Abstract
Summary: Diversity in adaptive responses is common within species and populations, especially when the heterogeneity of the frequently large populations found in environments is considered. By focusing on events in a single clonal population undergoing a single transition, we discuss how environmental cues and changes in growth rate initiate a multiplicity of adaptive pathways. Adaptation is a comprehensive process, and stochastic, regulatory, epigenetic, and mutational changes can contribute to fitness and overlap in timing and frequency. We identify culture history as a major determinant of both regulatory adaptations and microevolutionary change. Population history before a transition determines heterogeneities due to errors in translation, stochastic differences in regulation, the presence of aged, damaged, cheating, or dormant cells, and variations in intracellular metabolite or regulator concentrations. It matters whether bacteria come from dense, slow-growing, stressed, or structured states. Genotypic adaptations are history dependent due to variations in mutation supply, contingency gene changes, phase variation, lateral gene transfer, and genome amplifications. Phenotypic adaptations underpin genotypic changes in situations such as stress-induced mutagenesis or prophage induction or in biofilms to give a continuum of adaptive possibilities. Evolutionary selection additionally provides diverse adaptive outcomes in a single transition and generally does not result in single fitter types. The totality of heterogeneities in an adapting population increases the chance that at least some individuals meet immediate or future challenges. However, heterogeneity complicates the adaptomics of single transitions, and we propose that subpopulations will need to be integrated into future population biology and systems biology predictions of bacterial behavior.
INTRODUCTION
The adaptive success of bacteria needs explanation. Unfortunately from a reviewer's point of view, the evolutionary advantages of bacteria are diffusely distributed over many structural, metabolic, regulatory, microevolutionary, and genome rearrangement features as well as advantages of growth rate and population size. It is too large a task to integrate all the mechanistic ingredients of success into a single review, but at the same time we wish to reverse the extreme reductionist approach in which individual regulatory and mutational processes of bacteria are commonly considered in isolation. In addition, it is essential to reflect on the full complexity of adaptive possibilities as a note of realism when considering the emerging notion that evolution is predictable at an organismic systems level (251, 308). As a simple test of the science of bacterial adaptation, we wish to consider a single question: what are the ingredients of adaptation of a single bacterial population to a single transition between two environments? To our knowledge, there are no previous attempts to bring together all the population-level changes contributing to this kind of common transition. Given the current trend toward a systems approach to biological processes, it is important to start to integrate regulatory and mutational adaptation as well as to look at the effects of the inherent heterogeneity of these processes. Indeed the “adaptomic” description of regulatory and mutational responses to a single environmental change is a sensible step toward modeling the outcomes of multistep evolutionary change.
Even considering a single environmental challenge and a single population, there need to be exclusions to limit the scope of the discussion. We exclude very rapid structural, metabolic, or physiological changes (such as ion fluxes or changes in enzyme activity) at one end of the time scale for adaptation. Limiting our discussion to a single transition also allows us to ignore ecological specialization and speciation at the other extreme of the adaptational time scale. Thus, we discuss what can be described as regulatory and microevolutionary changes in a single, generic environmental transition. Finally, we also exclude the added complexities of multispecies communities and deal with a monoclonal population. This is less of an exclusion than it may seem, in that the tremendous level of heterogeneity within a single clonal population is one of the main themes in this review.
Three justifications can be offered for discussing regulatory and mutational responses together in a single environmental transition. First, one of the important trends in recent years has been a growing body of evidence linking regulatory and genomic adaptations. Examples include the cellular regulation of DNA repair processes leading to mutation rate variation and effects on evolvability (102, 112, 367) and the stochastic and deterministic effects leading to persister cells (179). Epistatic effects on regulatory circuits and their modulation during mutational adaptation are also increasingly evident as a component of fitness (66, 264). Indeed, the current trend indicates that regulatory adaptations inform the bacterium as to the genomic change that maximizes the chances of adaptation and evolutionary success.
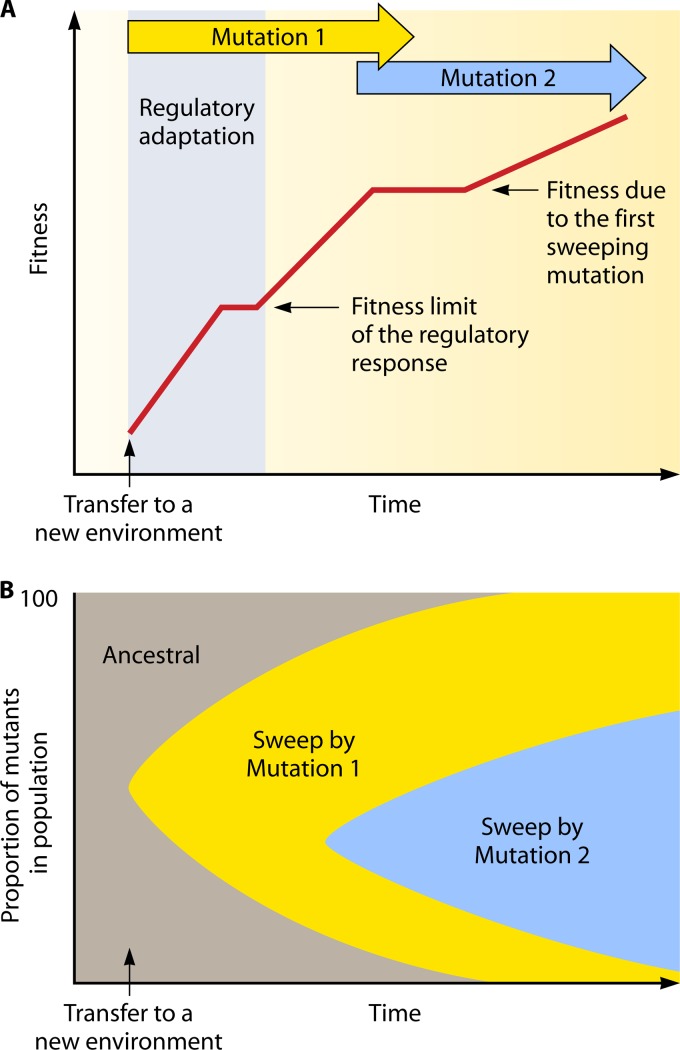
Second, a temporal perspective of adaptation in bacteria indicates the overlapping time frames of regulatory and mutational adaptations, as shown in Fig. 1. The synthesis of new transcripts and average-size proteins can be achieved within 30 s, and the time taken for induction of a large protein such as β-galactosidase is about 2.5 to 3 min (30). However, more complex developmental adaptations such as sporulation take considerably longer (up to 7 to 8 h) (70). Likewise, biofilm formation can be observed within an hour, but mature biofilms can take >24 h (214). Over such time scales, mutational selection can become significant. A mutant present in a large population with a growth advantage in the new environment can become highly enriched depending on its fitness increase (Fig. 1B). Given a large enough fitness difference, mutations in a gene can sweep a population within 24 to 48 h, and this has been demonstrated with regulatory mutations in experimental populations (97). In even more extreme selections, such as for high-level antibiotic resistance, survivors will be solely from the selection and multiplication of resistant mutants over all phases of adaptation. Resistant mutants, depending on their growth rate and other environmental constraints, can become large populations within 24 h.
Fig 1.
(A) Fitness increases through regulation and mutational processes in a population, relative to the prestressed state, upon a transition to a new environment. “Fitness” in this context is a qualitative measure of adaptation, and true competitive fitness in stress-induced versus uninduced cells is not available or normally measured. (B) The underlying, successive mutational sweeps result in ancestral types (gray) being replaced in the population by initially rare cells containing mutation 1, resulting in increased fitness to above that possible with the ancestor. Mutation 2 is enriched with continued selection and further increases population fitness.
Third, considering regulation and mutation together is consistent with historical views and definitions of adaptation. For bacteria, Ryan (286) noted 60 years ago: “Bacteria are remarkably plastic in their ability to undergo satisfactory adjustments in new environments. These adjustments are adaptations in the original biological sense of the word whether they are inherited or not, and irrespective of the underlying mechanisms.” Adaptation is thus a comprehensive process, and regulatory adaptations, epigenetic events, and inherited, mutational adaptations all contribute to fitness. We thus adopt the definition of “adaptation” to be “the process of change by which an organism or species becomes better suited to its environment” (however, a range of definitions of this much-discussed term exists [212]).
A further important consideration based on Fig. 1 is that the level of fitness obtained in an environmental transition through a regulated response may not be as high as that potentially available in a mutant present in the population. This was already demonstrated in early studies with the classic lac system, in that constitutive mutants outcompete wild-type, inducible bacteria in the presence of limiting lactose, resulting in the enrichment of mutants with better β-galactosidase levels than are available from induction; subsequently, gene dosage changes through amplification further increase enzyme levels (240). Hence, as suggested by Fig. 1, the selective advantage through mutation can be an important contributor to adaptation in populations, including in multiple steps if the selection is maintained. Altogether, these considerations suggest that the fitness increase available from a regulatory change fixed by the genetic blueprint is lower than that available from mutational change. Hence, regulation and microevolution through regulatory mutations provide a continuum of possibilities in adaptation (95).
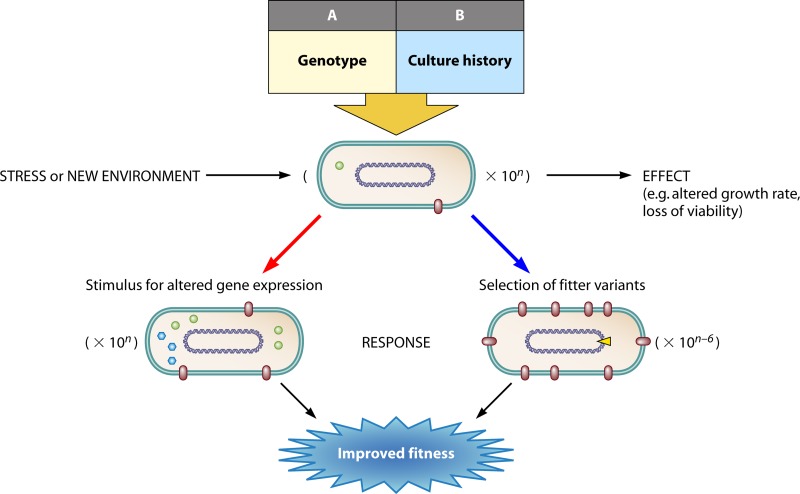
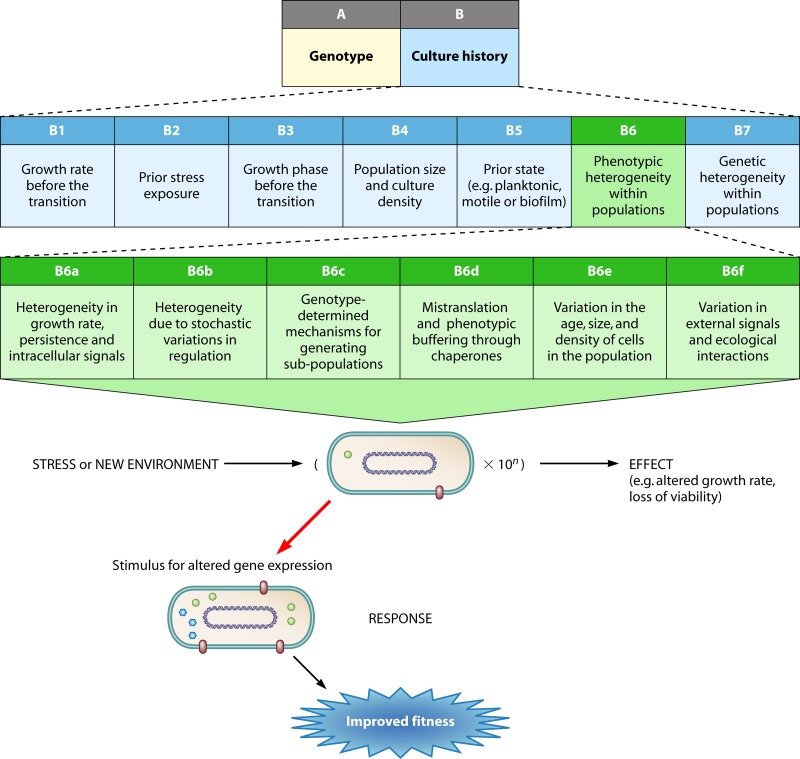
The overall scope of this review is outlined schematically in Fig. 2, which attempts to integrate bacterial adaptive pathways into a generic program during a single environmental transition, at the level of a bacterial population. The new environment in Fig. 2 can either reduce or increase the growth rate; in either case, the response in the new environment will change patterns of gene expression as well as the selection environment for fitter mutants. The regulatory response will be elicited in most but not all of the 10n cells in a population. The heterogeneities in the population arise from many sources (as described below) and complicate the quantitative prediction of the behavior of an organism such as Escherichia coli when it is transferred from one environment to another. Effects on both growth rate and viability should be calculable, but the effect of a transition is dependent on many variables in boxes A and B (Fig. 2), as further explained in Importance of the Genotype in Determining the Nature of Adaptive Responses and Importance of Culture History in Determining the Nature of Adaptive Responses below. We are far from being able to quantitatively estimate the contributions of all the inputs in Fig. 2 to 5. These include mutational adaptations, and mutational heterogeneities will certainly be dependent on the size of n in Fig. 2. This review will try to define the extent of this multifactorial problem. Complicating factors include not only the extensive population heterogeneity but also the overlapping interactions between mutational and regulatory responses that enrich the possibilities in adaptive pathways. The inputs A and B affecting the response of a bacterial population to an environmental transition are first discussed in turn.
Fig 2.
Adaptations from a prior state upon a transition to a new environment for bacteria (rods with DNA circle and cell components) through regulation (red arrow) and mutational processes (blue arrow). The number of bacteria in the population, 10n, nearly all undertake a regulatory transition, but the number gaining fitness through mutation is environment and function dependent, so the 10n − 6 shown can vary by orders of magnitude. The effects on cells depend on the genotype of the organism (A) and the history of the population (B), as discussed in the text.
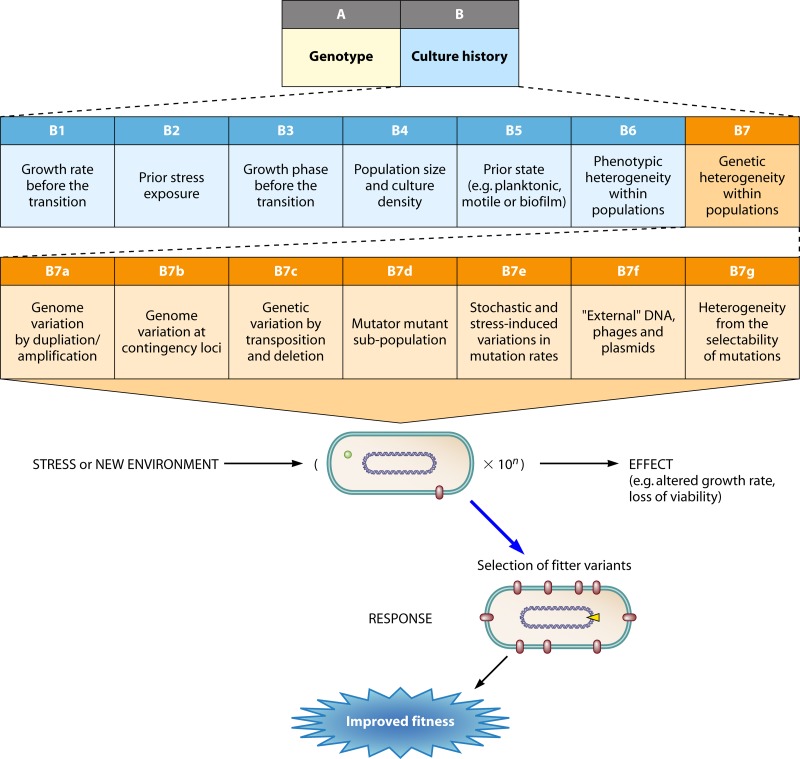
Fig 5.
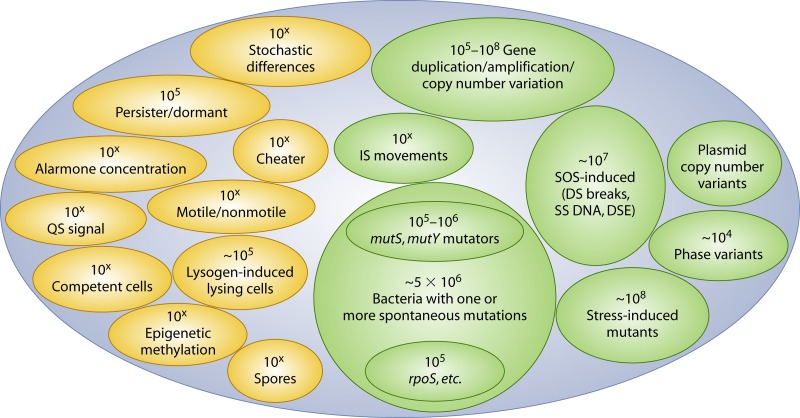
Aspects of genetic population heterogeneity relevant to adaptation, expanded upon from Fig. 3 in increasing detail. Each box is further discussed in the text. Other content and arrows are as in Fig. 2.
IMPORTANCE OF THE GENOTYPE IN DETERMINING THE NATURE OF ADAPTIVE RESPONSES
The genetic blueprint of an organism predetermines the adaptive pathways shown in Fig. 2 in two ways. The genome encodes the functional responses of the organism and how the genes are regulated. Bacterial genome sizes vary over a >10-fold range of DNA content because of various selection pressures (195), so their capacities to encode responses to environments also differ. In addition, the genome structure and the sequence itself influence the mutational possibilities in a chromosome.
The transcription factors and other regulatory components determine the response pattern of a bacterium, and these of course are genome encoded. Complex maps of the roles of the transcriptional regulators and their network organization are now available for some species, including E. coli (208). The size of the regulatory network varies from species to species and is related to genome size (219). Variation in regulation is evident not only from species to species, and the identity of genotypes in a population does not mean that transitions are identical in all of the 10n cells in a population (Fig. 2). The genome itself and the nature of the regulatory networks that it encodes create the scope for transcriptional noise and the possible bistable states in regulation (see “Heterogeneity due to stochastic variations in regulation” below for details). Phenotypic and regulatory variability within populations is thus influenced by several genome-determined processes associated with regulatory networks. Indeed, the ability to switch phenotypes within populations is under evolutionary selection and is also considered in detail below.
In considering regulation, we will focus largely on events and heterogeneities within populations of a single strain. Nevertheless, it is worth stressing that the inter- and intraspecies strain variations in regulation are extremely high according to recent data (52, 90, 151, 317). Although most of the genes involved in regulation are highly conserved, core elements in a genome, they are not necessarily operating in the same way, even in members of a species. The central elements in gene regulation, e.g., the concentrations of sigma factors and alarmones such as ppGpp, vary greatly under the same environmental conditions across a species (98, 304). The instability of such central elements of regulation is particularly evident within the lifetime of a clonal infection in a patient (177). Although variations are genotype specific, they do not always involve polymorphisms in the regulatory gene itself. The myriad of inputs into most central, high-level regulatory circuits allows many ways of influencing regulator levels. A prime example of this is with the E. coli sigma factor RpoS, which controls the expression of over 200 genes (357). The level of RpoS in the cell responds to external and internal stresses and is influenced by over 20 environmental signal inputs (142). The alternative input pathways controlling rpoS expression provide degenerate ways of affecting regulation, and indeed many alternative genes can change the particular regulatory circuit controlling RpoS levels (353). Even within a single adapting population, heterogeneities can arise, and regulatory mutations are the most frequent type in experimental evolution studies (52, 264, 353). Hence, it is not surprising that regulation across a species is not uniform.
The genotype-determined phenotypic effects of within-species variation are indeed profound, so one strain of E. coli can have a very different response to the same level of stress than another (24, 98) or a very different growth rate or competitive ability in particular environments (157). Genomics efforts have highlighted the relatively small core genome and the relatively large pan-genome within a species such as E. coli (193, 348), so it is highly likely that some of the pan-genome differences will affect adaptive responses. Plasmids and prophages add further variability to genomic capabilities (see “External DNA, genetic exchange, phages, and plasmids” below).
On the mutational side, the possibilities for change in a particular genome are influenced by the presence of genomic duplications, unstable elements such as microsatellites, transposable elements, repeats, and sequences contributing to high mutation rates, as discussed in “Genetic Heterogeneity within Populations” below. The great variation in the number and position of insertion sequence (IS) elements within a species must mean that the probability of particular mutations and genome rearrangements is strain specific. For example, the tendency to frequently acquire a mutator mutation in one laboratory strain of E. coli K-12 is linked to a specific IS5 movement. E. coli B strains do not have an IS at the same position and so do not generate mutators in the same way or with the same high frequency (110). Short repeats and the possibility of slipped-strand errors also differ between genomes (341). Such evolved strain differences could have considerable effects on mutation availability at various positions in the genome within strains. For example, the presence of a run of 8 G bases can potentiate a frameshift in a particular gene (109). Within a single population, we can ignore intraspecies differences, but we will point out where genome properties affect adaptive pathways. These influences are discussed in “Genetic Heterogeneity within Populations” below at a population level, in which mutations result in subpopulations with altered properties and an altered capacity for the transitions shown in Fig. 2.
The genotype also determines the mobility of genomic DNA and how recombinational mechanisms contribute to adaptation. This goes beyond the potential for rearrangements allowed by transposable elements and integrons, especially if the bacterium contains sex determinants such as the F factor or lysogenic phages that can facilitate cell-to-cell transfer of DNA. We consider the role of lateral gene transfer (LGT) in adaptation in “Genetic Heterogeneity within Populations” below.
