP is involved both in dampening the antiviral response and in promoting oncogenesis. On the other hand, the knocking down of TRBP expression in cells is likely to disrupt more than one pathway, which may impede cellular survival, and this may not be desirable in all circumstances. The identification of a molecule that specifically binds TRBP and restores a normal miRNA profile in cancer cells may help to elucidate the role of TRBP in other pathways and to explain the downregulation of certain miRNAs but not others (100, 131). Further elucidation of these pathways will help to identify therapies to target TRBP and restore normal pathways that have become dysfunctional in various pathologies.
ACKNOWLEDGMENTS
The work done in our laboratory is supported by grants from the Canadian Institutes of Health Research (A.G.) and by a Vanier Canada graduate scholarship (S.M.D.).
We are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Biographies

Sylvanne M. Daniels (M.Sc., 2006) obtained her undergraduate diploma in Cellular and Molecular Biology from the Université de Rennes 1, Rennes, France. She completed her M.Sc. in Functional Genomics and Health in 2006 at the Université de Rennes 1 and is currently conducting Ph.D. research at McGill University, Montréal, Québec, Canada. She is working on the role of TRBP in gene expression, RNA interference, and HIV-1 replication. She is the recipient of a CIHR Vanier Canada Graduate Scholarship.

Anne Gatignol (Ph.D., 1988) is a senior investigator at the Lady Davis Institute for Medical Research and an associate professor at the McGill University Departments of Experimental Medicine and Microbiology-Immunology, Montréal, Québec, Canada. She obtained her Ph.D. from the University of Toulouse, France. She did her postdoctoral training at George Washington University, Washington, DC, and the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, where she originally cloned the cDNA coding for TRBP. She started as an independent researcher of the Institut National de la Santé et de la Recherche Médicale at Cochin Institute, Paris, France, and moved to the Lady Davis Institute and McGill University, Montréal, Québec, Canada. She works on virus-cell interactions in the regulation of HIV replication. Her projects relate to understanding the role of ribonucleoprotein complexes during HIV replication, the interplay between RNA interference and viruses, as well as the use of RNA-based compounds against HIV.
REFERENCES
- 1. Arnaud N, et al. 2010. Hepatitis C virus controls interferon production through PKR activation. PLoS One 5:e10575 doi:10.1371/journal.pone.0010575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bannwarth S, Gatignol A. 2005. HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Curr. HIV Res. 3:61–71 [DOI] [PubMed] [Google Scholar]
- 3. Bannwarth S, et al. 2006. Cell-specific regulation of TRBP1 promoter by NF-Y transcription factor in lymphocytes and astrocytes. J. Mol. Biol. 355:898–910 [DOI] [PubMed] [Google Scholar]
- 4. Bannwarth S, et al. 2001. Organization of the human tarbp2 gene reveals two promoters that are repressed in an astrocytic cell line. J. Biol. Chem. 276:48803–48813 [DOI] [PubMed] [Google Scholar]
- 5. Barber GN. 2009. The NFAR's (nuclear factors associated with dsRNA): evolutionarily conserved members of the dsRNA binding protein family. RNA Biol. 6:35–39 [DOI] [PubMed] [Google Scholar]
- 6. Bass BL, Hurst SR, Singer JD. 1994. Binding properties of newly identified Xenopus proteins containing dsRNA-binding motifs. Curr. Biol. 4:301–314 [DOI] [PubMed] [Google Scholar]
- 7. Battisti PL, et al. 2003. Additive activity between the trans-activation response RNA-binding protein, TRBP2, and cyclin T1 on HIV type 1 expression and viral production in murine cells. AIDS Res. Hum. Retroviruses 19:767–778 [DOI] [PubMed] [Google Scholar]
- 8. Benkirane M, et al. 1997. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 16:611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bennasser Y, Le SY, Benkirane M, Jeang KT. 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22:607–619 [DOI] [PubMed] [Google Scholar]
- 10. Bennasser Y, Yeung ML, Jeang KT. 2006. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J. Biol. Chem. 281:27674–27678 [DOI] [PubMed] [Google Scholar]
- 11. Bennett RL, Blalock WL, May WS. 2004. Serine 18 phosphorylation of RAX, the PKR activator, is required for PKR activation and consequent translation inhibition. J. Biol. Chem. 279:42687–42693 [DOI] [PubMed] [Google Scholar]
- 12. Blair ED, et al. 1995. Expression of TAR RNA-binding protein in baculovirus and co-immunoprecipitation with insect cell protein kinase. J. Biomed. Sci. 2:322–329 [DOI] [PubMed] [Google Scholar]
