Summary
The DNA phage ΦX174 encodes the integral membrane protein E whose expression leads to host cell lysis by inhibition of the peptidoglycan synthesis enzyme MraY. Here we use mutagenesis to characterize the molecular details of the E lysis mechanism. We find that a minimal 18-residue region with the modified wild-type sequences of the conserved transmembrane helix of E is sufficient to lyse host cells and that specific residues within and at the boundaries of this helix are important for activity. This suggests that positioning of the helix in the membrane is critical for interactions with MraY. We further characterize the interaction site of the transmembrane helix with MraY demonstrating E forms a stable complex with MraY. Triggering cell lysis by peptidoglycan synthesis inhibition is a traditional route for antimicrobial strategies. Understanding the mechanism of bacterial cell lysis by E will provide insights into new antimicrobial strategies using re-engineered E peptides.
Keywords: MraY, alanine scanning, antibiotics, cell wall, peptidoglycan, microvirus
Introduction
Viruses of microbes, known as bacteriophages or more simply phages, make up the largest component of our biosphere (Bergh et al., 1989, Whitman et al., 1998, Clokie et al., 2011). Phages infect bacteria by injecting their genetic material resulting in expression and assembly of new particles. Lytic phages are released by causing breakdown of the host cell wall resulting in membrane rupture and cell death. The potential for phage antibacterial therapeutics based on lysis was recognized upon the very first phage discovery (d’Herelle, 1917). The efforts in developing phage therapies slowed upon the discovery of antibiotic drugs. In recent years, the increasing problem of drug resistant bacteria has rekindled the desire to utilize phage therapeutics, which may be more resistant to mutation (O’Flaherty et al., 2009). To accomplish this, understanding the mechanism of phage lysis will be useful to the engineering of novel compounds.
The lytic phage life cycle requires that particles escape the peptidoglycan shell, also known as murein. Based on current understanding, two general approaches achieve successful escape from the host cell (reviewed in Wang et al., 2000, Young & Blasi, 1995). The best understood method is found in double-stranded DNA phages. In the most basic case, these large phages encode a holin/endolysin system. Holins are cytoplasmic membrane proteins that assemble into large oligomers creating a pore in the lipid bilayer. This results in leakage of the cytoplasm releasing endolysins, which are muralytic enzymes that degrade the peptidoglycan layer (Young, 1992, Young et al., 2000).
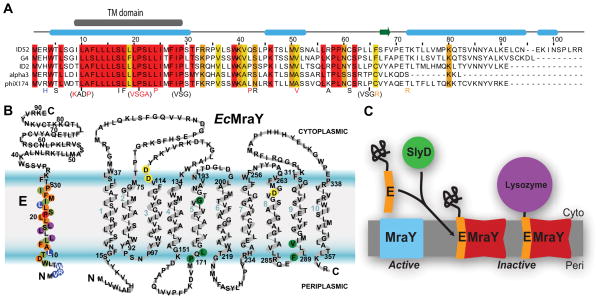
A different approach is utilized by small single-stranded genome phages. Although the mechanisms are not all understood, in studied cases the phage encodes a single lysis protein belonging to a family referred to broadly as ‘amurins’ (Bernhardt et al., 2002b). The most well characterized amurin is the protein encoded by gene E from the microvirus ΦX174, hereafter referred to simply as ‘E’. E is a small 91-residue protein (Sanger et al., 1977) (Fig. 1A) whose overexpression from a plasmid can cause host cell lysis (Sanger et al., 1977, Young & Young, 1982, Henrich et al., 1982). It contains a conserved N-terminal transmembrane domain and achieves lysis by inhibiting the transmembrane protein, MraY, which is an essential enzyme in peptidoglycan synthesis of bacteria (Fig. 1) (Buckley & Hayashi, 1986, Blasi & Lubitz, 1985, Bernhardt et al., 2000, Bernhardt et al., 2001).
Fig. 1.
ΦX174 lysis protein E and a model of its lysis mechanism.
A. Alignment of representative sequences of E isoforms available on NCBI. Five sequences were chosen based on diversity. Residue conservation for all E isoforms above 95%, 85%, and 75% are highlighted in red, orange, and yellow, respectively. The transmembrane (TM) domain is shown in a grey box above. Secondary structure was predicted based on the sequence alignment using Jpred (Cole et al., 2008) and indicated by light-blue box for α-helix and a green arrow for β-strand. Previous mutational studies are shown below the alignment: Non-lytic mutations in red; slow lysis-onset mutations in orange; mutations with no effect in lysis activity in black (Witte et al., 1997, Zheng et al., 2008b, Yu et al., 2011).
B. Topology of E and MraY based on the prediction by the TMHMM server. Sequences are overlaid. For MraY, E resistant mutations are shown in green circles while predicted catalytic aspartate residues are shown in yellow circles. Colored circles on E are based on Fig. 2 indicating the effect of alanine mutation. C. Model for the proposed lysis mechanism of E. MraY is active. E inserts into the cytoplasmic membrane with the help of the chaperone SlyD. MraY is inactive when E is bound. The soluble C-terminal domain of E can be replaced with another protein to retain lysis activity.
MraY catalyzes the first membrane-localized step of peptidoglycan precursor synthesis. The reaction transfers a phospho-N-acetylmuramyl pentapeptide from the carrier UDP- N-acetylmuramyl-pentapeptide to undecaprenol phosphate, generating Lipid I. MraY contains 10 transmembrane (TM) helices, with both termini oriented towards the periplasm (Fig. 1B) (Bouhss et al., 1999). Catalysis occurs in the cytoplasmic leaflet of the membrane. (van Heijenoort, 2001). MraY is essential in all bacteria making it an ideal candidate for antibiotic development; however, there are currently no clinical inhibitors of MraY (Dini, 2005, Bugg et al., 2006). E acts in vitro as a non-competitive inhibitor of the MraY reaction, suggesting that its mechanism of action could be used as a mode of antibiotic design (Zheng et al., 2009). Understanding the details of the E lysis mechanism will inform novel antimicrobial strategies using E or re-engineered peptides.
A notable feature of E-mediated lysis is a dependence on SlyD, a FKBP-type peptidyl-prolyl cis-trans isomerase, which is an abundant chaperone in E. coli (Roof et al., 1994, Roof et al., 1997, Bernhardt et al., 2002a). In E. coli, mutations that conferred resistance to E were found in the slyD gene. Furthermore, E fails to lyse cells in a slyD knockout strain of E. coli (Roof et al., 1994). Mutations in E (R3H and L19F) were identified that bypassed this requirement of SlyD by increasing the levels of E in the membrane. This suggested that turnover of wild-type E is relatively fast in the absence of SlyD (Bernhardt et al., 2002a). This leads to a model where SlyD stabilizes E, either directly or indirectly, preventing its degradation in the cell allowing inhibition of MraY (Fig. 1C) (Bernhardt et al., 2002a).
