Summary
Imprinting, parent-of-origin expression of alleles, plays an important role in regulating development in mammals and plants. DNA methylation catalyzed by DNA methyltransferases plays a pivotal role in regulating imprinting by silencing parental alleles. DEMETER (DME), a DNA glycosylase functioning in the base-excision DNA repair pathway, can excise 5-methylcytosine from DNA and regulate genomic imprinting in Arabidopsis. DME demethylates the maternal MEDEA (MEA) promoter in endosperm, thus resulting in expression of the maternal MEA allele. However, it is not known whether DME interacts with other proteins in regulating gene imprinting. Here we report the identification of histone H1.2 as a DME-interacting protein in a yeast two-hybrid screen and confirmation of their interaction by the in vitro pull-down assay. Genetic analysis of the loss-of-function histone h1 mutant shows that the maternal histone H1 allele is required for DME regulation of the MEA, FWA, and FIS2 imprinting in Arabidopsis endosperm while the paternal allele is dispensable. Furthermore, we show that mutations in histone H1 result in an increase of DNA methylation in the maternal MEA and FWA promoter in endosperm. Our results suggest that histone H1 is involved in the DME-mediated DNA methylation and gene regulation at imprinted loci.
Keywords: gene imprinting, DEMETER, histone H1, endosperm, DNA methylation, DNA demethylation, epigenetics
INTRODUCTION
Imprinting, the preferential expression of either maternal or paternal alleles, is an epigenetic phenomenon in plants and mammals (Reik and Walter, 2001; Feil and Berger, 2007; Huh et al., 2008). One of the widely accepted hypotheses of genomic imprinting is the parental conflict theory, which states that female and male parents have different interests in fitness of their progeny when one female can mate with multiple males (Ohad, 2007). The female parent wants all offspring to survive and thus prefers expression of genes that allocate limited resources equally to all offspring, whereas a male is only interested in its progeny, therefore favoring expression of genes that preferentially allocate resources to his offspring. Thus, different interests of female and male parents result in parent-of-origin expression of parental alleles of imprinted genes. DNA methylation, an addition of a methyl group at the 5-carbon position of cytosine, is an important epigenetic mechanism for regulating transposon silencing, gene expression, imprinting, and diseases (Feng et al., 2010; Law and Jacobsen, 2010). DNA methylation was thought to be permanent and unchanging after the methyl group is added by DNA methyltransferases. However, evidence has shown that DNA methylation is a dynamic process and the 5-methylcytosine can be actively excised by the DNA glycosylases, DEMETER (DME), REPRESSOR OF SILENCING 1 (ROS1), or DME-LIKE 2 (DML2), and replaced with a cytosine through the Base Excision Repair Pathway in Arabidopsis (Agius et al., 2006; Gehring et al., 2006; Huh et al., 2008; Niehrs, 2009). It had been debated whether 5-methylcytosine DNA glycosylases exist in mammals. Very recently, it has been reported that the DNA repair enzyme thymine DNA glycosylase can perform active demethylation by deamination-base excision repair in mammals (Cortellino et al., 2011).
Differing from mammals, there is haploid gametophytic growth in plants, i.e. postmeiotic cell divisions before fertilization. In Arabidopsis, a haploid megaspore at the end of meiosis undergoes three mitotic divisions to form a seven-cell, eight-nucleus female gametophyte containing the egg, central, synergid, and antipodal cells; the fusion of two haploid nuclei makes the nucleus of the diploid central cell. In the male gametophyte, a haploid microspore at the end of meiosis undergoes a mitotic cell division to give rise to one vegetative and one generative nucleus, and then the generative cell undergoes a mitotic cell division to result in two haploid sperm cells. There is also a double fertilization event that underlies gene imprinting in flowering plants. Fertilization of the egg cell by one sperm cell gives rise to a diploid embryo that ultimately generates the organs, tissues, and meristems of the plant (Goldberg et al., 1994). Fertilization of the central cell by the second sperm generates the triploid endosperm that supports embryo or seedling growth (Brown et al., 1999; Gehring et al., 2004). Imprinting mainly occurs in the endosperm tissue in plants (Hsieh et al., 2011), whereas imprinting can occur in embryos and in different adult tissues in mammals (Constancia et al., 2004; Gregg et al., 2010b).
MEDEA (MEA), a SET domain Polycomb group gene, was the first plant gene shown to be imprinted (maternally expressed and paternally silenced) in endosperm (Kinoshita et al., 1999), and its maternal expression is controlled by DNA methylation and demethylation (Xiao et al., 2003; Gehring et al., 2006; Jullien et al., 2006a). DNA METHYLTRANSFERASE 1 (MET1) is responsible for maintaining CpG methylation at the MEA locus (Xiao et al., 2003), whereas DME excises 5-methylcytosine of the maternal MEA allele, thus activating its expression (Gehring et al., 2006; Jullien et al., 2006b). Interestingly, silencing of the paternal MEA allele is not controlled by DNA methylation. Rather, the maternally expressed Polycomb group proteins, including MEA, maintain the paternal MEA allele silencing (Gehring et al., 2006). FLOWERING WAGENINGEN (FWA) homeodomain leucine zipper transcription factor and FERTILIZATION INDEPENDENT SEED 2 (FIS2) are also shown as imprinted genes in endosperm and regulated by DME (Kinoshita et al., 2004; Jullien et al., 2006a). Recently, it has been shown that STRUCTURE SPECIFIC RECOGNITION PROTEIN 1 (SSRP1), a high mobility group (HMG) domain containing component of a FACILITATES CHROMATIN TRANSCRIPTION/TRANSACTION (FACT) histone chaperone complex, is required for DNA demethylation and regulation of MEA imprinted genes, suggesting chromatin modification and DNA demethylation are linked in epigenetic reprogramming in Arabidopsis (Ikeda et al., 2011).
Chromatin, consisting of DNA and histone proteins, is the physiological template of genetic information in eukaryotes. Usually two molecules of each H2A, H2B, H3 and H4 form an octamer of nucleasome core and approximately two superhelical turns of DNA wrap around it to form a nucleasome (Jenuwein and Allis, 2001). Histone H1, a linker histone found between nucleosomes, is essential for organizing nucleosomes into the 30-nm chromatin by binding to the interior of the filament (Thoma and Koller, 1977; Graziano et al., 1994). Histone H1 has three domains in most species: a central globular domain, an N- terminal, and C-terminal tails (Hartman et al., 1977). Eukaryotic cells normally contain similar molar amounts of linker histone H1 and nucleosomes (Fan et al., 2003), i.e., nearly one H1 molecule for each core particle. Genetic evidence has shown that reduced amounts of H1 in cells result in shortening of the space between nucleosomes, altered chromatin structures, and changes in gene expression (Fan et al., 2003). Recent studies have shown that H1 cannot be simply defined as a global transcriptional repressor since histone H1 is also involved in transcriptional activation (Fan et al., 2005; Hashimoto et al., 2010). Reducing histone H1 can affect DNA methylation patterns of some imprinted genes in mammals and plants (Fan et al., 2005; Wierzbicki and Jerzmanowski, 2005). In Arabidopsis, it has been reported that 13 genes encode proteins containing domains similar to the conserved histone H1 globular domain, but only three genes AT1G06760, AT2G30620, and AT2G18050 were shown as linker histones through molecular and biochemical analysis, and were named as H1-1, H1-2, and H1-3, respectively (Gantt and Lenvik, 1991; Ascenzi and Gantt, 1999a; Fan et al., 2005; Wierzbicki and Jerzmanowski, 2005). Other H1-like genes are HMG and MYB domain-like DNA-binding proteins (Gantt and Lenvik, 1991; Ascenzi and Gantt, 1999a). H1-1 and H1-2 have 85% sequence identity at the nucleotide level in the globular domain while H1-3 shares less homology and can be induced by dehydration and abscisic acid (Ascenzi and Gantt, 1997, 1999b).
In this study, we performed a yeast two-hybrid screen and identified histone H1.2 (previously called H1-2) as a DME-interacting protein. We used a genetic approach to investigate the role of histone H1 in modulating gene imprinting and found that the maternal histone H1 allele is required for DME regulation of the imprinted expression of MEA, FWA and FIS2 while the paternal allele is dispensable in Arabidopsis endosperm. Our result also indicates that mutations in histone H1 result in an increase of DNA methylation in the maternal MEA and FWA promoter in endosperm, which suggests that histone H1 is involved in the DME-mediated DNA demethylation and gene expression at imprinted loci in Arabidopsis.