- 13. Blalock WL, Bavelloni A, Piazzi M, Faenza I, Cocco L. 2010. A role for PKR in hematologic malignancies. J. Cell. Physiol. 223:572–591 [DOI] [PubMed] [Google Scholar]
- 14. Blanchard D, et al. 2011. On the nature of in vivo requirements for rde-4 in RNAi and developmental pathways in C. elegans. RNA Biol. 8:458–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bourikas D, Stoeckli ET. 2003. New tools for gene manipulation in chicken embryos. Oligonucleotides 13:411–419 [DOI] [PubMed] [Google Scholar]
- 16. Braun RE. 2000. Temporal control of protein synthesis during spermatogenesis. Int. J. Androl. 23(Suppl 2):92–94 [DOI] [PubMed] [Google Scholar]
- 17. Bueno MJ, Malumbres M. 2011. MicroRNAs and the cell cycle. Biochim. Biophys. Acta 1812:592–601 [DOI] [PubMed] [Google Scholar]
- 18. Caballero OL, Chen YT. 2009. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 100:2014–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Castanotto D, et al. 2007. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 35:5154–5164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. 2010. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J. Mol. Biol. 404:392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang KY, Ramos A. 2005. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 272:2109–2117 [DOI] [PubMed] [Google Scholar]
- 22. Chendrimada TP, et al. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436:740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chi YH, Semmes OJ, Jeang KT. 2011. A proteomic study of TAR-RNA binding protein (TRBP)-associated factors. Cell Biosci. 1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Christensen HS, et al. 2007. Small interfering RNAs against the TAR RNA binding protein, TRBP, a Dicer cofactor, inhibit human immunodeficiency virus type 1 long terminal repeat expression and viral production. J. Virol. 81:5121–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Clerzius G, et al. 2009. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J. Virol. 83:10119–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clerzius G, Gélinas JF, Gatignol A. 2011. Multiple levels of PKR inhibition during HIV-1 replication. Rev. Med. Virol. 21:42–53 [DOI] [PubMed] [Google Scholar]
- 27. Cosentino GP, et al. 1995. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc. Natl. Acad. Sci. U. S. A. 92:9445–9449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Daher A, et al. 2009. TRBP control of PACT-induced phosphorylation of protein kinase R is reversed by stress. Mol. Cell. Biol. 29:254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Daher A, et al. 2001. Two dimerization domains in the trans-activation response RNA-binding protein (TRBP) individually reverse the protein kinase R inhibition of HIV-1 long terminal repeat expression. J. Biol. Chem. 276:33899–33905 [DOI] [PubMed] [Google Scholar]
- 30. Daniels SM, et al. 2009. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol. Biol. 10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Daviet L, et al. 2000. Analysis of a binding difference between the two dsRNA-binding domains in TRBP reveals the modular function of a KR-helix motif. Eur. J. Biochem. 267:2419–2431 [DOI] [PubMed] [Google Scholar]
- 32. Dehal P, et al. 2002. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298:2157–2167 [DOI] [PubMed] [Google Scholar]
- 33. Desper R, Gascuel O. 2004. Theoretical foundation of the balanced minimum evolution method of phylogenetic inference and its relationship to weighted least-squares tree fitting. Mol. Biol. Evol. 21:587–598 [DOI] [PubMed] [Google Scholar]
- 34. Dimitrova DI, et al. 2005. Lentivirus-mediated transduction of PKR into CD34(+) hematopoietic stem cells inhibits HIV-1 replication in differentiated T cell progeny. J. Interferon Cytokine Res. 25:345–360 [DOI] [PubMed] [Google Scholar]
- 35. Donzeau M, Winnacker EL, Meisterernst M. 1997. Specific repression of Tax trans-activation by TAR RNA-binding protein TRBP. J. Virol. 71:2628–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dorin D, et al. 2003. The TAR RNA-binding protein, TRBP, stimulates the expression of TAR-containing RNAs in vitro and in vivo independently of its ability to inhibit the dsRNA-dependent kinase PKR. J. Biol. Chem. 278:4440–4448 [DOI] [PubMed] [Google Scholar]
- 37. Doyle M, Jantsch MF. 2002. New and old roles of the double-stranded RNA-binding domain. J. Struct. Biol. 140:147–153 [DOI] [PubMed] [Google Scholar]
- 38. Duarte M, et al. 2000. Characterization of TRBP1 and TRBP2. Stable stem-loop structure at the 5′ end of TRBP2 mRNA resembles HIV-1 TAR and is not found in its processed pseudogene. J. Biomed. Sci. 7:494–506 [DOI] [PubMed] [Google Scholar]
