Abstract
In diffusion MRI, simultaneous multi-slice single-shot EPI acquisitions have the potential to increase the number of diffusion directions obtained per unit time, allowing more diffusion encoding in high angular resolution diffusion imaging (HARDI) acquisitions. Nonetheless, unaliasing simultaneously acquired, closely spaced slices with parallel imaging methods can be difficult, leading to high g-factor penalties (i.e., lower SNR). The CAIPIRINHA technique was developed to reduce the g-factor in simultaneous multi-slice acquisitions by introducing inter-slice image shifts and thus increase the distance between aliased voxels. Because the CAIPIRINHA technique achieved this by controlling the phase of the RF excitations for each line of k-space, it is not directly applicable to single-shot EPI employed in conventional diffusion imaging. We adopt a recent gradient encoding method, which we termed “blipped-CAIPI”, to create the image shifts needed to apply CAIPIRINHA to EPI. Here, we use pseudo-multiple replica SNR and bootstrapping metrics to assess the performance of the blipped-CAIPI method in 3× simultaneous multi-slice diffusion studies. Further, we introduce a novel image reconstruction method to reduce detrimental ghosting artifacts in these acquisitions. We show that data acquisition times for Q-ball and diffusion spectrum imaging (DSI) can be reduced 3-fold with a minor loss in SNR and with similar diffusion results compared to conventional acquisitions.
Keywords: Diffusion MRI, Simultaneous multi-slice, CAIPIRINHA, Parallel imaging
Introduction
Diffusion MRI is used for studying white matter structure in the brain, with diffusion tensor imaging (DTI) (Basser et al., 1994) being the most widely employed technique. More recently, high angular resolution diffusion imaging (HARDI) techniques have been developed to allow the detection of crossing major white matter fiber bundles. Some examples of these techniques include Q-ball Imaging (QBI) (Tuch et al., 2003), spherical deconvolution (SD) (Tournier et al., 2004) and Diffusion Spectrum Imaging (DSI) (Wedeen et al., 2005). A drawback of these techniques is their requirement for a greater number of diffusion encoded acquisitions compared to DTI, leading to an increase in acquisition time. In addition, these techniques often use a substantial amount of the acquisition sequence duration for the diffusion encoding gradients, resulting in long TR and scan times. For example, a typical 60 directions, 60 slices whole brain Q-ball acquisition can take up to 10 min to complete the diffusion and slice encoding, while a 257 direction whole-brain DSI scan lasts as long as 45 min. The length of these acquisitions limits their utility in clinical and research studies.
Diffusion MRI acquisitions typically rely on rapid single-shot 2D spin echo echo-planar imaging (EPI) sequences. Conventional accelerated 2D parallel imaging approaches (Griswold et al., 2002; Pruessmann et al., 1999; Sodickson and Manning, 1997) can greatly reduce the EPI readout or the echo train length by reducing the number of phase encoding steps by a factor of 2 to 4. This significantly reduces image distortion and blurring, resulting in an improved image quality of EPI acquisitions. However, this reduction in echo train length does not translate to a significant reduction in acquisition time because of the large fixed diffusion encoding time blocks. In comparison, accelerating data acquisition via simultaneous multi-slice approach for single-shot EPI can be very effective in decreasing scan time. Here, multiple slices are excited simultaneously, diffusion encoded with the same diffusion gradients, and readout simultaneously; thereby reducing total scan time by a factor equal to that of the number of simultaneously excited slices. Various methods have been proposed in the context of single-shot simultaneous multi-slice EPI, including Wideband imaging (Paley et al., 2006; Weaver, 1988), Simultaneous Image Refocusing (SIR) (Feinberg et al., 2002; Reese et al., 2009) and parallel image reconstruction based multi-slice imaging (Breuer et al., 2005; Larkman et al., 2001; Moeller et al., 2010; Nunes et al., 2006). However, these multi-slice techniques suffer from significant artifact and/or SNR loss issues. The Wideband approach results in a large voxel tilting artifact while the SIR technique necessarily lengthens the readout period of the EPI, thus increasing susceptibility induced image distortion in EPI and the minimal echo time (TE). On the other hand, multi-slice imaging techniques based on parallel image reconstruction can lead to a large SNR penalty related to the g-factor, since the aliased slices are generally close to each other due to a comparatively small field of view (FOV) in the slice direction (e.g. FOVslice =12 cm vs. FOVinplane =21 cm).
The controlled aliasing in parallel imaging results in higher acceleration (CAIPIRINHA) technique (Breuer et al., 2005) was developed to reduce the g-factor penalty of parallel imaging based simultaneous multi-slice acquisition. With this method, a different radio frequency (RF) pulse phase was used to excite each k-space line. For example if alternating 180° phase shifts are applied to successive ky lines for one of the simultaneously excited slices, that slice will be shifted by FOV/2 in y. This serves to increase the distance between aliasing pixels in the collapsed slices improving the ability of parallel imaging to unalias them. The CAIPIRINHA concept can greatly reduces the g-factor penalty of simultaneous multi-slice acquisition for many imaging techniques. However, the RF-pulse based CAIPIRINHA technique is not applicable to the single-shot EPI used in most diffusion imaging applications since only a single RF pulse is employed for all ky lines.
Recent improvements in simultaneous multi-slice EPI methods have renewed interest in their use for diffusion and functional MRI studies. A fusion of parallel imaging and SIR may allow acquisition of a large number of simultaneously excited slices with an acceptable artifact level and SNR loss (Feinberg et al., 2010; Setsompop et al., 2010). In addition, a large reduction in g-factor SNR penalty for parallel imaging based simultaneous multi-slice method has been achieved for single-shot EPI acquisition without detrimental blurring trade-off via the introduction of the blipped-CAIPI method (Setsompop et al., 2012), a CAIPIRINHA-based technique for single-shot EPI acquisition. We note that a CAIPIRINHA-based technique was first applied to a single-shot EPI acquisition by Nunes et al. (2006). With single-shot EPI acquisition, multiple RF pulses cannot be used to create the inter-slice image shift. Instead, Nunes et al. utilized a Wideband-like approach in the phase encoding and readout directions to create inter-slice shift in these directions, respectively. However, this results in pixel tilting (blurring) artifacts and limits the amount of inter-slice image shift that can be applied. The blipped-CAIPI method (Setsompop et al., 2012) is a modification of Nune’s approach, where a modified Wideband-like sequence is used to generate the desired interslice image shift in the phase encoding direction, but without the pixel tilting artifacts. With this method, it was shown that for a typical whole brain acquisition at 3T with a 32 channel coil, the average g-factor penalty of a 3-fold slice-accelerated acquisition can be reduced from 32% to 1% through the use of inter-slice image shift.