RESULTS
Identification of the histone H1 as a DME-interacting protein
DME, a DNA glycosylase that contains a conserved helix-hairpin-helix domain, can excise 5-methylcytosine residues at the imprinted MEA promoter in the Arabidopsis endosperm (Gehring et al., 2006). Are there other components involved in the DME-mediated DNA demethylation process? We hypothesized that there was a scaffold protein that interacted with DME and affected DNA demethylation at imprinted loci. To identify such a protein, we performed a yeast two-hybrid screen using the full-length DME as a bait. We screened cDNA libraries from Arabidopsis flowers as described (Fan et al., 1997; Wang et al., 2006). Approximately 179,000 clones were screened from the yeast two-hybrid cDNA libraries and 17 positive colonies were obtained. One of sequenced clones was identified as histone H1.2 (AT2G30620, called H1-2 in previous references). As shown in Figure 1a, H1.2 interacted with DME in yeast and activated the reporter genes His3 and LacZ. The quantitative assay showed that β – galactosidase activity in the yeast with constructs DME-pAS1 and H1.2-pACT (1.67 units) was 42-fold higher than that in the control yeast (0.04 units) (Figure 1a). To confirm the interaction between H1.2 and DME, an in vitro pull-down assay was performed. We fused H1.2 with Glutathione-S-Transferase (GST) epitope tag, expressed the fusion protein in bacteria, and purified it. When the fused maltose binding protein DME (MBP-DME) was purified and used as an input through GST-H1.2 bound column, the result showed that the fusion protein MBP-DME could be pulled down by GST-H1.2, but could not be pulled down by the control GST (Figure 1b). When MBP was used as an input in another control experiment, it could not be pulled down by either the fusion protein GST-H1.2 or GST itself (Figure 1b). These experiments demonstrate that H1.2 interacts with DME.
Figure 1. Histone H1 proteins interact with DME.
(a) Histone H1.2 interacts with DME in yeast. Histone H1.2 was isolated as a positive clone in a yeast two-hybrid screening of Arabidopsis cDNA library by using DME as a bait. Full-length DME fused with the GAL4 DNA binding domain, DME-pAS1 (DB), interacts with H1.2 fused with the GAL4 activation domain, H1.2-pACT (AD) (Fan et al., 1997). The concentration for 3-AT was 12.5 mM. The X-Gal qualitative and quantitative assays were as described (Ausubel, 2008). *The unit for galactosidase activity in the quantitative assay was 1000 × (A420−1.75 × A550)/(t (in minutes) × v (volume in ml) × OD600) in Figures 1a and 1c.
(b) H1.2 interacts with DME in vitro. GST or GST-H1.2 were immobilized on glutathione-superflow resin and used as bait to pull down MBP or MBP-DME. Proteins bound to the resin were separated by SDS-PAGE, blotted and probed with antibodies to MBP or GST. A partial DME with the N-terminal deletion (DMEΔN677) was used for the fusion protein (MBP-DME) as described (Gehring et al., 2006).
(c) H1.1 and H1.3 interact with DME in yeast. Full-length DME fused with the DNA binding domain, DME-pAS1 (DB), interacts with H1.1 and H1.3 fused with the GAL4 activation domain, H1.1-pACT(AD) and H1.3-pACT (AD), respectively. The concentration for 3-AT was 12.5 mM. The X-Gal assay was as described above in Figure 1a.
The histone H1 gene family in Arabidopsis
Histone H1 is a linker histone functioning in chromatin folding (Graziano et al., 1994; Bustin et al., 2005). It has been shown that there are several histone H1 variants in mammals (Fan et al., 2001; Wierzbicki and Jerzmanowski, 2005; Hashimoto et al., 2010). In Arabidopsis, there are three H1 linker histone genes (AT1G06760 [H1.1], AT2G30620 [H1.2], and AT2G18050 [H1.3]) (Gantt and Lenvik, 1991; Ascenzi and Gantt, 1999a; Fan et al., 2005; Wierzbicki and Jerzmanowski, 2005). To test if H1.1 and H1.3 also interact with DME, we cloned the H1.1 and H1.3 with Gal4 DNA activation domain (H1.1-pACT(AD) and H1.3-pACT(AD), respectively) and examined their interaction with DME fused with Gal4 DNA binding domain in pAS1 (DME-pAS1(DB)) in a yeast two-hybrid assay, the result showed that both H1.1 and H1.3 interact with DME, and activated the reporter genes His3 and LacZ in yeast (Figure 1c). The quantitative assay of β –galactosidase activity also indicated that H1.1 and H1.3 interact with DME in the yeast two-hybrid assay (Figure 1c). When yeast culture was serially diluted and spotted onto selection dropout medium, the result showed that H1.2, H1.1 and H1.3 interacted with DME (Figure S1).
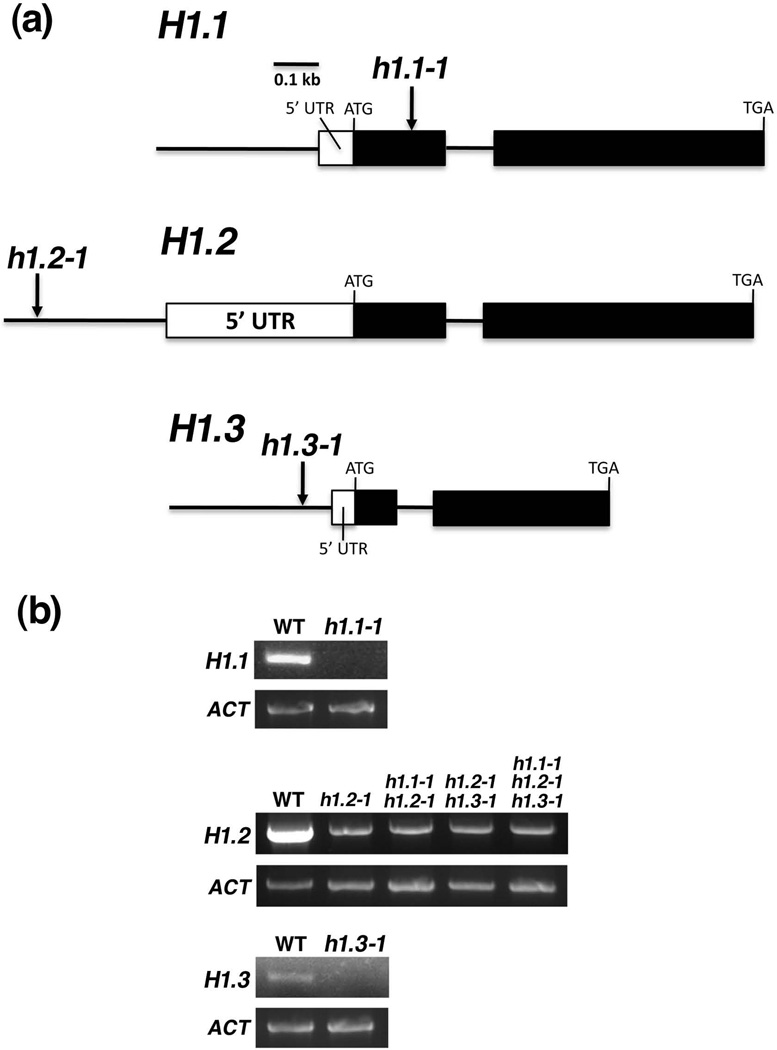
The transgenic RNAi line to silence histone H1.1, H1.2, and H1.3 was reported (Wierzbicki and Jerzmanowski, 2005). Since the RNAi approach using the transgene is dominant and cannot be used for imprinting studies, we searched for recessive T-DNA insertional mutants of the histone H1 genes in the Arabidopsis Biological Resource Center database. By genotyping T-DNA insertional lines (Figure 2a), we found h1.1-1 (Salk_128430C) has a T-DNA insertion in the first exon (215 bp downstream of transcription start), and h1.2-1 (Salk_002142) and h1.3-1 (Salk_025209) have a T-DNA insertion in the promoter (382 bp and 62 bp upstream of transcription start, respectively). To determine if the putative mutants are knockout or knockdown alleles, we examined expression of histone H1.1, H1.2, and H1.3 in their respective mutant endosperm tissues by RT-PCR. The results show that h1.1-1 is a knockout allele and h1.2-1 and h1.3-1 are knockdown alleles (Figure 2b).