- 39. Eekels JJ, Geerts D, Jeeninga RE, Berkhout B. 2011. Long-term inhibition of HIV-1 replication with RNA interference against cellular co-factors. Antiviral Res. 89:43–53 [DOI] [PubMed] [Google Scholar]
- 40. Erard M, Barker DG, Amalric F, Jeang KT, Gatignol A. 1998. An Arg/Lys-rich core peptide mimics TRBP binding to the HIV-1 TAR RNA upper-stem/loop. J. Mol. Biol. 279:1085–1099 [DOI] [PubMed] [Google Scholar]
- 41. Fabian MR, Sonenberg N, Filipowicz W. 2010. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79:351–379 [DOI] [PubMed] [Google Scholar]
- 42. Fabozzi G, Nabel CS, Dolan MA, Sullivan NJ. 2011. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J. Virol. 85:2512–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fire A, et al. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811 [DOI] [PubMed] [Google Scholar]
- 44. Förstemann K, et al. 2005. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 3:e236 doi:10.1371/journal.pbio.0030236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fu X, Xue C, Huang Y, Xie Y, Li Y. 2010. The activity and expression of microRNAs in prostate cancers. Mol. Biosyst. 6:2561–2572 [DOI] [PubMed] [Google Scholar]
- 46. Gale M, Jr, Tan SL, Katze MG. 2000. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 64:239–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garaigorta U, Chisari FV. 2009. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe 6:513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. García MA, et al. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70:1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. García MA, Meurs EF, Esteban M. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89:799–811 [DOI] [PubMed] [Google Scholar]
- 50. Garre P, Perez-Segura P, Diaz-Rubio E, Caldes T, de la Hoya M. 2010. Reassessing the TARBP2 mutation rate in hereditary nonpolyposis colorectal cancer. Nat. Genet. 42:817–818 [DOI] [PubMed] [Google Scholar]
- 51. Gatignol A, Buckler C, Jeang KT. 1993. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and Drosophila staufen. Mol. Cell. Biol. 13:2193–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gatignol A, Buckler-White A, Berkhout B, Jeang KT. 1991. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science 251:1597–1600 [DOI] [PubMed] [Google Scholar]
- 53. Gatignol A, Duarte M, Daviet L, Chang YN, Jeang KT. 1996. Sequential steps in Tat trans-activation of HIV-1 mediated through cellular DNA, RNA, and protein binding factors. Gene Expr. 5:217–228 [PMC free article] [PubMed] [Google Scholar]
- 54. Gatignol A, Jeang K-T. 1994. Expression cloning of genes encoding RNA binding proteins, p 18–28 In Adolph KW. (ed), Methods in molecular genetics: molecular virology techniques, part A, vol 4 Academic Press, San Diego, CA [Google Scholar]
- 55. Gatignol A, Lainé S, Clerzius G. 2005. Dual role of TRBP in HIV replication and RNA interference: viral diversion of a cellular pathway or evasion from antiviral immunity? Retrovirology 2:65 doi:10.1186/1742-4690-2-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gélinas JF, Clerzius G, Shaw E, Gatignol A. 2011. Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA-activated protein kinase. J. Virol. 85:8460–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gorry PR, et al. 1999. Diminished production of human immunodeficiency virus type 1 in astrocytes results from inefficient translation of gag, env, and nef mRNAs despite efficient expression of Tat and Rev. J. Virol. 73:352–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gorry PR, et al. 2003. Astrocyte infection by HIV-1: mechanisms of restricted virus replication, and role in the pathogenesis of HIV-1-associated dementia. Curr. HIV Res. 1:463–473 [DOI] [PubMed] [Google Scholar]
- 59. Gredell JA, Dittmer MJ, Wu M, Chan C, Walton SP. 2010. Recognition of siRNA asymmetry by TAR RNA binding protein. Biochemistry 49:3148–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123:631–640 [DOI] [PubMed] [Google Scholar]
- 61. Grishin NV. 1995. Estimation of the number of amino acid substitutions per site when the substitution rate varies among sites. J. Mol. Evol. 41:675–679 [DOI] [PubMed] [Google Scholar]
- 62. Grundhoff A, Sullivan CS. 2011. Virus-encoded microRNAs. Virology 411:325–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gu S, et al. 2011. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proc. Natl. Acad. Sci. U. S. A. 108:9208–9213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gupta V, Huang X, Patel RC. 2003. The carboxy-terminal, M3 motifs of PACT and TRBP have opposite effects on PKR activity. Virology 315:283–291 [DOI] [PubMed] [Google Scholar]