For most examples, the genome that we will discuss is that of the lab strains of E. coli, simply because most is known about it. Nevertheless, we will include adaptations such as competence or sporulation in Bacillus subtilis or biofilms in Pseudomonas aeruginosa where these provide better examples of adaptation or its intricacies. Of course readers will have other, possibly better examples of culture effects and population heterogeneity, and we apologize for such omissions.
IMPORTANCE OF CULTURE HISTORY IN DETERMINING THE NATURE OF ADAPTIVE RESPONSES
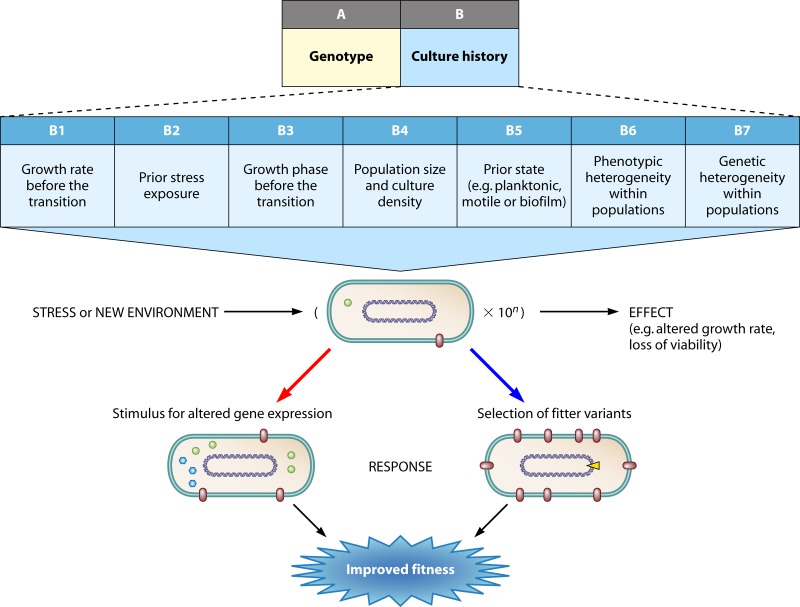
Besides the genotype, the immediate culture history of a population is an inevitable determinant of the overall effect of an environmental transition. In Fig. 3, we expand culture history effects into seven areas that can strongly influence the scope of the transition shown in Fig. 2. Perhaps surprisingly, the history of the culture is rarely discussed systematically where it comes to understanding adaptation and regulatory responses. Indeed, in laboratory studies when a simple, reproducible baseline is generally preferred, studies of adaptation try to eliminate variations due to history. A baseline population is often one that grows exponentially under near-optimal conditions and excess nutrient levels. What is known about transcriptional changes in model organisms such as E. coli is generally based on stresses and transitions applied to exponentially growing bacteria. This of course is extremely artificial, and in nature there is no single baseline; in each environmental transition, the baseline is that provided by the history of a population. In any case, there is extensive evidence that in bacteria grown in the same medium but at different growth rates (e.g., in chemostats) or into different growth states (e.g., exponential- versus stationary-phase or biofilm bacteria), the response to challenges differs greatly. The importance of culture history is widely recognized in food microbiology, where predictive methods for processing need to take into account the variations in bacterial properties, especially stress responses, due to prior culture conditions. It is therefore evident that we cannot hope to predict the adaptomics of a population and the effect of the same stress on the same organism unless we know how it was grown earlier. Therefore, in this section and in Fig. 3, we expand on seven different aspects of history that could or do affect adaptive responses.
Fig 3.
Aspects of culture history relevant to adaptation, expanded upon from Fig. 2 in increasing detail. Each box is further discussed in the text. Other content and arrows are as in Fig. 2.
Growth Rate before the Transition
Many constituents of bacterial cells as well as the cell size are strongly dependent on growth conditions (148, 196). For E. coli, the macromolecular composition is dependent more on the growth rate than on the nature of the growth medium used; media that support the same growth rate result in bacteria with similar macromolecular contents (34). Growth rate effects are not limited to batch cultures containing different substrates but are also found in nutrient-limited chemostats running at different dilution rates but with the same limiting substrate (94). In both situations, growth rate is a major influence on bacterial gene expression and regulation (96, 146, 159).
An underestimated consequence of bacteria having constituents that vary with growth rate is that their adaptive responses have different starting points upon a transition. Indeed, slow-growing bacteria (with doubling times of 3 to 4 h or more) have elevated levels of the alarmone ppGpp and the sigma factor RpoS, stimulating the general stress response as well as other stress proteins dependent on ppGpp (236, 325). The cellular content of stress protectants such as trehalose (whose synthesis is RpoS dependent) is much elevated during slow growth (336), so transitions into new environments with such bacteria are buffered by partly or highly expressed stress responses.
The ppGpp-controlled stringent response occurs during slow growth or when cells experience a nutritional downshift, e.g., become starved for an amino acid(s), nitrogen, or carbon, or when cells encounter various toxic agents (45). Elevated levels of ppGpp interact with the β and β′ subunits of RNA polymerase, affecting promoter binding preference and leading to the inhibition of transcription of stringently repressed genes, and induce transcription of stringently expressed genes (8). This results in a reduction of the synthesis of stable RNA (tRNA and rRNA), which restricts protein synthesis. This is accompanied by repression of other genes involved in normal growth and proliferation and induction of genes to relieve the stress, such as amino acid biosynthetic genes (for a review, see reference 197). rpoS is one of the genes that is induced by the stringent response, and many RpoS-controlled genes also require ppGpp for induction, so the general stress response and stringent response overlap each other (165, 332). Among the many changes that ppGpp plus RpoS elicits is a reduction in outer membrane permeability (185), probably accounting for antibiotic susceptibility of bacteria being a function of growth rate (39).
Making high levels of RpoS and ppGpp entails a cost to bacteria, because it diverts resources of the cell used in vegetative growth (304). Natural isolates of E. coli vary in growth rate (215), and part of the reason is that there is as much as a 10-fold variation in RpoS and/or ppGpp content between different E. coli strains growing under identical conditions. Individual populations are often a mix of rpoS mutants and rpoS wild-type subpopulations (98, 157). The levels and coexistence of rpoS mutant/wild-type subpopulations can reflect a trade-off between stress resistance (enhanced by RpoS) and nutritional competency (inhibited by RpoS), referred to as self-preservation and nutritional competence (SPaNC) balancing (95). Within a single genotype, the setting of the SPaNC balance is dependent on the growth rate and the culture history of the population (95). Therefore, depending on the strain type and culture history, there will be heterogeneity of RpoS levels within a population which could result in population heterogeneity in RpoS-dependent stress resistance.
Mutational processes are also affected by growth rate and stasis because of RpoS-induced mutagenesis (102, 112). In the same medium, a 6-fold decrease in growth rate in glucose-limited chemostats results in a 30-fold increase in the frequency of neutral mutations, with slow-growing cells having higher mutation rates (239). This is likely to be a regulated difference, and the high level of RpoS can induce error-prone DNA polymerase IV (166). In addition, it was shown recently that among starving cells, a small proportion exhibit heightened genomic instability during which multiple chromosomal structural changes can occur anywhere in the genome (181) (see “Genetic Heterogeneity within Populations” below).
It can also be envisaged that gene dosage varying with growth rate may affect mutational effects and DNA repair. Chromosomal copy number is growth rate dependent (34), with more copies at high growth rates. Although not tested, this could allow more opportunities for mutation elimination through homologous recombination under optimal growth conditions. These aspects will be expanded upon in “Genetic Heterogeneity within Populations” below when we discuss genetic variation.
The final growth rate effect on genomes that we consider is a question not of DNA change but of evolutionary selection acting on genomes. Adaptation involves not only genetic variation, which is growth rate dependent, but also selection, which is equally growth rate affected. There is mounting evidence that a mutation that is beneficial at one growth rate can be actually deleterious at another (198b, 239). This is again due to the different metabolic-regulatory contexts that a mutation meets in slow- and fast-growing bacteria. The fitness effects of many mutations may indeed turn out to be growth rate affected when further investigated, as also suggested by another example (53).
Prior Stress Exposure
Another aspect of bacterial history that affects the response of bacteria (Fig. 2) is prior exposure to a stress. This is partly for the same reasons to those discussed for growth rate effects in “Growth Rate before the Transition” above. In E. coli, the general stress response regulated by the sigma factor RpoS and ppGpp can be stimulated not only by growth rate but also by sublethal levels of acids, high osmolarity, or suboptimal temperatures (142). Induction of the general stress response results in global changes in metabolism that greatly increase the bacterium's resistance not only to the experienced stress but also to other stresses; e.g., exposure to carbon limitation increases resistance to oxidative stress, high temperature, and low pH (for a review, see reference 17). Induction by individual stresses affords cross-protection to other challenging transitions beyond the inducing stress. The best examples of cross-protection effects come from food microbiology studies on bacterial inactivation (170, 282, 359).
A second aspect of stress effects in the history of a culture is that, commonly, the cellular response may involve a stress-specific component as well as the general stress response. This is illustrated by acid tolerance in E. coli, where the defenses used to overcome sublethal levels of acid involve not only RpoS-dependent effects but also additional, acid-specific responses regulated in an RpoS-independent manner (101). This kind of dual regulatory response extends to other stresses, e.g., heat stress and oxidative stress (312). It is therefore an important point that the level of environmental resistance during a new transition as in Fig. 2 is a function of history regarding either specific or general responses or both.
The oxygen content of an environment and transitions from aerobiosis to anaerobiosis (and vice versa) can have a significant effect on adaptation. Oxygen presence potentiates oxygen detoxification mechanisms and elevates catalases and hydroperoxidases (254), and mutational processes are also affected differentially by aspects of aerobic/anaerobic metabolism (299). Hence, a population changing from an anaerobic to an aerobic environment may be more stressed by the same transition than one coming from aerobiosis.
As considered in more mechanistic detail below, high levels of stress exposure can change expression of the SOS response, DNA repair systems, and error-prone DNA polymerases and so can change mutation rates (see “Genetic Heterogeneity within Populations” below). If the stress is brought about by the presence of DNA-damaging chemicals or irradiation, the proportion of mutants in a population may increase and can potentially give different adaptive outcomes upon entry into a new environment because of the increased mutation supply in a population.
Growth Phase before the Transition
Improving survival at the cost of vegetative growth is a common bacterial strategy in response to adverse environments. Indeed, the whole advantage of differentiating into stationary growth phase or toward resistant forms (e.g., spores) is to induce a more generally protective state (160, 211, 345). Several reviews have discussed the properties of stationary-phase bacteria and the regulatory changes involved upon entry into, and maintenance in, stationary phase (100, 143, 160, 187). Microarray studies have revealed the extent of changes between exponential- and stationary-phase bacteria that induce the general stress response (256, 357) as well as other ppGpp-related responses (46). Given that approximately 10% of the genes in E. coli are differently regulated in stationary phase and that many of the genes expressed are involved in cell protection, it is evident why resistance to many stresses, such as acid stress or high osmolarity, is very different in exponential- and stationary-phase bacteria. Hence, the transition shown in Fig. 3 has a different baseline and result if exponentially growing or stationary-phase bacteria undertake a transition.
It is also possible to distinguish adaptive differences between early- and late-stationary-phase bacteria. During extended exposure to stationary phase, populations become more heterogeneous and complex. Diversity arises from incomplete sweeps by mutants that compete in the spent medium and have a growth advantage in stationary phase (GASP mutants) (100, 373). Some of these GASP mutants have mutations in global regulatory genes (rpoS or lrp), so the responses of these subpopulations are likely to be nonidentical if transferred to a new environment. Damaged cells are also a component of late-stationary-phase bacterial populations (241), so these can also be expected to respond differently to additional transitions.
In bacteria that can differentiate beyond stationary phase into spores, such as B. subtilis, the population heterogeneity is even more marked after starvation and induction of several coexisting responses and cell types (190). The simultaneous existence of spores, remaining vegetative bacteria, competent cells, toxin synthesis, and cannibalism gives rise to a mix of phenotypes (see “Genotype-determined mechanisms for generating multiple subpopulations” below); how such a mixture behaves in a new environmental transition is likely to be even more complex and is largely unexplored.
Genotypic stability in stationary phase also differ from that in exponential phase. Stationary phase is associated with altered regulation of DNA repair systems, most notably with reduced levels of MutSH (92) and reduced mismatch repair (MMR) (133, 134). GASP mutations are also stimulated by error-prone polymerase (372), which, as discussed in “Genetic Heterogeneity within Populations” below, is partly due to the higher levels of RpoS in stationary phase and elevated mutation rates due to stress-induced mutagenesis (SIM). Cell lysis and DNA release also occur in stationary phase, so the possibility of transformation is also potentially elevated in E. coli (249). Bacillus behavior upon starvation involves competence for DNA uptake, taking advantage of released DNA available from other lysed cells.
Population Size and Culture Density
Several of the determinants in adaptation are dependent on population size, so it matters whether a population undergoing an environmental transition is small (e.g., 103 to 104 cells) or large (108 to 1010 cells). The number of bacteria in a population determines the spontaneous mutation supply and thus the available range of mutations. The level of regulatory heterogeneity is also strongly affected by population size if dependent on the stochastic effects discussed in “Heterogeneity due to stochastic variations in regulation” below. A good example of how population size affects bacterial survival is that of a pathogen population inside a hostile host; large variation in the infectious dose of pathogens is a reflection of the importance of population size in overcoming host challenges.
Adaptations are in some cases impacted by intra- and interpopulation signaling in bacteria, and population density is an important factor in the local concentration and production of quorum signals (153, 315, 356, 362). A broad range of bacterial characteristics, including virulence, are affected by quorum sensing and signal molecules (169, 252), so it is highly likely that a population already subject to quorum signals will behave differently on moving to a new environment than a dilute, signal-less culture. Here again, a complicating factor is that heterogeneity occurs in the extent of the response to quorum signals in populations (259).
Rare beneficial mutations or particular gene duplications are unlikely to be present in small populations, and their fixation is also a function of population size. Adaptation in a small population has fewer sources of available variation, and mutator mutations are of particular secondary benefit in small populations (75). Hence, the same transition shown in Fig. 3 could result in different mutational possibilities with small or large populations of the same organism, simply because the right kind of variation is not present.
So far in this section we have been considering the population size before the environmental transition. If the transition involves a loss of viability (Fig. 2), then survivors may be a small proportion of the ancestral population. This can cause a bottleneck effect, and the full range of variation present in the prestress situation is not available in the survivors. The effective population size determined by the survivors in this scenario can affect the range of possible adaptive outcomes, especially with the rare genetic events considered in “Genetic Heterogeneity within Populations” below.
Prior State (e.g., Planktonic, Motile, or Biofilm)
Exponentially growing, fully mixed bacterial cultures are likely to be a rarity in nature. Bacteria not only frequently starve and enter stationary phase (as discussed above) but also often grow in an unmixed state subject to nutrient or stressor concentration gradients. In nonhomogenous environments, gradients are sensed by chemoreceptors and chemotaxis is a benefit, but motility is an expensive process (296). Motility is subject to regulation by many inputs from the environment (303), so the response shown in Fig. 3 may depend on the motile state of a population if swimming is an immediate benefit or cost in the new situation. Some bacteria hedge their bets by simultaneously producing both motile and sessile forms from the same genotype (154) (see “Phenotypic Variation within Populations” below).
Bacteria on solid surfaces can swarm and form aggregated cells based on intercellular signaling, so these prior states also set a different baseline for adaptation (294). A bacterium like P. aeruginosa is capable of three forms of motility: swimming, swarming, and twitching. Within swarming populations, because of complex signaling between members of a population, cells do not have common patterns of gene expression (333). In turn, this means that subpopulations in a swarming population have different baselines for further sudden changes of environment. Even more complex are bacterial aggregates that develop into mature biofilms with substructures and internal chemical gradients (191). In such scenarios, patterns of gene expression and cellular composition (especially in surface appendages and flagella) are widely different in subpopulations.
Biofilms show increased resistance to many types of environmental challenges and antibiotics (118). Several resistance mechanisms are involved in the same biofilm (198), and this is consistent with the view that a biofilm is not in a uniform physiological state (310). Antibiotic resistance of the biofilm subpopulations is aided by biofilm-inherent phenotypes, such as persister cells (see Phenotypic Variation within Populations” below), slow growth, and induction of rpoS-mediated stress responses (149). Biofilm subpopulations differ in gross growth characteristics as well (266). The main point here is that subpopulations in the history of a culture before a transition will need to be considered in the response to a transition. If the population in Fig. 3 comes from a biofilm rather than a homogeneous population, the responses to a new environment are inevitably going to be nonidentical in the subpopulations.
Phenotypic Variation within Populations
Importantly, most of our understanding of bacterial behavior is from bulk cultures, averaging results over large populations. The extensive descriptions of bacterial physiology, regulation, and metabolism in reference works such as Escherichia coli and Salmonella: Cellular and Molecular Biology (231) and most of our understanding of transcriptional regulatory networks in E. coli (15) are based on studied adaptations of averaged populations.
In the past 10 years, though, there has been an increasing interest in heterogeneity as an inherent feature of bacterial growth and behavior (69). As already touched on above, however, the finding of pure-culture heterogeneity is not new, and both growth rate and antibiotic susceptibility variations within populations are long known (26, 155). The kinetics of killing by stresses also pointed to heterogeneities in bacterial populations (31). Further evidence for culture heterogeneities has come from the application of flow cytometry, identifying four or more physiological states in some exponential-phase bacterial cultures (68, 355). With the advent of microfluidics and high-sensitivity detection methods, it is now possible to dissect population effects as happening in individual cells. What is revealed is the high level of noise in gene expression across the whole genome (323). Cell-to-cell variations in macromolecules are high even with abundant proteins, perhaps due to a variable metabolic capacity to produce proteins (337). The reasons underlying noise and heterogeneity are considered in the subsections below. At the single-cell level, the early findings on growth rate heterogeneity have been substantiated in aging bacteria (309), and population diversification of individual cells in spore formation has been identified (345). The single-cell studies of antibiotic persistence have also provided strong indications for population growth rate heterogeneities and dormant cells (12). The evidence for population complexity in any bacterial culture is thus overwhelming.
The way that the distinct subpopulations adapt in new environments is mostly unknown, unless, as in the case of antibiotic persistence, the phenotype of the subpopulation is importantly obvious (see “Heterogeneity in growth rate, persistence, and intracellular signals” below). Another factor is that the totality of the heterogeneity is seldom considered; persistent cells coexist with many other subpopulations differing in motility, gene expression, cell size, etc., in the same culture. Thus, the main emphasis in this section is on how culture history influences heterogeneities in a wide range of characteristics. There has been rapid progress in defining the cellular features and evolutionary benefits that contribute to nonuniformities in populations, but it will be a challenge for the future to determine the adaptomics of heterogeneous populations. We can only begin to define the scale of the problem, as shown in Fig. 4.
Fig 4.
Aspects of phenotypic heterogeneity relevant to adaptation, expanded upon from Fig. 3 in increasing detail. Each box is further discussed in the text. Other content and arrows are as in Fig. 2.
Heterogeneity in growth rate, persistence, and intracellular signals.
As noted in “Growth Rate before the Transition” above, the growth rate affects many characteristics of a bacterial cell. Given this importance in defining cellular characteristics, an important question is whether all the 10n bacteria undertaking a transition are growing uniformly. Even for exponentially growing cultures, this question has already been answered in the negative; several independent approaches (12, 155, 309) all suggest that resting or slow-growing bacteria are present in growing populations.
The clearest indication that populations contain slow-growing or dormant subpopulations comes from the phenomenon of bacterial persistence, which is defined as the capacity of some antibiotic-sensitive bacteria to survive lethal concentrations of bactericidal antibiotics (179). This property was initially demonstrated when Staphylococcus aureus cultures treated with penicillin always contained a small fraction of survivors, on the order of 10−6 or fewer (26). More than 40 years later, Moyed and his group isolated high-persister (hipAB) mutants of E. coli (29, 223, 224), in which the frequency of persisters after ampicillin exposure could reach as high as 10−2 of the population. Another high-persister locus (hipQ) was later identified (364). Direct observations of hip mutants growing in a specially designed microfluidic chamber identified two kind of subpopulations: the type I persisters, which are a preexisting subpopulation of nongrowing cells generated at the stationary phase, and the type II persisters, which are generated during exponential growth (12). Type II persisters are slow growers but are not as growth arrested as type I persisters. Fluorescence-activated cell sorting and differences in gene expression of inactive cells suggested that a third physiological state, distinct from exponential- and stationary-phase forms, is also possible (293). Therefore, a wild-type culture consists of three or four subpopulations of bacteria growing at different rates.
Persistence is strongly influenced by genetic elements called toxin-antitoxin (TA) modules present in the genomes of prokaryotes, which typically consist of two genes expressed as an operon (114). One of the genes encodes a stable toxin (which inhibits some important cellular functions) and the other an unstable antitoxin which neutralizes the toxin and also acts as an autoregulator of expression. TA modules found on plasmids constitute a maintenance mechanism (115, 140). TA modules are also commonly found in bacterial chromosomes. During experiments on gene expression in persistent cells of E. coli, it was observed that some well-characterized TA modules (relBE, mazEF, and dinJ/yafQ) were among the overexpressed genes (156). Overexpression of the plasmid-borne relE toxin gene, coding for a translation inhibitor, resulted in a 10- to 10,000-fold increase in persisters (54). Single-cell methodologies revealed that the hipBA (TA) module determines the onset as well as duration of a transient growth arrest (281). Successive deletion of all the TA loci of E. coli progressively reduced the level of persisters, showing that persistence is a phenotype common to TA loci. These results support a simple model according to which TA loci are activated in a small fraction of growing bacteria, which in turn induces dormancy and persistence (204).