The role of the C-terminal cytoplasmic domain of E has been investigated. Truncation from the C-terminus of E, as few as a 17-residue deletion, abolishes its lysis activity. Interestingly, attaching various proteins to truncated E rescues lysis activity (Fig. 1C) (Maratea et al., 1985, Buckley & Hayashi, 1986). The activity of E was retained when the C-terminal region was replaced with either β-galactosidase or chloramphenicol acetyl transferase but not when it was fused to an anthranilate synthase subunit. It was suggested that E oligomerizes through its C-terminus stabilizing E in the membrane (Buckley & Hayashi, 1986). A variant of protein E containing the first 29 residues was the shortest demonstrated that retained the lysis activity as a fusion (Buckley & Hayashi, 1986). Moreover, in vitro a similar peptide was demonstrated to be a non-competitive inhibitor of MraY (Mendel et al., 2006, Zheng et al., 2009).
A variety of genetic studies on E identified several mutations that were unable to cause lysis (Fig. 1A) (Witte et al., 1997, Zheng et al., 2008b, Yu et al., 2011). The most notable of these is that any tested change of Pro21 (Gly, Ala, Ser, Val) inactivated E. Though limited in the positions tested, these experiments suggested that there were critical features of E involved in function.
Here we use a series of mutational analysis to identify the importance of each residue to the lysis function of E. Initially, we use alanine-scanning mutagenesis (residues 2–29 including the TM domain) to define important residues and identify additional groups for more specific mutations. We next use an E fusion to determine the minimal region required for lysis and characterize residues involved in direct interactions with MraY. Finally, we show binding of E directly to MraY. This allows us to determine the minimal components of an E-peptide for inhibition of MraY
Results
Alanine-scanning of the conserved N-terminal region of E
We performed a thorough analysis of the amino acid sequence requirements for E using alanine-scanning (Cunningham & Wells, 1989). All non-alanine residues in the conserved N-terminal domain (residues 2–29) were replaced with alanine to assay lysis activity with the exception of Ala11 which was changed to a serine. A construct was generated in an inducible vector that contained E followed by a 6x-His and FLAG tag. Protein production in a DH5α variant was induced using the T7 promoter. Post-induction lysis was monitored as a drop in OD550 over time, with lysis onset defined as the time-point at which the OD550 began to decrease. Both 6xHis-tagged and 6xHis-FLAG tagged E constructs showed the same lysis onset as the non-tagged wild-type E, suggesting neither the 6xHis-tag nor FLAG-tag affected the lysis activity of E (Fig. S1A). Wild-type E resulted in lysis onset at ~10 min with complete lysis after ~30–40 min in the DH5α variant (Fig. S1A). This is consistent with previous reported results (Bernhardt et al., 2002a) and supports a model wherein lysis occurs during cell division. Surprisingly, for the majority of variants that insert properly into the membrane, only the previously identified P21A variant resulted in a complete loss of lysis (Fig. S1B) (Witte et al., 1997, Zheng et al., 2008b). We were concerned that high levels of expression by the T7 promoter might mask phenotypes; therefore, it was necessary to establish a minimum expression level for lysis that might better reflect the protein levels seen during the phage life cycle.
We identified an expression strain that would allow us to reproducibly control the levels of mRNA production. The strain Lemo21(DE3) expresses lysozyme on a titratable rhamnose promoter, resulting in rhamnose concentration dependent inhibition of the T7 RNA polymerase, and reducing linearly the subsequent expression level of a protein using a T7 promoter (Wagner et al., 2008). Testing a range of rhamnose concentrations showed that even at the maximum recommended concentration (2mM rhamnose) we could observe lysis (Fig. S1C). The reduction in expression resulted in a significant delay in lysis onset beyond the previously observed 10 min to 25–35 min. We reasoned that this level of E expression was more suitable to detect phenotypes that were not obvious in the high-level expression system. All of subsequent alanine-scanning experiments were performed with 2 mM rhamnose.
All of the alanine variants were re-tested revealing a variety of new phenotypes. Eight of the mutants (residues 2–4, 10, and 24) resulted in an earlier lysis onset (Fig. 2A and B and Fig. S2A). Based on our rhamnose titrations, it is clear that an increase in expression levels affects lysis onset. We compared protein E levels after 20 minutes by Western blot (Fig. S2B). Each of these mutants had higher levels of protein relative to wild-type. Earlier lysis onset due to increased protein levels had previously been observed (R3H/L19F) (Bernhardt et al., 2002a). Six of the mutants (W7A, T9A, L13A, S17A, S22A, I25A, and I28A) resulted in a longer lysis onset phenotype likely due to observed lower levels of protein (Fig. S2).
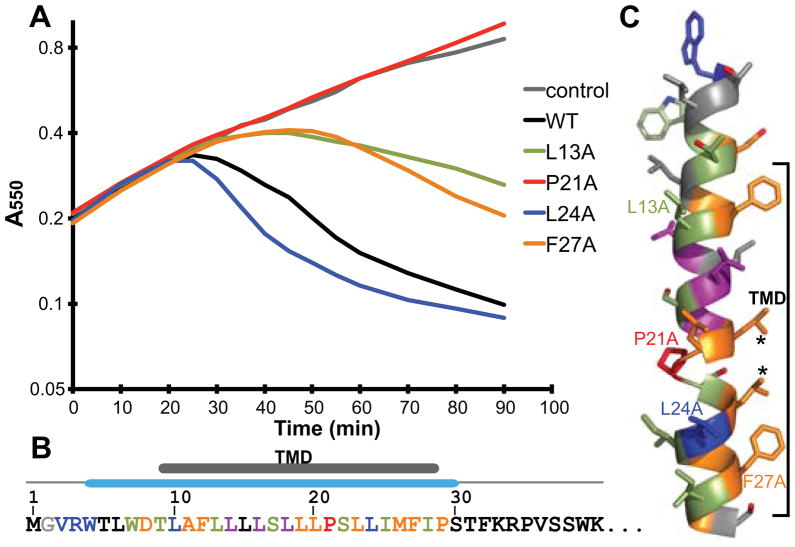
Fig. 2.
Alanine-scanning of the N-terminal domain of E.
A. Representative growth curves of alanine-scanning mutants in Lemo21 cells. Lines are based on the average curve from Fig. S2. Expression of each E construct was induced at time= 0. Various phenotypes and protein expression levels of each mutation are categorized by color: black, no obvious change in lysis onset; blue, faster lysis onset due to higher protein levels; lightgreen, delayed lysis onset due to lower protein levels; purple, colony-to-colony variation in lysis onset; orange, delayed lysis onset with no obvious protein level change; red, no lytic activity with no protein level change.
B. Summary of alanine-scanning results. The first 40 residues of E are shown. The TM domain and the secondary structure are indicated above. Colors of each residue are according to A.
C. A model for the first α-helix. Colors are according to A. Residues from growth curves in A are labeled. Asterisks indicate residues changed to phenylalanine later in the text.