- 65. Haase AD, et al. 2005. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 6:961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hitti EG, Sallacz NB, Schoft VK, Jantsch MF. 2004. Oligomerization activity of a double-stranded RNA-binding domain. FEBS Lett. 574:25–30 [DOI] [PubMed] [Google Scholar]
- 67. Hock J, Meister G. 2008. The Argonaute protein family. Genome Biol. 9:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Huang X, Hutchins B, Patel RC. 2002. The C-terminal, third conserved motif of the protein activator PACT plays an essential role in the activation of double-stranded-RNA-dependent protein kinase (PKR). Biochem. J. 366:175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hutvagner G, Simard MJ. 2008. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9:22–32 [DOI] [PubMed] [Google Scholar]
- 70. Ito T, Yang M, May WS. 1999. RAX, a cellular activator for double-stranded RNA-dependent protein kinase during stress signaling. J. Biol. Chem. 274:15427–15432 [DOI] [PubMed] [Google Scholar]
- 71. Jaskiewicz L, Filipowicz W. 2008. Role of Dicer in posttranscriptional RNA silencing. Curr. Top. Microbiol. Immunol. 320:77–97 [DOI] [PubMed] [Google Scholar]
- 72. Jha BK, et al. 2011. Inhibition of RNase L and RNA-dependent protein kinase (PKR) by sunitinib impairs antiviral innate immunity. J. Biol. Chem. 286:26319–26326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]
- 74. Ketting RF. 2011. The many faces of RNAi. Dev. Cell 20:148–161 [DOI] [PubMed] [Google Scholar]
- 75. Kharrat A, Macias MJ, Gibson TJ, Nilges M, Pastore A. 1995. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 14:3572–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim DH, Villeneuve LM, Morris KV, Rossi JJ. 2006. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 13:793–797 [DOI] [PubMed] [Google Scholar]
- 77. Kim MS, et al. 2010. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. J. Pathol. 221:139–146 [DOI] [PubMed] [Google Scholar]
- 78. Kini HK, Walton SP. 2009. Effect of siRNA terminal mismatches on TRBP and Dicer binding and silencing efficacy. FEBS J. 276:6576–6585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kok KH, Ng MH, Ching YP, Jin DY. 2007. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J. Biol. Chem. 282:17649–17657 [DOI] [PubMed] [Google Scholar]
- 80. Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N. 1992. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science 257:1685–1689 [DOI] [PubMed] [Google Scholar]
- 81. Koscianska E, Starega-Roslan J, Krzyzosiak WJ. 2011. The role of Dicer protein partners in the processing of microRNA precursors. PLoS One 6:e28548 doi:10.1371/journal.pone.0028548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kozak CA, Gatignol A, Graham K, Jeang KT, McBride OW. 1995. Genetic mapping in human and mouse of the locus encoding TRBP, a protein that binds the TAR region of the human immunodeficiency virus (HIV-1). Genomics 25:66–72 [DOI] [PubMed] [Google Scholar]
- 83. Krol J, Loedige I, Filipowicz W. 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11:597–610 [DOI] [PubMed] [Google Scholar]
- 84. Laraki G, et al. 2008. Interactions between the double-stranded RNA-binding proteins TRBP and PACT define the Medipal domain that mediates protein-protein interactions. RNA Biol. 5:92–103 [DOI] [PubMed] [Google Scholar]
- 85. Lau PW, Potter CS, Carragher B, MacRae IJ. 2009. Structure of the human Dicer-TRBP complex by electron microscopy. Structure 17:1326–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lecellier CH, et al. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308:557–560 [DOI] [PubMed] [Google Scholar]
- 87. Lee JY, et al. 2004. Merlin, a tumor suppressor, interacts with transactivation-responsive RNA-binding protein and inhibits its oncogenic activity. J. Biol. Chem. 279:30265–30273 [DOI] [PubMed] [Google Scholar]
- 88. Lee JY, et al. 2006. Merlin facilitates ubiquitination and degradation of transactivation-responsive RNA-binding protein. Oncogene 25:1143–1152 [DOI] [PubMed] [Google Scholar]
- 89. Lee K, Fajardo MA, Braun RE. 1996. A testis cytoplasmic RNA-binding protein that has the properties of a translational repressor. Mol. Cell. Biol. 16:3023–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lee Y, et al. 2006. The role of PACT in the RNA silencing pathway. EMBO J. 25:522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Li S, Sen GC. 2003. PACT-mediated enhancement of reporter gene expression at the translational level. J. Interferon Cytokine Res. 23:689–697 [DOI] [PubMed] [Google Scholar]
- 92. Lima WF, et al. 2009. Binding and cleavage specificities of human Argonaute2. J. Biol. Chem. 284:26017–26028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liu Q, Paroo Z. 2010. Biochemical principles of small RNA pathways. Annu. Rev. Biochem. 79:295–319 [DOI] [PubMed] [Google Scholar]