A potential link between the environment, stress, and persistence comes from the role of ppGpp. This alarmone is elevated in stressed cells, but inactivation of the relA-spoT gene pair, involved in the synthesis of ppGpp, brought the persister level of the hip mutant to that of the hip+ wild-type strain (162). This defect could be complemented in trans by the wild-type relA gene. Based on these observations, Korch et al. (162) proposed a model of persister formation invoking a causative role for ppGpp, and indeed the mazEF TA is under ppGpp control (207). Another stress-related connection to generating dormant populations comes from the induction of persisters by DNA damage through the SOS-induced TisB toxin (78). Strategies of survival through dormancy and persistent subpopulations may be linked into sensing stressful environments.
The implication of ppGpp in persisters raises another interesting question. If subpopulations with different growth rates occur in any population, then these cells will also have different ppGpp levels. This is because the intracellular concentration of the alarmone ppGpp is central to responding to growth rate changes in the cell (197, 268). The phenotypes of bacteria growing at different rates are significantly different for the reasons already discussed in “Growth Rate before the Transition” above; ppGpp levels are key to the global regulation of ribosome synthesis, cell size, and global gene regulation (197). If a large population has slow-growing bacteria, through either TA effects or stochastic variations such as cells with a temporary deficit of ribosomes or RNA polymerases (see “Heterogeneity due to stochastic variations in regulation” below), then the alarmone levels within a population may also be heterogeneous. Transitions such as nutrient exhaustion also cause very rapid, drastic increase in ppGpp levels; ppGpp was shown to change in E. coli within a minute of starvation or addition of glucose (232). This was studied in bulk populations, but the rapidity of change suggests that if a large population has regions of nonconstant nutrient levels, then the alarmone levels within a population will be heterogeneous. This is more likely in structured environments such as on an agar plate or biofilms, but it may occur with imperfect mixing in liquid culture.
The likely heterogeneities in ppGpp levels in a population resulting from growth rate differences may be mirrored by other intracellular signaling molecules. Growth rate strongly affects the levels of cyclic AMP (cAMP) in bacteria (23, 237). In turn, cAMP controls a large number of catabolic and anabolic processes and metabolic fluxes (230). Thus, populations with growth rate heterogeneities may have significant effects on metabolism. Experimentally, there was indeed considerable heterogeneity in respiration in bacterial cultures (274); only 90 to 95% are respiration active in growing populations, and heterogeneity increased with starvation. Whether this is indeed due to variation in cAMP or ppGpp remains to be established.
Growth rate can alter other forms of regulation involving small molecules. Intracellular inducer and repressor compounds are responsible for controlling a large number of relatively specific responses encoded within the genome. The majority of regulation in bacteria like E. coli involves so-called local transcription factors and inducers/repressors that control a few genes (15). This includes the lac system, whose bistability is caused largely by differences in intracellular inducer concentrations between cells due to differences in transporter function (235). A piece of evidence linking growth rates to heterogeneity is the demonstration that the bistable switch responsible for coexisting bacteria with distinct levels of lac operon expression is affected by the history of the population and indeed its growth rate (277). Bistability and stochastic variations in populations are considered in the next section, but it is important to realize that the growth rate acts as an epigenetic factor in the generation of population heterogeneity.
The intracellular pool sizes of other regulon inducers such as galactose or maltotriose are a function of growth rate (93), so the expression of genes (additional to those regulated by ppGpp) is also a function of growth rate. There are some important compounds whose variation with growth rate is less well understood; other alarmones such as bis-(3′,5′)-cyclic dimeric GMP (c-di-GMP) are also important in defining bacterial phenotypes (141). Metabolomics has not yet advanced to a single-cell level except for individual molecules (22), but advances in this area will reveal possible heterogeneities in the pool sizes of inducers and alarmones such as cAMP, ppGpp, and c-di-GMP. However, given the above evidence, it would not be surprising if pool size variations resulted in phenotypic differences due to heterogeneity in growth rates or vice versa. In addition, the effect of prior growth rate on stochastic variation needs more thorough investigation.
Given the importance of environmental signals and stationary phase in persisters, culture history before a transition as shown in Fig. 4 is likely to have a strong influence on the overall population and subpopulation frequencies. The evolutionary advantage of having dormant subpopulations is very evident from clinical settings. For example, P. aeruginosa from chronic cystic fibrosis infections shows a much higher frequency of high-persister types in isolates from a patient in later years than in the earlier phase of infection (225). This provides a clear demonstration of the benefit (to the bacterium) of persistence as well as its evolvability. The fitness loss due to reduced growth rate in persisters pays off as a risk-reducing or “insurance” strategy in catastrophic situations (179).
Heterogeneity due to stochastic variations in regulation.
As well as the lactose operon bistability and persistence mentioned above, some of the better characterized examples of nonheritable variation in properties include the lysis-lysogeny switch of phage lambda, chemotaxis in E. coli, phase variation in a number of pathogens, and sporulation and competence in B. subtilis (9, 69, 85, 277, 300). In all these cases, variation can occur due to the fact that signals are not discrete because of random fluctuations inherent to the biochemical reactions occurring in the cell. This stochastic variation is called “noise.” Phenotypic variability in isogenic populations arises from regulatory mechanisms that exploit this signal noise, particularly when the number of molecules involved in biological processes happens to be small. This signal variability can be transformed into phenotypic variability by utilizing a regulatory network designed to suppress or amplify the effects of noise. For example, negative feedback can dampen the effects of noise (21), whereas positive feedback and other network motifs can exploit biochemical noise to generate population-level variability (85).
Experimental analyses of single-cell expression illustrated the extent of noise in transcription and translation in individual bacteria, resulting in the observed differences in protein concentrations (121, 320, 323). There are two kinds of noise linked to the expression process: the “intrinsic” noise, determined by the gene sequence and the properties of the protein it encodes, and the “extrinsic” noise which describes the fluctuations determined by variation in the number of polymerases and ribosomes in the cell, which are dependent on the metabolic state of the cell, cell cycle phase, or cell age (297, 337). Both sources of noise are hypothesized to contribute to individuality; however, it is not clear which source is dominant, and this will likely vary between regulatory systems (85).
Noise signal modulation can transform a graded response into a binary state, in which cells express a certain gene at low or high levels. At the population level, this switch-like behavior can result in a bimodal distribution in gene expression. It is this kind of pattern that produces two different subpopulations and is called “bistability.” Experimentally, some bistable gene expression patterns rely on positive feedback as well as double-negative feedback (toggle switch) (347).
Not all genes are equally noisy, so some phenotypes are more variable than others. A screen for noisy promoters in Salmonella enterica showed that some proteins, especially flagellar gene products, show high stochasticity (105). By broadening the range of characters such as motility and environmental stress resistance across a population, high gene expression noise can increase the likelihood that some cells within the population are better able to endure environmental transitions (9, 31). Experimental results providing support for this hypothesis were obtained in a study that demonstrated a competitive advantage of stress-resistant yeast mutants under high stress due to increased phenotypic heterogeneity (27). Experimental evolution has also shown that selection conditions can favor altered switching in bacteria (20). There is thus increasing evidence that changing noise levels, like other genome-encoded traits, are heritable and evolvable, being subject to selective pressures during the course of adaptation.
However, a high variability in the expression of individual genes does not always confer a selective advantage and may significantly increase the fitness disadvantage under low-stress conditions. The noise would decrease the likelihood of generating viable progeny, as better-fit cells would stochastically change their expression levels and become unfit prior to cell division (104). Consequently, noise reduction mechanisms preserving the fidelity of regulatory signals may evolve in some cellular functions (21, 83). This is borne out by the difference in noise between genes in a genome (105).
Genotype-determined mechanisms for generating multiple subpopulations.
Probably all bacteria can lower risks from environmental transitions by participating in stochastic bet-hedging through creating diversity (347). A particularly clear example of noise-related diversification comes from studies with B. subtilis. The complex subpopulation structure of course makes trying to predict the outcome of a single environmental transition with B. subtilis even more interesting and complex than with E. coli, because B. subtilis differentiates into several distinct, specialized cell types, depending on the culture history (189). There are conditions where a culture may consist of four or more subpopulations, each with individual properties and fitness advantages. The topic of Bacillus differentiation and the multiple signals and environmental influences affecting it is well reviewed and so will not be repeated here (85, 189, 345, 347), but from the point of view of adaptation and a subpopulation structure, it is instructive to speculate how a population of mixed types undertakes a transition as shown in Fig. 4. Clearly, the proportion of each type and its distinct benefit will determine which of the subtypes emerges with a selective advantage upon a transition.
One form of intrapopulation divergence is in motility. Populations of B. subtilis may have coexisting sessile and motile cells even in exponential phase (154). In terms of a single transition, the adaptomics in a new environment will depend on whether motility is an advantage and movement toward nutrients is required.
If B. subtilis is starved, about half the cells morph into spores. Heterogeneity of sporulating populations includes at least two cell types: sporulating cells, in which the master regulator of sporulation Spo0A is active, and nonsporulating cells, in which Spo0A is inactive (123). If this mixture ends up in a highly stressed environment, the passive resistance of dormant spores provides a major advantage. Nonsporulating cells can also undertake different pathways to acquiring new properties. About 10% differentiate into competent cells able to accept foreign DNA (85, 189, 347). The competence state allows Bacillus to generate diversity through the interchange of genetic information (60). Indeed, using mathematical models, Wylie et al. (371) predicted that if the total population size is approximately constant, cells which stochastically switch between the competent and vegetative phenotypes will prevail in competition experiments against otherwise isogenic cells that either are competence negative or fully commit to competence.
This differentiation into the competent state is a process driven not by genetic differences among cells but rather by a stochastic regulatory mechanism (371). Competence in B. subtilis is also a transient process, limited to a short time during the stationary growth phase (347). A complex regulatory network that integrates signals from various pleiotropic regulators controls competence (173) and the expression of all genes that encode the DNA uptake and integration machinery (344). Although subject to various regulatory inputs, the key factor is the stochastic fluctuation in conjunction with the positive-feedback loop that amplifies the signal such that the concentration of the competence regulator ComK exceeds a threshold in some cells, activating the positive loop that drives these cells into the competent state (346).
Another source of heterogeneity in B. subtilis is that in some situations, bacteriocins are used to kill members of the same clonal population. Experimental data showed that at the onset of sporulation, bacteria secrete extracellular killing factors that lyse sibling nonsporulating cells, releasing nutrients into the medium on which “cannibal” cells can feed (124). Cannibalism might be an additional way to minimize survival risks by allowing the mobilization of the last nutrients within a niche—the “last supper” (314)—to delay spore formation until the prolonged depletion of nutrients forces the cells to sporulate (124). In B. subtilis, killing of nonsporulating cells is controlled by two independent gene clusters that are activated at low concentrations of Spo0A (55). At an early stage of sporulation when the level of activated Spo0A is low, genes involved in auxiliary roles such as cannibalism or building of multicellular aerial structures are turned on (108). Then, if harsh conditions continue, there is a progressive increase in the intracellular concentration of activated Spo0A that promotes activation of genes directly linked to the sporulation (123).
The strategies used by Bacillus are also adopted by other bacteria, although the regulatory mechanisms may be different. Competence and DNA uptake are found in taxonomically diverse bacteria (84) and are linked to cannibalistic strategies in Streptococcus pneumoniae (128, 139). Cells that are competent for natural genetic transformation lyse noncompetent cells, and virulence factors are released. In contrast to sporulation, nutritional stress does not trigger the development of the competence state in S. pneumoniae, which is induced in rich medium during early logarithmic growth phase and develops in response to different environmental signals (58), such as changes in pH or the presence of antibiotics (60, 269). In addition, it has been suggested that fratricide may generate diversity by genetic transformation (57, 59), and experimental evidence strongly supports a role for fratricide in lateral gene transfer (152).
Cannibalism is also associated with yet another subpopulation of B. subtilis, namely, that involved in biofilm development (192). Environmental, but not laboratory domesticated, strains of this bacterium form aerial structures in which sporulation occurs (33). In Spo0A-active cells, the cannibal subpopulation involved in killing nonsporulating siblings and delaying the sporulation process promotes at the same time the growth of specialized matrix-producing cells that induce the secretion of these extracellular components (192, 349). This adds yet another ecologically specialized subpopulation to the armory of B. subtilis.
In summary, there may be up to six types of cells in a B. subtilis population, depending on culture history: motile bacteria, sessile cells, bacteriocin producers, residual vegetative cells, competent cells, and biofilm formers. The behavior of each subpopulation in response to a new transition needs to be considered for the full adaptomic description of such bacteria.
Mistranslation and phenotypic buffering through chaperones.
The same genotype can give rise to different phenotypes by a cell making mistranslated proteins from the same coding sequence. Amino acid misincorporation during translation occurs once in every 1,000 to 10,000 codons translated (243, 253), which means that around 15% of average-length proteins will contain at least one misincorporated amino acid. Therefore, exactly replicated genomes are common, but perfectly synthesized proteomes never occur in a bacterium (81). In addition, transcription errors, premature termination, faulty posttranslational modifications, and kinetic missteps during folding are some of the common translational errors over which a plethora of quality control systems act to correct (81, 221). At the same time, the fidelity of protein synthesis is also influenced by intracellular and extracellular factors such as amino acid starvation (221). Therefore, it is important to note that translational error is under a dynamic equilibrium and its alteration directly affects bacterial fitness.
It is intuitively assumed that high mistranslation causes reduced growth rate and fitness because it results in altered proteins with less overall activity. Indeed, the presence of misacylated aminoacyl-tRNA (aa-tRNA) in a protease-deficient strain caused a significant reduction of the growth rate (285). However, under selection pressure, bacteria not only tolerate the presence of misacylated aa-tRNA but also can even require it for growth (285). For instance, E. coli can tolerate up to 10% of faulty protein, coping with mistranslation by triggering the heat shock response, which stimulates nonoptimized polypeptides to achieve a native conformation or be degraded (285). In this way, bacterial cells ensure the presence of sufficient functional protein even though at a considerable energetic cost (285). Under certain conditions, increased levels of mistranslation can even be advantageous: in Acinetobacter baylyi, substitutions in isoleucyl-tRNA synthetase that allow the mischarging of tRNAIle with Val confer an increased growth rate compared to that of wild-type bacteria under conditions of limiting Ile and excess Val (11).
In evolution, organisms can theoretically take advantage of beneficial phenotypes generated by error (209, 360). Mistranslation errors may cause individuals in a population to be beneficially prepared for a transition to a new environmental challenge, but advantageous phenotypes caused by mistranslation are not obviously heritable. However, phenotypic diversity generated by mistranslation can potentially allow heritable adaptation. The discovery in E. coli and other bacteria of a hypermutagenesis phenotype associated with codon ambiguity, the “translational stress mutagenesis” (TSM) phenotype, provides a potential solution to the problem of heritable transmission of protein translation errors (1–3, 77). Ambiguous codon decoding expands the proteome and generates new phenotypes. Selection of advantageous phenotypes creates positive feedback pressure that maintains ambiguous codon decoding, leading to synthesis of mutant DNA polymerases and DNA repair enzymes and emergence of hypermutagenic clones with an increase in genome mutational load. This accelerates the fixation of advantageous phenotypes and their transmission to the progeny (1–3, 77). Therefore, the high cellular tolerance to mistranslation opens the possibility that evolution of proteomic and phenotypic novelty could be fixed through hypermutagenesis (221). The possible importance of transient mutators, defined as wild-type bacteria that, due to occasional transcription or translation errors, display a mutator phenotype, was already proposed theoretically by Ninio (234), who suggested that transient mutators produce at least 10% of the single mutations and more than 95% of the simultaneous double mutations in an E. coli population; this needs experimental validation.
Another source of possible heterogeneity comes from chaperones, or heat shock proteins, that are capable of buffering the negative effect of mutations causing protein defects (202, 329). The level of the heat shock chaperones DnaK and GroEL was increased in lineages that had accumulated many mutations, and experimental overproduction of GroEL further increased the fitness of lineages containing deleterious mutations (329). In transitions such as those shown in Fig. 4, the availability of buffered mutations deleterious in the original environment could lead to the acquisition of neutral genetic diversity but accelerate the rate of adaptation in a new environment where the available mutations are beneficial.
Variation in the size, age, and density of cells in the population.
The fate and response of bacteria are subject to yet other population-level heterogeneities. One of these is in the size of the bacteria, because asymmetry in cell partitioning can generate cells both shorter and longer than the mean (67). Size can affect physical properties relevant to an environment (e.g., protist predation is dependent on size [131]), but other fitness effects are also size dependent. For example, if the environmental challenge is through phage infection, the size of cells in a growing population may intriguingly influence the lysis-lysogeny decision in E. coli with phage lambda (313). Also, infection of persister cells and vegetative cells can have different outcomes and produce different decisions in the lytic-lysogenic choice in phage-bacterial interaction (257).
The physical cell density of bacteria in a population growing in liquid culture is also heterogeneous (with up to 15 discrete fractions!) (205). The glycogen content of cells is largely responsible for density differences; the presence of very different glycogen levels in cells in a population is related to the metabolic characteristics of individuals as well as their regulation (363). However, there is no systematic study available to describe the fates of all the possible combinations of age, size, and density subpopulations in a single culture or whether they respond differently to stress.
Of course, heterogeneity is also seen in biofilms, and different cell types can be observed and recovered from biofilms (265, 368). Even colonies on agar plates contain subpopulations with distinct properties (288). In structured environments such as these, a series of local differences (e.g., nutrient gradients and cell-cell signaling, considered in “Variation in external signals and ecological interactions” below) are likely to contribute to this heterogeneity.
Bacterial cell composition is affected by the production of faulty proteins, as discussed in “Mistranslation and phenotypic buffering through chaperones” above. Recent research suggests that aggregates formed by denatured proteins are themselves a signal for cellular divergence. In a normal growing culture of E. coli, the evidence suggests that protein aggregation is predominantly at one end of a cell at old cell poles (184, 203). Stewart et al. (309), using cell imaging and tracking technology with dividing E. coli, observed that despite the morphological identity of daughter cells arising from symmetrical cell fission, one of the progeny cells inherits preexisting components (old pole) of the mother cell while the other inherits “de novo”-synthesized elements (new pole) (309). Notably, cells which successively accumulated old-pole elements showed a reduced growth rate, decreased numbers of offspring, and higher chances of death. Finally, upon reaching around 100 divisions, the older-pole-containing cells ceased to grow (309). Therefore, using an asymmetrical partitioning strategy, some cells would accrue irreparably damaged components, compromising their own reproductive potential in order to create fitter, rejuvenated offspring with maximal cellular capacities (307). The relevance of an asymmetric distribution of bacterial damage in a population in adaptomics is that aged and active bacteria presumably react differently to secondary transitions, as shown in Fig. 4.
Variation in external signals and ecological interactions.
The image of bacterial populations as consisting of isolated, noninteracting individuals has given way in recent years to one in which they interact extensively (161). Cannibalism and fratricide, discussed above in “Genotype-determined mechanisms for generating multiple subpopulations,” are examples of interactive behavior in bacteria, in which groups of genetically identical or closely related organisms synchronize their patterns of gene expression to achieve specific goals that are unattainable for single cells acting on their own. Such collaborative behavior, which depends on cell-cell communication with secreted signaling (quorum-sensing) molecules, is widespread among bacteria.
Intrapopulation signaling through quorum-sensing systems influences many bacterial properties, such as the expression of virulence genes, biofilm formation, antibiotic susceptibility, cellular differentiation, and genetic competence (16, 161, 168). Interactions between bacterial cells involving other diffusible factors, i.e., secreted antibiotics, bacteriocins (44), or toxins (89), may also result in antagonistic interactions, which may kill nonresistant bacteria, nonimmune closely related species, or siblings, respectively (161). Whether quorum signals are present just prior to a transition determines the presence or otherwise of these phenotypes.
An important factor in the production of signal molecules and the local concentration of quorum sensor molecules or autoinducers is the density of the culture (321). As mentioned in “Population Size and Culture Density” above, high-density cultures undertaking an environmental transition may be in a state affected by the presence of signal molecules. For example, in E. coli, the response to challenges such as acid stress is modulated by population-level signaling. The release of indole can influence the behavior of neighboring cells (144). This signal molecule also affects antibiotic sensitivity in populations, in which subpopulations release indole (168). The history-dependent changes described in “Growth Rate before the Transition,” “Prior Stress Exposure,” and “Growth Phase before the Transition” above can also be elicited by population density effects. For example, bacterial characteristics similar to those in stationary phase can be elicited by high population density, even when growth is exponential. Bacteria growing at the same rate in chemostat cultures but at high densities exhibit many of the properties of stationary-phase bacteria (186) and so are likely to be more resistant to secondary challenges than low-density cultures. The concentrations of both the cells themselves and released signal molecules will therefore determine the transitions and responses shown in Fig. 3. The above results also suggest that population-level susceptibility, as against individual cell susceptibility, may be different.
Bacteria release a surprisingly wide range of molecules, especially in stationary phase (276). Released compounds include nucleotides and amino acids, and recycling of metabolites and nutrients from lysed cells provides a new ecological background to stationary-phase cultures, as it does in starving B. subtilis (see “Genotype-determined mechanisms for generating multiple subpopulations” above). Released nutrients provide new resources to compete for, and in long-term stationary phase, several mutations that enable E. coli to better compete for low concentrations of amino acids sweep (378). The heterogeneity of stationary-phase cultures (see “Growth Phase before the Transition” above) is at least partly due to adaptation to this more complex environment. In addition, released compounds such as amino acids can act to signal aggregative or motility-related changes (220). The profile of a culture is thus determined by the presence or absence of these signals and alternative released nutrients.