Here, we show that the blipped-CAIPI based simultaneous multi-slice method can reduce the acquisition time of Q-ball and DSI acquisitions 3-fold. We also propose a novel modification for blipped-CAIPI image reconstruction to reduce inter-slice image ghost artifact that is specific to this type of acquisition. The performance of the method is compared to conventional acquisitions using image-based metrics based on pseudo-multiple replica (Robson et al., 2008) and diffusion-based metrics based on bootstrapping techniques (Jones, 2003; Pajevic and Basser, 2003). Using these metrics, we show that the data acquisition times for Q-ball and DSI can be reduced 3-fold with minimal loss in SNR or diffusion information, thereby providing an important gain in SNR per unit time. As a final part of this work, a 1418-direction DSI dataset was collected with the blipped-CAIPI based simultaneous multi-slice acquisition on a novel MR system equipped with a high performance CONNECTOM gradient system to demonstrate the potential in using such acquisition schemes to obtain a very large, high quality diffusion dataset.
Methods
Blipped-CAIPI sequence
With the blipped-CAIPI sequence, Gz encoding gradient blips are applied simultaneously with the phase encoding blips of the EPI readout train to impart phase differences between simultaneously excited imaging slices and provide an inter-slice shift between them. With this method, an amplitude cycling scheme on the Gz gradient is employed to generate the desired phase differences, but prevents undesirable voxel tilting artifacts. For details of this sequence please refer to Setsompop et al. (2012).
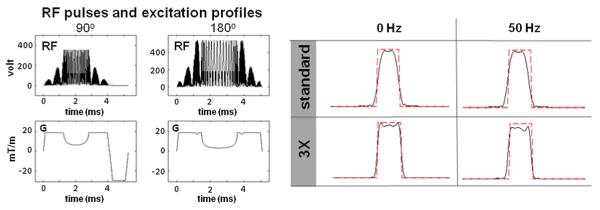
To achieve simultaneous multi-slice excitation, conventional slice-selective RF pulses are frequency-modulated and summed. Fig. 1 left shows the modulated 90° and 180° RF pulses and the corresponding slice-selective gradients for the 3× simultaneous multi-slice excitation. The Shinnar-Le Roux (SLR) algorithm (Pauly et al., 1991) was used to design the RF pulses while the Variable Rate Selective Excitation (VERSE) method (11) was applied to reduce the peak RF voltage and thus the specific absorption rate (SAR). To compensate for the degradation in the slice selection profile at off-resonance frequencies due to VERSE, the Time BandWidth (TBW) product of the RF pulses was increased compared to the conventional pulses. For 90° excitation, the TBW product was 6, VERSE factor 3×, and pulse length 4.8 ms. For 180° refocusing pulses, these parameters were 6, 6×, and 5.38 ms. Bloch simulation was employed to assess the slice selection performance of the multi-slice RF pulses in comparison to the standard RF pulses normally used in conventional diffusion sequences. Fig. 1 right shows a comparison of the excitation profiles from a 1× standard and a 3× simultaneous multi-slice 90°-180°-180° excitations at on resonance and at 50 Hz off-resonance. The normalized root mean square error (nRMSE) from the ideal profile (normalized by the area under the ideal profile) for the 90°-180°-180° sequence was 17.6% at 0 Hz and 28.5% at ±50 Hz off-resonance for our designed RF pulses. This was an improvement over the standard RF pulse train (27.4% and 32.1% for spins off-resonant by 0 Hz and 50 Hz, respectively).
Fig. 1.
Left: Modulate SLR 90° and 180° RF pulses for simultaneous multi-slice excitation. Right: comparison of the resulting excitation profiles of 1× standard and a 3× simultaneous multi-slice 90°–180°–180° excitation at on resonance and at 50 Hz.
Acquisitions
To compare the performance of the simultaneous multi-slice acquisition with conventional acquisition, the SE-EPI diffusion-weighted imaging was performed with a twice-refocused sequence (Feinberg and Jakab, 1990; Reese et al., 2003) using either conventional single-slice imaging or with a 3× slice-accelerated simultaneous multi-slice acquisition. Acquisitions were obtained from 3 healthy subjects after obtaining informed consent using an institutionally approved protocol. Imaging was performed using a 3T Siemens whole-body TIM Trio scanner and the commercially available 32-channel head array coil (Siemens Medical Solutions, Erlangen, Germany).
Three experiments were carried out to compare the performance of the 3× slice-accelerated blipped-CAIPI acquisitions to their equivalent conventional acquisitions with no slice-acceleration. The first experiment assessed SNR and diffusion metrics after applying slice acceleration factor (Rsl)=3 to Q-ball imaging without in-plane acceleration. Acquisition parameters were: resolution=2 mm isotropic; FOV=208 mm×208 mm×126 mm; Partial Fourier=6/8; Bandwidth=1658 Hz/pixel, b=3000 s/mm2, 64 directions, one b=0 image, 63 axial slices, TE=125 ms. For the conventional (Rsl=1) acquisition, TR=11.3 s and the total acquisition time Tacq=12.2 min. For the simultaneous multi-slice acquisition, 3 slices separated by 4.2 cm were simultaneously excited with a FOV/2 inter-slice image shift. The Rsl=3 slice accelerated sequence resulted in TR=3.8 s and Tacq=4.1 min. Five repetitions of the Rsl=1 and Rsl=3 acquisitions were collected to assess diffusion-based metrics via bootstrap analysis. Additionally, to evaluate the effect of TR reduction, 5 datasets with a reduced FOVz 1× acquisition (21 slices) and a TR matching the 3× acquisition (TR=3.8 s) were collected. Finally, to evaluate the effect of time averaging, 5 extra repetitions of the 3× acquisitions were collected to allow for a synthesis of 5 repetitions of 2 averages of 3× acquisitions (Tacq=8.2 min). A 1.5 averages of 3× acquisition (Tacq=6.2 min) was also created from this dataset by performing averaging from 2 repetitions for every other diffusion gradient direction.