- 94. Liu Q, et al. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301:1921–1925 [DOI] [PubMed] [Google Scholar]
- 95. Ma E, MacRae IJ, Kirsch JF, Doudna JA. 2008. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 380:237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Maas S, Kawahara Y, Tamburro KM, Nishikura K. 2006. A-to-I RNA editing and human disease. RNA Biol. 3:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. 2008. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. U. S. A. 105:512–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Mallanna SK, et al. 2006. Inhibition of anatid herpes virus-1 replication by small interfering RNAs in cell culture system. Virus Res. 115:192–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Maniataki E, Mourelatos Z. 2005. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 19:2979–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Melo S, et al. 2011. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc. Natl. Acad. Sci. U. S. A. 108:4394–4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Melo SA, et al. 2009. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat. Genet. 41:365–370 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 102. Meurs E, et al. 1990. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62:379–390 [DOI] [PubMed] [Google Scholar]
- 103. Meurs EF, Galabru J, Barber GN, Katze MG, Hovanessian AG. 1993. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. U. S. A. 90:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Mounir Z, Koromilas AE. 2010. Uncovering the PKR pathway's potential for treatment of tumors. Future Oncol. 6:643–645 [DOI] [PubMed] [Google Scholar]
- 105. Murphy D, Dancis B, Brown JR. 2008. The evolution of core proteins involved in microRNA biogenesis. BMC Evol. Biol. 8:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Nie Y, Hammond GL, Yang JH. 2007. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J. Virol. 81:917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Nishikura K. 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79:321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Noland CL, Ma E, Doudna JA. 2011. siRNA repositioning for guide strand selection by human Dicer complexes. Mol. Cell 43:110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ong CL, et al. 2005. Low TRBP levels support an innate human immunodeficiency virus type 1 resistance in astrocytes by enhancing the PKR antiviral response. J. Virol. 79:12763–12772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Papadopoulos JS, Agarwala R. 2007. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23:1073–1079 [DOI] [PubMed] [Google Scholar]
- 111. Pare JM, et al. 2009. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol. Biol. Cell 20:3273–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Park H, et al. 1994. TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc. Natl. Acad. Sci. U. S. A. 91:4713–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Parker GS, Maity TS, Bass BL. 2008. dsRNA binding properties of RDE-4 and TRBP reflect their distinct roles in RNAi. J. Mol. Biol. 384:967–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Paroo Z, Ye X, Chen S, Liu Q. 2009. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell 139:112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Patel CV, Handy I, Goldsmith T, Patel RC. 2000. PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J. Biol. Chem. 275:37993–37998 [DOI] [PubMed] [Google Scholar]
- 116. Patel RC, Sen GC. 1998. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 17:4379–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Perdiguero B, Esteban M. 2009. The interferon system and vaccinia virus evasion mechanisms. J. Interferon Cytokine Res. 29:581–598 [DOI] [PubMed] [Google Scholar]
- 118. Perron MP, Provost P. 2009. Protein components of the microRNA pathway and human diseases. Methods Mol. Biol. 487:369–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Peters GA, Hartmann R, Qin J, Sen GC. 2001. Modular structure of PACT: distinct domains for binding and activating PKR. Mol. Cell. Biol. 21:1908–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Peters GA, Li S, Sen GC. 2006. Phosphorylation of specific serine residues in the PKR activation domain of PACT is essential for its ability to mediate apoptosis. J. Biol. Chem. 281:35129–35136 [DOI] [PubMed] [Google Scholar]
- 121. Pindel A, Sadler A. 2011. The role of protein kinase R in the interferon response. J. Interferon Cytokine Res. 31:59–70 [DOI] [PubMed] [Google Scholar]