As mentioned in “Prior State (e.g., Planktonic, Motile, or Biofilm)” above, chemical signals play an important role in the formation of biofilms. The aggregates themselves create gradients of oxygen and nutrient availabilities, causing differences in gene expression depending on nutrient levels (291). A range of self-produced autoinducers as well as external molecules contribute to biofilm formation (191). In P. aeruginosa, along with many other Gram-negative organisms, quorum-sensing systems respond to autoinducers termed acyl homoserine lactones. In B. subtilis, surfactin acts as a unidirectional signal so that one subpopulation of cells produces the molecule, whereas other cells respond to it and produce matrix (191). All these kinds of interactions cause heterogeneities in a population.
Bacterial cells also interact by means of contact-dependent systems. As in other processes described above, contact-dependent interactions may have positive or negative effects on bacterial cells. Particularly, sharing of components through contact-dependent structures such as nanotubes within and between species has been suggested (82). The new identification of specific systems involved in contact-dependent activities as well as their wide conservation strongly suggests that contact-dependent interactions are a widespread process and form part of the general interaction of the bacterial cell with its environment (161). A phenomenon called “contact-dependent growth inhibition” (CDI) was described, in which a uropathogenic E. coli strain produced a significant reduction in the number of viable E. coli K-12 cells (7). Other contact-dependent phenomena in different bacteria have been described (82, 128, 174, 226, 228, 331).
Finally, members of microbial populations are heterogeneous in terms of cheating to obtain commonly produced goods such as siderophores (iron chelators) or externally located enzymes such as β-lactamase (56, 135, 350). Some organisms stop producing the shared product and live off the rest of the population, which still bears the cost of production. The proportion of cheaters in a population is subject to environmental influences (36), so in the context of the single environmental transition that we are dealing with, the proportion of nonproducers and producers of shared resources will depend on culture history and the structure of the population (e.g., biofilm versus planktonic) (37).
Even given our narrow focus on clonal populations and a single transition, the heterogeneities and interactions discussed in “Phenotypic Variation within Populations” clearly define the formidable scale of the problem in trying to assess the makeup of a population at a single time point, before the transfer to a new environment. So far, we have considered mainly phenotypic variation, but we now turn to the added dimension of genomic variation and selection in the adapting population.
Genetic Heterogeneity within Populations
Despite the depth of regulatory solutions available for bacteria to survive a change in environmental conditions, the circumstances that a bacterial population may face in nature are many and can be varied and unpredictable, so it is not possible to have an effective regulatory response to every environment. Mainly due to costs and trade-offs, there are no superfit organisms in nature. In the absence of an appropriate phenotypic response in a new environment, there will be selection pressure to adapt via genetic change, which is the subject we focus on in the following subsections.
In considering how an organism adapts via heritable changes, we are concerned with variations that produce either altered gene expression or expression of an altered gene(s). The way by which this occurs is through mutational events, which encompasses a diverse array of mechanisms and frequencies of occurrence, including amplifications, deletions, point mutation, insertions, etc. (106). In large populations, as often occur, mutational variation is considerable: in each generation, due to errors in replication, a spontaneous mutation arises on average in 5 × 10−3 of the population, and after n generations, the proportion of ancestral organisms will be, statistically, 0.995n (68). If n = 109 in Fig. 2, then about 5 × 106 bacteria in that population potentially carry a mutation. As discussed below, processes such as amplification and phase variation are even more frequent (5). Adaptation, though, is not just the generation of genomic variability, and we also consider how selection acts upon genomic variation to result in a diversity of fitter offspring. In the following sections we discuss how both genomic variation and selectional processes increase the richness of adaptive outcomes.
In order to maintain genetic stability from generation to generation, the process of DNA replication should be carried out with very high fidelity; this is important since mutations are much more likely to be deleterious than beneficial (79, 260). To this end, the mutation rate per base pair per generation (μb) is maintained at a very low level; for example, in E. coli, μb is estimated to be around 5 × 10−10 (80, 194), although other estimates are lower still (242). The majority of these mutations are neutral or deleterious, while the frequency of beneficial mutations in E. coli has been estimated at around 10−8 to 10−9 per genome (147, 283). This may be an underestimate due to clonal interference (competition in populations between multiple bacteria with independent mutations) that will exist, particularly in larger populations (116). However, the most optimistic estimate in the absence of clonal interference is still only 2 × 10−5 per genome, which is orders of magnitude below the overall mutation rate (260).
The hypothesis that mutations are kept to a minimum in evolution is not altogether convincing, however. A survey of the spontaneous mutations arising in 787 natural isolates of E. coli did not indicate a species-wide trend toward a uniform, low rate of spontaneous mutagenesis; in contrast, there was a broad distribution of frequencies of spontaneous mutation to rifampin resistance, with many strains differing by several orders of magnitude (28). A high mutation rate may result in faster adaptation to an environmental change by providing a greater number of alternative genetic solutions from which beneficial solutions can be selected. Of course, the balance between mutation rate and genetic stability represents a trade-off, i.e., the ability to evolve to adapt to change balanced with the need to maintain the integrity of the organism.
There are several ways in which a bacterial genome can change the costs and benefits of this trade-off. In other words, bacteria shift this balance such that when conditions are favorable, the bulk of the population undertakes high-fidelity genome replication (i.e., “stick with what is working”), but when conditions change, there is the capacity within the population to produce new genetic solutions. Several identified mechanisms change mutation rates conditionally. These mechanisms include the following: (i) increasing gene copy number by amplification, which can increase the dosage of a beneficial gene or provide redundancy to increase the target(s) for adaptive mutation while simultaneously decreasing the risk of loss of gene function through deleterious mutation (5); (ii) limiting mutations spatially by possessing contingency loci, i.e., regions of the genome that are more susceptible to mutation, as exemplified by the mechanism(s) of phase variation (341); (iii) taking advantage of a subpopulation of constitutive mutator mutants with an increased mutation rate that can be enriched by secondary selection in times of need; (iv) limiting mutations temporally, i.e., increasing mutation rates under stress (a process called stress-induced mutagenesis) (28, 102, 112), including increased mutation rate during stationary phase (100); and (v) acquiring new genes by lateral transfer (42). All of the above can provide a great deal of genomic variation within a population that can be exploited to adapt to environmental change. The following sections and Fig. 5 examine each of the above mechanisms in more detail and discuss how they contribute to genetic heterogeneity.
Genome variation by duplication/amplification.
Gene duplication/amplification (GDA) is one of the most common types of genome change. It encompasses both simple duplications and higher-level amplifications of up to 40 copies of a gene(s), with sizes of the amplified region of up to several megabases (6, 328). GDA can affect practically any gene and is inherently unstable/reversible. The ease of reversibility of GDA means that the cost of the duplication can be lost in a nonselective environment.
The high frequency of GDA in bacteria means that large populations will be heterogeneous before and during the transitions shown in Fig. 4. GDA occurs in a growing population of bacteria at a frequency of >10−2 to around 10−4 per genome (4, 272, 302). From these frequencies and the average size of the amplified regions, it has been estimated that around 10% of all cells in a nonselectively growing culture contain a gene duplication somewhere in their genome (5). If the duplications are spread evenly throughout the chromosome, then around 2 × 104 in a population of 109 E. coli bacteria contain a duplication of a particular gene. The culture history also influences the frequency of GDA, which can be induced by prior exposure to cellular stressors (136) and may be part of the strategy for stress-induced mutagenesis (138), as further considered below.
Much of our understanding of GDAs had been obtained from studies of S. enterica, in which GDAs have been found in all chromosomal regions except the terminus of replication (4). GDA has been proposed to occur by several mechanisms, and it is likely that different ones account for simple duplications and higher-level amplifications (5, 113, 172, 279, 290). The most frequently observed mechanism for formation of simple duplications occurs by RecA-dependent nonequal homologous recombination between regions bounded by directly repeated sequences such as rRNA (rrn) operons, transposable elements, and other repetitive sequences (4, 5, 129, 182, 273, 290). However, other duplications that are both repeat sequence and RecA independent can also occur (290). It has also been proposed that stress might induce repair of broken replication forks to switch from high-fidelity homologous recombination to nonhomologous repair, thus promoting copy number change (137). Higher-level, multiple amplifications can then form from duplications via RecA-dependent recombination between the tandem repeat generated by the initial duplication (273). Multiple amplifications can also form via a rolling-circle replication mechanism in the absence of preexisting duplications (261, 290).
GDA can increase fitness in a selective environment by increasing the copy number (gene dosage) of a beneficial gene, effectively increasing its expression, much like a positive regulatory change. GDA therefore provides a quick and reversible way of adapting, which is in some respects closer to regulatory changes than the more permanent genetic change normally considered heritable. As mentioned above, GDAs are unstable and are easily lost under nonselective conditions by homologous recombination between the repeated amplified sequences (263, 290).
GDA can also act as a facilitator of more permanent genetic changes. Increasing the copy number of a gene permits “prospecting” for beneficial mutations in one replica while still maintaining one or more wild-type copies, abating the risk of loss or reduced gene function because of deleterious mutations. Once a beneficial mutation is acquired, the GDA may be lost (5).
Genome variation at contingency loci.
Contingency loci are hypermutable regions of genomic DNA that typically mediate the high-frequency, reversible on-off switching of gene expression, as exemplified by the phenomenon of phase variation. Contingency loci allow an organism to confine an increased rate of change to particular genes whose heterogeneity of expression in a population is of advantage and to reduce the deleterious cost of increased mutation rate throughout the genome. Rates of switching at contingency loci can be much higher than the spontaneous mutation rate, ranging from 10−2 to 10−5 per genome per generation (222). This, considering the stochastic nature of the switching and the fact that there are often several phase-varied genes in a genome, results in considerable genotypic heterogeneity within a population.
The process of phase variation in bacteria is most studied in bacterial pathogens, where there is heterogeneity in the expression of genes whose products (usually a cell surface component) are visible to the host's immune system. Variation provides a mechanism for bacterial populations to survive clearance by host immune responses (for reviews, see references 222 and 289). There are three common mechanisms for local genome variation, namely, (i) through changes in short sequence repeats (SSRs) in or near contingency genes, (ii) through specific inversions of particular parts of the genome, and (iii) through epigenetic regulation mediated by DNA methylation (341).
Some contingency genes contain SSRs made up of 1- to 6-bp units, either within the promoter region or in the coding sequence of the gene. This facilitates phase variation via the mechanism of slipped-strand mispairing (SSM) (222, 338). SSM occurs during DNA replication or repair requiring strand separation of the parental DNA duplex, giving rise to single-stranded DNA, and synthesis of new daughter DNA, which must then reanneal to the template. The presence of a repeat sequence facilitates the ability of the template and daughter strands to slip in relation to one another, resulting in loss or gain of a repeat unit sequence and leading to frameshift mutations or changes in promoter sequence spacing, which can then result in altered gene expression. Conversely, the same mechanism can switch gene expression on again following further rounds of DNA synthesis and slipped-strand mispairing (222). SSM is important in many pathogens, particularly Neisseria meningitidis and Helicobacter pylori, in which 19 and 65 SSM-mediated phase variation loci have been identified, respectively (275, 339).
Another mechanism that can give rise to phase variation of contingency gene expression is site-specific recombination. This involves the inversion of a short DNA segment flanked by sequences recognized by specific recombinases, which contains the contingency gene or its promoter sequence. The gene(s) is then either expressed or not depending on the orientation of the invertible sequence (343). A well-known example of phase variation controlled by site-specific recombination is phase variation of type 1 fimbriae in E. coli (343). Flagellar phase variation in S. enterica is also controlled coordinately by an inversion event (298). Another related mechanism of phase variation involves the precise excision or incorporation of insertion sequences (343). This phase variation mechanism is restricted to a few IS elements, e.g., IS2561 in Staphylococcus epidermidis (343).
Methylation of DNA is involved in a number of examples of phase variations. Unlike the previous two, this mechanism is epigenetic, since the DNA coding is not altered. Methylation can result in phase variation through a transcriptional regulator, as occurs in interdependent expression of fimbrial and flagellar genes in E. coli (reviewed in reference 342). Two Dam methyltransferase sites and pap DNA methylation patterns are involved in the regulation of the phase variation switch between the on and off states in the synthesis of Pap pili in E. coli (32). Dam-dependent phase variation in E. coli also accounts for changes in another antigen, Ag43 (130). A third example of a surface component regulated in this way involves Salmonella lipopolysaccharide modification genes (35). Hence, a population of bacteria can consist of subpopulations whose methylation pattern and gene expression are determined by epigenetic processes.
As noted in the next section, phase variation may even play a role in regulating the mutation rate, since it has recently been shown that the expression of mutL, a gene involved in the mismatch repair system in S. enterica serovar Typhimurium, can be reversibly switched on or off via SSM-mediated changes in a number of small tandem repeats (49, 50). Thus, such a genetic switch would enable a subset of a population to readily convert to a hypermutator phenotype, which would be of potential benefit in times of stress, as discussed below. The authors went on to show that the tandem repeat sequences were present in MMR genes from many other bacteria, suggesting that on-off switching of hypermutation may be a prevalent mechanism.
In all of the mechanisms affecting contingency loci, the rate of generation of phase variants is controlled by cellular components, be they methylases, polymerases, or DNA inversion mechanisms. Culture history can change the frequency of switching and contribute environmental regulation to switching rates (18). Stochastic effects discussed above can also contribute to the many factors that affect contingency heterogeneity within a species and a population.
Variation due to transpositions and deletions.
In bacterial genomes, transposable elements and especially insertion sequences (IS elements) are widely and diversely distributed (201). Transposable elements in a genome can aid adaptation and fitness (47, 48). Many examples of transposition arise in adapted strains in experimental evolution experiments, and these generate a high level of genetic diversity within E. coli (110, 250) and lead to adaptive mutations (375). Several of the IS insertions in experimental evolution studies are beneficial under the selection conditions (110). Movement of IS elements contributes to bacterial adaptation by increasing the mutation supply, in turn enabling the organisms to overcome an environmental challenge (318, 361, 379).
The rates of transposition vary from element to element; for example, under nutrient limitation of E. coli, no changes were associated with either IS30 or IS150 but IS1, IS3, and IS5 movements were common (110). The precise numbers of transpositions in any individual population are not thoroughly understood, but methods for quantitating transposition frequency are being developed (248). Nevertheless, based on the high number of transpositions in experimental selections (110, 250), a large population undertaking a transition shown in Fig. 4 undoubtedly includes some genomes with IS movements. The possibility of transpositions in large resistance gene islands on chromosomes and plasmids in multiple drug-resistant isolates is even greater, especially under antibiotic selection (330).
There are examples of organisms in which the frequency of transpositions is also affected by stressful environments, especially when DNA is damaged (87) and perhaps also under extended starvation (227). Thus, culture history may have an effect on these classes of mutation; for example, the prior UV exposure of a population shown in Fig. 3 could affect the proportion of mutations caused by transposable elements.
IS elements also provide a means of obtaining deletion mutations. Repeat copies of IS elements provide homologies permitting deletions to form, as of course do other repeats in genomes (25). An interesting example in E. coli is provided by the means of generating mutator mutations through an IS5-mediated transposition followed by deletion of the intervening region (110). Several additional mechanisms of deletion formation have been proposed, and some of these are present in stationary-phase bacteria (13, 14). Hence, this form of mutation, like other forms of mutation discussed in “Mutator mutant subpopulations” below, may be under a culture history influence.
Mutator mutant subpopulations.
When a bacterial population faces a change in environmental conditions, it may be worth the risk of raising the mutation rate in order to increase the occurrence of beneficial mutations. One way to achieve this in a population or at the species level is via the existence of subpopulations of constitutive mutator or hypermutator mutants (119).
A major factor in the basal mutation rate is the capacity for DNA repair; mutations in DNA repair genes generally result in increased mutation rates (216). There are many components of DNA repair that influence bacterial mutability (145). The most common and best understood mutations causing a heritable, constitutive mutator phenotype involve genes of the DNA mismatch repair (MMR) system (mutS, mutL, mutH, and uvrD), which are responsible for proofreading after DNA replication as well as the inhibition of interspecies recombination events (49, 180, 271, 319). Mutations in the MMR system can elevate mutation rates by more than 100-fold (319).
When starting with a pure nonmutator E. coli clone, by the time 109 cells are present through division in a population like that shown in Fig. 5, about 105 mutator mutants are present in this population through spontaneous mutations in mutator genes (206). If beneficial mutations are selected in such populations (such as mutations for antibiotic resistance or better growth under nutrient limitation), the MMR mutator cell is 100 times more likely to contain a beneficial mutation. The cell containing a combination of an MMR mutation plus a beneficial mutation is then enriched through selection. Hitchhiking with beneficial mutations allows MMR mutant bacteria to reach high proportions in rapidly evolving cultures (217, 238). This process of secondary selection is most likely responsible for the high proportion of mutators in nature (72, 119).
Indeed, MMR constitutive mutator mutants are present at a surprisingly high frequency in natural populations of pathogenic and commensal E. coli strains, making up to 1% or more of the population (167). Many of the mutations in naturally occurring mutator isolates are deletions (164). The frequency of mutators can reach very high levels in uropathogenic E. coli (72) or P. aeruginosa populations chronically infecting the cystic fibrosis lung, where mutators can make up 40% of the population (91, 244). P. aeruginosa cystic fibrosis lung infections are usually lifelong and often are aggressively treated with prolonged antibiotic therapy, and the high frequency may reflect the repeated selection of antibiotic resistance associated with mutators (91, 244, 365).
In addition to increasing the rate of point mutations, a deficient MMR system also results in an increased incorporation of foreign DNA via lateral transfer, owing to the role of the MMR system as an inhibitor of this process (210). E. coli mutants without MutS or MutL recombine Salmonella DNA into E. coli at about 1,000-fold-higher rates than cells with an intact MMR system (305). This is obviously of importance for adaptation as well as the lateral acquisition of antibiotic resistance from other organisms (327). The heterogeneity and mosaicism of mutator gene sequences suggest that changes of mutation and recombination rates by frequent loss and reacquisition of MMR functions occur in E. coli genomes (38, 73).
Mutation of the MMR system may be subject to a more deliberate adaptive strategy akin to phase variation. As discussed above, the MMR system is potentially phase variable, perhaps across a range of bacterial species (49, 50). The evidence suggests that repeat sequences present in MMR genes may act as “on-off” switches facilitating the easy conversion to a mutator phenotype to better respond to stressful conditions.
Stochastic and stress-induced variation in mutation rates.
In contrast to acquiring a constitutive mutator phenotype via mutation of the MMR system, bacteria can also express a transient elevated mutation rate in two ways, although the two mechanisms may overlap. Temporary mutators or transient mutators (218) can arise for the same stochastic reasons as the phenotypic variation discussed in “Heterogeneity due to stochastic variations in regulation” above. As predicted on theoretical grounds (234), transcriptional and translational errors or cell divisions that result in low levels of MMR components can result in transient mutagenesis. In a large population like that we are considering, all these stochastic events can give rise to subpopulations with elevated mutation rates. The second mechanism causing temporarily increasing mutation rates, known as stress-induced mutagenesis (SIM), is less stochastic and has elements of determinism; several regulatory genes associated with stress resistance are associated with SIM (102, 112, 326). A control mechanism with stress inputs temporally isolates increased mutation to times when the bacteria are maladapted to their environment.
SIM appears to be a common feature of E. coli populations, with up to 80% of isolates of E. coli showing a stress-inducible mutagenesis phenotype in aged colonies (28; see reference 369 for an alternative view). This potentially provides a large increase in mutation supply in a population. However, large variations in mutation rate increases exist, ranging from a few fold to >1,000-fold (28), so the extent of SIM appears to be strain specific and may reflect differences in recent evolutionary selective pressures.
Several types of genetic alterations can be involved in SIM, including base substitutions, small deletions and insertions, gross chromosomal rearrangements, copy number variations, and movement of mobile elements (102, 112, 136, 319). A common theme, though, is the requirement for one or more stress responses to induce mutagenesis, e.g., the SOS DNA damage response, the RpoS general stress response, and the stringent response (102, 111, 112, 319, 326). The link between stress and mutagenesis extends to extracytoplasmic stressors (e.g., those causing membrane damage) through another sigma factor, RpoE (117). RpoE, aside from its better-known role in membrane protein protection, is thought to promote double-strand breaks and feed into SIM induction by affecting genome integrity (117). Multiple inputs ensure temporal control of mutagenesis to times of stress, while ensuring genetic stability when cells are well adapted.
The type of stress that is intuitively the easiest to link with increased mutagenesis is one that causes DNA damage, either from exposure to DNA-damaging agents or from uncompleted DNA repair/replication. DNA damage induces the derepression of SOS genes, many of which are involved in DNA repair and recombination (99, 255). The initial SOS response involves induction of enzymes that repair DNA with high fidelity; however, with greater damage, a more mutagenic response is initiated, involving three alternative DNA polymerases, i.e., Pol II, the Y family polymerase Pol IV (encoded by dinB), and Pol V (encoded by umuDC), which, in contrast to normal replicative Pol III, can replicate past DNA lesions (99). Two of these, Pol IV and Pol V, are both low-fidelity polymerases and therefore replicate DNA in an error-prone manner. In dividing cells, it is the induction of Pol V-dependent replication during the SOS response that is largely responsible for up to a 100-fold increase in chromosome-wide mutagenesis (292, 306, 366). In stationary-phase cells, Pol IV is involved in higher mutation rates (40). RecA also plays a central role, since in addition to upregulating dinB and umuDC transcription, RecA also interacts directly with Pol IV and Pol V to modulate their mutagenic activity and is involved in the cleavage of nascent UmuD to form functional Pol V (120, 150, 322). The SOS-induced mutagenesis is probably a by-product of a survival-at-any-cost mechanism to overcome otherwise fatal DNA lesions. Nevertheless, it also increases genetic diversity within the population at a time of stress, which may then be of advantage to adaptation.