The second experiment focused on SNR and diffusion metrics in Q-ball acquisition with 2× in-plane acceleration and imaging parameters similar to the first experiment. The TE was reduced to 118 ms due to the 2× in-plane acceleration (which shortens the EPI readout) and with 60 axial slices instead of 63 slices. The TR and total acquisition time of this protocol were 9.2 s and 10 min for conventional (Rsl=1) acquisition, and 3.1 s and 3.4 min for 3× slice accelerated acquisition. Due to the 2× in-plane acceleration, a FOV/2 shift between the slices would result in no net voxel shift between the slices. Thus, an inter-slice image shift of FOV/4 was employed to prevent voxels in adjacent slices with same (x,y) locations from overlapping in the collapsed image. The inter-slice image shift was achieved by applying the FOV/2 blipped-CAIPI Gz train to the reduced FOV acquisition. For bootstrap analysis, 5 repetitions of the 1× and 3× data were collected. An additional 10 repetitions of the 3× data were acquired for use to assess the performance of 2 averages and 3 averages of 3× acquisitions (Tacq=6.8 and 10.2 min). The robustness of the bootstrap method was also evaluated from the 3× data by computing bootstrap metrics for three independent sets, 5 repetitions each.
The third experiment was for a DSI acquisition protocol where the imaging parameters were: resolution=2.5 mm isotropic; FOV= 200 mm×200 mm×127.5 mm; Partial Fourier=6/8; Bandwidth= 2083 Hz/pixel, bmax=7000 s/mm2, 256 directions, with additional b=0 image every 20 TRs (for motion correction), 51 sagittal slices, and TE=157 ms. No in-plane acceleration was used. The TR and total acquisition time for this protocol were 10 s and 45 min for the conventional acquisition and 3.4 s and 15 min for the 3× slice accelerated acquisition.
In addition to the aforementioned experiments that will be use to compare the performance of simultaneous multi-slice acquisition with conventional acquisition, a high quality DSI dataset was also collected with simultaneous multi-slice acquisition to demonstrate the potential of using such acquisition scheme to obtain a very large, high quality diffusion dataset. For this, Stejskal–Tanner based diffusion EPI acquisitions were obtained from a healthy volunteer using a novel 3T system (MAGNETOM Skyra CONNECTOM†, Siemens Healthcare, Erlangen, Germany) equipped with the AS302 CONNECTOM gradient with Gmax = 300 mT/m and Slew=200 T/m/s. A custom-built 64-channel RF head array was used for reception. 3× slice-accelerated simultaneous multi-slice with 2× in-plane acceleration was employed (with an intra-slice image shift of FOV/4). Imaging parameters were: resolution=2 mm isotropic; FOV=200 mm×200 mm×126 mm; no Partial Fourier; Bandwidth=2273 Hz/pixel (effective echo spacing=0.27 ms), bmax=10500 s/mm2, 1418 directions, with additional b=0 image every 20 TRs (for motion correction purpose), 63 axial slices, and TE=64 ms. The TR and total acquisition time for this protocol were 2.1 s and 52 min. The maximum gradient strength for this acquisition was limited to 200 mT/m to reduce eddy-current distortions.
Reconstruction
The slice-GRAPPA algorithm (Setsompop et al., 2012) was used to unalias the multiple slices. In this algorithm, a GRAPPA-like kernel was fit to each slice of a pre-scan calibration dataset acquired one slice at a time and then applied to the aliased data to estimate the k-space points of each individual imaging slice. Thus, for the 3-fold slice-accelerated acquisition, 3 separate sets of GRAPPA kernels were fitted and applied, one for each imaging slice. For the acquisitions with both in-plane and slice acceleration, the reconstructions were performed in sequential steps: first the slice-GRAPPA was applied to separate the aliased slices, then conventional GRAPPA was used to generate the missing k-space lines for the in-plane under-sampled slices.
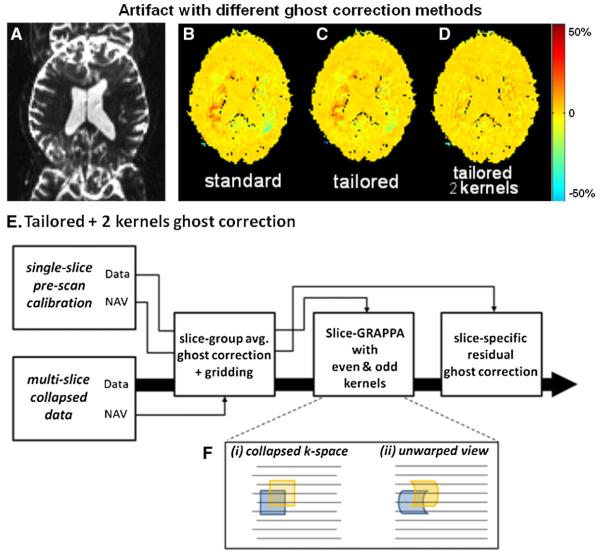
Field inhomogeneities and eddy currents can create slice-specific ghosting artifacts during the reconstruction of multi-slice acquisitions. If the ghost correction is performed on the collapsed slices (using the phases of the odd and even reference lines acquired before reading out the collapsed slices), then only the slice-group average ghost is corrected. We call this the “standard” correction. To correct for slice-specific ghosts, Moeller et al. (2010) applied a static residual even/odd phase correction to each slice after slice separation via a SENSE/GRAPPA-based reconstruction (Blaimer et al., 2006). This phase correction was estimated from pre-scan calibration data, which was acquired one slice at a time. We used a similar scheme where the phase correction from the single slice reference data was applied to the slice unaliased by the slice-GRAPPA method. We refer to this as the “tailored” ghost correction method. We evaluated this even/odd phase correction on a 3× slice-accelerated acquisition with FOV/2 inter-slice image shift. Fig. 2 shows a percentage signal error image of the unaliased simultaneous multi-slice image of the middle slice. In the standard ghost correction scheme the middle slice (shown in Fig. 2B) shows a significant inter-slice ghost artifact originating from the top slice. The artifact is slightly reduced by the use of the tailored ghost correction (Fig. 2C), but further improvement to the reconstruction is required to reduce the ghost artifact to an acceptable level. This illustrates that a small level of ghosting from a high intensity region of the top slice can cause a large signal change in the voxels with lower intensity in the center slice. This is particularly prominent for blipped CAIPI acquisitions with FOV/2 inter-slice shift, where the ghost from the top imaging slice is directly in the middle of the FOV of the center slice.