Figure 2. Three histone H1 genes H1.1, H1.2, and H1.3 in Arabidopsis.
(a) Gene structures of H1.1, H1.2, and H1.3 as well as their respective mutant alleles are shown. Filled and open rectangles represent exons and 5’ untranslated region (UTR), respectively. Lines represent introns or promoter. ATG and TGA indicate the translational start and stop codons, respectively. Arrows indicate the locations of T-DNA insertion. h1.1-1 has a T-DNA insertion in the first exon (+ 215 bp relative to transcription start). h1.2-1 and h1.3-1 have a T-DNA insertion in the promoter (− 382 bp and − 62 bp relative to transcription start, respectively).
(b) Expression of histone H1.1, H1.2, and H1.3 in their respective mutants. Endosperm tissues from seed at 8-DAP of respective genotypes were collected and total RNA was isolated. Gene expression was examined by using semi-quantitative RT-PCR. ACTIN (ACT) was used as a control for the experiment.
Mutations in histone H1 affect the MEA, FWA and FIS2 imprinting
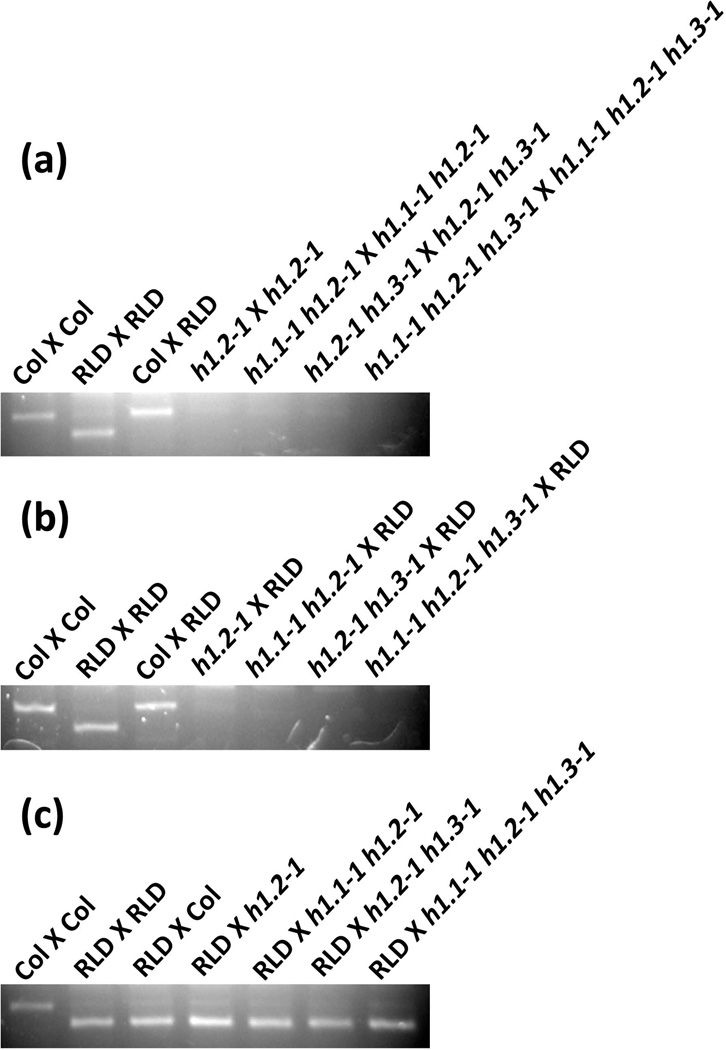
It has been shown that histone H1 depletion in mouse embryonic stem cells affects expression of imprinted genes (Fan et al., 2005). Since histone H1 interacts with DME (Figure 1) and DME is required for the maternal MEA allele expression in endosperm (Choi et al., 2002), we speculated that mutations in histone H1 might affect the MEA imprinting in Arabidopsis. To test this hypothesis, we utilized the polymorphism of the MEA locus between ecotypes Col-0 and RLD, performed reciprocal genetic crosses between the single mutant h1.2-1, double mutant h1.1-1 h1.2-1 and h1.2-1 h1.3-1, and triple mutant h1.1-1 h1.2-1 h1.3-1 in Col-0 background with RLD plants, and examined allele specific expression of MEA in the crossed silique endosperm (Figure 3). In the self-pollinated single mutant h1.2-1 (h1.2-1 X h1.2-1), expression of the maternal MEA allele was reduced compared with that in the control crosses (Col X Col and Col X RLD), and MEA expression is further reduced in the self-pollinated double mutants (h1.1-1 h1.2-1 X h1.1-1 h1.2-1 and h1.2-1 h1.3-1 X h1.2-1 h1.3-1) and undetectable in the triple mutant (h1.1-1 h1.2-1 h1.3-1 X h1.1-1 h1.2-1 h1.3-1) (Figure 3a). When the maternal histone h1 single, double and triple mutants were crossed with wild type RLD male (Figure 3b), we obtained a similar result as that in self-pollinated mutants: the maternal MEA expression was reduced in the h1.2-1 cross (h1.2-1 X RLD), was again reduced in the double mutant crossed with RLD (h1.1-1 h1.2-1 X RLD and h1.2-1 h1.3-1 X RLD), and undetectable in the triple mutant cross (h1.1-1 h1.2-1 h1.3-1 X RLD). Interestingly, when the histone h1 single, double and triple mutants as the male parent were crossed with wild type RLD female (Figure 3c), the maternal MEA allele from RLD ecotype was expressed to a similar level as the wild type control cross (RLD X RLD or RLD X Col). This suggests that the histone h1 mutations from the male parent did not affect expression of the maternal MEA allele in the crosses. Together, these results indicate that mutations in the histone H1 affect expression of the imprinted MEA gene in Arabidopsis.
Figure 3. Mutations in histone H1 affect MEA imprinting in Arabidopsis endosperm.
(a) MEA expression is reduced or undetected in self-pollinated histone h1 mutants.
(b) The maternal histone h1 mutation diminishes or abolishes MEA expression in endosperm.
(c) The paternal histone h1 mutation has no effect on expression of MEA in endosperm.
Endosperm tissues from seed at 8-DAP of respective crossed siliques were collected and total RNA was isolated. All h1 mutants are in Col-0 background and the maternal parent was shown first in the crosses (♀ X ♂). The MEA alleles from Col-0 and RLD were distinguished by a dCAPS marker. MEA expression was examined by using semi-quantitative RT-PCR.