Three leucine-to-alanine mutants in the TM domain (L14A, L16A, and L18A) exhibited drastic colony-to-colony variation in lysis onset (Fig. S2). In the high expression experiments, the L18A mutant failed to insert into the membrane (Fig. S1A). In the lower expression experiments, insertion of the L18A mutant into the membrane is improved resulting in lysis. These results demonstrate that some residue changes from leucine to alanine within the TM likely cause defects in membrane insertion.
The remaining mutants all had levels of E that were similar or higher than wild-type. Nine of these (D8A, A11S, F12A, L19A, L20A, L23A, M26A, F27A and P29A) resulted in a longer lysis onset phenotype. Residues Asp8 and Pro29 are at the ends of the predicted TM helix (Fig. 2B). These residues, one being charged and the other breaking the alpha helix, could be important in establishing the position of the helix in the lipid bilayer. Changing these residues to alanine may alter the relative position of the TM helix. Residues Ala11, Phe12, Leu19, Leu20, Leu23, Met26 and Phe27 are all located on the same face of the TM helix (Fig. 2C). Thus, they may be involved in direct interactions with MraY. Again, only the previously described alanine mutant at Pro21 resulted in complete loss of lysis function.
Leucine mutants of the TM domain
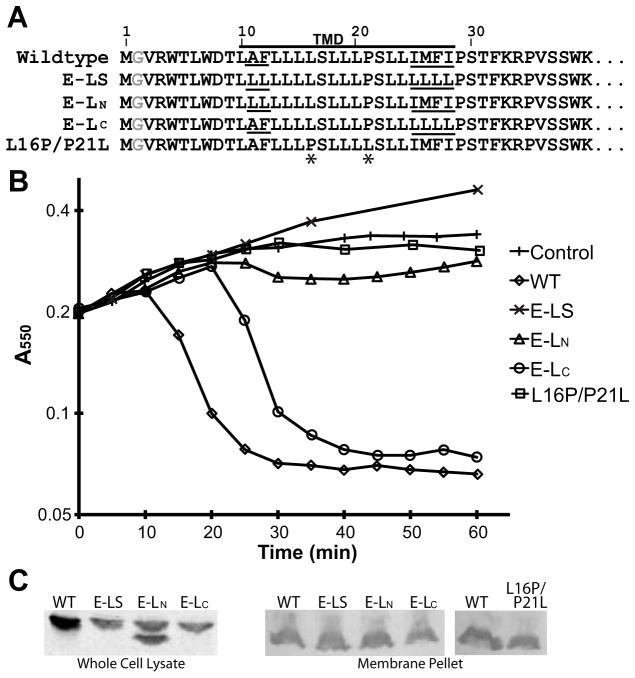
The importance of the length/positioning and potential distortions within the TM for the lysis activity of E was demonstrated in alanine-scanning. To test whether these factors are the only requirement for E function, we built a construct in which residues 10–28 in the predicted TM domain, except for Ser17, Pro21, and Ser22, are changed into leucines (referred to as ‘E-LS’). In the sequence of E-LS, both N-terminal and C-terminal ends of the TM domain differ from the wild-type sequence (Fig. 3A). E-LS was expressed in cells but did not retain lysis activity (Fig. 3B and C). Protein levels of E-LS were lower compared to wild-type suggesting that changes in the TM may have affected the rate of membrane insertion. Nonetheless, total abolishment of lysis activity indicates that other residues in the TM domain besides the kink at 21 are important in lysis activity.
Fig. 3.
Effect of leucine mutations.
A. Sequences of the wild-type, leucine variants and the L16P/P21L mutant. Underlines indicate positions of residues that vary in the leucine variants. Asterisks indicate the position of mutated residues in the L16P/P21L mutant.
B. Growth curves of the mutants in TOP10 cells.
C. (left) Western blot of whole cells for the leucine mutants. An unknown secondary protein product is evident in E-LN. (right) Western blot of membrane fractions.
To consider other residue requirements, two constructs were generated focused on either side of the TM, E-LN (mutation of N-terminal residues Ala11 and Phe12 to leucines) and E-LC (mutation of C-terminal residues Ile125, Met126, Phe127, and Ile25 to leucines) (Fig. 3A). E-LC retained lysis function despite a lower level of protein expression that resulted in longer lysis onset (Fig. 3B and C). Similarly, when expression was repressed in Lemo21 cells, the lysis onset was further delayed, although there was no lysis activity with maximum repression (Fig. S3). E-LN is expressed at a comparable level as E-LC yet lost its lysis activity completely under all conditions (Fig. 3B, C and Fig. S3). To confirm that all three mutants are successfully integrated into the bilayer, the membrane fraction was isolated. All the variants of E are found in this fraction (Fig. 3C) An secondary protein product is evident in the whole cell of E-LN (Fig. 3C). This is likely a soluble cleavage product as it is missing in the membrane fraction (Fig. 3C). Nevertheless, protein levels of all three constructs in the membrane are comparable, suggesting that the cleavage product does not affect lysis activity of E-LN. Altogether, these results suggest the N-terminal residues of the TM domain, which are in the periplasmic leaflet, play a direct role in E lysis function.
We additionally tested the importance of the position of a kink within the TM by changing Leu16 into a proline and Pro21 into a leucine (Fig. 3A). This L16P/P21L mutant was integrated into membrane (Fig. 3C) but failed to lyse cells, suggesting that having a kink at any position within the TM domain is not sufficient for lysis activity (Fig 3B).
Defining the minimal length of E required for lysis activity
Truncation of the cytoplasmic C-terminal domain of E is deleterious to E function but can be rescued by fusion to another protein. One suggested role of the C-terminal domain is to facilitate oligomerization, as previous functional fusions involved proteins that formed oligomers (Buckley & Hayashi, 1986). To test this hypothesis, we chose two known monomeric proteins, a maltose binding protein (MBP) and an inactive lysozyme, for testing as E C-terminal fusions. The N-terminal 37 residues of E were used in the initial fusion construct. Expression of either the MBP or the lysozyme fusion resulted in lysis onset similar to wild type (Fig. S4A). Therefore, oligomerization of E by the soluble domain is not necessary for lysis function. The lysozyme fusion provides a useful tool for exploring the minimal length of E required for function.