Fig. 2.
Comparison of FOV/2 ghost correction methods for blipped-CAIPI acquisition. A) Collapsed 3 slice-accelerated acquisition with FOV/2 inter-slice image shift, B)–D) image artifact in the separated center slice as a percent signal change, for standard, tailored, and tailored with two GRAPPA kernels reconstruction methods. Significant inter-slice ghost artifact (mainly from the top slice) can be observed with standard ghost correction reconstruction. Minor reduction of this artifact is achieved via the tailored ghost correction method, while a major improvement is provided by the addition of the two GRAPPA kernels method. E) Flow diagram of the tailored ghost+two GRAPPA kernels method. F) Shows the justification for the two GRAPPA kernel method, where (i) shows the application of a GRAPPA kernel to the even (blue) and odd (yellow) lines of the collapsed k-space data in the presence of even/odd phase imperfection, and (ii) illustrates the differences in k-space coverage in the aligned k-space co-ordinate for the even and odd line application of the kernel. With these differences in k-space coverage, a different kernel should be use for the odd and even line. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The inter-slice ghost artifact is caused by imperfect separation of the single slice data from the collapsed data through the use of a single GRAPPA kernel set in the slice-GRAPPA algorithm. This algorithm fails to simultaneously remove the top slice image and its ghost during the reconstruction of the center slice. To reduce inter-slice ghost artifact, we proposed the use of separate slice-GRAPPA kernels for the odd and even k-space lines (in addition to the tailored ghost correction). The overall scheme of this ghost correction is shown in Fig. 2E. To illustrate how separate even and odd kernels can reduce inter-slice ghost, we examine how slice-GRAPPA kernel is applied to the slice-collapsed data (after slice-group average ghost correction). Fig. 2F(i) shows the application of a GRAPPA kernel to the even and odd lines (blue and yellow) of the slice-collapsed data. Note that the k-space data of the collapsed image is not perfectly ghost corrected since a different correction is needed for the signal of each slice in this collapsed data. Fig. 2F(ii) shows the differences in the coverage in the unwarped k-space coordinate that this kernel would have for the odd and even line application (note that the kernel coverage will actually be different for each imaging slice of the collapsed image since the amount of warping is different in each slice). Due to this difference, we use a different GRAPPA kernel for the odd and the even lines to effectively unalias the imaging slices and eliminate inter-slice ghost artifacts. The first kernel set is estimated using the even lines fitting of the pre-scan calibration data and then applied on the even lines of the collapsed data to generate the even lines data of the individual slice. The second kernel set is generated and applied in a similar way but on the odd data lines. Both kernel sets must be estimated from the pre-scan dataset after slice-group average phase corrections from the reference lines of the simultaneous multi-slice acquisition.
The even/odd phase correction is commonly performed prior to the gridding operation. Here, the slice-group average ghost correction based on the navigator of the collapsed data was applied prior to the gridding process (this was done for all the ghost correction cases in Fig. 2). However, the slice specific even/odd phase correction can only be applied after the slice-GRAPPA operation, which needs to be performed on gridded k-space data.
In low SNR acquisitions, sum-of-square (SoS) combination of coil channel data represents a poor approximation to the optimal coil combination (weighting by the coil sensitivity profiles) since weighting the pixel intensity of each coil by itself is essentially a random weighting for low SNR data. Additionally, the magnitude operation generates a non-central Chi-squared distribution (Constantinides et al., 1997) noise distribution and introduces significant bias. This is an important concern for diffusion weighted acquisitions, particularly at high b-values where the image SNR is low and has a detrimental effect on the estimation of the diffusion metrics (Jones and Basser, 2004). To avoid this problem, we used a combination based on the sensitivity profile of the coil array, where the complex sensitivity profiles were estimated from polynomial fitting/smoothing of the b=0 pre-scan data. A bootstrap based comparison between sensitivity combined and SoS coil combinations for HARDI diffusion data set is also provided in the Supplemental material section.
Image-based SNR and residual aliasing artifact quantification
The image SNR of the simultaneous multi-slice technique was compared to that of the unaccelerated acquisition using the pseudo-multiple replica method (Robson et al., 2008) with 1000 image pseudo time series of the coil-combined images, as described in Setsompop et al. (2012). Using this method, the SNR ratio maps (slice-accelerated to conventional acquisition) were created. For acquisitions without inplane acceleration, this represents the g-factor penalty of slice acceleration (1/gsl). For acquisitions with in-plane acceleration, this represents the ratio of g-factor penalties ginplane/ginplane + sl. Additionally, the normalized mean square error (nRMSE) measure of the residual aliasing artifact was calculated. The SNR ratios and residual aliasing artifact were evaluated over a brain-sized region of interest.
Quality assessment of Q-ball data using bootstrap
The pseudo-multiple replica method allows the estimation of the variability of the reconstruction. Non-reconstruction related (or acquisition instability related, e.g., subject motion) effects can also have an impact on the data. Such effects are likely to be dependent on many acquisition parameters such as b-value and diffusion direction of the acquired image. In this work, to capture both the reconstruction related and the non-reconstruction related variability in a given diffusion acquisition, bootstrap analysis was performed. The quality of the simultaneous multi-slice (3×) and conventional (1×) data for Q-ball reconstruction was assessed using regular bootstrap with 5 acquired repetitions using the methodology of Cohen-Adad et al. (2011). The evaluation was performed for in-plane acceleration factors (Rinplane) of 1 and 2. Each set of HARDI data (Rsl=3× and 1×) was corrected for motion using FSL FLIRT (Jenkinson and Smith, 2001) where the motion was estimated directly from the acquired diffusion weighted images. The data were then re-shuffled using regular bootstrapping with jackknife sampling to synthesize 500 bootstrapped repetitions. For each bootstrapped dataset, the diffusion orientation distribution function (ODF) was estimated voxel-wise using Q-ball reconstruction based on spherical harmonics functions (Descoteaux et al., 2007). The reproducibility of the ODF was assessed using the Jensen-Shannon Divergence (JSD) and the 95% angular confidence interval was derived for the first and the second ODF maxima (CI1 and CI2).