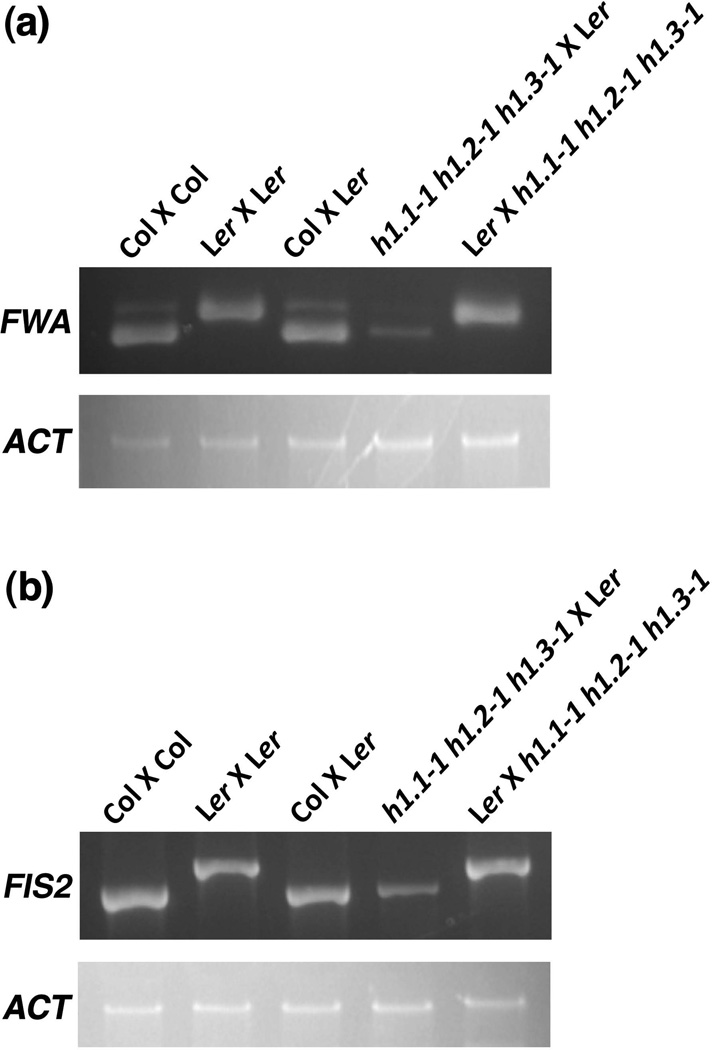
DME also regulates imprinted expression of FWA and FIS2 in endosperm (maternally expressed and paternally silenced) (Kinoshita et al., 2004; Jullien et al., 2006a). To examine whether mutations in H1 affects imprinting of FWA and FIS2, we used RT-PCR to measure allele specific expression of FWA and FIS2 in endosperm of the reciprocal crosses between the triple mutant h1.1-1 h1.2-1 h1.3-1 in Col-0 and wild type Ler. The result showed that expression of the maternal FWA and FIS2 alleles were significantly reduced when the triple mutant female was crossed with wild type male, whereas expression of the maternal alleles was not affected in the reciprocal cross with the triple mutant h1.1-1 h1.2-1 h1.3-1 as male (Figure 4). This result suggests that histone H1 might have a broad effect on expression of imprinted genes in Arabidopsis.
Figure 4. Mutations in histone H1 affect FWA and FIS2 imprinting in Arabidopsis endosperm.
(a) The maternal histone h1 mutation diminishes maternal FWA expression in endosperm.
(b) The maternal histone h1 mutation reduces maternal FIS2 expression in endosperm.
Endosperm tissues from seed at 8-DAP of respective crossed siliques were collected and total RNA was isolated. All h1 mutants are in Col-0 background and the maternal parent was shown first in the crosses (♀ X ♂). The FWA and FIS2 alleles from Col-0 and Ler were distinguished as described (Kinoshita et al., 2004; Jullien et al., 2006a). FWA and FIS2 expression was examined by using semi-quantitative RT-PCR. ACTIN (ACT) was amplified from the cDNA as a control.
Effects of the histone H1 mutation on methylation of the maternal MEA and FWA promoter in endosperm
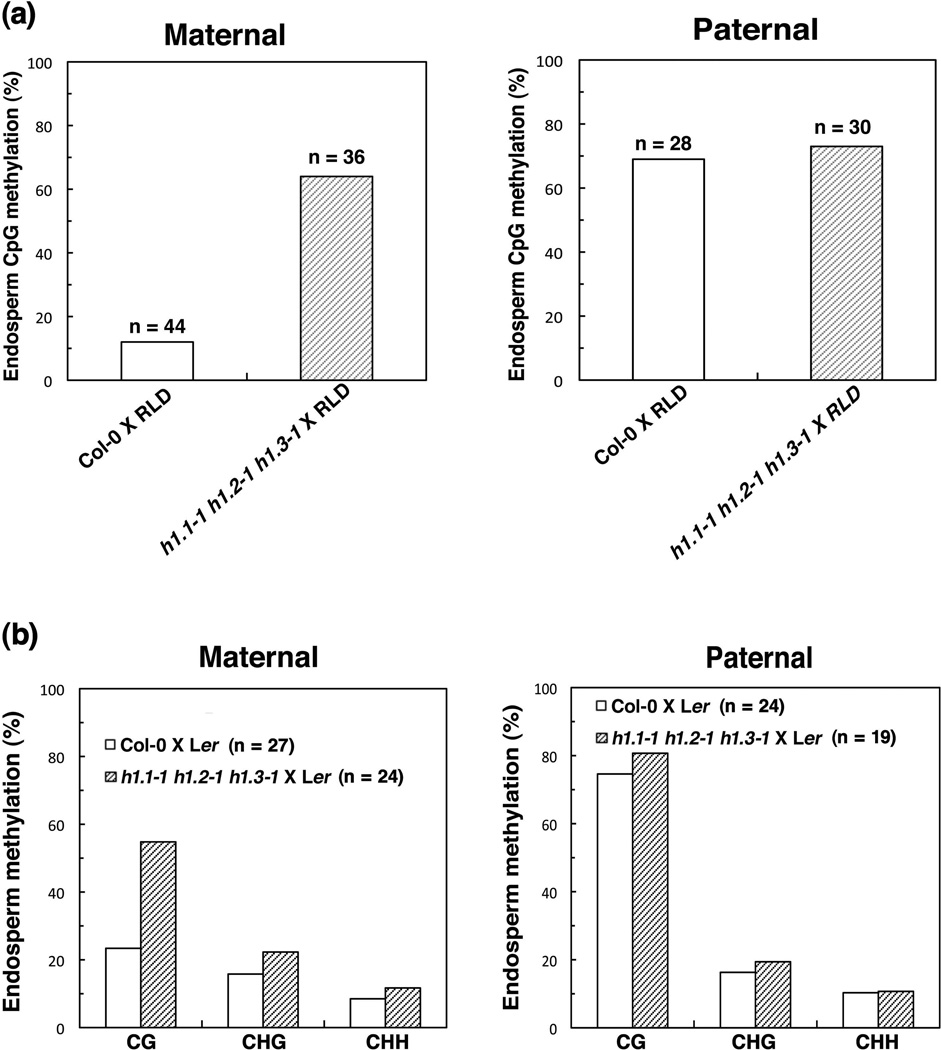
It has been reported that the maternal MEA promoter, as well as the 3’ repeats, are methylated and its expression is antagonistically controlled by MET1 and DME (Xiao et al., 2003; Gehring et al., 2006). In the Arabidopsis RNAi lines with reduced expression of H1, there is a significant increase in CpG methylation in the 183-bp intergenic sequence repeats adjacent to the imprinted MEA gene (MEA-ISR) in leaf tissues (Wierzbicki and Jerzmanowski, 2005). We have shown that the histone H1 interacts with the DME protein in vitro (Figure 1). If H1 is required for the DME-mediated DNA demethylation, mutations in H1 would result in an increase in DNA methylation in the maternal MEA promoter in endosperm. To test this hypothesis, we isolated endosperm tissues from the F1 seed of the histone h1 triple mutant (h1.1-1 h1.2-1 h1.3-1) female crossed with wild type RLD male as well as the endosperm of the control cross (Col-0 female X RLD male), extracted endosperm genomic DNA, and performed bisulfite sequencing of the – 500 bp MEA promoter, which is important for regulating MEA expression (Choi et al., 2002; Choi et al., 2004; Gehring et al., 2006). The result showed that there was 12% methylated cytosine in the maternal MEA promoter in – 500 bp region of the control cross endosperm (Col-0 female X RLD male) (Figure 5a). However, there was 64% methylated cytosine in the maternal MEA promoter in – 500 bp region in the endosperm of the histone h1 triple mutant (h1.1-1 h1.2-1 h1.3-1) female crossed with wild type RLD male (Figure 5a), a significant increase in DNA methylation. For the paternal MEA allele, the DNA methylation levels were similar in wild type control and the triple mutant cross. This result shows that mutations in histone H1 from the maternal parent cause an increase in DNA methylation of the maternal MEA promoter in the endosperm, which might in turn suppress the maternal MEA expression in the endosperm.
Figure 5. Histone H1 mutations result in hypermethylation of the maternal MEA and FWA allele in endosperm.
(a) The maternal histone h1 mutation causes hypermethylation of the maternal MEA allele in endosperm. DNA methylation profile of the maternal and paternal MEA allele in −500 bp in endosperm of crossed seeds was shown. DNA was isolated from endosperm, treated with bisulfite, PCR amplified, cloned, and sequenced. Bisulfite sequencing results are from the genomic endosperm DNA of the crossed siliques of four different triple mutant h1.1-1 h1.2-1 h1.3-1 plants. “n” represents the number of total clones that were sequenced in the bisulfite sequencing.