The population history, as discussed in “Growth Rate before the Transition,” “Prior Stress Exposure,” and “Growth Phase before the Transition” above, has a major influence on mutation rates through a general stress-sensing mechanism; there is strong evidence that the general stress response induces the error-prone Pol IV (40, 166). The induction of Pol IV is mediated by the sigma factor RpoS and occurs when cells enter stationary phase or experience the other stresses discussed above in “Growth Rate before the Transition” and “Prior Stress Exposure.” There are thought to be two ways in which the general stress response increases the mutation rate, both of which are RpoS dependent. The first is by upregulation of the expression of the error-prone DNA polymerase Pol IV (111, 166), and the second is by downregulating expression of the MMR proteins MutS and MutH (28, 334). The RpoS-dependent nature of both of these mechanisms can give rise to heterogeneity in mutagenesis levels in a population concurrent with heterogeneity in RpoS levels. This indeed explains the heterogeneity in mutation rates across the species E. coli (287).
Another player in the control of RpoS/Pol IV mutagenesis is polyphosphate [poly(P)], which consists of chains of phosphate tens to hundreds of residues long and is produced by the enzyme polyphosphate kinase (PpK) (for a review, see reference 163). Poly(P) accumulates in cells entering stationary phase and is required for induction of RpoS and of RecA (295, 335). Thus, poly(P) can influence SIM through its regulation of RpoS; however, poly(P) may also play a more direct role by regulating the activity of Pol IV (316).
The stringent response mediated by ppGpp is a third stress signal that contributes to SIM, partly through RpoS induction (165, 332). This overlap means that the RpoS-induced mutagenesis mentioned above will also occur during the stringent response and, similarly, that heterogeneity in RpoS expression (see “Prior Stress Exposure” above) will result in heterogeneity in stringent response-induced mutagenesis. Like for RpoS, the levels of expression of ppGpp can vary greatly across different strains of E. coli, and mutations affecting ppGpp production readily occur (98), which provides further heterogeneity in SIM within a population.
As mentioned above, the stringent response induces transcription of amino acid biosynthetic genes and other genes to relieve stress. Transcription requires the formation of loops of single-stranded DNA, which has been proposed to be more susceptible to damage (370). Potentially, actively transcribed genes can acquire more damage than nontranscribed genes; consistent with this is the fact that DNA repair enzymes are recruited to transcribed genes (213). In particular, error-prone Pol IV associates with RNA polymerase that is stalled at lesions in DNA that it is transcribing, to facilitate translesion DNA synthesis to enable transcription to proceed (62). This provides a potential mechanism by which genes that are being actively transcribed are more prone to mutation via increased error-prone DNA synthesis. This would target mutation to genes that are actually required to overcome stress (370). Indeed, this mechanism has been shown to be involved in SIM in both lacZ and tetR mutation assay systems, increasing mutagenesis by more than 400-fold (63).
It is tempting to think that SIM is an evolved mechanism to increase evolvability under stress, and evidence is indeed accumulating to suggest that this is the case (112). As mentioned above, the increase in mutation rate may even be targeted to genes that are actively being transcribed, which would target adaptation to genes and pathways that are most likely to lead to an advantage in overcoming a particular stress (370). The mutation rate increase is indeed also restricted to subpopulations of cells, because it appears the hypermutation is mostly restricted to about 1% of stressed cells that have double-strand breaks in DNA (258). Also, the stress-induced GDA changes may be in subpopulations of bacteria that show repeated template switching during DNA replication (181). All this mutational heterogeneity in subpopulations adds to the rich variation from which adaptive strategies can emerge in bacteria.
External DNA, genetic exchange, phages, and plasmids.
As discussed in “Phenotypic Variation within Populations” above, a factor subject to bacterial phenotypic variation is the competence to take up DNA. Besides competence, bacteria have other DNA uptake mechanisms involving conjugation and phage-mediated transduction (10, 43). The ability to access foreign DNA leads to a huge increase in the potential for genetic variation, and indeed the analysis of bacterial genomes points to the importance of lateral gene transfer (LGT) in bacterial evolution (176, 376). The core genome shared by all E. coli strains is fewer than 1,000 genes, so even at the species level, a great number of genes get moved around (193). However, in the system we are dealing with, in a clonal population with similar genomes, the possibilities for major variation through LGT are significant only if the new environment has other bacteria as a source of external DNA. In many natural environments, this may indeed be the case. It is extremely difficult to estimate the frequency of such LGT-type events in single transitions, and these will be very variable depending on the environment, sex factors, ability to take up DNA, and ability to recombine foreign DNA. In any case, this lateral source of variation in microevolution is probably rare relative to the other processes discussed here, such as contingency gene changes, GDAs, or even base change mutations. However, it does seem that some species change more often by LGT than others, so the capacity for LGT is also a question of genotype (376).
Nevertheless, there are circumstances when genetic exchange is advantageous even within an E. coli population. Experimental evolution studies of pure cultures in the presence of the F plasmid (promoting conjugation in E. coli) showed that the fixation of beneficial mutations was enhanced through plasmid-mediated gene transfer (65). Conjugation allows beneficial mutations that arise in different lineages to recombine, hence increasing the number of possibilities in adaptation. Hence, the transition shown in Fig. 2 will depend on the genotype (presence of conjugative plasmids) and the proportion of the population that carries such elements (depending on plasmid stability).
Besides conjugation and genetic exchange, plasmids and phages add another dimension to bacterial variability. The genes carried in extragenomic elements have the potential to alter not only the physiology and metabolism of a cell but also its capacity to generate mutations. The error-prone DNA polymerases can be encoded in plasmids and contribute to mutational adaptation (122, 324). Together with the variation in plasmid content itself as a result of environmental stresses (358), a bacterial population may be heterogeneous because of these plasmid-encoded factors.
The well-known dependence of prophage induction by stresses, coupled to the SOS response in phages such as lambda (278), also has the potential to influence heterogeneity in a population. The basal level of lambda induction, in about 1 in 104 cell divisions, is increased by a variety of stresses (41). Hence, in a large population of lysogenic bacteria, heterogeneity and lysis due to released phage is a likely scenario.
Heterogeneity from the selectability of mutations.
Selection and within-population competition occur constantly in evolution. With a change of environment as shown in Fig. 2, altered fitness characteristics that can benefit a subpopulation of mutants in the old population are selected. In the above sections we have discussed several mechanisms whereby such fitter mutants can arise from GDA, LGT, spontaneous mutations, or elevated mutation rates. Still, there can be situations where mutations are limiting; no major adaptive shifts in the population are likely if the beneficial mutations in the particular environment are absent, or particularly rare, or if the population is very small. In these cases of limiting mutation supply, mutational adaptation is unlikely, but elevated mutation rates can overcome this limitation, as discussed above. Selection can then result in changing ratios of subpopulations within the population. There are further scenarios, however, in which the composition of a population may not change appreciably as a result of selection. The fixation of mutations, especially weakly beneficial mutations, is also a limitation, and drift can eliminate these before fixation (245).
More frequently, however, significant population changes can be associated with mutational events. First, a strongly beneficial mutation can sweep through a population in a new environment, replacing the ancestral bacteria. This occurs in lethal selections such as for antibiotic resistance among a sensitive population. Such sweeps result in a new but genetically homogeneous population. Other, more complex, types of population shift can also occur. In large populations or if the mutation supply is high, several different beneficial mutations may be present simultaneously. This scenario is likely to be common in the large populations that occur in nature. In laboratory evolution experiments that use large populations (109 to 1010 bacteria), the appearance of several beneficial mutations is more the norm. Experiments have revealed a range of conditions under which bacterial diversity is generated in a relatively short period (200, 270, 280). The classic scenario for such divergence is that if the new environment has alternative niches and is itself heterogeneous, then different bacteria may adapt toward the different niches (270). As shown in Table 1, if alternative carbon sources are present upon the transition, then some bacteria can specialize on one and some on the other nutrient (171, 377). This ecological explanation of diversification entirely depends on the new environment shown in Fig. 2. If the new environment results in a biofilm with its distinct zones for example, diversity can later arise in this niche-dependent manner (265).
Table 1.
Proposed models of diversification leading to heterogeneity in populations
| Proposed mechanism | Example(s) | Reference(s) |
|---|---|---|
| Alternative niches or resources in the new environment | Specialization on one of several carbon sources or one of several locations (e.g., air-water interface) in the environment | 171, 377 |
| Mutation-selection balance | Can occur when a high mutation rate and the slow fixation of mutations lead to several coexisting genotypes in a population | 74, 374 |
| Negative frequency-dependent selection | The competitive fitness of a type increases as it becomes rarer so does not eliminate competitor | 178 |
| Trade-offs | With a SPaNC trade-off, a type with higher stress resistance but a lower growth rate coexists with a variant with less resistance but a better growth rate; trade-offs can also lead to coexistence of rate yield variants | 126, 127 |
| Trade-offs plus frequency-dependent selection | Theoretical, no example available | 76 |
| Trade-offs plus mutation rate differences | Theoretical, no example available | 19 |
| Convergent evolution of equally fit types | Different genotypes result from the same regulatory change and provide the same benefit in the selection environment | 125, 353 |
| Niche creation | Cross-feeding between a producer of a fermentation product and an evolved user organism; cannibalism | 107, 280, 284 |
In recent years, the diversification of populations through mutational processes has also been demonstrated even in new environments that are not structured and without multiple nutrient sources. How diversity arises in microevolutionary adaptation is still not resolved, but it can lead to divergence to the point of individuality in less than 100 generations (200). Heterogeneity obtained in this way, together with the lack of complete elimination of competitors, can have several causes and explanations, as further summarized in Table 1.
It is not yet clear which of the proposed mechanisms in Table 1 has a dominant or even contributing role in the diversification of adapting populations. Some, like coexistence based on trade-offs (127), do contribute to the diversity in E. coli chemostat culture populations (199), and negative frequency dependence is also shown by some isolates in the same environment (200). Further experimental evidence for coexistence based on cannibalism or cross feeding also exists (158, 284). Less experimental support (and testing) is currently available for the contributions of more theoretical models (mutation-selection balance, trade-offs, and trade-offs plus frequency-dependent selection in Table 1). Altogether, the evidence is that in large populations a combination of several mechanisms in Table 1 contributes to the overall diversity (198a). None of the mechanisms in Table 1 is disproven, so there is potentially a rich array of selection pathways available to evolving bacteria. It is tempting to conclude that diversity in bacterial populations arises from the wide pool of selection mechanisms as well as the flexible mutation supply.
CONCLUSIONS ON CULTURE HISTORY AS A DETERMINANT OF ADAPTIVE POSSIBILITIES
A common theme within the many experimental examples described in Importance of Culture History in Determining the Nature of Adaptive Responses above is that culture history changes the responses and heterogeneity of bacterial populations. The predictability of adaptive outcomes with a single environmental transition is hence very much dependent on the baseline population history, for both regulatory and mutational responses. The prior growth rate, growth phase, population density, population structure, prior stress exposure, and all the environmental conditions in the history of the organism (e.g., O2 exposure) determine the possibilities for further adaptation. The behavior of bacteria upon transition to a new environment, such as resistance characteristics and capacity to use new resources, is certainly dependent on the baseline set by the culture history.
Most existing results on environmental effects come from exposing bacteria growing exponentially in rich medium to a stress. These experiments use a more or less identical baseline for adaptation. We know very little about what happens to bacteria that are not optimally growing when further stressed or transferred to a better medium, except for early upshift experiments on composition (196). Indeed, to fully understand the adaptomics of bacteria, a whole new series of experiments is required, where adaptation starts not from exponentially growing, unstressed bacteria with excess nutrients but with the many nonoptimal situations and the heterogeneous populations that we discussed above and which are more likely to occur in nature.
The influence of history and many kinds of stresses on mutational processes means that the extent of genetic variation and hence paths to fitness can also differ in stressed and unstressed baseline populations. As shown in Table 2, it is already possible to identify many links between environmental sensing and the mutational processes we have dealt with, and it would not be surprising if more links emerge. Altogether, the cross-interactions between regulation and mutation supply make the adaptomics of populations and microevolutionary outcomes extremely complex and sensitive to small changes in the environment.
Table 2.
Cross-interactions between phenotypic and mutational effects in populations
| Mutational process or effect | Example(s) | Reference(s) |
|---|---|---|
| SIM | DNA damage inducing error-prone polymerases; starvation/stress-mediated increase in RpoS inducing error-prone polymerase | 188, 267, 311 |
| Phase variation | Environmental regulation of switching rates | 18 |
| GDA | Stress-induced amplification | 181 |
| Competence/LGT | DNA uptake regulated by starvation and other stresses | 345 |
| Transient mutation | Mistranslation of DNA repair proteins and stochastic variations | 218 |
| Buffering phenotypes of mutations | Mutational effects altered by chaperones | 202, 329 |
CONCLUSIONS ON THE ADAPTOMIC INGREDIENTS OF EVOLUTIONARY SUCCESS IN BACTERIA
Undoubtedly, microbes in general and bacteria in particular are extremely successful in filling the niches available in the biosphere. Here we consider the importance of the adaptive mechanisms that we have discussed to the global success of bacteria.
Dykhuizen (86) argued that a contributing factor to the evolutionary survival of bacteria is an ability to avoid mass extinctions over geological time, owing to their capacity to withstand rapid changes in environmental conditions. In turn, such survival in fluctuating environments requires highly developed adaptive capabilities. As discussed in “Phenotypic Variation within Populations” above, free-living bacteria indeed have a wide range of environmental sensing and regulatory response mechanisms. In addition to this, we have seen in “Genetic Heterogeneity within Populations” above that mutational variation and selection are well developed in populations of bacteria, which ensures that the confined limits imposed by the blueprint of regulatory responses do not limit bacterial survival. The capacities for adaptation defined above indeed allow evolution of microorganisms to occur rapidly, particularly under strong selective pressures. With mutations that can sweep large populations within days (97), there is a convergence of ecological and evolutionary time scales. In this section, we identify various adaptomic principles underlying the contribution of regulatory and heritable adaptations leading to fitness.
Population Heterogeneity as an Ingredient of Evolutionary Success
A number of characteristics of bacterial populations provide sources of adaptability underpinning the long-term evolutionary success defined by Dykhuizen (86). An essential component of bacterial success is diversity itself. Biodiversity in an ecological sense has long been seen as a stabilizing force and may represent a form of biological insurance to more common environmental responses (229). The Bacteria and Archaea are the most genetically diverse superkingdoms of life (103, 247). Bacteria exhibit not only species diversity but also huge intraspecies variation. In addition to this, populations of individual strains exhibit phenotypic heterogeneity and diversification strategies, as discussed above. Even in genetically similar populations, a large fraction of bacteria exhibit states that differ from that of the majority. The subpopulations may have lower fitness in the extant conditions, even extending to stasis, but may have an advantage upon transition into a new environment. Evolutionary bet-hedging involves a trade-off between the mean and variance of fitness, such that phenotypes with reduced mean fitness may be at a selective advantage under certain conditions (71). Diversity and bet-hedging at every level, from populations to species to superkingdoms, are inherent ingredients to bacterial success.
Whether population heterogeneities contribute to bacterial fitness or not, the experimental evidence for the contribution of heterogeneity to phenotypes is overwhelming; for example, death curves when bacteria are challenged by stresses or antibiotics indicate nonuniform responses. Generally, these do not follow the kinetics expected from homogeneous populations (31). The heterogeneity and presence of more resistant bacteria in populations affect the shape of killing curves or even the growth pattern under stress, as well as phenomena such as persistence with antibiotics. Another example is with subpopulation dynamics at superoptimal temperatures; irregular growth curves were explained by postulating the coexistence of two subpopulations: a more resistant, growing population and a temperature-sensitive, inactivating population (340). Also importantly, the proportion of the more resistant forms is itself a function of medium composition and culture history (31), so the course of adaptation shown in Fig. 3 to 5 is generally not likely to be a uniform response with different culture histories.
In this review, we have focused on the adaptomics of pure clonal populations. To underline the variation even in these individual populations, it is worth bringing together all the heterogeneities operating even at this taxonomic level of bacterial population biology. In Fig. 6, we attempt to collate all the subpopulations we have discussed. We also include, where available, the proportions of populations likely to exist in the subpopulations. We use a hypothetical, composite organism to emphasize the range of variations. Figure 6 undoubtedly contains inaccuracies, but the likelihood is that we underestimate the extent because of yet-undiscovered subpopulations. Also, we ignore subsubgroups with properties due to overlaps in groupings (e.g., cells with both double-strand breaks and GDA) that would also be possible in large populations.
Fig 6.
The background shading represents 109 bacteria, and the inner shapes represent subpopulations with different phenotypic (yellow) or genetic (green) characteristics. Included are characteristics from several species, so not all bacterial populations will have all subpopulations and the proportions are history dependent, as discussed in the text. In mildly stressed cells (e.g., at the onset of starvation or with mild DNA damage), the numbers in the subpopulations are from the closest published values but are indicative rather than absolute; these too are mostly history and genotype dependent. Where these numbers are extremely variable or unknown, 10x is used.
Based on Fig. 6 and the heterogeneities described above in “Phenotypic Variation within Populations” and “Genetic Heterogeneity within Populations,” a conclusion of this review has to be that adaptation upon an environmental transition can be considered a whole-population change only as a gross simplification. Indeed, an important message from this compilation is that the subpopulations in any large bacterial population need to be considered in adaptomics or other areas of microbiology where population behavior is important. Even if all of the transition rates and frequencies of various states could be measured precisely, it is likely that in adaptomics the best one can hope for is a probabilistic understanding of what will occur in any given transition. For high-frequency transitions (such as sporulation, persisters, etc.), the probability of transitions is likely to be predictable, but for rarer events, such as mutations, a Luria-Delbrück jackpot distribution is more likely. Hence, the results of rare outcomes will have a greater variance and influence how a mixed population like that shown in Fig. 6 behaves.
It is likely that we have not covered all sources of heterogeneity in bacterial populations or indeed the important question whether all the heterogeneity is adaptive. For example, some heterogeneity may be linearly derived in daughter cells from parental cells with differing regulator concentrations or mistranslation errors. We have generally assumed that the heterogeneity is adaptively useful, although the detailed evidence for this is incomplete. A case in point is the mistranslation heterogeneity in cells, which may be nonadaptive accidents of errors in cellular processes. A future need is to obtain evidence for or against the adaptive nature of heterogeneities at every level.
Population Evolvability as an Ingredient of Evolutionary Success
Another identifiable component of bacterial success is evolvability, the capacity of bacteria to change mutational and selectional processes under stress (112, 301). The availability of mutations in populations is far from fixed, and the processes leading to altered mutation rates are increasingly understood. In several examples discussed above, the settings of regulatory processes, determined by the environment, inform and direct the cell toward mutational changes. The theme of diversity or population heterogeneity also impacts evolvability. Mutator subpopulations can represent several percent of many pathogen populations and are a source of beneficial mutations. In addition, some explanations of elevated mutation rates in nonmutator bacteria depend on SIM subpopulations and limiting mutations in space and time (112). The high availability of genome amplifications, both in gene dosage effects and perhaps in generating “adaptive” mutations (262), is another ingredient in generating genetic variation. In any case, mutation supply is not constant across a population that has constitutive mutators, cells with elevated error-prone polymerases, and genome-amplified subpopulations on top of a bulk population with “normal” spontaneous mutation rates. Evolvability can hence vary with the proportion of genome-altered subpopulations. Evolution involves a balance between bacterial integrity maintained by minimizing deleterious mutations under healthy vegetative conditions and switching to greater variation in times of stress.
An essential link in evolvability is the ability to translate regulatory responses sensitive to the environment to the generation of genetic heterogeneity. Mutation supply becomes significant especially in stressed situations when beneficial mutations are most useful. A clear pattern of regulatory and mutational adaptations being linked emerges. Genetic heterogeneity arising via genome duplications or error-prone DNA replication, as well as the other mechanisms mentioned in “Genetic Heterogeneity within Populations” above, is linked to stress responses in partly defined ways. It really does not matter for bacteria whether “adaptive” mutations during starvation occur via duplications or SOS- plus RpoS-mediated stress responses, but it is environmental inputs that stimulate genetic variation in both.
Metabolic and Regulatory Redundancy as an Ingredient of Evolutionary Success
A considerable evolutionary advantage of adapting bacteria is the degeneracy in function and robustness that bacteria encode (352). It is common for bacteria to have more than one protein with the capacity to influence the same metabolic step or the regulation of the same genes. This degeneracy aids adaptation because the capacity to lose or change by mutation some component of function or regulation does not kill the cell. Redundancy and robustness built into bacteria not only help survival in the presence of deleterious mutations (183, 351, 354) but could actually increase the number of loci for beneficial, adaptive changes (175, 353). In this respect, degeneracy contributes to the selectability of diversity that can evolve during adaptation, as discussed in “Genetic Heterogeneity within Populations” above.
In multicomponent regulatory networks or signal transduction pathways, degeneracy allows mutations in different genes to result in the same phenotype. A good example of this is within the general stress response controlled by RpoS (17) and the cost that this imposes on the SPaNC balance (95). Recent evidence of degenerate mechanisms to obtain mutational adaptive shifts in the SPaNC balance and RpoS levels comes from population changes in adaptation to nutrient limitation in the laboratory (353) or in pathogen populations adapting to an extraintestinal environment (177). A similar phenotype in terms of SPaNC balance can come from mutations in rpoS itself, hfq, spoT, and rssB (198a, 353) and others that indirectly influence RpoS levels (177). The mutational degeneracy of changing RpoS levels reinforces the huge diversity in RpoS across the species E. coli (51, 98). The similar effect on SPaNC is accompanied by very different phenotypic changes with, for example, hfq and spoT mutations (353). These secondary differences provide the ingredients for phenotypic diversification seen in populations and across the species.