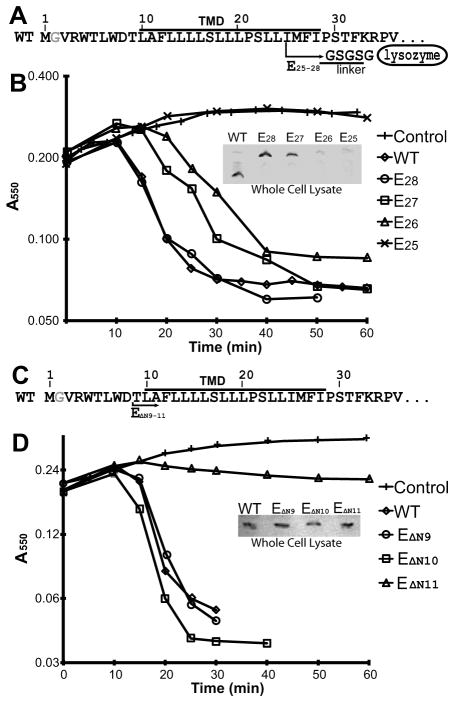
Using our inactive lysozyme fusion, we generated a series of truncation variants in E to identify the shortest region required for function. Constructs containing the N-terminal 25 to 28 residues are referred to as E25, E26, E27 and E28 (Fig. 4A). Of these, only E28 retained wild-type lysis rates in the DH5alpha strain (Fig. 4B). Both E27 and E26 showed a delay in lysis onset while E25 lost lysis activity, likely due to a lower level of protein expression (Fig. 4B). Thus, the full TM domain (E28) is required for efficient lysis. To further compare to wild-type, we tested E28 in a slyD knockout strain to see if, like wild-type, the truncation/fusion would be unable to lyse. Intriguingly, the expressed E28 fusion resulted in lysis in the slyD knockout strain (Fig. S4B). This suggests that the lysozyme domain fusion performs a similar role to the function of SlyD in stabilizing E in the membrane. A possible explanation for this is that, in wild-type E. coli, SlyD may interact directly with the C-terminal domain of E stabilizing the TM in the membrane.
Fig. 4.
Defining the minimal length of E required for lysis activity.
A. Illustration of the lysozyme fusion constructs. e.g. E25 contains the first 25 residues followed by the linker and an inactive lysozyme shown after the arrow.
B. Growth curves of wild-type and the E25–28 fusion constructs in TOP10 cells. Inset, Western blot of whole cells after 10 min of induction.
C. Illustration of the N-terminal deletion constructs.
D. Growth curves of deletion constructs in TOP10 cells. Inset, Western blot of whole cells after 10 min of induction.
There are no reports on the effect of truncations N-terminal to the TM of E. To determine the minimal region of E required for lysis activity, we created several N-terminal deletion constructs starting with removal of the first nine residues, EΔN9 through EΔN11. In each construct, the deleted residues are replaced by Met-Gly (Fig. 4C). Both EΔN9 and EΔN10 showed similar lysis onsets as wild-type and retained similar to slightly lower protein levels (Fig. 4D). On the contrary, the EΔN11 construct lost lysis activity despite having similar protein levels to the EΔN10 construct (Fig. 4D). Thus, the first 10 residues of E are not required for lysis activity.
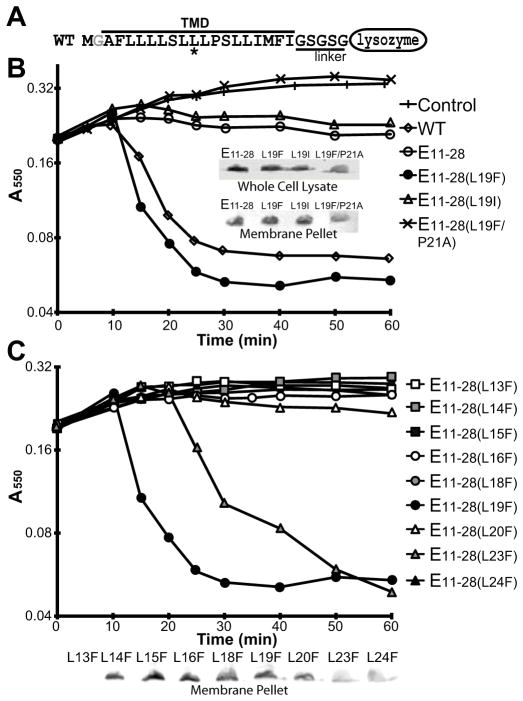
To test the minimal requirement defined above for lysis activity, we generated a construct of the minimal region of E containing residues 11 through 28 fused to lysozyme, referred to as E11–28 (Fig. 5A). This minimal construct displayed a slight decrease in cell density but did not show a complete lysis despite its successful insertion into the membrane (Fig. 5B), showing that the combined N- and C-terminal truncations abolished lysis activity while individually having no apparent defect.
Fig. 5.
Lysis activity of the minimal region.
A. Illustration of the minimal region constructs that contains residues 11 to 28 of E followed by the linker and an inactive lysozyme. The asterisk indicates the position of Leu19.
B. Growth curves of wild-type and various minimal region constructs in TOP10 cells. Insert, Western blot of both whole cells and the membrane fraction after 10 min of induction.
C. Growth curves of all Leu-to-Phe mutants of the minimal region in TOP10 cells. Inset, Western blot of the membrane fraction after 10 min of induction. L13F failed to insert into the membrane.
Restoring lysis activity of the minimal region
ΦX174 phages containing mutations that bypass the requirement of SlyD were identified as plaques on a lawn of E. coli carrying the slyD null allele. Three mutations were identified (R3H, L19F, and the combination of R3H/L19F) (Bernhardt et al., 2002a). The R3H/L19F mutant was shown to bypass the requirement of SlyD by increasing levels of E (Bernhardt et al., 2002a). The separate effects of the R3H and L19F mutation on the synthesis of E was not shown. The change from arginine to histidine in the periplasmic N-terminus may facilitate correct topology in the membrane by enhancing the “positive-inside” rule of TM insertion (Andersson et al., 1992). The L19F enhancement is harder to rationalize, therefore, we introduced the L19F mutation, located in the middle of TM domain, into E11–28 to test if this residue change can recover the lysis activity of the minimal region. The E11–28(L19F) mutant indeed restored lysis activity even though the protein level both in the cells and the membrane was comparable to E11–28 (Fig. 5B). To further investigate if the recovery of lysis activity by the L19F mutation is due to efficieny of membrane insertion, Leu19 was also changed to a Ile residue that is energetically more favored for membrane insertion than Leu and Phe residues (Table S2) (Hessa et al., 2007). This E11–28(L19I) mutant displayed a behavior similarly to E11–28 and did not lyse the cells completely (Fig. 5B). These results show that a restored lysis activity of the minimal region by the L19F mutation is likely due to some improvement in interactions with MraY. In addition, we introduced a P21A mutation into E11–28(L19F). This E11–28(L19F/P21A) mutant did not show any lysis activity (Fig. 5B), suggesting that the L19F mutation does not complement the loss of lysis activity by the P21A mutaiton.
Leu19 is one of the residues proposed to be involved in direct interaction with MraY in the alanine-scanning study (Fig. 2). According to the TM model in Fig. 2C, Leu23 is on the same face as Leu19 and had a similar phenotype to Leu19 in alanine-scanning. Based on this, we suspected that a L23F mutation may have the same effect as L19F on the minimal region. To test this hypothesis, we made L23F mutant along with changing each of the other leucines in the minimal region into phenylalanines. E11–28(L23F) regained the lysis activity while all the other phenylalanine mutants failed to show any lysis activity (Fig. 5C). The lysis onset of E11–28(L23F) was delayed compared to L19F likely due to less efficient membrane insertion (Fig. 5C). These results confirm that the Leu19 and Leu23 are involved in direct interaction with MraY and suggest that the phenylalanine mutations introduce beneficial interactions with MraY.