(b) The maternal histone h1 mutation results in increased CG methylation of the maternal 5’ repeats of FWA in endosperm. DNA methylation profile of the maternal and paternal FWA allele in 5’ repeats in endosperm of crossed seeds were shown. Bisulfite sequencing was done as described above in Figure 5a. “n” represents the number of total clones that were sequenced in the bisulfite sequencing.
FWA is imprinted and regulated by DME demethylation in Arabidopsis endosperm (Kinoshita et al., 2004). Since the H1 mutation affected FWA imprinting (Figure 4a), we speculated that the H1 mutation could have an effect on methylation of the 5’ direct repeats of FWA (Soppe et al., 2000). To test this, we examined methylation status of the 5’ repeats of FWA in the F1 endosperm of the mutant and wild type crosses. The result showed that there was 23% methylated CpG, 16% CpHpG, and 9% CpHpH in the maternal 5’ direct repeats of FWA in the control cross endosperm (Col-0 female X Ler male) (Figure 5b). However, there was a substantial increase in DNA methylation: 55% methylated CpG, 22% CpHpG, and 11% CpHpH in the maternal 5’ repeat region in the endosperm of the histone h1 triple mutant (h1.1-1 h1.2-1 h1.3-1) female crossed with wild type Ler male (Figure 5b). For the paternal FWA allele, the DNA methylation levels were similar in the triple mutant cross compared with wild type control (Figure 5b). This result indicated that mutations in histone H1 from the maternal parent result in an increase in CpG methylation of the maternal 5’ repeats of FWA in the endosperm.
Effects of the histone h1 mutation on plant growth and development
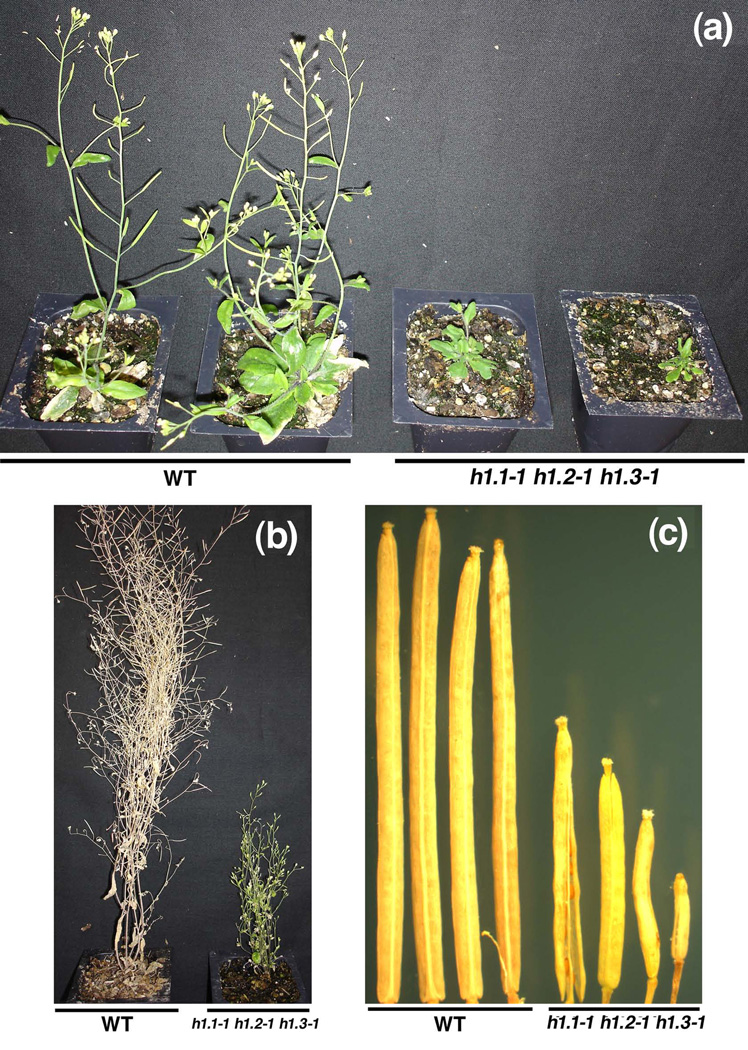
The self-pollinated heterozygous MEA/mea mutant plant gives rise to approximately 50% aborted seed and the homozygous mea/mea is usually not viable (Kinoshita et al., 1999; Vielle-Calzada et al., 1999; Luo et al., 2000). If the H1 mutation completely abolishes the maternal MEA expression in the triple mutant h1.1-1 h1.2-1 h1.3-1 plants, we would observe nearly 100% seed abortion in the self-pollinated triple mutant. However, we did not see the expected 100% seed abortion, but 16.7% seed abortion in the self-pollinated F3 triple mutant h1.1-1 h1.2-1 h1.3-1 plants (74 aborted seeds among total 444 seeds, Table 1). We observed 13.4% aborted seed (82 aborted seeds among total 611 seeds) when the F3 triple h1.1-1 h1.2-1 h1.3-1 mutant female was crossed with wild type Col-0 male, and 4.8% aborted seed (27/562) in the reciprocal cross (Table 1). It seemed that the seed abortion phenotype caused by the h1 mutation was different from that in mea (i.e. endosperm over proliferation followed by seed collapse). The lack of mea-like seed abortion could be due to very low expression of MEA in the triple h1.1-1 h1.2-1 h1.3-1 mutant. The reason might be that the h1.2-1 and h1.3-1 mutants are not null alleles although we did not detect the maternal MEA expression in the triple mutant under our RT-PCR conditions (Figure 3). In Arabidopsis, suppression of histone H1 using RNAi approaches caused developmental abnormalities (Wierzbicki and Jerzmanowski, 2005). In the F1 to F3 progeny, we did not observe any developmental phenotype except some aborted seeds in the h1 mutant. However, in the self-pollinated F4 triple mutant h1.1-1 h1.2-1 h1.3-1 progeny, we observed some mutant plants showing small stature (Figure 6a), delayed flowering (Figure 6b), and shorter siliques (Figure 6c). Since the abnormal developmental defects were observed in the self-pollinated F4 generation and there was no genomic hypomethylation in the h1 mutant (Figure S2), we speculate that sporadic developmental abnormalities can be due to epigenetic alteration in the h1 mutant plants – an epiallele that is not associated with the H1 locus.
Table 1.
Seed abortion caused by the H1 mutation
| Parental Genotype (Female X Male)a |
Viable Seed |
Aborted Seed | Total Seed | Seed Abortion (%) |
|---|---|---|---|---|
| h1.1-1 h1.2-1 h1.3-1 X h1.1-1 h1.2-1 h1.3-1 | 370 | 74 | 444 | 16.7 |
| h1.1-1 h1.2-1 h1.3-1 X Col-0 | 529 | 82 | 611 | 13.4 |
| Col-0 X h1.1-1 h1.2-1 h1.3-1 | 535 | 27 | 562 | 4.8 |
The F3 progeny of the h1.1-1 h1.2-1 h1.3-1 mutant plants were used in the genetic crosses.
Figure 6. The triple mutant h1.1-1 h1.2-1 h1.3-1 shows abnormal developmental phenotypes.
(a) Wild type Col-0 and the F4 generation triple mutant h1.1-1 h1.2-1 h1.3-1 plants at 6 weeks old. The h1.1-1 h1.2-1 h1.3-1 mutant plant shows delay in flowering and small stature.
(b) Wild type Col-0 and the F4 generation triple mutant h1.1-1 h1.2-1 h1.3-1 plants at 10 weeks old. The h1.1-1 h1.2-1 h1.3-1 mutant plant shows delayed flowering, small stature and is almost infertile.
(c) Siliques of wild type Col-0 and the F4 generation triple mutant h1.1-1 h1.2-1 h1.3-1 plant. The mutant siliques are shorter and have fewer or no seeds.
The histone H1 is not an imprinted gene in Arabidopsis
Since only the maternal H1 mutation affects expression of the maternal alleles of MEA, FWA, and FIS2, whereas the H1 mutation from the paternal side has no effect (Figure 3), we were curious whether the parent-of-origin effect of H1 on expression of MEA, FWA, and FIS2 in endosperm was due to the imprinted feature of H1 itself. To test this hypothesis, we crossed Col-0 with RLD, isolated embryo and endosperm RNA from crossed seed at 8 DAP, and designed a dCAPS marker to distinguish the parental H1.2 allele. The result showed that the histone H1.2 is not imprinted in either embryo or endosperm (Figure 7a).