Quality assessment of DSI data using tractography
Each DSI dataset was corrected for motion using FSL FLIRT. Motion was assessed from the b=0 images interspersed every 20 volumes using FSL FLIRT (6° of freedom). Each diffusion weighted data was then corrected using the transformation matrix associated to the closest b=0 image (closest in time).
For the data acquired with the CONNECTOM gradient system, we also corrected for non-rigid distortions induced by eddy-currents. In contrast to the other DSI data, this dataset was more affected by eddycurrent distortions due to the use of the Stejskal-Tanner sequence (Stejskal and Tanner, 1965) (as opposed to the twice refocused sequence). To correct for eddy-current distortions we employed the method proposed in Bodammer et al. (2004), where each diffusion-encoding gradient was also applied in the opposite direction, producing 709 (1418/2) pairs of directions. FLIRT was used to estimate a transformation matrix to register each pair of images acquired with opposite gradient directions. Registration was constrained to translation along Y, stretching along Y and shearing in the X-Y plane (therefore 3° of freedom). Based on the coefficients, an average transformation matrix was generated and applied to both images (one positive, one negative).
ODF and tractography were conducted using the Diffusion ToolKit (Wang et al., 2007). The within-voxel angular threshold for tractography was 45° and minimum track length was 2.5 cm. The number of tracks and the average track length±standard deviation (SD) were analyzed. Additionally, for the 1× and 3× data comparison, we labeled 18 major white-matter pathways following the protocol described in Wakana et al. (2007), which defines two regions of interest (ROIs) per pathway in parts of the anatomy that each pathway is known to traverse. To eliminate variability due to manual labeling in the two data sets and make our comparison as unbiased as possible, the ROIs used here were not drawn manually on the 1× and 3× data. Instead we obtained the ROIs from a different data set of 33 healthy subjects, where we had previously labeled the same pathways (Yendiki et al., 2011). We averaged the respective ROIs from the 33 subjects in MNI space and we mapped the average ROIs to the native space of the 1× and 3× data sets using affine registration. In each dataset, we isolated the tractography streamlines going through the respective ROIs to identify the 18 pathways. The average fractional anisotropy (FA) and the volume (number of voxels) of each of the pathways were assessed for both the 1× and 3× data.
Results
Fig. 2 assesses the amount of artifacts for the simultaneous multi-slice acquisitions with 3 different ghost correction methods. Figs. 2B–D show a ratio image of the conventional single-slice prescan data and the unaliased simultaneous multi-slice image of the center slice. Either the standard ghost correction (2B), the tailored ghost correction (2C), or the tailored ghost correction+two GRAPPA kernel approach (2D) was used. Table 1 lists the mean and SD of the artifact, and the percentage of pixels that has more than 10% signal error for all three ghost correction techniques. The artifact level in the tailored ghost correction+two GRAPPA kernel approach is significantly lower than in the other two methods. Compared to the new tailored ghost+two GRAPPA approach, the standard ghost correction method results in 30% higher mean artifact level and increases the number of pixels with more than 10% signal error by more than a factor of 3.
Table 1.
Tabulation of (i) the mean and standard deviation of the image artifact and (ii) percentage of pixels that have more than 10% signal error, separately for the three different ghost correction methods.
| %Error: mean±Std | Pixels with >10% error | |
|---|---|---|
| Standard | 2.6±3.5 | 4.5% |
| Tailored-ghost | 2.4±3.26 | 3.5% |
| Tailored-ghost with 2 kernels | 1.97±2.3 | 1.4% |
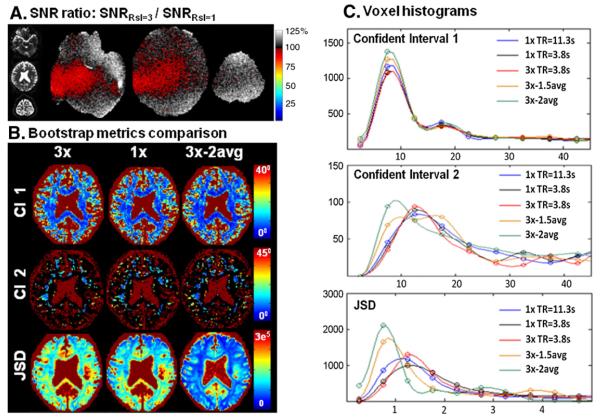
Fig. 3 shows SNR and bootstrap metrics for the Q-ball acquisition with no in-plane acceleration. Panel A shows the unfolded images from 3× slice accelerated blipped-CAIPI acquisition with FOV/2 inter-slice shift and the corresponding Monte-Carlo generated SNR ratio maps (comparing the SNR of this acquisition to the standard Rsl=1× acquisition). The average SNR ratio±SD across the three slices was 1.04±0.07 and the minimum SNR ratio value was 0.89 (after smoothing the SNR ratio map with a 5×5 voxel square kernel). In some regions the retained SNR was greater than unity indicating some noise cancelation in the reconstruction process as well as the changes in the noise coupling across channels that affects the SNR of the coil combination process. This increase in retained SNR was previously demonstrated in low acceleration GRAPPA acquisitions (Polimeni et al., 2008) and the effect of noise cancelation was also previously described by Sodickson (2000) in the context of SMASH theory. The nRMSE of the aliasing artifact for this acquisition is 1.3%. Panel B shows the bootstrap uncertainty metrics for the center slice where the average SNR ratio was 1.02±0.06. The left side of Panel B contains the maps of the 95% confidence interval of the primary (CI 1) and the secondary (CI 2) fiber directions and JSD of the ODF for i) 3× slice-accelerated (4 min), ii) non-accelerated (12 min) and iii) two averages of 3× slice-accelerated (8 min) acquisitions. Panel C shows the corresponding voxel histograms of the confidence intervals and JSD of the three acquisition schemes and of iv) 1.5 averages of 3× slice-accelerated acquisition (6 min) and v) non-accelerated acquisition with reduced FOVz (21 slices, 4 min). The reproducibility in the 3× slice-accelerated acquisition is slightly lower than in the standard 1× acquisition, which has a longer TR and thus slightly higher SNR (note that the higher the JSD, the lower the reproducibility of the ODF). However, bootstrap metrics from the 3× acquisition are very similar to that of the reduced FOVz non-accelerated acquisition with the same TR. The 1.5 and 2 averages of 3× slice-accelerated acquisitions result in markedly higher reproducibility; illustrating the gain in SNR per unit time of the blipped-CAIPI acquisition.