Both E28 and E11–28(L19F) did not show any defect in lysis in the DH5α variant. We tested E11–28(L19F) for lysis activity in the slyD knockout strain. Surprisingly, although E11–28(L19F) showed lysis activity, the lysis onset was delayed compared to E28 (Fig. 5B). This may imply that SlyD plays an additional role to stablizing E through the C-terminus. One possibilites could be promoting membrane targeting and insertion through the N-terminal residues preceding the TM domain.
Interactions of E and MraY in vitro
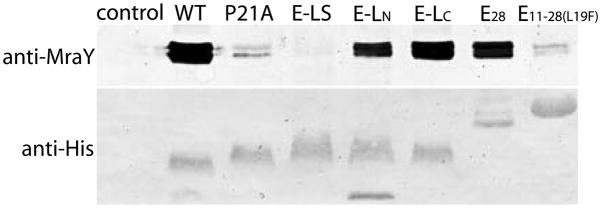
Previous studies speculated that E inhibits MraY via direct interaction between their TM domains (Bernhardt et al., 2000, Bernhardt et al. 2001, Zheng et al., 2008a). To prove a direct E-MraY interaction, a pulldown experiment was designed. Cell cultures expressing the 6xHis-tagged E were lysed, solubilized in detergent and then E was bound on a Ni-affinity resin. The protein was released from the resin after extensive washing, and the presence in elution of E and native MraY was confirmed by Western blot against the tag on E and an antibody against E. coli MraY (Zheng et al., 2008a). In order to maximize the protein yield, cell cultures were induced at OD550 of ~1.5 instead of ~0.5. This prevented the cells from rapid lysis providing sufficient quantities of cells for purification of the E-MraY complex. A Western blot confirmed that E captured significant quantities of MraY, i.e. the proteins form a stable complex (Fig. 6).
Fig. 6.
Protein E forms a stable complex with MraY.
Tagged wild-type E and E variants were expressed and purified on the Ni-NTA column along with a control (an empty pRSFDuet vector). The presence of the E variants and captured native MraY were confirmed by western blot using anti-penta-His and anti-MraY. Molecular weights of wild-type, P21A, E-LS, E-LN, and E-LC are comparable, but all variants have different mobility on the gel. Similarly, E11–28(L19F) runs slower than E28 on the gel despite its smaller molecular weight likely differences in charge. Two bands of native MraY are visible under these conditions.
Based on our earlier results, we chose to test several E mutant constructs for the ability to bind to MraY. The non-lytic E-LS mutant completely failed to capture MraY (Fig. 6). The non-lytic P21A variant significantly reduced the amount of MraY captured (Fig. 6). The lytic E-LC captured slightly reduced amounts of MraY (Fig. 6). Interestingly, the non-lytic E-LN showed similar to slightly reduced protein levels of MraY compared to E-LC (Fig. 6). This supports the hypothesis that the N-terminal residues of the TM play a key role in E lysis function. Moreover, the results suggest that binding and lysis are not completely correlated. The total amount of purified E28 is significantly less than the wild-type; nevertheless, E28 captured a significant amount of MraY (Fig. 6). The lytic E11–28(L19F) captured significantly less MraY similar to the levels as the non-lytic P21A (Fig. 6).
Discussion
The single-stranded DNA phage ΦX174 has evolved a simple lysis system encoding the transmembrane protein E (Hutchison & Sinsheimer, 1966), which inhibits peptidoglycan synthesis (Bernhardt et al., 2000, Bernhardt et al., 2001). Here we report an extensive study of the elements of E that are required for lysis using site-specific mutagenesis of the first 29 amino acids. Somewhat surprisingly, only the single alanine mutation at Pro21 completely abolished lysis activity.
Using a tightly regulated promoter, we identified several mutations that exhibited a change in lysis onset. Most that showed a delay of lysis onset also had reduced levels of E consistent with the notion that the timing of lysis correlates directly to the amount of available E. Two mutants, D8A and P29A, had significantly delayed lysis onset despite levels of E similar to wild-type. Asp8 is a charged residue preceding the start of the predicted TM domain. Changing this residue into a hydrophobic alanine may affect TM length and the relative position of Pro21 in the membrane. Similarly, Pro29 is located at the end of the TM. Changing this residue into an alanine may shift the end of the TM domain from residue 28 to residue 32 again affecting the length and position in the membrane. The results suggest that the length of the TM helix and the position of Pro21 are important for the lysis function of E.
The reason for the essential role of the kink/distortion at Pro21 in the TM remains a question. Prolines are common in membrane proteins mediating helical kinks that allow for tightly packed structures (Cordes et al., 2002, Yohannan et al., 2004). We reasoned that if this is the case the position of the kink might not be critical. We tested this by moving the proline approximately one turn of the helix closer to the periplasm (L16P/P21L). The inability of this variant to facilitate lysis suggests the position of the kink is crucial. This is further illustrated by the inability of P21A to form a stable complex with MraY. Pro21 presumably generates the critical shape necessary for interaction with MraY.
A number of other mutants (A11S, F12A, L19A, L20A, L23A, M26A, and F27A) also exhibited delayed lysis onset in various degrees despite similar protein levels to wild-type. These side chains are all predicted to be on the same face of the TM helix (Fig. 2C). This suggests that these residues are involved in direct interaction with MraY upon inhibition.
We further investigated more subtle phenotypes by looking specifically at the role of the hydrophobic residues in the TM. This resulted in three constructs in which residues were replaced by leucine. Changes at the C-terminus of the TM, E-LC, had no effect; however, changes at the N-terminal side, E-LS, E-LN, resulted in a loss of function. The implication of this is that interactions in the periplasmic leaflet at the N-terminal end of the TM are important for MraY inhibition.
To date, five mutations on MraY were found to have resistance to E. All of those mutations were mapped onto TM helices 5 and 9 (Bernhardt et al., 2000, Zheng et al., 2008a), suggesting the conserved TM domain of E interacts with these TM domains of MraY upon inhibition (Fig. 1B). Mutations in MraY at positions P170L, ΔL172 (a deletion of a leucine), F288L, and V291M all locate at the periplasmic end of the TM helices. Our findings also suggest that N-terminal residues of the E TM helix at the periplasmic side are crucial for lysis activity. The action of E as a non-competitive inhibitor of MraY (Zheng et al., 2009) would be consistent with binding in the periplasmic leaflet to allosterically stabilize a non-functional conformation of MraY.
Current inhibitors against MraY are thought to mimic catalytic intermediates (Bugg et al., 2006) and bind at the active site that is likely on the cytoplasmic side of the membrane (Lloyd et al., 2004). These inhibitors have not found clinical use due to low activity in vivo (Dini, 2005, Bugg et al., 2006). There appears to be a requirement that these drugs are lipophilic; therefore, the lack of success of these compounds may partially be due to inefficient delivery across the cytoplasmic membrane. The ability of E to act as an effector on the periplasmic side suggests that allosteric inhibition may be possible using small molecules. If true, this would increase the likelihood of identifying clinically relevant compounds.