CONCLUSIONS ON THE PREDICTABILITY OF ADAPTATION: FROM SINGLE TRANSITIONS TO EVOLUTIONARY OUTCOMES
Evolution is deceptively simple in Darwinian terms, consisting of fitness based on genetic variation and selection. For example, these ingredients can be analyzed and modeled by simplifying reality, by assuming particular mutation rates and fixed selection coefficients that allow a description of the fixation of fitter organisms (132). In some cases, adjusting for biological variation by assuming distributions of character values, as done in population biology, is undertaken (245). However, this is also mistaken, as the heterogeneities described in this review are generally not a continuum but often occur with distinctly different patterns (as in bimodal on-off stochastic variation, persister growth/nongrowth, or stress-induced/basal mutation rates differing by orders of magnitude) and with different effects on parts of the chromosome or sets of genes. This consideration of population heterogeneity reinforces the problem of “frail hypotheses” in population biology identified by Ninio (233). To further quote Ninio: “I suggest that population geneticists should invest more effort in refining the numerical values of the critical parameters used in their models. They should take into account the recent proposals on how mutations arise. They should also pay more attention to phenotypic variations, and develop criteria to discriminate between proposed evolutionary mechanisms that can actually work, and others that cannot.” We fully concur.
To extrapolate single-transition adaptations to long-term evolutionary outcomes is probably premature. To understand how populations evolve over several rounds of mutation and selection, it needs to be realized that population heterogeneities increase with continued growth and continued selection, even in simple environments. Many studies in the past 10 years have used bacteria in experimental evolution, in which an ancestor is subjected to a particular environment(s) for extended periods (64, 88). These may be through continued subculturing in the same medium or through continuous cultures kept over many days. An unstated assumption in studies of evolution, intrinsic to many experimental evolution studies, including our own (110, 200, 353), is that we start with an ancestor population that is genotypically and phenotypically homogeneous. As discussed above, this overlooks the heterogeneities in bacterial populations. Thus, a more realistic description of experiments started with a clonal ancestral population is that at the start of the evolution experiments, we already start with many distinct subpopulations. The problem is that the mutation rate and benefit of mutations may be different in each subpopulation. There may well be several subexperiments going on, with lineages appearing at very early times in adaptation. This interpretation is entirely consistent with the extremely rapid divergence of evolving populations even in the simplest media (200), which may also depend on a multitude of selection mechanisms (Table 1) that provide alternative pathways to fitness.
Future adaptomics studies will need to investigate whether some of the adaptational divergence seen in effectively all experimental evolution studies of bacterial populations has its seeds in ancestral heterogeneity. Additionally, a deeper analysis of the selection steps in a single environmental transition is needed.
Models of growth and organismic function are targets of systems biology (64, 246), and here again, it is essential to question whether population-level behavior is fully covered by existing models. Heterogeneity and growth history features set challenges in metabolic networks even if stochastic features are incorporated. As in adaptation/evolution studies, systems biology involving mutational or environmental perturbations needs to take into account the history and heterogeneity of the populations studied.
As a final thought, we suggest that progress in the adaptomics of populations can contribute to the science of microbiology in further areas beyond systems biology and population biology. First, our grasp of bacterial behavior will expand by using baseline cultures other than exponential batch cultures as the norm in physiological adaptation studies. Second, research on how heterogeneity contributes to infection processes will influence our thinking about the diagnosis and combating of infectious diseases. Third, the era of “pure culture” microbiology is nearing an end, and heterogeneity is going to be integral to our understanding of microbial processes, not just in ecosystems but also in pure clone experiments. Even in the genomics of microbes, a higher accuracy in sequencing technology and more careful analysis will undoubtedly reveal the metagenomics of intrapopulation clonal diversity suggested by Fig. 6.
ACKNOWLEDGMENTS
We thank current and previous colleagues, especially Ram Maharjan, for helping us to obtain a broad understanding of adaptive processes, although they are not to be blamed for our errors and omissions.
This work was supported by U.S. Army Research Office grant W911NF1010045 and grants from the Australian Research Council to T.F.
REFERENCES
- 1. Al Mamun AA, Gautam S, Humayun MZ. 2006. Hypermutagenesis in mutA cells is mediated by mistranslational corruption of polymerase, and is accompanied by replication fork collapse. Mol. Microbiol. 62:1752–1763 [DOI] [PubMed] [Google Scholar]
- 2. Al Mamun AA, Marians KJ, Humayun MZ. 2002. DNA polymerase III from Escherichia coli cells expressing mutA mistranslator tRNA is error-prone. J. Biol. Chem. 277:46319–46327 [DOI] [PubMed] [Google Scholar]
- 3. Al Mamun AA, Rahman MS, Humayun MZ. 1999. Escherichia coli cells bearing mutA, a mutant glyV tRNA gene, express a recA-dependent error-prone DNA replication activity. Mol. Microbiol. 33:732–740 [DOI] [PubMed] [Google Scholar]
- 4. Anderson P, Roth J. 1981. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between ribosomal RNA (Rrn) cistrons. Proc. Natl. Acad. Sci. U. S. A. 78:3113–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andersson DI, Hughes D. 2009. Gene amplification and adaptive evolution in bacteria. Annu. Rev. Genet. 43:167–195 [DOI] [PubMed] [Google Scholar]
- 6. Andersson DI, Slechta ES, Roth JR. 1998. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science 282:1133–1135 [DOI] [PubMed] [Google Scholar]
- 7. Aoki SK, et al. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 309:1245–1248 [DOI] [PubMed] [Google Scholar]
- 8. Artsimovitch I, et al. 2004. Structural basis for transcription regulation by alarmone ppGpp. Cell 117:299–310 [DOI] [PubMed] [Google Scholar]
- 9. Avery SV. 2006. Microbial cell individuality and the underlying sources of heterogeneity. Nat. Rev. Microbiol. 4:577–587 [DOI] [PubMed] [Google Scholar]
- 10. Babic A, Lindner AB, Vulic M, Stewart EJ, Radman M. 2008. Direct visualization of horizontal gene transfer. Science 319:1533–1536 [DOI] [PubMed] [Google Scholar]
- 11. Bacher JM, Waas WF, Metzgar D, de Crecy-Lagard V, Schimmel P. 2007. Genetic code ambiguity confers a selective advantage on Acinetobacter baylyi. J. Bacteriol. 189:6494–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625 [DOI] [PubMed] [Google Scholar]
- 13. Balbinder E. 1993. Multiple pathways of deletion formation in Escherichia coli. Mutat. Res. 299:193–209 [DOI] [PubMed] [Google Scholar]
- 14. Balbinder E. 2001. Stationary phase deletions in Escherichia coli. I. Evidence for a new deletion pathway. Mutat. Res. 479:19–36 [DOI] [PubMed] [Google Scholar]
- 15. Balleza E, et al. 2009. Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol. Rev. 33:133–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bassler BL, Losick R. 2006. Bacterially speaking. Cell 125:237–246 [DOI] [PubMed] [Google Scholar]
- 17. Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65:189–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bayliss CD. 2009. Determinants of phase variation rate and the fitness implications of differing rates for bacterial pathogens and commensals. FEMS Microbiol. Rev. 33:504–520 [DOI] [PubMed] [Google Scholar]
- 19. Beardmore RE, Gudelj I, Lipson DA, Hurst LD. 2011. Metabolic trade-offs and the maintenance of the fittest and the flattest. Nature 472:342–346 [DOI] [PubMed] [Google Scholar]
- 20. Beaumont HJE, Gallie J, Kost C, Ferguson GC, Rainey PB. 2009. Experimental evolution of bet hedging. Nature 462:90–97 [DOI] [PubMed] [Google Scholar]
- 21. Becskei A, Serrano L. 2000. Engineering stability in gene networks by autoregulation. Nature 405:590–593 [DOI] [PubMed] [Google Scholar]
- 22. Bermejo C, Haerizadeh F, Takanaga H, Chermak D, Frommer WB. 2011. Optical sensors for measuring dynamic changes of cytosolic metabolite levels in yeast. Nat. Protoc. 6:1806–1817 [DOI] [PubMed] [Google Scholar]
- 23. Bettenbrock K, et al. 2007. Correlation between growth rates, EIIA(Crr) phosphorylation, and intracellular cyclic AMP levels in Escherichia coli K-12. J. Bacteriol. 189:6891–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bhagwat AA, et al. 2006. Functional heterogeneity of RpoS in stress tolerance of enterohemorrhagic Escherichia coli strains. Appl. Environ. Microbiol. 72:4978–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bichara M, Wagner J, Lambert IB. 2006. Mechanisms of tandem repeat instability in bacteria. Mutat. Res. 598:144–163 [DOI] [PubMed] [Google Scholar]
- 26. Bigger JW. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet ii:497–500 [Google Scholar]
- 27. Bishop AL, Rab FA, Sumner ER, Avery SV. 2007. Phenotypic heterogeneity can enhance rare-cell survival in ‘stress-sensitive’ yeast populations. Mol. Microbiol. 63:507–520 [DOI] [PubMed] [Google Scholar]
- 28. Bjedov I, et al. 2003. Stress-induced mutagenesis in bacteria. Science 300:1404–1409 [DOI] [PubMed] [Google Scholar]
- 29. Black DS, Kelly AJ, Mardis MJ, Moyed HS. 1991. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 173:5732–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boezi JA, Cowie DB. 1961. Kinetic studies of β-galactosidase induction. Biophys. J. 1:639–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Booth IR. 2002. Stress and the single cell: intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Int. J. Food Microbiol. 78:19–30 [DOI] [PubMed] [Google Scholar]
- 32. Braaten BA, Nou X, Kaltenbach LS, Low DA. 1994. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell 76:577–588 [DOI] [PubMed] [Google Scholar]
- 33. Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 98:11621–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bremer H, Dennis PP. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p 1553–1569 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 35. Broadbent SE, Davies MR, van der Woude MW. 2010. Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Mol. Microbiol. 77:337–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brockhurst MA, Habets M, Libberton B, Buckling A, Gardner A. 2010. Ecological drivers of the evolution of public-goods cooperation in bacteria. Ecology 91:334–340 [DOI] [PubMed] [Google Scholar]
- 37. Brockhurst MA, Hochberg ME, Bell T, Buckling A. 2006. Character displacement promotes cooperation in bacterial biofilms. Curr. Biol. 16:2030–2034 [DOI] [PubMed] [Google Scholar]
- 38. Brown EW, LeClerc JE, Li BG, Payne WL, Cebula TA. 2001. Phylogenetic evidence for horizontal transfer of mutS alleles among naturally occurring Escherichia coli strains. J. Bacteriol. 183:1631–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brown MRW, Collier PJ, Gilbert P. 1990. Influence of growth rate on susceptibility to antimicrobial agents: modification of the cell envelope and batch and continuous culture studies. Antimicrob. Agents Chemother. 34:1623–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bull HJ, Lombardo MJ, Rosenberg SM. 2001. Stationary-phase mutation in the bacterial chromosome: recombination protein and DNA polymerase IV dependence. Proc. Natl. Acad. Sci. U. S. A. 98:8334–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Campbell AM. 1996. Bacteriophages, p 2325–2338 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed American Society for Microbiology, Washington, DC [Google Scholar]
- 42. Campbell AM. 2000. Lateral gene transfer in prokaryotes. Theor. Popul. Biol. 57:71–77 [DOI] [PubMed] [Google Scholar]
- 43. Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brussow H. 2003. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6:417–424 [DOI] [PubMed] [Google Scholar]
- 44. Cascales E, et al. 2007. Colicin biology. Microbiol. Mol. Biol. Rev. 71:158–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cashel M, Gentry D, Hernandez VJ, Vinella D. 1996. The stringent response, p 1458–1496 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed American Society for Microbiology, Washington, DC [Google Scholar]
- 46. Chang DE, Smalley DJ, Conway T. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45:289–306 [DOI] [PubMed] [Google Scholar]
- 47. Chao L, McBroom SM. 1985. Evolution of transposable elements: an IS10 insertion increases fitness in Escherichia coli. Mol. Biol. Evol. 2:359–369 [DOI] [PubMed] [Google Scholar]
- 48. Chao L, Vargas C, Spear BB, Cox EC. 1983. Transposable elements as mutator genes in evolution. Nature 303:634–635 [DOI] [PubMed] [Google Scholar]
- 49. Chen F, et al. 2010. Multiple genetic switches spontaneously modulating bacterial mutability. BMC Evol. Biol. 10:277 doi:10.1186/1471-2148-10-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen F, et al. 2010. mutL as a genetic switch of bacterial mutability: turned on or off through repeat copy number changes. FEMS Microbiol. Lett. 312:126–132 [DOI] [PubMed] [Google Scholar]
- 51. Chiang SM, Dong T, Edge TA, Schellhorn HE. 2011. Phenotypic diversity caused by differential RpoS activity among environmental Escherichia coli isolates. Appl. Environ. Microbiol. 77:7915–7923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chiang SM, Schellhorn HE. 2010. Evolution of the RpoS regulon: origin of RpoS and the conservation of RpoS-dependent regulation in bacteria. J. Mol. Evol. 70:557–571 [DOI] [PubMed] [Google Scholar]
- 53. Chou HH, Berthet J, Marx CJ. 2009. Fast growth increases the selective advantage of a mutation arising recurrently during evolution under metal limitation. PLoS Genet. 5:652 doi:10.1371/journal.pgen.1000652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. 2001. ReIE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. U. S. A. 98:14328–14333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chung JD, Stephanopoulos G, Ireton K, Grossman AD. 1994. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J. Bacteriol. 176:1977–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Clark DR, et al. 2009. Evolution of altruists and cheaters in near-isogenic populations of Escherichia coli. Front. Biosci. 14:4815–4824 [DOI] [PubMed] [Google Scholar]
- 57. Claverys JP, Havarstein LS. 2007. Cannibalism and fratricide: mechanisms and raisons d'etre. Nat. Rev. Microbiol. 5:219–229 [DOI] [PubMed] [Google Scholar]
- 58. Claverys JP, Havarstein LS. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7:d1798–d1814 [DOI] [PubMed] [Google Scholar]
- 59. Claverys JP, Martin B, Havarstein LS. 2007. Competence-induced fratricide in streptococci. Mol. Microbiol. 64:1423–1433 [DOI] [PubMed] [Google Scholar]
- 60. Claverys JP, Prudhomme M, Martin B. 2006. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 60:451–475 [DOI] [PubMed] [Google Scholar]
- 61. Reference deleted.
- 62. Cohen SE, Godoy VG, Walker GC. 2009. Transcriptional modulator NusA interacts with translesion DNA polymerases in Escherichia coli. J. Bacteriol. 191:665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cohen SE, Walker GC. 2010. The transcription elongation factor NusA is required for stress-induced mutagenesis in Escherichia coli. Curr. Biol. 20:80–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Conrad TM, Lewis NE, Palsson BO. 2011. Microbial laboratory evolution in the era of genome-scale science. Mol. Syst. Biol. 7:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cooper TF. 2007. Recombination speeds adaptation by reducing competition between beneficial mutations in populations of Escherichia coli. PLoS Biol. 5:1899–1905 doi:10.1371/journal.pbio.0050225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cooper TF, Remold SK, Lenski RE, Schneider D. 2008. Expression profiles reveal parallel evolution of epistatic interactions involving the CRP regulon in Escherichia coli. PLoS Genet. 4:e35 doi:10.1371/journal.pgen.0040035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cullum J, Vicente M. 1978. Cell-growth and length distribution in Escherichia coli. J. Bacteriol. 134:330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Davey HM, Kell DB. 1996. Flow cytometry and cell sorting of heterogeneous microbial populations—the importance of single-cell analyses. Microbiol. Rev. 60:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Davidson CJ, Surette MG. 2008. Individuality in bacteria. Annu. Rev. Genet. 42:253–268 [DOI] [PubMed] [Google Scholar]
- 70. Dawes IW, Kay D, Mandelst J. 1969. Sporulation in Bacillus subtilis. Establishment of a time scale for morphological events. J. Gen. Microbiol. 56:171. [DOI] [PubMed] [Google Scholar]
- 71. de Jong IG, Haccou P, Kuipers OP. 2011. Bet hedging or not? A guide to proper classification of microbial survival strategies. Bioessays 33:215–223 [DOI] [PubMed] [Google Scholar]
- 72. Denamur E, et al. 2002. High frequency of mutator strains among human uropathogenic Escherichia coli isolates. J. Bacteriol. 184:605–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Denamur E, et al. 2000. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell 103:711–721 [DOI] [PubMed] [Google Scholar]
- 74. Desai MM, Fisher DS. 2007. Beneficial mutation selection balance and the effect of linkage on positive selection. Genetics 176:1759–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. de Visser JAGM, Zeyl CW, Gerrish PJ, Blanchard JL, Lenski RE. 1999. Diminishing returns from mutation supply rate in asexual populations. Science 283:404–406 [DOI] [PubMed] [Google Scholar]
- 76. Doebeli M, Ispolatov I. 2010. Complexity and diversity. Science 328:494–497 [DOI] [PubMed] [Google Scholar]
- 77. Dorazi R, Lingutla JJ, Humayun MZ. 2002. Expression of mutant alanine tRNAs increases spontaneous mutagenesis in Escherichia coli. Mol. Microbiol. 44:131–141 [DOI] [PubMed] [Google Scholar]
- 78. Dorr T, Vulic M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8:317. 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Drake JW. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. U. S. A. 88:7160–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Drake JW, Charlesworth B, Charlesworth D, Crow JF. 1998. Rates of spontaneous mutation. Genetics 148:1667–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Drummond DA, Wilke CO. 2009. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 10:715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dubey GP, Ben-Yehuda S. 2011. Intercellular nanotubes mediate bacterial communication. Cell 144:590–600 [DOI] [PubMed] [Google Scholar]
- 83. Dublanche Y, Michalodimitrakis K, Kummerer N, Foglierini M, Serrano L. 2006. Noise in transcription negative feedback loops: simulation and experimental analysis. Mol. Syst. Biol. 2:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dubnau D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217–244 [DOI] [PubMed] [Google Scholar]
- 85. Dubnau D, Losick R. 2006. Bistability in bacteria. Mol. Microbiol. 61:564–572 [DOI] [PubMed] [Google Scholar]
- 86. Dykhuizen DE. 1998. Santa Rosalia revisited—why are there so many species of bacteria. Antonie Van Leeuwenhoek 73:25–33 [DOI] [PubMed] [Google Scholar]
- 87. Eichenbaum Z, Livneh Z. 1998. UV light induces IS10 transposition in Escherichia coli. Genetics 149:1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Elena SF, Lenski RE. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4:457–469 [DOI] [PubMed] [Google Scholar]
- 89. Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. 2006. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell 124:549–559 [DOI] [PubMed] [Google Scholar]
- 90. Fay JC, Wittkopp PJ. 2008. Evaluating the role of natural selection in the evolution of gene regulation. Heredity 100:191–199 [DOI] [PubMed] [Google Scholar]
- 91. Feliziani S, et al. 2010. Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLoS One 5:e12669 doi:10.1371/journal.pone.0012669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Feng G, Tsui HCT, Winkler ME. 1996. Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. J. Bacteriol. 178:2388–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ferenci T. 1996. Adaptation to life at micromolar nutrient levels: the regulation of Escherichia coli glucose transport by endoinduction and cAMP. FEMS Microbiol. Rev. 18:301–317 [DOI] [PubMed] [Google Scholar]
- 94. Ferenci T. 2008. Bacterial physiology, regulation and mutational adaptation in a chemostat environment. Adv. Microb. Physiol. 53:169–229 [DOI] [PubMed] [Google Scholar]
- 95. Ferenci T. 2005. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol. Microbiol. 57:1–8 [DOI] [PubMed] [Google Scholar]
- 96. Ferenci T. 1999. Regulation by nutrient limitation. Curr. Opin. Microbiol. 2:208–213 [DOI] [PubMed] [Google Scholar]
- 97. Ferenci T. 2008. The spread of a beneficial mutation in experimental bacterial populations: the influence of the environment and genotype on the fixation of rpoS mutations. Heredity 100:446–452 [DOI] [PubMed] [Google Scholar]
- 98. Ferenci T, Galbiati H, Betteridge T, Phan K, Spira B. 2011. The constancy of global regulation across a species: the concentrations of ppGpp and RpoS are strain-specific in Escherichia coli. BMC Microbiol. 11:62 doi:10.1186/1471-2180-11-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Fernandez de Henestrosa AR, et al. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35:1560–1572 [DOI] [PubMed] [Google Scholar]
- 100. Finkel SE. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat. Rev. Microbiol. 4:113–120 [DOI] [PubMed] [Google Scholar]
- 101. Foster JW, Spector MP. 1995. How Salmonella survive against the odds. Annu. Rev. Microbiol. 49:145–174 [DOI] [PubMed] [Google Scholar]
- 102. Foster PL. 2007. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42:373–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. 2009. The bacterial species challenge: making sense of genetic and ecological diversity. Science 323:741–746 [DOI] [PubMed] [Google Scholar]
- 104. Fraser D, Kaern M. 2009. A chance at survival: gene expression noise and phenotypic diversification strategies. Mol. Microbiol. 71:1333–1340 [DOI] [PubMed] [Google Scholar]
- 105. Freed NE, et al. 2008. A simple screen to identify promoters conferring high levels of phenotypic noise. PLoS Genet. 4:e1000307 doi:10.1371/journal.pgen.1000307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Friedberg EC, et al. 2006. DNA repair and mutagenesis, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 107. Friesen ML, Saxer G, Travisano M, Doebeli M. 2004. Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution 58:245–260 [PubMed] [Google Scholar]