Fig. 3.
Results from 3× slice-accelerated blipped-CAIPI acquisition with FOV/2 inter-slice shift. A) Unfolded images of an aliased slice group and the corresponding Monte-Carlo generated SNR ratio maps, where the SNR retention is close to 100% in all locations B) bootstrap metrics comparison. Left: 95% confidence interval of the primary (CI 1) and the secondary (CI 2) fiber directions and Jenson-Shannon Divergence (JSD) of the ODF of the Q-ball reconstruction for i) 3× slice-accelerated (4 min), ii) non-accelerated (12 min) and iii) two averages of 3× slice-accelerated (6 min) acquisitions. C) The corresponding voxel histograms of the confidence intervals and JSD of the three acquisition schemes and of iv) 1.5 averages of 3× slice-accelerated acquisition (5.5 min) and v) non-accelerated acquisition with reduced FOVz (with 21 slices to achieve a matching TR to the 63 slices 3× slice-accelerated acquisition, 4 min). The angular uncertainties and JSD measures of the 3× slice-accelerated acquisition were very similar to that of the reduced FOVz non-accelerated acquisition. The performances of these acquisitions were marginally worse than that of the standard non-accelerated acquisition which a longer TR and slightly higher signal. The 1.5 and 2 averages of 3× slice-accelerated acquisitions provided progressively better performance; illustrating the gain in SNR per unit time of the blipped-CAIPI acquisition.
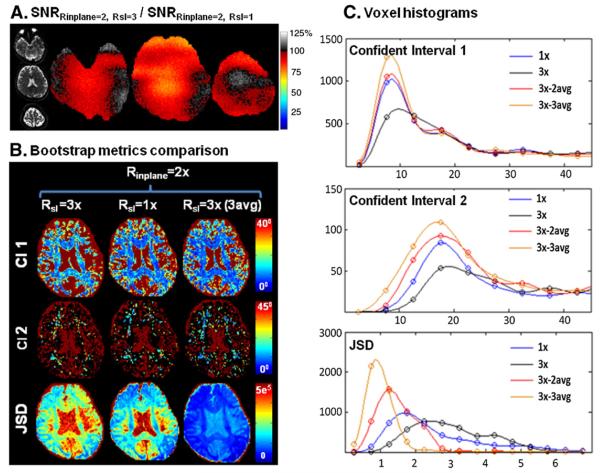
Fig. 4 shows the SNR ratios and diffusion metrics for the Q-ball acquisition with Rinplane =2. Thus, conventional (Rsl×Rinplane =1×2) and simultaneous multi-slice (Rsl×Rinplane =3×2) acquisitions are compared. Fig. 4A shows the unfolded images from the blipped-CAIPI acquisition slice group and the Monte Carlo generated SNR ratio maps. The average SNR ratio±SD across the three slices was 0.9±0.08 with a minimum SNR ratio value of 0.73 (after smoothing the ratio maps with a 5×5 voxel square kernel). The nRMSE of the aliasing artifact for this acquisition is 3%. Fig. 4B shows the bootstrap uncertainty metrics (95% confidence interval for fiber directions 1 and 2, and JSD) for the center slice, which had the lowest average SNR ratio (0.85±0.08). From left to right, these metrics are shown for total acquisition times of 3.3 min, 10 min, and 10 min. Thus we assessed the metrics for Rsl×Rinplane =3×2 with both accelerated acquisition time (3.3 min) as well as with matched acquisition time to the Rsl=1 acquisition by taking 3 averages of the Rsl=3 acquisition. Fig. 4C shows the voxel histograms of the uncertainty measures for the aforementioned recordings and for a two averages (6.6 min) of the Rsl×Rinplane =3×2 acquisition. The uncertainty of the 3.3 min acquisition is higher than that of the conventional 10 min acquisition, highlighting the SNR reduction per shot. The 2 and 3 averages data (6.6 and 10 min acquisitions) both provide superior performance in comparison to the conventional 10 min acquisition, with further improvements with more averages. This illustrates the gain in SNR per unit time of the blipped-CAIPI acquisition.
Fig. 4.
Results from 3× slice and 2× in-plane accelerated blipped-CAIPI acquisition with FOV/4 inter-slice shift. A) Unfolded images of an aliased slice group and the corresponding Monte-Carlo generated SNR ratio. B) Bootstrap metrics comparison left: 95% confidence interval of the primary (CI 1) and the secondary (CI 2) fiber directions and JSD of the ODF of the Q-ball reconstruction i) 3× slice and 2× in-plane accelerated (3.3 min), ii) 2× in-plane accelerated (10 min) and iii) three averages of 3× slice and 2× in-plane accelerated (10 min) acquisitions. C) The corresponding voxel histograms of the confident intervals and JSD of the three acquisition schemes and of iv) two averages of the 3× slice and 2× in-plane accelerated (6.6 min) acquisition. The uncertainty measures of the 3× slice and 2× in-plane accelerated (3.3 min) acquisition are higher than that of the 2× in-plane accelerated (10 min) acquisition; highlighting an SNR reduction per shot. The 2 and 3 averages of the 3× slice and 2× in-plane accelerated acquisition (6.6 and 10 min) both provide superior performances in comparison to the 2× in-plane accelerated acquisition, with performance clearly improving with more averages. This illustrates the gain in SNR per unit time of the blipped-CAIPI acquisition.