Previous work suggested that protein E is rapidly turned over in the absence of the chaperone SlyD. This can be rescued by overexpression, which is supported by work here that shows the level of expression is critical to function. This suggests that binding and inhibition of MraY by E is established in a concentration-dependent manner. However, since single mutations can suppress the slyD knockout, it is surprising that E evolved to require SlyD for its stabilization instead of optimizing its expression levels. This suggests that SlyD may be important in regulating the timing of lysis.
The role of the C-terminus of E has not been fully elucidated. Prior work had demonstrated that the C-terminus was critical to function as somewhat minimal truncations result in loss of function. Surprisingly, this could be rescued by appending a wide range of soluble proteins, which we demonstrate can include monomeric proteins. Furthermore, the lysozyme fusion construct was shown to bypass the SlyD requirement. We can speculate from those results that SlyD stabilizes E through some interactions at the C-terminus and that the C-terminal truncations disrupts those interactions, causing loss of function.
The C-terminal truncation/fusion studies revealed that the first 28 residues are required for sufficient membrane insertion, and we have gone further to show that the first 10 residues are not required for lysis function of E. The evidence that the first 10 residues of E are not required appears to contradict a recent study using gene shuffling that showed the first 25 residues are not essential for E-mediated lysis function (Yu et al., 2011). In that work, a possible alternate start codon (TTG) at nucleotide 16 was not discussed. With that possibility, the tested protein E variant would only miss the first 6 residues agreeing with the results described here.
The reason for the absence of lysis activity in E11–28 may be explained by reduced binding affinity to MraY. We were able to restore the lysis activity of this minimal region by changing a Leu residue to a Phe residue at position 19 and 23. Those 2 residues are positioned on the face of the TM helix that we propose to be involved in interactions with MraY. Intriguingly, this face of the TM helix contains two phenylalanine residues at position 12 and 27 that show reduced lysis when changed to alanine (Fig. 2C). Phenylalanine residues were previously shown to promote interactions between TM domains (Unterreitmeier et al. 2007). Thus, changing Leu19 and Leu23 into phenylalanines are likely stabilizing the interaction with MraY. The capture assay showed that the binding affinity of E11–28(L19F) to MraY is greatly reduced compared to E28, implying that some of the first 10 residues may also be involved in interactions with MraY although deleting those residues from the wild-type E did not affect lysis activity.
MraY remains one of the great hopes for new targets that inhibit peptidoglycan synthesis (Dini, 2005, Bugg et al., 2006). Protein E from ΦX174 is an amurin representing a unique class of inhibitors that may suggest a novel route for drug design. Here, we have characterized the minimal elements of protein E required for efficient lysis to further elucidate the mechanism of this unusual antibiotic. While the precise details of the E/MraY interaction are not yet understood, the overall picture is becoming clearer. We defined the face of the TM helix involved in interactions with MraY, the N-terminal residues that are important in lysis function, and the minimal region with modified sequences sufficient for lysis activity. Today, more and more efforts are put into developing high-throughput MraY functional assay (Shapiro et al., 2012, Tanino et al. 2011, Mravljak et al, 2011). This work will allow for more refined in vitro studies testing short peptide sequences established here for MraY inhibition. Moreover, our work will help to inform the design of drugs targeting MraY.
Experimental Procedures
Construction of plasmids
The bacteriophage ΦX174 lysis protein E gene was amplified by recursive PCR (Prodromou & Pearl, 1992) using the primers shown in Table S1 based on the first isolate sequenced (NCBI: NP_040709) (Sanger et al., 1977). The 5′ and 3′ primers contain NcoI and XhoI restriction sites, respectively. An artifact of cloning resulted in the insertion of a Gly after the first Met in the wild-type E sequence. An additional 6x His-tag was added at the C-terminus for Western Blotting. PCR product was digested with NcoI and XhoI and ligated into the multiple cloning site of pRSFDuet (Novagen) that was modified to introduce FLAG-tag at the C-terminus of the inserted gene. Mutants were generated by the Quickchange method (Qiagen). E-LS, E-LN, and E-LC variants were amplified by recursive PCR using the primers shown in Table S1 cloned in the same manner as the wild-type construct.
Various lengths of the TM domain of E (25–28) and an inactive T4 lysozyme containing a 6x-His tag at the C-terminus were amplified separately. The TM domain of E and lysozyme were fused by another round of PCR, generating E25–28 constructs (e.g. E25 is a fusion of the first 25 residues of E to T4 lysozyme with the linker Gly-Ser-Gly-Ser-Gly in between. PCR product was digested with NcoI and NotI and ligated into the multiple cloning site of pRSFDuet (Novagen). The inactive T4 lysozyme (T26Q (Poteete et al., 1991)) was generated by standard mutagenesis.
Lysis Assay
The pRSFDuet-Ewt and mutant plasmids were transformed into TOP10(DE3) (TOP10 variant from Invitrogen that includes the viral polymerase) or Lemo21(DE3) (Novagen). For TOP10 cultures, 2 mL of an overnight culture were inoculated into 50 mL of LB media containing 35 μg ml−1 kanamycin then grown by shaking at 225 rpm at 37 °C. For Lemo21 cultures, growths were the same except that the media additionally contained 34 μg ml−1 chloramphenicol and 2 mM of L-rhamnose unless otherwise specified. Cells were induced at an optical density (OD550) of ~0.2 with either 1 mM IPTG for TOP10 cultures or 0.4 mM IPTG for Lemo21. Growths were monitored by periodic OD measurements. To confirm that toxicity was not contributing to our phenotypes, plasmids for two variants that had delayed onset, W7A and T9A, were confirmed to have the correct inserted gene sequence after growth overnight.
Membrane Fractionation
Growth was done under the same conditions as the lysis assay in TOP 10(DE3) except that cells were induced at an OD550 of ~0.5 and 10 ml aliquots were taken after 10 minutes. Samples were pelleted by centrifugation at 4,000g for 10 min at 4 °C. Cell pellets were resuspended in 1 mL of 20 mM Tris, pH 7.5, 300 mM NaCl, 10% glycerol, 5 mM of β-mercaptoethanol, 0.1 mM PMSF, and 0.1 mg ml−1 chicken egg lysozyme and subjected to sonication to lyse cells. Cell debris was pelleted by centrifugation at 20,000g for 30 min at 4 °C and discarded. The supernatant was then centrifuged at ~400,000g for 20 min 4 °C to pellet the membrane fraction. Membrane pellets were resuspended in SDS sample buffer in volumes normalized to the final OD550, and western blots were carried out as described below.