Figure 7. H1.2 is not imprinted in embryo or endosperm and the H1 mutation does not affect expression of DME in ovules.
(a) Both maternal and paternal H1.2 alleles were detected in embryo or endosperm of 8-DAP seed in the cross between Col-0 and RLD. A fragment of H1.2 cDNA was amplified by PCR, and digested with PstI to distinguish a dCAPS marker between Col-0 and RLD at the H1.2 locus.
(b) DME expression in wild type Col-0 and the triple mutant h1.1-1 h1.2-1 h1.3-1. RNA was isolated from the ovule tissues of wild type and the mutant plants and used for RT-PCR examination of DME expression.
We have shown that mutations in the maternal H1 affect methylation status of the maternal MEA promoter and expression of the maternal MEA (Figure 3 and 4). We thought it was because H1 is involved, directly or indirectly, in the DME-mediated DNA demethylation. To exclude the possibility that DME expression was affected by the H1 mutation, we examined expression of DME in ovules of the triple mutant h1.1-1 h1.2-1 h1.3-1 as well as wild type. The result showed that DME expression in the triple mutant h1.1-1 h1.2-1 h1.3-1 was similar to that in wild type (Figure 7b), suggesting that the H1 mutation did not affect the DME activity.
DISCUSSION
Histone H1 affects gene imprinting
Genomic imprinting is an evolutionary conserved epigenetic phenomenon in mammals and plants (Feil and Berger, 2007). Through genomic approaches, many imprinted genes have recently been identified in Arabidopsis (Gehring et al., 2011; Hsieh et al., 2011; McKeown et al., 2011; Wolff et al., 2011), rice (Luo et al., 2011), and mammals (Gregg et al., 2010a; Gregg et al., 2010b), but detailed molecular mechanisms regulating genome imprinting is still elusive. Our study shows that histones H1.1, H1.2, and H1.3 interact with DME in vitro (Figure 1). Loss-of-function mutations in histone H1 result in a significant increase in DNA methylation of the maternal MEA and FWA promoter (Figure 5) and cause a reduction in the maternal MEA and FWA expression in endosperm (Figures 3 and 4). Interestingly, only the maternal H1 mutation has effects on MEA and FWA imprinting whereas the H1 mutation from the paternal side has no effect (Figures 3 and 4). We found that the histone H1.2 is not imprinted in either embryo or endosperm (Figure 7a). Differing from mammals in gametogenesis, there is a haploid gametophytic growth stage: postmeiotic cell divisions before fertilization in the flowering plants. DME expression is restricted to the central cell in the female gametophyte, the progenitor of the endosperm (Choi et al., 2002). DME is also expressed in the vegetative cell of pollen that is not involved in fertilization, but not in sperm cells (Schoft et al.). When the maternal h1 mutant plant was crossed with a wild type male, it is likely that the h1 mutation in the female gametophyte influences targeting of DME to the imprinted MEA locus, thus affecting maternal MEA allele expression. Since only the vegetative cell in pollen expresses DME and sperm cells involved in fertilization does not express DME, the paternal wild type allele cannot rescue the defect caused by the maternal h1 mutation. The result that h1 mutation from the male has no effect on MEA imprinting suggests that H1 expression and its interaction with DME in the female gametophyte is sufficient for sustaining the maternal MEA expression in the central cell and endosperm.
Role of histone H1 in the DME-mediated DNA demethylation in the Arabidopsis genome
How is a DNA glycosylase like DME targeted to a specific locus or transposon to perform its demethylase function in the genome? Does the plant with the h1 mutation fail to target DME into the MEA locus in the central cell and lead to hypermethylation of the maternal MEA promoter, thus resulting in reduced maternal MEA expression? It is likely that DME interacts with some scaffold proteins, like H1 variants, to facilitate its localization to the imprinted locus, MEA. One possibility is that mutations in histone H1 can abolish DME targeting or reduce its efficiency, and result in an increase in DNA methylation in the MEA promoter (Figure 5a), thus leading to reduced MEA expression in endosperm (Figure 3). It has been reported that there is a significant increase of CpG methylation in the MEA intergenic subtelomeric repeat (MEA-ISR) region in leaf tissues of the H1 RNAi plants (Wierzbicki and Jerzmanowski, 2005), which is conceptually consistent with our result (Figure 5a). How can histone H1 affect DNA methylation? Our results suggest histone H1 plays a direct or indirect role in the DME-mediated DNA demethylation. Little is known what is the exact role of histone H1 in the DME-mediated DNA demethylation in the genome, and we speculate the following possible mechanisms. First, histone H1 has basic lysine-rich regions that interact with DNA (Hendzel et al., 2004; Fang et al., 2011) and this might enhance DME binding to DNA in vivo. H1 variants have direct effects on chromatin structure (Bustin et al., 2005; Fan et al., 2005), which might allow DME and/or transcription factors to access the chromatin, thus activating MEA expression in the central cell. Second, it has been reported that other proteins can replace or exclude a nucleosome-specific histone H1 variant and affect chromatin structure (Swindle and Engler, 1998; Ju et al., 2006). It is possible that some histone H1 variants associated with the methylated locus like MEA or transposable element can bring DME to DNA through their interaction, and then DME can replace histone H1 variants and open up chromatin, thus demethylating the MEA locus and activating its expression. Third, in our original yeast two-hybrid screen for DME-interacting proteins (Figure 1 and as described in the result), we also uncovered other nuclear proteins including transcription factors and proteins with motifs mediating protein-protein or DNA-protein interactions, which can play a more direct role in targeting DME into a specific locus in the genome. Characterization of other DME-interacting proteins might reveal molecular mechanisms of how the DME DNA glycosylase performs its demethylation in the genome.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
T-DNA lines were ordered from ABRC for each of the three H1 genes, H1.1, H1.2, and H1.3. The T-DNA insertion in h1.1-1 (SALK_128430C) was detected in the first exon by amplifying a 493-bp PCR fragment with primers: SALK-128430-2, 5’-GAATAAACGAAATCGAATCTC-3’ and LBa1, 5’-TGGTTCACGTAGTGGGCCATCG-3’. The T-DNA insertion in h1.2-1 (SALK_002142) was detected in the promoter 382-bp upstream of transcription start by amplifying a 633-bp PCR fragment with primers: H1.2 promoter TDNA, 5’-CGTATATTTCAGTAGAGGAAAACTTGCC-3’ and LBa1 (see above). The T-DNA insertion in h1.3-1 (SALK_025209) was detected in the promoter 62-bp upstream of transcription start by amplifying a 552-bp PCR fragment with primers: H1.3 promoter F2, 5’-CTCTGGTTTAAACTTTCTCCCATC-3 and LBa1. Once T-DNA insertions were found in a plant, PCR amplification was used to determine homozygosity by amplifying an endogenous gene fragment across the T-DNA insertion. A 930-bp endogenous fragment was amplified in H1.1 with primers: 5’-CGGGATCCATGTCAGAGGTGGAAATAGAG-3’ and 5’-CCGCTGGAGTCACTTCTTAACCCTAGAAGA-3’; A 2074-bp endogenous fragment was amplified in H1.2 with primers: H1.2 promoter TDNA (see above) and 5’- GCGAGCTCTCACTTCTTAGCCTCCCTAG-3’; A 1405-bp endogenous fragment was amplified in H1.3 with primers: 5’-GCCTCTCGGTAAATGTGGTAAA-3’ and 5’-CGGGATCCTCAAGCAGCGGAAGCTT-3’. Three homozygous single mutants h1.1-1, h1.2-1, and h1.3-1 were backcrossed to wild type for three times and then used to create the double mutants (h1.1-1 h1.2-1, h1.1-1 h1.3-1, H1.2-1 h1.3-1) and the triple mutant (h1.1-1 h1.2-1 h1.3-1). Arabidopsis plants were grown in growth chambers or greenhouse under 16-hour light and 8-hour dark at 23 °C.