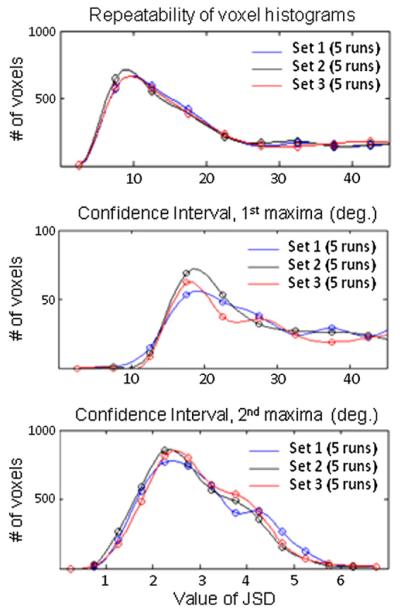
Fig. 5 shows the repeatability of the bootstrap metrics by evaluating three independent bootstrap data sets (5 runs each) of the Rsl×R-inplane =3×2 accelerated acquisition. The voxel histogram plots of the confidence intervals and JSD of three independent datasets are shown. Good repeatability of the uncertainty measures from the three independent data sets is observed, with confidence interval of the primary fiber direction (CI 1) plots having the smallest variation, and the confidence interval of the secondary fiber direction (CI 2) plots having the largest variation.
Fig. 5.
Voxel histogram plots of the confident intervals and JSD of three independent bootstrap datasets of the 3× slice and 2× in-plane accelerated acquisition. Good agreement between the uncertainty measures from the three independent datasets can be observed.
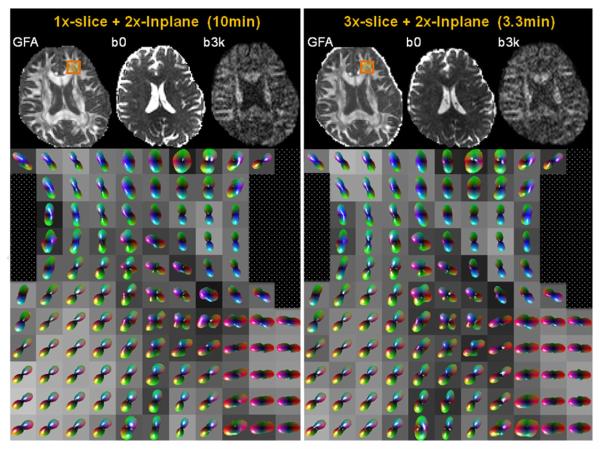
Fig. 6 provides a qualitative comparison of the diffusion weighted images and metrics of the Rsl×Rinplane =1×2 accelerated (10 min), and Rsl×Rinplane =3×2 accelerated (3.3 min) acquisitions. The GFA, b=0 image, b=3000 image and Q-ball based ODFs of a region (orange square) are shown for both acquisitions. Visual comparison suggests similar diffusion ODF between acquisition with and without simultaneous multi-slice in the white matter. Largest differences are observed in the gray matter where the ODF is expected to be highly variable due to the issues of reliably estimating the ODF in low anisotropy areas. These regions have been masked to focus the comparison on white matter. Although the two acquisitions were aligned prior to comparison, it is also clear that the alignment is not perfect. It is difficult to interpret the significance of the differences in the glyphs without knowledge of the test-retest statistics.
Fig. 6.
Comparison of the general fractional isotropy (GFA), b=0 image, b=3000 image, and Q-ball based orientation distribution function of a zoomed in region (orange square) of the 2× in-plane accelerated (10 min), and 3× slice and 2× in-plane accelerated (3.3 min) acquisitions. Similar results are observed.
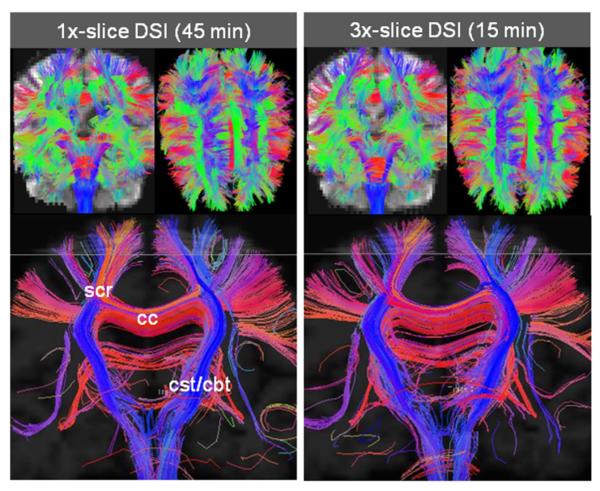
Fig. 7 shows the tractography results from the 256 directions DSI acquisition for the Rsl×Rinplane =1×1 (45 min), and Rsl×Rinplane =3×1 accelerated (15 min) acquisitions with very similar tract distributions. The top part of the figure shows whole-brain tractography results, in which one can appreciate the similarity between the two reconstructions. The number of tracks and their average lengths were 42,420 and 68±39 mm for the 1× acquisition and 42,500 and 65±37 mm for the 3× multi-slice accelerated acquisition. The bottom part of Fig. 7 shows the tracts that reside within a 17.5 mm coronal slab. Again, the 1× and the 3× reconstruction are visually very similar. Major tracts are reconstructed in both datasets, such as the corpus callosum (cc), corticospinal tract (cst), corticobulbar tract (cbt) and the superior corona radiata (scr). These results also highlight the benefit of using DSI reconstruction to be able to reconstruct multiple fiber orientation within voxels. The corresponding Monte-Carlo generated SNR ratio maps for the 3× acquisition (not shown) is very similar to that of the Rsl ×Rinplane =3×1 accelerated Q-ball acquisition, with an average SNR ratio of ~1.
Fig. 7.
Comparison of the tractography results of 1× (45 min) vs. 3× (15 min) slice accelerated DSI acquisitions (256 directions). The top panel shows whole-brain tractography results while the bottom panel shows the tracts that reside within a 17.5 mm coronal slab. The 1× and 3× tractography results appear to be very similar. These results also highlight the benefit of DSI acquisition in the reconstruction of multiple fiber orientations within voxels.