MraY Capture Assay
The pRSFDuet-Ewt and –mutant, and an empty pRSFDuet vector were transformed into TOP10(DE3). 25 mL of an overnight culture were inoculated into 1L of 2X YT media containing 35 μg ml−1 kanamycin then grown by shaking at 225 rpm at 37 °C. Cells were induced at an optical density (OD550) of ~1.5 with 1 mM IPTG. Each 1L culture was harvested after 20 minutes by centrifugation at 4,000g at 4 °C. Cell pellets were resuspended in 50 mL of 20 mM Tris, pH 7.5, 300 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, and 0.1 mM PMSF and subjected to the M-110L microfluidizer (Microfluidics) to lyse cells. Cell debris was pelleted by centrifugation at 20,000g for 30 min 4 °C and discarded. The supernatant was then centrifuged at 250,000 g for 1 hour 4 °C to pellet the membrane fraction. Membrane pellets were resuspended in 5 mL of 20 mM Tris, pH 7.5, 300 mM NaCl, 5% glycerol, 2 % n-Dodecyl-β-D-maltoside (DM), 30 mM imidazole, 5 mM β-mercaptoethanol, and 0.1 mM PMSF and incubated at 4 °C for 1 hour and then centrifuged at ~400,000g for 30 min. The supernatant was mixed with 200 μl of Ni-NTA agarose (Qiagen) equilibrated in the wash buffer (20 mM Tris, pH 7.5, 300 mM NaCl, 5% glycerol, 0.15 % DM, 30 mM imidazole, and 5 mM β-mercaptoethanol) and incubated at 4 °C for 1 hour. The sample was then flowed through a gravity column. The Ni-NTA agarose on the column was washed with 4 mL of the wash buffer, and the proteins were eluted with 200 μl of 20 mM Tris, pH 7.5, 300 mM NaCl, 5% glycerol, 0.15 % DM, 300 mM imidazole, and 5 mM β-mercaptoethanol 3 times. Each elution fraction was then mixed with 66 μl of 4X SDS sample buffer, and western blots were carried out as described below.
SDS-PAGE and Western blotting
SDS-PAGE and Western blotting were performed generally as described previously (Zheng et al., 2008b). Quantification of expressed E mutant protein levels in whole cells was estimated by Western blot. Growth was done under the same conditions as the Lysis Assay except that cells were induced at an OD550 of ~0.5 and 10 ml aliquots were taken after 10 minutes for TOP10 cultures and after 20 minutes for Lemo21 cultures. Samples were pelleted by centrifugation at 4,000g for 10 min at 4 °C. Pellets were resuspended in SDS sample buffer in volumes normalized to the final OD550. Western blots were prepared by standard methods on nitrocellulose membranes, except for native MraY detection, which used PVDF membranes. Membranes were washed and incubated with appropriate antibodies (for FLAG-tagged mutants: anti-FLAG (Rockland) 1:5,000 dilution and anti-rabbit-IRDye800CW (LI-COR Bioscience) at 1:10,000 dilution; or His-tagged mutants, anti-penta-His (Qiagen) 1:2,000 and anti-mouse-IRDye800CW (LI-COR Bioscience) at 1:10,000 and visualized in a 800 nm channel by Odyssey Infrared Imaging System. For membrane pellets and E variants from the pull-down assay, we used anti-mouse-alkaline phosphatase (Rockland) at 1:8,000 as a secondary antibody, and protein bands were chemically visualized with a NBT/BCIP solution (Thermo Scientific). For detection of MraY, anti-MraY (provided by Ry Young, Texax A&M) at 1:1,000 was used as a primary antibody, and anti-mouse-alkaline phosphatase (Rockland) at 1:1,000 was used as a secondary antibody, and protein bands were chemically visualized with a NBT/BCIP solution.
Supplementary Material
Acknowledgments
We are grateful to Jaeyoon Chung and Nadia Iqbal for help in generating the mutant constructs. We thank R. Young and T.O. Yeates for critical reading of the manuscript. We thank members of the laboratory for support and useful discussion. The slyD knockout strain and the anti-MraY antibody were kindly provided by Ry Young, Texas A&M University. This work is supported by a Jane Coffin Childs Memorial Fund fellowship to S.T. and a Searle Scholar fellowship, a Burroughs-Wellcome Fund Career Award and a NIH Pioneer Award (Grant 1DP1OD008304-01) to W.M.C.
References
- Andersson H, Bakker E, von Heijne G. Different positively charged amino acids have similar effects on the topology of a polytopic transmembrane protein in Escherichia coli. J Biol Chem. 1992;267:1491–1495. [PubMed] [Google Scholar]
- Bergh O, Borsheim KY, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, Roof WD, Young R. Genetic evidence that the bacteriophage phi X174 lysis protein inhibits cell wall synthesis. P Natl Acad Sci USA. 2000;97:4297–4302. doi: 10.1073/pnas.97.8.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, Roof WD, Young R. The Escherichia coli FKBP-type PPIase SlyD is required for the stabilization of the E lysis protein of bacteriophage phi X174. Mol Microbiol. 2002a;45:99–108. doi: 10.1046/j.1365-2958.2002.02984.x. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, Struck DK, Young R. The lysis protein E of phi X174 is a specific inhibitor of the MraY-catalyzed step in peptidoglycan synthesis. J Biol Chem. 2001;276:6093–6097. doi: 10.1074/jbc.M007638200. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, I, Wang N, Struck DK, Young R. Breaking free: “protein antibiotics” and phage lysis. Res Microbiol. 2002b;153:493–501. doi: 10.1016/s0923-2508(02)01330-x. [DOI] [PubMed] [Google Scholar]
- Blasi U, Lubitz W. Influence of C-terminal modifications of phi X174 lysis gene E on its lysis-inducing properties. J Gen Virol. 1985;66(Pt 6):1209–1213. doi: 10.1099/0022-1317-66-6-1209. [DOI] [PubMed] [Google Scholar]
- Bouhss A, Mengin-Lecreulx D, Le Beller D, Van Heijenoort J. Topological analysis of the MraY protein catalysing the first membrane step of peptidoglycan synthesis. Mol Microbiol. 1999;34:576–585. doi: 10.1046/j.1365-2958.1999.01623.x. [DOI] [PubMed] [Google Scholar]
- Buckley KJ, Hayashi M. Lytic activity localized to membrane-spanning region of phi X174 E protein. Mol Gen Genet. 1986;204:120–125. doi: 10.1007/BF00330198. [DOI] [PubMed] [Google Scholar]
- Bugg TD, Lloyd AJ, Roper DI. Phospho-MurNAc-pentapeptide translocase (MraY) as a target for antibacterial agents and antibacterial proteins. Infect Disord Drug Targets. 2006;6:85–106. doi: 10.2174/187152606784112128. [DOI] [PubMed] [Google Scholar]
- Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1:31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes FS, Bright JN, Sansom MS. Proline-induced distortions of transmembrane helices. J Mol Biol. 2002;323:951–960. doi: 10.1016/s0022-2836(02)01006-9. [DOI] [PubMed] [Google Scholar]
- Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- d’Herelle F. An invisible microbe that is antagonistic to the dysentery bacillus. Compres rendus Acad Sciences. 1917;165:373–375. [Google Scholar]
- Dini C. MraY Inhibitors as Novel Antibacterial Agents. Curr Top Med Chem. 2005;5:1221–1236. doi: 10.2174/156802605774463042. [DOI] [PubMed] [Google Scholar]
- Henrich B, Lubitz W, Plapp R. Lysis of Escherichia coli by induction of cloned phi X174 genes. Mol Gen Genet. 1982;185:493–497. doi: 10.1007/BF00334146. [DOI] [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- Hutchison CA, 3rd, Sinsheimer RL. The process of infection with bacteriophage phi-X174. X. Mutations in a phi-X Lysis gene. J Mol Biol. 1966;18:429–447. doi: 10.1016/s0022-2836(66)80035-9. [DOI] [PubMed] [Google Scholar]
- Lloyd AJ, Brandish PE, Gilbey AM, Bugg TD. Phospho-N-acetyl-muramyl-pentapeptide translocase from Escherichia coli: catalytic role of conserved aspartic acid residues. J Bacteriol. 2004;186:1747–1757. doi: 10.1128/JB.186.6.1747-1757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maratea D, Young K, Young R. Deletion and fusion analysis of the phage phi X174 lysis gene E. Gene. 1985;40:39–46. doi: 10.1016/0378-1119(85)90022-8. [DOI] [PubMed] [Google Scholar]
- Mendel S, Holbourn JM, Schouten JA, Bugg TD. Interaction of the transmembrane domain of lysis protein E from bacteriophage phiX174 with bacterial translocase MraY and peptidyl-prolyl isomerase SlyD. Microbiology. 2006;152:2959–2967. doi: 10.1099/mic.0.28776-0. [DOI] [PubMed] [Google Scholar]
- Mravljak J, Monasson O, Al-Dabbagh B, Crouvoisier M, Bouhss A, Gravier-Pelletier C, Le Merrer Y. Synthesis and biological evaluation of a diazepanone-based library of liposidomycins analogs as MraY inhibitors. European Journal of Medicinal Chemistry. 2011;46:1582–1592. doi: 10.1016/j.ejmech.2011.02.006. [DOI] [PubMed] [Google Scholar]
- O’Flaherty S, Ross RP, Coffey A. Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol Rev. 2009;33:801–819. doi: 10.1111/j.1574-6976.2009.00176.x. [DOI] [PubMed] [Google Scholar]
- Poteete AR, Sun DP, Nicholson H, Matthews BW. Second-site revertants of an inactive T4 lysozyme mutant restore activity by restructuring the active site cleft. Biochemistry. 1991;30:1425–1432. doi: 10.1021/bi00219a037. [DOI] [PubMed] [Google Scholar]
- Prodromou C, Pearl LH. Recursive PCR: a novel technique for total gene synthesis. Protein Eng. 1992;5:827–829. doi: 10.1093/protein/5.8.827. [DOI] [PubMed] [Google Scholar]
- Roof WD, Fang HQ, Young KD, Sun J, Young R. Mutational analysis of slyD, an Escherichia coli gene encoding a protein of the FKBP immunophilin family. Mol Microbiol. 1997;25:1031–1046. doi: 10.1046/j.1365-2958.1997.5201884.x. [DOI] [PubMed] [Google Scholar]
- Roof WD, Horne SM, Young KD, Young R. slyD, a host gene required for phi X174 lysis, is related to the FK506-binding protein family of peptidyl-prolyl cis-trans-isomerases. J Biol Chem. 1994;269:2902–2910. [PubMed] [Google Scholar]
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Shapiro AB, Jahic H, Gao N, Hajec L, Rivin O. A High-Throughput, Homogeneous, Fluorescence Resonance Energy Transfer-Based Assay for Phospho-N-acetylmuramoyl-pentapeptide Translocase (MraY) J Biomol Screen. 2012;17:662–672. doi: 10.1177/1087057112436885. [DOI] [PubMed] [Google Scholar]
- Tanino T, Al-Dabbagh B, Mengin-Lecreulx D, Bouhss A, Oyama H, Ichikawa S, Matsuda A. Mechanistic analysis of muraymycin analogues: a guide to the design of MraY inhibitors. J Med Chem. 2011;54:8421–8439. doi: 10.1021/jm200906r. [DOI] [PubMed] [Google Scholar]
- Unterreitmeier S, Fuchs A, Schaffler T, Heym RG, Frishman D, Langosch D. Phenylalanine promotes interaction of transmembrane domains via GxxxG motifs. J Mol Biol. 2007;374:705–718. doi: 10.1016/j.jmb.2007.09.056. [DOI] [PubMed] [Google Scholar]
- van Heijenoort J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat Prod Rep. 2001;18:503–519. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- Wagner S, Klepsch MM, Schlegel S, Appel A, Draheim R, Tarry M, Hogbom M, van Wijk KJ, Slotboom DJ, Persson JO, de Gier JW. Tuning Escherichia coli for membrane protein overexpression. P Natl Acad Sci USA. 2008;105:14371–14376. doi: 10.1073/pnas.0804090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. P Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte A, Schrot G, Schon P, Lubitz W. Proline 21, a residue within the alpha-helical domain of phiX174 lysis protein E, is required for its function in Escherichia coli. Mol Microbiol. 1997;26:337–346. doi: 10.1046/j.1365-2958.1997.5781941.x. [DOI] [PubMed] [Google Scholar]
- Yohannan S, Faham S, Yang D, Whitelegge JP, Bowie JU. The evolution of transmembrane helix kinks and the structural diversity of G protein-coupled receptors. P Natl Acad Sci USA. 2004;101:959–963. doi: 10.1073/pnas.0306077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD, Young R. Lytic action of cloned phi X174 gene E. J Virol. 1982;44:993–1002. doi: 10.1128/jvi.44.3.993-1002.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R, Blasi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]
- Young R, Wang I, Roof WD. Phages will out: strategies of host cell lysis. Trends Microbiol. 2000;8:120–128. doi: 10.1016/s0966-842x(00)01705-4. [DOI] [PubMed] [Google Scholar]
- Yu SY, Peng W, Si W, Yin L, Liu SG, Liu HF, Zhao HL, Wang CL, Chang YH, Lin YZ. Enhancement of bacteriolysis of shuffled phage PhiX174 gene E. Virol J. 2011;8:206. doi: 10.1186/1743-422X-8-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Struck DK, Bernhardt TG, Young R. Genetic analysis of MraY inhibition by the phiX174 protein E. Genetics. 2008a;180:1459–1466. doi: 10.1534/genetics.108.093443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Struck DK, Dankenbring CA, Young R. Evolutionary dominance of holin lysis systems derives from superior genetic malleability. Microbiology. 2008b;154:1710–1718. doi: 10.1099/mic.0.2008/016956-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Struck DK, Young R. Purification and functional characterization of phiX174 lysis protein E. Biochemistry. 2009;48:4999–5006. doi: 10.1021/bi900469g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.