- 108. Fujita M, Gonzalez-Pastor JE, Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187:1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Funchain P, et al. 2000. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics 154:959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Gaffe J, et al. 2011. Insertion sequence-driven evolution of Escherichia coli in chemostats. J. Mol. Evol. 72:398–412 [DOI] [PubMed] [Google Scholar]
- 111. Galhardo RS, et al. 2009. DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics 182:55–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Galhardo RS, Hastings PJ, Rosenberg SM. 2007. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 42:399–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Galitski T, Roth JR. 1997. Pathways for homologous recombination between chromosomal direct repeats in Salmonella typhimurium. Genetics 146:751–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Gerdes K, Christensen SK, Lobner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371–382 [DOI] [PubMed] [Google Scholar]
- 115. Gerdes K, Rasmussen PB, Molin S. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. U. S. A. 83:3116–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Gerrish PJ, Lenski RE. 1998. The fate of competing beneficial mutations in an asexual population. Genetica 103:127–144 [PubMed] [Google Scholar]
- 117. Gibson JL, et al. 2010. The sigma E stress response is required for stress-induced mutation and amplification in Escherichia coli. Mol. Microbiol. 77:415–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Gilbert P, Maira-Litran T, McBain AJ, Rickard AH, Whyte FW. 2002. The physiology and collective recalcitrance of microbial biofilm communities. Adv. Microb. Physiol. 46:203–256 [PubMed] [Google Scholar]
- 119. Giraud A, Radman M, Matic I, Taddei F. 2001. The rise and fall of mutator bacteria. Curr. Opin. Microbiol. 4:582–585 [DOI] [PubMed] [Google Scholar]
- 120. Godoy VG, et al. 2007. UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol. Cell 28:1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Golding I, Paulsson J, Zawilski SM, Cox EC. 2005. Real-time kinetics of gene activity in individual bacteria. Cell 123:1025–1036 [DOI] [PubMed] [Google Scholar]
- 122. Goldsmith M, Sarov-Blat L, Livneh Z. 2000. Plasmid-encoded MucB protein is a DNA polymerase (pol RI) specialized for lesion bypass in the presence of MucA′, RecA, and SSB. Proc. Natl. Acad. Sci. U. S. A. 97:11227–11231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Gonzalez-Pastor JE. 2011. Cannibalism: a social behavior in sporulating Bacillus subtilis. FEMS Microbiol. Rev. 35:415–424 [DOI] [PubMed] [Google Scholar]
- 124. Gonzalez-Pastor JE, Hobbs EC, Losick R. 2003. Cannibalism by sporulating bacteria. Science 301:510–513 [DOI] [PubMed] [Google Scholar]
- 125. Gresham D, et al. 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 4:e1000303 doi:10.1371/journal.pgen.1000303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Gudelj I, Beardmore RE, Arkin SS, Maclean RC. 2007. Constraints on microbial metabolism drive evolutionary diversification in homogeneous environments. J. Evol. Biol. 20:1882–1889 [DOI] [PubMed] [Google Scholar]
- 127. Gudelj I, et al. 2010. An integrative approach to understanding microbial diversity: from intracellular mechanisms to community structure. Ecol. Lett. 13:1073–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Guiral S, Mitchell TJ, Martin B, Claverys JP. 2005. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc. Natl. Acad. Sci. U. S. A. 102:8710–8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Haack KR, Roth JR. 1995. Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics 141:1245–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Haagmans W, Van Der Woude M. 2000. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol. Microbiol. 35:877–887 [DOI] [PubMed] [Google Scholar]
- 131. Hahn MW, Höfle MG. 1999. Flagellate predation on a bacterial model community: interplay of size-selective grazing, specific bacterial cell size, and bacterial community composition. Appl. Environ. Microbiol. 65:4863–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Haldane JBS. 1927. The mathematical theory of natural and artificial selection. V. Selection and mutation. Proc. Camb. Philos. Soc. 23:838–844 [Google Scholar]
- 133. Harris RS, et al. 1999. Mismatch repair is diminished during stationary-phase mutation. Mutat. Res. 437:51–60 [PubMed] [Google Scholar]
- 134. Harris RS, et al. 1997. Mismatch repair protein MutL becomes limiting during stationary-phase mutation. Genes Dev. 11:2426–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Harrison F, Browning LE, Vos M, Buckling A. 2006. Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol. 4:21 doi:10.1186/1741-7007-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Hastings PJ. 2007. Adaptive amplification. Crit. Rev. Biochem. Mol. Biol. 42:271–283 [DOI] [PubMed] [Google Scholar]
- 137. Hastings PJ, Lupski JR, Rosenberg SM, Ira G. 2009. Mechanisms of change in gene copy number. Nat. Rev. Genet. 10:551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hastings PJ, Slack A, Petrosino JF, Rosenberg SM. 2004. Adaptive amplification and point mutation are independent mechanisms: evidence for various stress-inducible mutation mechanisms. PLoS Biol. 2:e399 doi:10.1371/journal.pbio.0020399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Havarstein LS, Martin B, Johnsborg O, Granadel C, Claverys JP. 2006. New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol. Microbiol. 59:1297–1307 [DOI] [PubMed] [Google Scholar]
- 140. Hayes F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496–1499 [DOI] [PubMed] [Google Scholar]
- 141. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 142. Hengge-Aronis R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hengge-Aronis R. 1993. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72:165–168 [DOI] [PubMed] [Google Scholar]
- 144. Hirakawa H, Hayashi-Nishino M, Yamaguchi A, Nishino K. 2010. Indole enhances acid resistance in Escherichia coli. Microb. Pathog. 49:90–94 [DOI] [PubMed] [Google Scholar]
- 145. Horst JP, Wu T, Marinus MG. 1999. Escherichia coli mutator genes. Trends Microbiol. 7:29–36 [DOI] [PubMed] [Google Scholar]
- 146. Hua Q, Yang C, Oshima T, Mori H, Shimizu K. 2004. Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures. Appl. Environ. Microbiol. 70:2354–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Imhof M, Schlotterer C. 2001. Fitness effects of advantageous mutations in evolving Escherichia coli populations. Proc. Natl. Acad. Sci. U. S. A. 98:1113–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Ingraham JL, Maaloe O, Neidhardt FC. 1983. Growth of the bacterial cell. Sinauer Associates, Sunderland, MA [Google Scholar]
- 149. Ito A, Taniuchi A, May T, Kawata K, Okabe S. 2009. Increased antibiotic resistance of Escherichia coli in mature biofilms. Appl. Environ. Microbiol. 75:4093–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Jarosz DF, Beuning PJ, Cohen SE, Walker GC. 2007. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 15:70–77 [DOI] [PubMed] [Google Scholar]
- 151. Jishage M, Ishihama A. 1997. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J. Bacteriol. 179:959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Johnsborg O, Eldholm V, Bjornstad ML, Havarstein LS. 2008. A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae and related commensal species. Mol. Microbiol. 69:245–253 [DOI] [PubMed] [Google Scholar]
- 153. Kaiser D, Losick R. 1993. How and why bacteria talk to each other. Cell 73:873–885 [DOI] [PubMed] [Google Scholar]
- 154. Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 19:3083–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Kelly CD, Rahn O. 1932. The growth rate of individual bacterial cells. J. Bacteriol. 23:147–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. King T, Ishihama A, Kori A, Ferenci T. 2004. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J. Bacteriol. 186:5614–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Kinnersley MA, Holben WE, Rosenzweig F. 2009. E unibus plurum: genomic analysis of an experimentally evolved polymorphism in Escherichia coli. PLoS Genet. 5:713 doi:10.1371/journal.pgen.1000713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Klumpp S, Zhang ZG, Hwa T. 2009. Growth rate-dependent global effects on gene expression in bacteria. Cell 139:1366–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Kolter R, Siegele DA, Tormo A. 1993. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47:855–874 [DOI] [PubMed] [Google Scholar]
- 161. Konovalova A, Sogaard-Andersen L. 2011. Close encounters: contact-dependent interactions in bacteria. Mol. Microbiol. 81:297–301 [DOI] [PubMed] [Google Scholar]
- 162. Korch SB, Henderson TA, Hill TM. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50:1199–1213 [DOI] [PubMed] [Google Scholar]
- 163. Kornberg A, Rao NN, Ault-Riche D. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89–125 [DOI] [PubMed] [Google Scholar]
- 164. Kotewicz ML, Brown EW, Leclerc JE, Cebula TA. 2003. Genomic variability among enteric pathogens: the case of the mutS-rpoS intergenic region. Trends Microbiol. 11:2–6 [DOI] [PubMed] [Google Scholar]
- 165. Kvint K, Farewell A, Nystrom T. 2000. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of sigma(s). J. Biol. Chem. 275:14795–14798 [DOI] [PubMed] [Google Scholar]
- 166. Layton JC, Foster PL. 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 50:549–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Leclerc JE, Li BG, Payne WL, Cebula TA. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208–1211 [DOI] [PubMed] [Google Scholar]
- 168. Lee HH, Molla MN, Cantor CR, Collins JJ. 2010. Bacterial charity work leads to population-wide resistance. Nature 467:82–U113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Lee JH, Lee J. 2010. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 34:426–444 [DOI] [PubMed] [Google Scholar]
- 170. Leenanon B, Drake MA. 2001. Acid stress, starvation, and cold stress affect poststress behavior of Escherichia coli O157:H7 and nonpathogenic Escherichia coli. J. Food Prot. 64:970–974 [DOI] [PubMed] [Google Scholar]
- 171. Le Gac M, et al. 2008. Metabolic changes associated with adaptive diversification in Escherichia coli. Genetics 178:1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Lehner AF, Hill CW. 1980. Involvement of ribosomal ribonucleic acid operons in Salmonella typhimurium chromosomal rearrangements. J. Bacteriol. 143:492–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Leisner M, Stingl K, Frey E, Maier B. 2008. Stochastic switching to competence. Curr. Opin. Microbiol. 11:553–559 [DOI] [PubMed] [Google Scholar]
- 174. Lemonnier M, et al. 2008. The evolution of contact-dependent inhibition in non-growing populations of Escherichia coli. Proc. R. Soc. B Biol. Sci. 275:3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Lenski RE, Barrick JE, Ofria C. 2006. Balancing robustness and evolvability. PLoS Biol. 4:2190–2192 doi:10.1371/journal.pbio.0040428 [Google Scholar]
- 176. Lerat E, Daubin V, Ochman H, Moran NA. 2005. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 3:807–814 doi:10.1371/journal.pbio.0030130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Levert M, et al. 2010. Molecular and evolutionary bases of within-patient genotypic and phenotypic diversity in Escherichia coli extraintestinal infections. PLoS Pathog. 6:1125 doi:10.1371/journal.ppat.1001125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Levin BR. 1988. Frequency dependent selection in bacterial populations. Philos. Trans. R. Soc. B 319:459–472 [DOI] [PubMed] [Google Scholar]
- 179. Lewis K. 2010. Persister cells. Annu. Rev. Microbiol. 64:357–372 [DOI] [PubMed] [Google Scholar]
- 180. Li GM. 2008. Mechanisms and functions of DNA mismatch repair. Cell Res. 18:85–98 [DOI] [PubMed] [Google Scholar]
- 181. Lin DX, et al. 2011. Global chromosomal structural instability in a subpopulation of starving Escherichia coli cells. PLoS Genet. 7:2223 doi:10.1371/journal.pgen.1002223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Lin RJ, Capage M, Hill CW. 1984. A repetitive DNA-sequence, Rhs, responsible for duplications within the Escherichia coli K-12 chromosome. J. Mol. Biol. 177:1–18 [DOI] [PubMed] [Google Scholar]
- 183. Lind PA, Berg OG, Andersson DI. 2010. Mutational robustness of ribosomal protein genes. Science 330:825–827 [DOI] [PubMed] [Google Scholar]
- 184. Lindner AB, Madden R, Dernarez A, Stewart EJ, Taddei F. 2008. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. U. S. A. 105:3076–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Liu XQ, Ferenci T. 2001. An analysis of multifactorial influences on the transcriptional control of ompF and ompC porin expression under nutrient limitation. Microbiology 147:2981–2989 [DOI] [PubMed] [Google Scholar]
- 186. Liu XQ, Ng C, Ferenci T. 2000. Global adaptations resulting from high population densities in Escherichia coli cultures. J. Bacteriol. 182:4158–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Llorens JMN, Tormo A, Martinez-Garcia E. 2010. Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 34:476–495 [DOI] [PubMed] [Google Scholar]
- 188. Lombardo MJ, Aponyi I, Rosenberg SM. 2004. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics 166:669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Lopez D, Kolter R. 2010. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol. Rev. 34:134–149 [DOI] [PubMed] [Google Scholar]
- 190. Lopez D, Vlamakis H, Kolter R. 2009. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 33:152–163 [DOI] [PubMed] [Google Scholar]
- 191. López D, Vlamakis H, Kolter R. 2010. Biofilms. Cold Spring Harb. Perspect. Biol. 2:a000398 doi:10.1101/cshperspect.a000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Lopez D, Vlamakis H, Losick R, Kolter R. 2009. Cannibalism enhances biofilm development in Bacillus subtilis. Mol. Microbiol. 74:609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Lukjancenko O, Wassenaar TM, Ussery DW. 2010. Comparison of 61 sequenced Escherichia coli genomes. Microb. Ecol. 60:708–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Lynch M. 2010. Evolution of the mutation rate. Trends Genet. 26:345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Lynch M. 2006. Streamlining and simplification of microbial genome architecture. Annu. Rev. Microbiol. 60:327–349 [DOI] [PubMed] [Google Scholar]
- 196. Maaloe O, Kjeldgaard NO. 1966. Control of macromolecular synthesis. WA Benjamin, New York, NY [Google Scholar]
- 197. Magnusson LU, Farewell A, Nystrom T. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13:236–242 [DOI] [PubMed] [Google Scholar]
- 198. Mah TFC, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39 [DOI] [PubMed] [Google Scholar]
- 198a. Maharjan R, et al. 2012. The multiplicity of divergence mechanisms in a single evolving population. Genome Biol. 13:R41 doi:10.1186/gb-2012-13-6-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198b. Maharjan R, McKenzie C, Yeung A, Ferenci T. The basis of antagonistic pleiotropy in hfq mutations that have opposite effects on fitness at slow and fast growth rates. Heredity, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Maharjan R, Seeto S, Ferenci T. 2007. Divergence and redundancy of transport and metabolic rate-yield strategies in a single Escherichia coli population. J. Bacteriol. 189:2350–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Maharjan R, Seeto S, Notley-McRobb L, Ferenci T. 2006. Clonal adaptive radiation in a constant environment. Science 313:514–517 [DOI] [PubMed] [Google Scholar]
- 201. Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Maisnier-Patin S, et al. 2005. Genomic buffering mitigates the effects of deleterious mutations in bacteria. Nat. Genet. 37:1376–1379 [DOI] [PubMed] [Google Scholar]
- 203. Maisonneuve E, Ezraty B, Dukan S. 2008. Protein aggregates: an aging factor involved in cell death. J. Bacteriol. 190:6070–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U. S. A. 108:13206–13211 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 205. Makinoshima H, Nishimura A, Ishihama A. 2002. Fractionation of Escherichia coli cell populations at different stages during growth transition to stationary phase. Mol. Microbiol. 43:269–279 [DOI] [PubMed] [Google Scholar]
- 206. Mao EF, Lane L, Lee J, Miller JH. 1997. Proliferation of mutators in a cell population. J. Bacteriol. 179:417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Marianovsky I, Aizenman E, Engelberg-Kulka H, Glaser G. 2001. The regulation of the Escherichia coli mazEF promoter involves an unusual alternating palindrome. J. Biol. Chem. 276:5975–5984 [DOI] [PubMed] [Google Scholar]
- 208. Martinez-Antonio A, Janga SC, Thieffry D. 2008. Functional organisation of Escherichia coli transcriptional regulatory network. J. Mol. Biol. 381:238–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Masel J. 2006. Cryptic genetic variation is enriched for potential adaptations. Genetics 172:1985–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Matic I, Rayssiguier C, Radman M. 1995. Interspecies gene exchange in bacteria—the role of SOS and mismatch repair systems in evolution of species. Cell 80:507–515 [DOI] [PubMed] [Google Scholar]
- 211. Matin A. 1991. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol. Microbiol. 5:3–10 [DOI] [PubMed] [Google Scholar]
- 212. Mayr E. 1983. How to carry out the adaptationist program? Am. Nat. 121:324–334 [Google Scholar]
- 213. Mellon I. 2005. Transcription-coupled repair: a complex affair. Mutat. Res. 577:155–161 [DOI] [PubMed] [Google Scholar]
- 214. Merritt JH, Kadouri DE, O'Toole GA. 2011. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 1B:1.1–1.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Mikkola R, Kurland CG. 1992. Selection of laboratory wild-type phenotype from natural isolates of Escherichia coli in chemostats. Mol. Biol. Evol. 9:394–402 [DOI] [PubMed] [Google Scholar]
- 216. Miller JH. 1996. Spontaneous mutators in bacteria—insights into pathways of mutagenesis and repair. Annu. Rev. Microbiol. 50:625–643 [DOI] [PubMed] [Google Scholar]
- 217. Miller JH, et al. 1999. Direct selection for mutators in Escherichia coli. J. Bacteriol. 181:1576–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Miller JH, et al. 2000. Temporary and permanent mutators lacking the mismatch repair system: the enhancement of mutators in cell populations. Cold Spring Harbor Symp. Quant. Biol. 65:241–252 [DOI] [PubMed] [Google Scholar]
- 219. Minezaki Y, Homma K, Nishikawa K. 2005. Genome-wide survey of transcription factors in prokaryotes reveals many bacteria-specific families not found in Archaea. DNA Res. 12:269–280 [DOI] [PubMed] [Google Scholar]
- 220. Mittal N, Budrene EO, Brenner MP, van Oudenaarden A. 2003. Motility of Escherichia coli cells in clusters formed by chemotactic aggregation. Proc. Natl. Acad. Sci. U. S. A. 100:13259–13263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Moura GR, Carreto LC, Santos MA. 2009. Genetic code ambiguity: an unexpected source of proteome innovation and phenotypic diversity. Curr. Opin. Microbiol. 12:631–637 [DOI] [PubMed] [Google Scholar]
- 222. Moxon R, Bayliss C, Hood D. 2006. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 40:307–333 [DOI] [PubMed] [Google Scholar]
- 223. Moyed HS, Bertrand KP. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Moyed HS, Broderick SH. 1986. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 166:399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 192:6191–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Murat D, Goncalves L, Dassa E. 2008. Deletion of the Escherichia coli uup gene encoding a protein of the ATP binding cassette superfamily affects bacterial competitiveness. Res. Microbiol. 159:671–677 [DOI] [PubMed] [Google Scholar]
- 227. Naas T, Blot M, Fitch WM, Arber W. 1995. Dynamics of IS-related genetic rearrangements in resting Escherichia coli K-12. Mol. Biol. Evol. 12:198–207 [DOI] [PubMed] [Google Scholar]
- 228. Nackerdien ZE, Keynan A, Bassler BL, Lederberg J, Thaler DS. 2008. Quorum sensing influences Vibrio harveyi growth rates in a manner not fully accounted for by the marker effect of bioluminescence. PLoS One 3:e1671 doi:10.1371/journal.pone.0001671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Naeem S, Li S. 1997. Biodiversity enhances ecosystem reliability. Nature 390:507–509 [Google Scholar]
- 230. Nanchen A, Schicker A, Revelles O, Sauer U. 2008. Cyclic AMP-dependent catabolite repression is the dominant control mechanism of metabolic fluxes under glucose limitation in Escherichia coli. J. Bacteriol. 190:2323–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Neidhardt FC, et al. (ed). 1996. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 232. Neubauer P, Ahman M, Tornkvist M, Larsson G, Enfors SO. 1995. Response of guanosine tetraphosphate to glucose fluctuations in fed-batch cultivations of Escherichia coli. J. Biotechnol. 43:195–204 [DOI] [PubMed] [Google Scholar]
- 233. Ninio J. 2010. Frail hypotheses in evolutionary biology. PLoS Genet. 6:e1001067 doi:10.1371/journal.pgen.1001067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Ninio J. 1991. Transient mutators: a semiquantitative analysis of the influence of translation and transcription errors on mutation rates. Genetics 129:957–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Noel JT, Pilyugin SS, Narang A. 2009. The diffusive influx and carrier efflux have a strong effect on the bistability of the lac operon in Escherichia coli. J. Theor. Biol. 256:14–28 [DOI] [PubMed] [Google Scholar]
- 236. Notley L, Ferenci T. 1996. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli? J. Bacteriol. 178:1465–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Notley-McRobb L, Death A, Ferenci T. 1997. The relationship between external glucose concentration and cAMP levels inside Escherichia coli—implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiology 143:1909–1918 [DOI] [PubMed] [Google Scholar]
- 238. Notley-McRobb L, Seeto S, Ferenci T. 2002. Enrichment and elimination of mutY mutators in Escherichia coli populations. Genetics 162:1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Notley-McRobb L, Seeto S, Ferenci T. 2003. The influence of cellular physiology on the initiation of mutational pathways in Escherichia coli populations. Proc. R. Soc. Lond. Ser. B Biol. Sci. 270:843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Novick A, Horiuchi T. 1961. Hyper-production of beta-galactosidase by Escherichia coli bacteria. Cold Spring Harbor Symp. Quant. Biol. 26:239–245 [DOI] [PubMed] [Google Scholar]
- 241. Nystrom T. 2004. Stationary-phase physiology. Annu. Rev. Microbiol. 58:161–181 [DOI] [PubMed] [Google Scholar]
- 242. Ochman H, Elwyn S, Moran NA. 1999. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. U. S. A. 96:12638–12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Ogle JM, Ramakrishnan V. 2005. Structural insights into translational fidelity. Annu. Rev. Biochem. 74:129–177 [DOI] [PubMed] [Google Scholar]
- 244. Oliver A, Canton R, Campo P, Baquero F, Blazquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1253 [DOI] [PubMed] [Google Scholar]