Yeast two-hybrid screen and assay
The full-length DME was generated by PCR amplification, and fused with the GAL4 DNA binding domain (DB) in pAS1 (Fan et al., 1997) and pGBKT7 (Wang et al., 2006). DME - DB was used as a bait to screen cDNA libraries fused with the GAL4 DNA activation domain (AD) in pACT as described (Fan et al., 1997; Wang et al., 2006). H1.2 was a positive clone identified in the above yeast two-hybrid library screening. For experiments of H1.1 and H1.3 interact with DME in yeast, Arabidopsis H1.1 and H1.3 were amplified by PCR and fused with the GAL4 activation domain in pACT, H1.1-pACT (AD) and H1.3- pACT (AD). The constructs were then transformed into the yeast containing DME-pAS1 (DB) and interaction clones were selected on yeast dropout media. The concentration of 3-AT used in the selection medium was 12.5 mM if not specified. To measure β-galactosidase activity, the qualitative assay using a filter lift and quantitative assay using liquid culture were as described (Ausubel, 2008).
Protein expression and purification
The construct for DME protein expression, c2x-DMEΔN677 was provided by Dr. Huh (Mok et al., 2010). Expression of DMEΔN677 was performed as described (Gehring et al., 2006). For the expression of MBP protein, BL21 cells containing pMAL-c2x (NEB) were induced at 0.3 mM IPTG in LB medium at 37°C for 3 hours. MBP and MBP-tagged DMEΔN677 fusion proteins were purified with amylose resin (NEB). To make the expression construct for histone H1.2, the coding region of H1.2 gene was amplified by PCR with primers H1.2Ex-F (5’-CGCGGATCCATGTCTATAGAGGAAGAAAACGTT-3’) and H1.2Ex-R (5’-CGCCTCGAGTCACTTCTTAGCCTTCCTAGTCGA-3’) and cloned into pGEX 4T-1 expression vector. The resulting plasmid was transformed into BL21 cells and induced with 1 mM IPTG in LB medium at 24°C for 3 hours. GST and GST-H1.2 fusion proteins were purified with glutathione-superflow resin (Clontech).
In vitro pull-down assay
GST or GST-tagged histone H1 proteins were immobilized on glutathione-superflow resin and used as bait. The purified MBP or MBP-DMEΔN677 proteins as input were added to the resin and incubated in TBS (25 mM Tris-HCl, 150 mM NaCl, pH7.2) on nutator at 4°C for 3 hours. After four times of wash with TBS, the beads were resuspended in TBS and protein loading buffer. The beads were boiled for 5 min and centrifuged at 12,000 rpm for 3 min. Input proteins and the supernatants were separated on 10% SDS-PAGE, blotted, and detected by antibodies to MBP or GST.
Gene expression and imprinting analysis
For examining expression of histone H1.1, H1.2, and H1.3 in the respective mutant plants, endosperm tissues from seed at 8-DAP of respective plants were isolated and total RNA was extracted. Gene expression was examined by using semi-quantitative RT-PCR using 28–30 PCR amplification cycles. Primer pairs for amplifying cDNA of H1.1, H1.2 and H1.3 were: H1.1-F1 and H1.1-R1; H1.2-Forward and H1.2-R1; H1.3-NcoI Y2H-pACT2 and H1.3-Y2H-R1 BamHI, respectively. The primers used for histone H1 expression cover the entire coding region of the genes, which distinguished cDNA from any possible genomic DNA contamination in the assay. ACTIN (ACT) was used as a control for the experiment. For investigating effects of histone H1 mutations on MEA imprinting in Arabidopsis, endosperm tissues from seed of respective crosses at 8-DAP were collected and total RNA was isolated. The MEA alleles from Col-0 and RLD were distinguished by a dCAPS marker and MEA expression was examined by using RT-PCR as described (Kinoshita et al., 1999). Expression of FWA and FIS2 was examined by using primers and RT-PCR as described (Kinoshita et al., 2004; Jullien et al., 2006a).
dCAPS marker for H1-2
dCAPS marker was designed with the polymorphism PERL0376855 between Col-0 and Ler ecotypes. PCR products amplified by primers dCAPS-F (5’-GTTCCAACGACTGTTGACTCAGGAGCAGC-3’) and dCAPS-R (5’-GTATTG ACTAGATCCAGTTCTCTCCTTC-3’) were digested with Pst I restriction enzyme and separated on 3% agarose gel. The PCR product from Col-0 contains the Pst I site but the Ler allele does not.
Bisulfite sequencing
Four independent triple mutant h1.1-1 h1.2-1 h1.3-1 plants were used in the genetic cross for the bisulfite sequencing assay. Endosperm tissues from seed of respective crosses (Col-0 X RLD and h1.1-1 h1.2-1 h1.3-1 X RLD) at 8-DAP were isolated and genomic DNA was extracted as described (Rea et al., 2011). Genomic DNA was then digested with Xho I and Pst I followed by bisulfite treatment. The bisulfite conversion was performed using Methylcode Bisulfite Conversion Kit (Invitrogen). After conversion, the MEA promoter at - 500 bp region was PCR-amplified by using primers mea7520TFc and mea7933TRc for the top strand, mea7529F and mea7935R for the bottom strand, PCR fragments were then cloned into pCR2.1-TOPO vector (Invitrogen) and positive colonies were selected for sequencing as described (Xiao et al., 2003). The sequences from the maternal MEA allele from Col-0 background were differentiated from those from the paternal RLD allele based on the polymorphism in the region between Col-0 and RLD ecotypes (Gehring et al., 2006). For bisulfite sequencing 5’ repeats of FWA, bisulfite treatment was the same as described above, but two rounds of semi-nested PCR were used to amplified the fragment of 5’ repeats. The first round PCR cycling condition was as described (Pappas et al., 2009). 0.5 µl of first round PCR product was used as template for the second round PCR. The PCR cycling condition for second round PCR was 94°C for 1 min; 30 cycles of 94°C for 30 sec, 55°C for 30 sec, 72 °C for 1 min; 72°C for 5 min. To amplifying the top strand of 5’ repeats of FWA, primers used in the first round PCR were FWA-top F (5’-GGTTTTATATTAATATTAAAGAGTTATGGGTYGAAGTTT-3’) and FWA-top R (5’-CAAAATACTTTACACATAAACRAAAAACAAACAAATCRAA-3’) as described (Chan et al., 2006), and primers for the second round were FWA-top F and FWA-TR2 (5’-AACCAAAATCATTCTCTAAACAAAATRTAAAAAAATC-3’). To amplifying the bottom strand of 5’ repeats of FWA, primers FWA-bottom F (5’-TAAAGTATTTTATATATAAGYGAAAAATAGATAAATTGG-3’) and FWA-bottom R (5’-AATTCTATACTAATATCAAAAAATTATAAACCRAAACCC-3’) were used for the first round PCR, and primers FWA-BF1 (5’-TTCTATACTAATATCAAARARTTATRRRCCRAARCCC-3’) and FWA-BR2 (5’-TAAAYAAAATGTAAAAAAATYTGATTTTTGGYTGA-3’) were used for the second round PCR. The maternal FWA allele (Col-0 ecotype) and paternal FWA allele (Ler ecotype) in the cross were distinguished based on the DNA sequence polymorphisms (Soppe et al., 2000).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Robert L. Fischer, in whose laboratory the original yeast two-hybrid screen was performed and W.X. greatly appreciates R.L. Fischer’s generosity of letting him to carry on the project and his valuable comments on the manuscript. We thank Eric Odle and Kaushal Patel for maintenance of Arabidopsis plants, thank the ABRC at The Ohio State University for providing seeds and libraries, Ramin Yadegary for providing the yeast two-hybrid cDNA library, Jin Hoe Huh for providing DME clone, Andrzej Jerzmanowski for providing the H1.1 H1.2 H1.3 RNAi construct, and Daniel Zilberman for providing his putative h1 mutant seed. This work was supported by National Institutes of Health grants 1R15GM086846-01 and 3R15GM086846-01S1 to W. X.