- 122. Rossi JJ. 2005. Mammalian Dicer finds a partner. EMBO Rep. 6:927–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Rouleau GA, et al. 1993. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature 363:515–521 [DOI] [PubMed] [Google Scholar]
- 124. Rowe TM, Rizzi M, Hirose K, Peters GA, Sen GC. 2006. A role of the double-stranded RNA-binding protein PACT in mouse ear development and hearing. Proc. Natl. Acad. Sci. U. S. A. 103:5823–5828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sadler AJ, Williams BR. 2007. Structure and function of the protein kinase R. Curr. Top. Microbiol. Immunol. 316:253–292 [DOI] [PubMed] [Google Scholar]
- 126. Sakurai K, et al. 2011. A role for human Dicer in pre-RISC loading of siRNAs. Nucleic Acids Res. 39:1510–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Sanghvi VR, Steel LF. 2011. The cellular TAR RNA binding protein, TRBP, promotes HIV-1 replication primarily by inhibiting the activation of double-stranded RNA-dependent kinase PKR. J. Virol. 85:12614–12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Sanghvi VR, Steel LF. 2011. A re-examination of global suppression of RNA interference by HIV-1. PLoS One 6:e17246 doi:10.1371/journal.pone.0017246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Saunders LR, Barber GN. 2003. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 17:961–983 [DOI] [PubMed] [Google Scholar]
- 130. Scoles DR. 2008. The merlin interacting proteins reveal multiple targets for NF2 therapy. Biochim. Biophys. Acta 1785:32–54 [DOI] [PubMed] [Google Scholar]
- 131. Shan G, et al. 2008. A small molecule enhances RNA interference and promotes microRNA processing. Nat. Biotechnol. 26:933–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Siffroi JP, et al. 2001. Expression of the TAR RNA binding protein in human testis. Mol. Hum. Reprod. 7:219–225 [DOI] [PubMed] [Google Scholar]
- 133. Singh M, Castillo D, Patel CV, Patel RC. 2011. Stress-induced phosphorylation of PACT reduces its interaction with TRBP and leads to PKR activation. Biochemistry 50:4550–4560 [DOI] [PubMed] [Google Scholar]
- 134. Soifer HS, et al. 2008. A role for the Dicer helicase domain in the processing of thermodynamically unstable hairpin RNAs. Nucleic Acids Res. 36:6511–6522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. St Johnston D, Brown NH, Gall JG, Jantsch M. 1992. A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. U. S. A. 89:10979–10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Suh N, Blelloch R. 2011. Small RNAs in early mammalian development: from gametes to gastrulation. Development 138:1653–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Suk K, et al. 2011. Reconstitution of human RNA interference in budding yeast. Nucleic Acids Res. 39:e43 doi:10.1093/nar/gkq1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Tabara H, Yigit E, Siomi H, Mello CC. 2002. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109:861–871 [DOI] [PubMed] [Google Scholar]
- 139. Tebit DM, Arts EJ. 2011. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect. Dis. 11:45–56 [DOI] [PubMed] [Google Scholar]
- 140. Trofatter JA, et al. 1993. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell 72:791–800 [DOI] [PubMed] [Google Scholar]
- 141. Vorburger SA, et al. 2005. Gene therapy with E2F-1 up-regulates the protein kinase PKR and inhibits growth of leiomyosarcoma in vivo. Mol. Cancer Ther. 4:1710–1716 [DOI] [PubMed] [Google Scholar]
- 142. Wang HW, et al. 2009. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 16:1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wang S, Chen AJ, Shi LJ, Zhao XF, Wang JX. 2012. TRBP and eIF6 homologue in Marsupenaeus japonicus play crucial roles in antiviral response. PLoS One 7:e30057 doi:10.1371/journal.pone.0030057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Wang S, Liu N, Chen AJ, Zhao XF, Wang JX. 2009. TRBP homolog interacts with eukaryotic initiation factor 6 (eIF6) in Fenneropenaeus chinensis. J. Immunol. 182:5250–5258 [DOI] [PubMed] [Google Scholar]
- 145. Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L. 1999. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell Biol. 19:2220–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Yamashita S, et al. 2011. Structures of the first and second double-stranded RNA-binding domains of human TAR RNA-binding protein. Protein Sci. 20:118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Zhong J, Edelhoff S, Disteche C, Braun RE. 1998. The gene encoding PRBP, the mouse homolog of human TRBP, maps to distal chromosome 15. Mamm. Genome 9:413–414 [DOI] [PubMed] [Google Scholar]
- 148. Zhong J, Peters AH, Lee K, Braun RE. 1999. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat. Genet. 22:171–174 [DOI] [PubMed] [Google Scholar]