- 245. Orr HA. 2009. Fitness and its role in evolutionary genetics. Nat. Rev. Genet. 10:531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Orth JD, et al. 2011. A comprehensive genome-scale reconstruction of Escherichia coli metabolism—2011. Mol. Syst. Biol. 7:535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Pace NR. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734–740 [DOI] [PubMed] [Google Scholar]
- 248. Pajunen MI, et al. 2010. Universal platform for quantitative analysis of DNA transposition. Mobile DNA 1:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Palchevskiy V, Finkel SE. 2006. Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J. Bacteriol. 188:3902–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Papadopoulos D, et al. 1999. Genomic evolution during a 10,000-generation experiment with bacteria. Proc. Natl. Acad. Sci. U. S. A. 96:3807–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Papp B, Notebaart RA, Pal C. 2011. Systems-biology approaches for predicting genomic evolution. Nat. Rev. Genet. 12:591–602 [DOI] [PubMed] [Google Scholar]
- 252. Parker CT, Sperandio V. 2009. Cell-to-cell signalling during pathogenesis. Cell. Microbiol. 11:363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Parker J. 1989. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 53:273–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Partridge JD, Scott C, Tang Y, Poole RK, Green J. 2006. Escherichia coli transcriptome dynamics during the transition from anaerobic to aerobic conditions. J. Biol. Chem. 281:27806–27815 [DOI] [PubMed] [Google Scholar]
- 255. Patel M, Jiang QF, Woodgate R, Cox MM, Goodman MF. 2010. A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V. Crit. Rev. Biochem. Mol. Biol. 45:171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Patten C, Kirchhof M, Schertzberg M, Morton R, Schellhorn H. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272:580–591 [DOI] [PubMed] [Google Scholar]
- 257. Pearl S, Gabay C, Kishony R, Oppenheim A, Balaban NQ. 2008. Nongenetic individuality in the host-phage interaction. PLoS Biol. 6:957–964 doi:10.1371/journal.pbio.0060120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Pennington JM, Rosenberg SM. 2007. Spontaneous DNA breakage in single living Escherichia coli cells. Nat. Genet. 39:797–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Pérez PD, Hagen SJ. 2010. Heterogeneous response to a quorum-sensing signal in the luminescence of individual Vibrio fischeri. PLoS One 5:e15473 doi:10.1371/journal.pone.0015473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Perfeito L, Fernandes L, Mota C, Gordo I. 2007. Adaptive mutations in bacteria: high rate and small effects. Science 317:813–815 [DOI] [PubMed] [Google Scholar]
- 261. Petit MA, Mesas JM, Noirot P, Moreldeville F, Ehrlich SD. 1992. Induction of DNA amplification in the Bacillus subtilis chromosome. EMBO J. 11:1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Pettersson ME, Andersson DI, Roth JR, Berg OG. 2005. The amplification model for adaptive mutation: simulations and analysis. Genetics 169:1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Pettersson ME, Sun S, Andersson DI, Berg OG. 2009. Evolution of new gene functions: simulation and analysis of the amplification model. Genetica 135:309–324 [DOI] [PubMed] [Google Scholar]
- 264. Philippe N, Crozat E, Lenski RE, Schneider D. 2007. Evolution of global regulatory networks during a long-term experiment with Escherichia coli. Bioessays 29:846–860 [DOI] [PubMed] [Google Scholar]
- 265. Poltak SR, Cooper VS. 2011. Ecological succession in long-term experimentally evolved biofilms produces synergistic communities. ISME J. 5:369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Ponciano JM, La HJ, Joyce P, Forney LJ. 2009. Evolution of diversity in spatially structured Escherichia coli populations. Appl. Environ. Microbiol. 75:6047–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Ponder RG, Fonville NC, Rosenberg SM. 2005. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19:791–804 [DOI] [PubMed] [Google Scholar]
- 268. Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:35–51 [DOI] [PubMed] [Google Scholar]
- 269. Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313:89–92 [DOI] [PubMed] [Google Scholar]
- 270. Rainey PB, Travisano M. 1998. Adaptive radiation in a heterogeneous environment. Nature 394:69–72 [DOI] [PubMed] [Google Scholar]
- 271. Rayssiguier C, Thaler DS, Radman M. 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342:396–401 [DOI] [PubMed] [Google Scholar]
- 272. Reams AB, Kofoid E, Savageau M, Roth JR. 2010. Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics 184:1077–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Reams AB, Neidle EL. 2004. Gene amplification involves site-specific short homology-independent illegitimate recombination in Acinetobacter sp. strain ADP1. J. Mol. Biol. 338:643–656 [DOI] [PubMed] [Google Scholar]
- 274. Rezaeinejad S, Ivanov V. 2011. Heterogeneity of Escherichia coli population by respiratory activity and membrane potential of cells during growth and long-term starvation. Microbiol. Res. 166:129–135 [DOI] [PubMed] [Google Scholar]
- 275. Richardson AR, Stojiljkovic I. 2001. Mismatch repair and the regulation of phase variation in Neisseria meningitidis. Mol. Microbiol. 40:645–655 [DOI] [PubMed] [Google Scholar]
- 276. Rinas U, Hellmuth K, Kang RJ, Seeger A, Schlieker H. 1995. Entry of Escherichia coli into stationary phase is indicated by endogenous and exogenous accumulation of nucleobases. Appl. Environ. Microbiol. 61:4147–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Robert L, et al. 2010. Pre-dispositions and epigenetic inheritance in the Escherichia coli lactose operon bistable switch. Mol. Syst. Biol. 6:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Rokney A, et al. 2008. Host responses influence on the induction of lambda prophage. Mol. Microbiol. 68:29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Romero D, Palacios R. 1997. Gene amplification and genomic plasticity in prokaryotes. Annu. Rev. Genet. 31:91–111 [DOI] [PubMed] [Google Scholar]
- 280. Rosenzweig RF, Sharp RR, Treves DS, Adams J. 1994. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics 137:903–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Rotem E, et al. 2010. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. U. S. A. 107:12541–12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Rowe MT, Kirk R. 1999. An investigation into the phenomenon of cross-protection in Escherichia coli O157:H7. Food Microbiol. 16:157–164 [Google Scholar]
- 283. Rozen DE, DE Visser J, Gerrish PJ. 2002. Fitness effects of fixed beneficial mutations in microbial populations. Curr. Biol. 12:1040–1045 [DOI] [PubMed] [Google Scholar]
- 284. Rozen DE, Philippe N, Arjan DE Visser J, Lenski RE, Schneider D. 2009. Death and cannibalism in a seasonal environment facilitate bacterial coexistence. Ecol. Lett. 12:34–44 [DOI] [PubMed] [Google Scholar]
- 285. Ruan B, et al. 2008. Quality control despite mistranslation caused by an ambiguous genetic code. Proc. Natl. Acad. Sci. U. S. A. 105:16502–16507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Ryan FJ. 1952. Adaptation to use lactose in Escherichia coli. J. Gen. Microbiol. 7:69–88 [DOI] [PubMed] [Google Scholar]
- 287. Saint-Ruf C, Sopta JPM, Matic I. 2007. Causes and consequences of DNA repair activity modulation during stationary phase in Escherichia coli. Crit. Rev. Biochem. Mol. Biol. 42:259–270 [DOI] [PubMed] [Google Scholar]
- 288. Saint-Ruf C, Taddei FO, Matic I. 2004. Stress and survival of aging Escherichia coli rpoS colonies. Genetics 168:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Salaun L, Snyder LAS, Saunders NJ. 2003. Adaptation by phase variation in pathogenic bacteria. Adv. Appl. Microbiol. 52:263–301 [DOI] [PubMed] [Google Scholar]
- 290. Sandegren L, Andersson DI. 2009. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat. Rev. Microbiol. 7:578–588 [DOI] [PubMed] [Google Scholar]
- 291. Schembri MA, Kjaergaard K, Klemm P. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253–267 [DOI] [PubMed] [Google Scholar]
- 292. Schlacher K, Goodman MF. 2007. Timeline—lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat. Rev. Mol. Cell Biol. 8:587–594 [DOI] [PubMed] [Google Scholar]
- 293. Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53 doi:10.1186/1471-2180-6-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Shapiro JA. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52:81–104 [DOI] [PubMed] [Google Scholar]
- 295. Shiba T, et al. 1997. Inorganic polyphosphate and the induction of rpoS expression. Proc. Natl. Acad. Sci. U. S. A. 94:11210–11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Sibona GJ. 2007. Evolution of microorganism locomotion induced by starvation. Phys. Rev. E 76:1919 10.1103/PhysRevE.76.011919 [DOI] [PubMed] [Google Scholar]
- 297. Silva-Rocha R, de Lorenzo V. 2010. Noise and robustness in prokaryotic regulatory networks. Annu. Rev. Microbiol. 64:257–275 [DOI] [PubMed] [Google Scholar]
- 298. Silverman M, Zieg J, Hilmen M, Simon M. 1979. Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc. Natl. Acad. Sci. U. S. A. 76:391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Singh A, Karimpour-Fard A, Gill RT. 2010. Increased mutation frequency in redox-impaired Escherichia coli due to RelA- and RpoS-mediated repression of DNA repair. Appl. Environ. Microbiol. 76:5463–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Smits WK, Kuipers OP, Veening JW. 2006. Phenotypic variation in bacteria: the role of feedback regulation. Nat. Rev. Microbiol. 4:259–271 [DOI] [PubMed] [Google Scholar]
- 301. Sniegowski PD, Murphy HA. 2006. Evolvability. Curr. Biol. 16:R831–R834 [DOI] [PubMed] [Google Scholar]
- 302. Sonti RV, Roth JR. 1989. Role of gene duplications in the adaptation of Salmonella typhimurium to growth on limiting carbon sources. Genetics 123:19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Soutourina OA, Bertin PN. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol. Rev. 27:505–523 [DOI] [PubMed] [Google Scholar]
- 304. Spira B, Hu X, Ferenci T. 2008. Strain variation in ppGpp concentration and RpoS levels in laboratory strains of Escherichia coli K-12. Microbiology 154:2887–2895 [DOI] [PubMed] [Google Scholar]
- 305. Stambuk S, Radman M. 1998. Mechanism and control of interspecies recombination in Escherichia coli. I. Mismatch repair, methylation, recombination and replication functions. Genetics 150:533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Steinborn G. 1978. Uvm mutants of Escherichia coli K-12 deficient in UV Mutagenesis. 1. Isolation of uvm mutants and their phenotypical characterization in DNA-repair and mutagenesis. Mol. Gen. Genet. 165:87–93 [DOI] [PubMed] [Google Scholar]
- 307. Stephens C. 2005. Senescence: even bacteria get old. Curr. Biol. 15:R308–R310 [DOI] [PubMed] [Google Scholar]
- 308. Stern DL, Orgogozo V. 2009. Is genetic evolution predictable? Science 323:746–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Stewart EJ, Madden R, Paul G, Taddei F. 2005. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 3:e45 doi:10.1371/journal.pbio.0030045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6:199–210 [DOI] [PubMed] [Google Scholar]
- 311. Storvik KAM, Foster PL. 2010. RpoS, the stress response sigma factor, plays a dual role in the regulation of Escherichia coli's error-prone DNA polymerase IV. J. Bacteriol. 192:3639–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Storz G, Hengge-Aronis R. 2000. Bacterial stress responses. ASM Press, Washington, DC [Google Scholar]
- 313. St-Pierre F, Endy D. 2008. Determination of cell fate selection during phage lambda infection. Proc. Natl. Acad. Sci. U. S. A. 105:20705–20710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Stragier P. 2006. To kill but not be killed: a delicate balance. Cell 124:461–463 [DOI] [PubMed] [Google Scholar]
- 315. Straight PD, Kolter R. 2009. Interspecies chemical communication in bacterial development. Annu. Rev. Microbiol. 63:99–118 [DOI] [PubMed] [Google Scholar]
- 316. Stumpf JD, Foster PL. 2005. Polyphosphate kinase regulates error-prone replication by DNA polymerase IV in Escherichia coli. Mol. Microbiol. 57:751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Suiter AM, Banziger O, Dean AM. 2003. Fitness consequences of a regulatory polymorphism in a seasonal environment. Proc. Natl. Acad. Sci. U. S. A.:2134994100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Sun X, Dennis JJ. 2009. A novel insertion sequence derepresses efflux pump expression and preadapts Pseudomonas putida S12 for extreme solvent stress. J. Bacteriol. 191:6773–6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Sundin GW, Weigand MR. 2007. The microbiology of mutability. FEMS Microbiol. Lett. 277:11–20 [DOI] [PubMed] [Google Scholar]
- 320. Swain PS, Elowitz MB, Siggia ED. 2002. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl. Acad. Sci. U. S. A. 99:12795–12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Swift S, Throup JP, Williams P, Salmond GPC, Stewart GSAB. 1996. Quorum sensing—a population-density component in the determination of bacterial phenotype. Trends Biochem. Sci. 21:214–219 [PubMed] [Google Scholar]
- 322. Tang MJ, et al. 1999. UmuD′ C-2 is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. U. S. A. 96:8919–8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Taniguchi Y, et al. 2010. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329:533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Tark M, et al. 2005. A DNA polymerase V homologue encoded by TOL plasmid pWW0 confers evolutionary fitness on Pseudomonas putida under conditions of environmental stress. J. Bacteriol. 187:5203–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Teich A, et al. 1999. Growth rate related concentration changes of the starvation response regulators sigma(S) and ppGpp in glucose-limited fed-batch and continuous cultures of Escherichia coli. Biotechnol. Prog. 15:123–129 [DOI] [PubMed] [Google Scholar]
- 326. Tenaillon O, Denamur E, Matic I. 2004. Evolutionary significance of stress-induced mutagenesis in bacteria. Trends Microbiol. 12:264–269 [DOI] [PubMed] [Google Scholar]
- 327. Tenaillon O, Taddei F, Radman M, Matic I. 2001. Second-order selection in bacterial evolution: selection acting on mutation and recombination rates in the course of adaptation. Res. Microbiol. 152:11–16 [DOI] [PubMed] [Google Scholar]
- 328. Tlsty TD, Albertini AM, Miller JH. 1984. Gene amplification in the lac region of E. coli. Cell 37:217–224 [DOI] [PubMed] [Google Scholar]
- 329. Tokuriki N, Tawfik DS. 2009. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 459:668–U671 [DOI] [PubMed] [Google Scholar]
- 330. Toleman MA, Walsh TR. 2011. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol. Rev. 35:912–935 [DOI] [PubMed] [Google Scholar]
- 331. Tong H, et al. 2007. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol. Microbiol. 63:872–880 [DOI] [PubMed] [Google Scholar]
- 332. Traxler MF, Chang DE, Conway T. 2006. Guanosine 3′,5′-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 103:2374–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Tremblay J, Deziel E. 2010. Gene expression in Pseudomonas aeruginosa swarming motility. BMC Genomics 11:587 doi:10.1186/1471-2164-11-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Tsui HCT, Feng G, Winkler ME. 1997. Negative regulation of mutS and mutH repair gene expression by the hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 179:7476–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Tsutsumi K, Munekata M, Shiba T. 2000. Involvement of inorganic polyphosphate in expression of SOS genes. Biochim. Biophys. Acta 1493:73–81 [DOI] [PubMed] [Google Scholar]
- 336. Tweeddale H, Notley-McRobb L, Ferenci T. 1998. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“metabolome”) analysis. J. Bacteriol. 180:5109–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Tyagi S. 2010. E. coli, what a noisy bug. Science 329:518–519 [DOI] [PubMed] [Google Scholar]
- 338. van Belkum A, Scherer S, van Alphen L, Verbrugh H. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. van Belkum A, et al. 1997. Variable number of tandem repeats in clinical strains of Haemophilus influenzae. Infect. Immun. 65:5017–5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Van Derlinden E, Boons K, Van Impe JF. 2011. Escherichia coli population heterogeneity: subpopulation dynamics at super-optimal temperatures. Food Microbiol. 28:667–677 [DOI] [PubMed] [Google Scholar]
- 341. van der Woude MW. 2011. Phase variation: how to create and coordinate population diversity. Curr. Opin. Microbiol. 14:205–211 [DOI] [PubMed] [Google Scholar]
- 342. van der Woude MW. 2006. Re-examining the role and random nature of phase variation. FEMS Microbiol. Lett. 254:190–197 [DOI] [PubMed] [Google Scholar]
- 343. van der Woude MW, Baumler AJ. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. van Sinderen D, et al. 1995. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol. 15:455–462 [DOI] [PubMed] [Google Scholar]
- 345. Veening J-W, et al. 2008. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc. Natl. Acad. Sci. U. S. A. 105:4393–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346. Veening JW, Smits WK, Hamoen LW, Kuipers OP. 2006. Single cell analysis of gene expression patterns of competence development and initiation of sporulation in Bacillus subtilis grown on chemically defined media. J. Appl. Microbiol. 101:531–541 [DOI] [PubMed] [Google Scholar]
- 347. Veening JW, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62:193–210 [DOI] [PubMed] [Google Scholar]
- 348. Vieira G, et al. 2011. Core and panmetabolism in Escherichia coli. J. Bacteriol. 193:1461–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349. Vlamakis H, Aguilar C, Losick R, Kolter R. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 22:945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350. Vulic M, Kolter R. 2001. Evolutionary cheating in Escherichia coli stationary phase cultures. Genetics 158:519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Wagner A. 2005. Distributed robustness versus redundancy as causes of mutational robustness. Bioessays 27:176–188 [DOI] [PubMed] [Google Scholar]
- 352. Wagner A. 2008. Robustness and evolvability: a paradox resolved. Proc. R. Soc. Lond. Ser. B. 275:91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353. Wang L, et al. 2010. Divergence involving global regulatory gene mutations in an Escherichia coli population evolving under phosphate limitation. Genome Biol. Evol. 2:478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354. Wang Z, Zhang JZ. 2009. Abundant indispensable redundancies in cellular metabolic networks. Genome Biol. Evol. 1:23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355. Want A, et al. 2011. Multi-parameter flow cytometry and cell sorting reveal extensive physiological heterogeneity in Bacillus cereus batch cultures. Biotechnol. Lett. 33:1395–1405 [DOI] [PubMed] [Google Scholar]
- 356. Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319–346 [DOI] [PubMed] [Google Scholar]
- 357. Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: sigma(S)-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358. Wegrzyn G, Wegrzyn A. 2002. Stress responses and replication of plasmids in bacterial cells. Microb. Cell Fact. 1:2 doi:10.1186/1475-2859-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359. Wesche AM, Gurtler JB, Marks BP, Ryser ET. 2009. Stress, sublethal injury, resuscitation, and virulence of bacterial foodborne pathogens. J. Food Prot. 72:1121–1138 [DOI] [PubMed] [Google Scholar]
- 360. Whitehead DJ, Wilke CO, Vernazobres D, Bornberg-Bauer E. 2008. The look-ahead effect of phenotypic mutations. Biol. Direct 3:18 doi:10.1186/1745-6150-3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361. Whiteway J, et al. 1998. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J. Bacteriol. 180:5529–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362. Williams P, et al. 1992. Small molecule-mediated density-dependent control of gene expression in prokaryotes: bioluminescence and the biosynthesis of carbapenem antibiotics. FEMS Microbiol. Lett. 79:161–167 [DOI] [PubMed] [Google Scholar]
- 363. Wilson WA, et al. 2010. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol. Rev. 34:952–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364. Wolfson JS, Hooper DC, McHugh GL, Bozza MA, Swartz MN. 1990. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and beta-lactam antimicrobial agents. Antimicrob. Agents Chemother. 34:1938–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Woodford N, Ellington MJ. 2007. The emergence of antibiotic resistance by mutation. Clin. Microbiol. Infect. 13:5–18 [DOI] [PubMed] [Google Scholar]
- 366. Woodgate R. 1992. Construction of a umuDC operon substitution mutation in Escherichia coli. Mutat. Res. 281:221–225 [DOI] [PubMed] [Google Scholar]
- 367. Woods RJ, et al. 2011. Second-order selection for evolvability in a large Escherichia coli population. Science 331:1433–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368. Workentine ML, et al. 2010. Phenotypic and metabolic profiling of colony morphology variants evolved from Pseudomonas fluorescens biofilms. Environ. Microbiol. 12:1565–1577 [DOI] [PubMed] [Google Scholar]
- 369. Wrande M, Roth JR, Hughes D. 2008. Accumulation of mutants in “aging” bacterial colonies is due to growth under selection, not stress-induced mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 105:11863–11868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Wright BE. 2004. Stress-directed adaptive mutations and evolution. Mol. Microbiol. 52:643–650 [DOI] [PubMed] [Google Scholar]
- 371. Wylie CS, Trout AD, Kessler DA, Levine H. 2010. Optimal strategy for competence differentiation in bacteria. PLoS Genet. 6:e1001108 doi:10.1371/journal.pgen.1001108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372. Yeiser B, Pepper ED, Goodman MF, Finkel SE. 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. U. S. A. 99:8737–8741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Zambrano MM, Kolter R. 1996. Gasping for life in stationary phase. Cell 86:181–184 [DOI] [PubMed] [Google Scholar]
- 374. Zhang X-S, Hill WG. 2005. Genetic variability under mutation selection balance. Trends Ecol. Evol. 20:468–470 [DOI] [PubMed] [Google Scholar]
- 375. Zhang ZG, Saier MH. 2009. A mechanism of transposon-mediated directed mutation. Mol. Microbiol. 74:29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Zhaxybayeva O, Doolittle WF. 2011. Lateral gene transfer. Curr. Biol. 21:R242–R246 [DOI] [PubMed] [Google Scholar]
- 377. Zhong S, Khodursky A, Dykhuizen DE, Dean AM. 2004. Evolutionary genomics of ecological specialization. Proc. Natl. Acad. Sci. U. S. A. 101:11719–11724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378. Zinser ER, Kolter R. 1999. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 181:5800–5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379. Zinser ER, Schneider D, Blott M, Kolter R. 2003. Bacterial evolution through the selective loss of beneficial genes: trade-offs in expression involving two loci. Genetics 164:1271–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]