Footnotes
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
REFERENCES
- Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci U S A. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascenzi R, Gantt JS. A drought-stress-inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants. Plant Mol Biol. 1997;34:629–641. doi: 10.1023/a:1005886011722. [DOI] [PubMed] [Google Scholar]
- Ascenzi R, Gantt JS. Molecular genetic analysis of the drought-inducible linker histone variant in Arabidopsis thaliana. Plant Mol Biol. 1999a;41:159–169. doi: 10.1023/a:1006302330879. [DOI] [PubMed] [Google Scholar]
- Ascenzi R, Gantt JS. Subnuclear distribution of the entire complement of linker histone variants in Arabidopsis thaliana. Chromosoma. 1999b;108:345–355. doi: 10.1007/s004120050386. [DOI] [PubMed] [Google Scholar]
- Ausubel F, Brent R, Kingston R, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. 2008;3:13.16.11–13.16.15. [Google Scholar]
- Brown RC, Lemmon BE, Nguyen H, Olsen O-A. Development of endosperm in Arabidopsis thaliana. Sex. Plant Reprod. 1999;12:32–42. [Google Scholar]
- Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 2006;4:e363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Harada JJ, Goldberg RB, Fischer RL. An invariant aspartic acid in the DNA glycosylase domain of DEMETER is necessary for transcriptional activation of the imprinted MEDEA gene. Proc Natl Acad Sci U S A. 2004;101:7481–7486. doi: 10.1073/pnas.0402328101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. DEMETER, a DNA Glycosylase Domain Protein, Is Required for Endosperm Gene Imprinting and Seed Viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- Constancia M, Kelsey G, Reik W. Resourceful imprinting. Nature. 2004;432:53–57. doi: 10.1038/432053a. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Hu Y, Tudor M, Ma H. Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 1997;12:999–1010. doi: 10.1046/j.1365-313x.1997.12050999.x. [DOI] [PubMed] [Google Scholar]
- Fan Y, Sirotkin A, Russell RG, Ayala J, Skoultchi AI. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol Cell Biol. 2001;21:7933–7943. doi: 10.1128/MCB.21.23.7933-7943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Fang H, Clark DJ, Hayes JJ. DNA and nucleosomes direct distinct folding of a linker histone H1 C-terminal domain. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Berger F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 2007;23:192–199. doi: 10.1016/j.tig.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt JS, Lenvik TR. Arabidopsis thaliana H1 histones. Analysis of two members of a small gene family. Eur J Biochem. 1991;202:1029–1039. doi: 10.1111/j.1432-1033.1991.tb16466.x. [DOI] [PubMed] [Google Scholar]
- Gehring M, Choi Y, Fischer RL. Imprinting and Seed Development. Plant Cell. 2004;16 doi: 10.1105/tpc.017988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Missirian V, Henikoff S. Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One. 2011;6:e23687. doi: 10.1371/journal.pone.0023687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R. Plant embryogenesis: Zygote to seed. Science. 1994;266:605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- Graziano V, Gerchman SE, Schneider DK, Ramakrishnan V. Histone H1 is located in the interior of the chromatin 30-nm filament. Nature. 1994;368:351–354. doi: 10.1038/368351a0. [DOI] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010a;329:682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010b;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D, Westoby M. Parent-specific gene expression and the triploid endosperm. American Naturalist. 1989;134:147–155. [Google Scholar]
- Haig D, Westoby M. Genomic imprinting in endosperm: its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Phil. Trans. R. Soc. Lond. B. 1991;333:1–14. [Google Scholar]
- Hartman PG, Chapman GE, Moss T, Bradbury EM. Studies on the role and mode of operation of the very-lysine-rich histone H1 in eukaryote chromatin. The three structural regions of the histone H1 molecule. Eur J Biochem. 1977;77:45–51. doi: 10.1111/j.1432-1033.1977.tb11639.x. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Takami Y, Sonoda E, Iwasaki T, Iwano H, Tachibana M, Takeda S, Nakayama T, Kimura H, Shinkai Y. Histone H1 null vertebrate cells exhibit altered nucleosome architecture. Nucleic Acids Res. 2010;38:3533–3545. doi: 10.1093/nar/gkq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel MJ, Lever MA, Crawford E, Th'ng JP. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- Hsieh TF, Shin J, Uzawa R, Silva P, Cohen S, Bauer MJ, Hashimoto M, Kirkbride RC, Harada JJ, Zilberman D, Fischer RL. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci U S A. 2011;108:1755–1762. doi: 10.1073/pnas.1019273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JH, Bauer MJ, Hsieh TF, Fischer RL. Cellular programming of plant gene imprinting. Cell. 2008;132:735–744. doi: 10.1016/j.cell.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Kinoshita Y, Susaki D, Iwano M, Takayama S, Higashiyama T, Kakutani T, Kinoshita T. HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis. Dev Cell. 2011;21:589–596. doi: 10.1016/j.devcel.2011.08.013. [DOI] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Jullien PE, Kinoshita T, Ohad N, Berger F. Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell. 2006a;18:1360–1372. doi: 10.1105/tpc.106.041178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien PE, Katz A, Oliva M, Ohad N, Berger F. Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr Biol. 2006b;16:486–492. doi: 10.1016/j.cub.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Arabidopsis MET1 Cytosine Methyltransferase Mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Yadegari R, Harada JJ, Goldberg RB, Fischer RL. Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell. 1999;11:1945–1952. doi: 10.1105/tpc.11.10.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–523. doi: 10.1126/science.1089835. Published online 510.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci U S A. 2000;97:10637–10642. doi: 10.1073/pnas.170292997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Taylor JM, Spriggs A, Zhang H, Wu X, Russell S, Singh M, Koltunow A. A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002125. e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown PC, Laouielle-Duprat S, Prins P, Wolff P, Schmid MW, Donoghue MT, Fort A, Duszynska D, Comte A, Lao NT, Wennblom TJ, Smant G, Kohler C, Grossniklaus U, Spillane C. Identification of imprinted genes subject to parent-of-origin specific expression in Arabidopsis thaliana seeds. BMC Plant Biol. 2011;11:113. doi: 10.1186/1471-2229-11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok YG, Uzawa R, Lee J, Weiner GM, Eichman BF, Fischer RL, Huh JH. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proc Natl Acad Sci U S A. 2010;107:19225–19230. doi: 10.1073/pnas.1014348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends in Genetics. 1991;7:45–48. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Niehrs C. Active DNA demethylation and DNA repair. Differentiation. 2009;77:1–11. doi: 10.1016/j.diff.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Ohad N. Plant development: parental conflict overcome. Nature. 2007;447:275–276. doi: 10.1038/447275a. [DOI] [PubMed] [Google Scholar]
- Pappas JJ, Toulouse A, Bradley WE. A modified protocol for bisulfite genomic sequencing of difficult samples. Biol Proced Online. 2009;11:99–112. doi: 10.1007/s12575-009-9010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea M, Chen M, Luan S, Bhangu D, Braud M, Xiao W. Determination of DNA methylation of imprinted genes in Arabidopsis endosperm. J Vis Exp. 2011 doi: 10.3791/2327. http://www.jove.com/index/Details.stp?ID=2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: Parental influence on the genome. Nature Rev. Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Schoft VK, Chumak N, Choi Y, Hannon M, Garcia-Aguilar M, Machlicova A, Slusarz L, Mosiolek M, Park JS, Park GT, Fischer RL, Tamaru H. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc Natl Acad Sci U S A. 2011;108:8042–8047. doi: 10.1073/pnas.1105117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJM. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- Swindle CS, Engler JA. Association of the human papillomavirus type 11 E1 protein with histone H1. J Virol. 1998;72:1994–2001. doi: 10.1128/jvi.72.3.1994-2001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F, Koller T. Influence of histone H1 on chromatin structure. Cell. 1977;12:101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- Thomas JO. The higher order structure of chromatin and histone H1. J Cell Sci Suppl. 1984;1:1–20. doi: 10.1242/jcs.1984.supplement_1.1. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Thomas J, Spillane C, Coluccio A, Hoeppner MA, Grossniklaus U. Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 1999;13:2971–2982. doi: 10.1101/gad.13.22.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Tyson MD, Jackson SS, Yadegari R. Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:13244–13249. doi: 10.1073/pnas.0605551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Jerzmanowski A. Suppression of histone H1 genes in Arabidopsis results in heritable developmental defects and stochastic changes in DNA methylation. Genetics. 2005;169:997–1008. doi: 10.1534/genetics.104.031997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MT, Spillane C, Nordborg M, Rehmsmeier M, Kohler C. High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002126. e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Gehring M, Choi Y, Margossian L, Pu H, Harada JJ, Goldberg RB, Pennell RI, Fischer RL. Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Developmental Cell. 2003;5:891–901. doi: 10.1016/s1534-5807(03)00361-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.