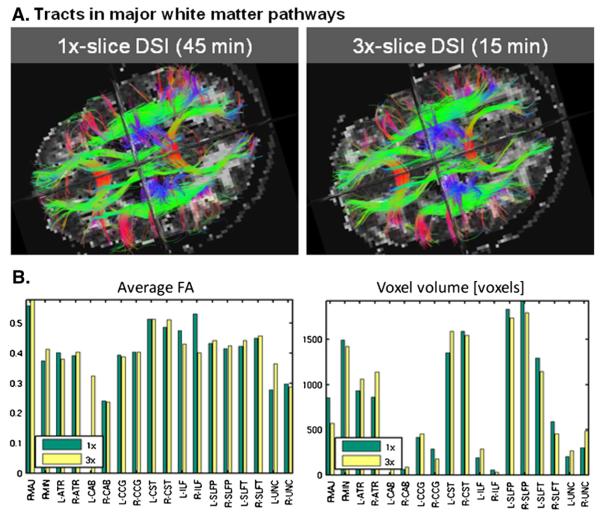
Fig. 8A shows images of the labeled pathways in the 1× and 3× DSI datasets. Fig. 8B shows plots of the corresponding average FA and volume of each pathway. In general we found good agreement between the 1× and 3× results. The most notable differences were in weaker pathways that only consisted of very few streamlines and thus were more sensitive to noise and had lower test-retest reliability than the stronger pathways. This was the case especially for the left cingulum-angular bundle (CAB), which did not have any streamlines in the 1× but was observed in the 3× data, and the right inferior longitudinal fasciculus (ILF), which had very few streamlines in both data sets. Also, we found some reduction in the volume of the corpus callosum — forceps major (FMAJ) pathway in the 3× data. A one-way ANOVA testing the difference in mean FA within each tract between the 1× and 3× DSI dataset did not show any significant differences (F<0.0001, p=0.98).
Fig. 8.
A) Axial view of white-matter pathways labeled from streamline DSI tractography in 1× (45 min) and 3× (15 min) data. Visible in this view are the forceps minor and major of the corpus callosum, the anterior thalamic radiations, the cingulum, the superior longitudinal fasciculus, and the superior endings of the corticospinal tract. B) Average FA (left) and volume in number of voxels (right) for each of the 18 labeled pathways, as obtained from the 1× (green) and 3× (yellow) data sets. Intra-hemispheric pathways are indicated by “L-” (left) or “R-” (right). The pathways are: corpus callosum — forceps major (FMAJ), corpus callosum — forceps minor (FMIN), anterior thalamic radiation (ATR), cingulum-angular (infracallosal) bundle (CAB), cingulum-cingulate gyrus (supracallosal) bundle (CCG), corticospinal tract (CST), inferior longitudinal fasciculus (ILF), superior longitudinal fasciculus-parietal bundle (SLFP), superior longitudinal fasciculus-temporal bundle (SLFT), uncinate fasciculus (UNC). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
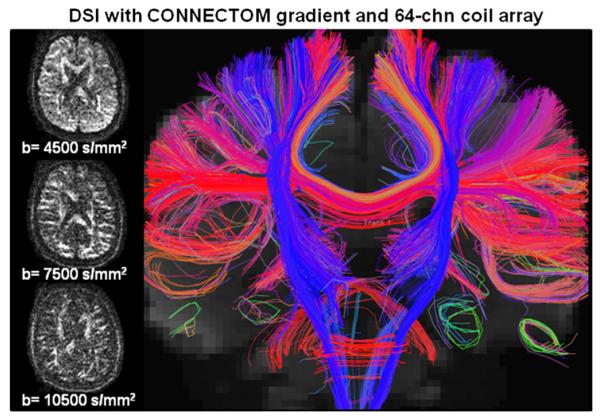
Fig. 9 shows diffusion weighted images (DWI) and tractography result of the 1418-direction DSI acquisition (Rsl×Rinplane =3×2), where the average SNR ratio±SD across the three slices was 1.05±0.08 and the minimum SNR ratio value was 0.87. With the short TE achieved through the use of the CONNECTOM gradient, high resolution DWIs with good SNR and contrast were achieved in a single shot at b=10500 s/mm2. The right of Fig. 9 shows the corresponding tractography result of tracts that reside within a 16 mm coronal slab.
Fig. 9.
Results from the 1418-direction DSI acquisition (Rsl×Rinplane =3×2) acquired using the CONNECTOM gradient and a 64-channel coil array. Left: diffusion weighted images at different b values, illustrating good SNR and contrast in a single shot up to b=10,500 s/mm2 right: tractography result of tracts that reside within a 16 mm coronal slab.
Discussion and conclusion
In this study, performance of the blipped-CAIPI method in 3× simultaneous multi-slice diffusion imaging was assessed using a pseudo-multiple replica SNR measure. In addition, reproducibility of the Q-ball ODFs was assessed via bootstrapping metrics. DSI tractography was qualitatively assessed and the average FA and volume of the major white matter pathways were compared. Through these quantitative and qualitative assessments, we show that the data acquisition times for Q-ball and DSI can be reduced 3-fold using simultaneous multislice with small loss in SNR or diffusion information compared to conventional acquisitions, thereby providing large gains in sensitivity per unit time.
The bootstrap metrics performance was found to be sensitive and repeatable. For example, the measure of Q-ball ODF reproducibility (JSD) was sensitive to minor SNR differences in the raw data. This is illustrated via the results in the Q-ball measures of Fig. 3. These data contained minor differences in SNR level between the Rsl×Rinplane=1×1 acquisition acquired with TR=11.8 s and again at TR=3.8 s. The expected SNR difference due to the TR differences for white matter (T1~850 ms) was around ~1%. Some additional SNR differences can also result from imperfect slice profiles and slice interleaving. For TR=3.8 s, the saturation-recovery factor of the unintentionally crushed signal due to imperfect excitation of spatially adjacent slice (excited TR/2 prior) was ~11% (e-(TR/2)/T1). Overall, the expected SNR loss was small. Nonetheless, this small SNR loss still translated to visible differences in the Q-ball bootstrap metrics. In addition, the bootstrap metrics were shown to be repeatable in three independent bootstrap datasets with identical acquisition parameters (Fig. 5). Here we note that the angular confidence interval is more robust for the 1st (CI 1) than for the 2nd maximum (CI 2), and that JSD shows good reproducibility across the three bootstrap datasets. The reduction in robustness of the 2nd maxima is likely to be from the lower SNR of this metric.
The bootstrap results of the slice-accelerated acquisitions agree well with the predicted SNR loss from the pseudo-multiple replica simulation and the expected time averaging SNR gain from averaging across multiple repetitions. This is highlighted in Fig. 4 (Q-ball acquisition with Rsl×Rinplane=3×2 acceleration). Based on the pseudo-multiple replica simulation, the average reduction in SNR for the Rsl×Rinplane=3×2 accelerated acquisition was 0.85 (for the center slice). This SNR loss could, in principle, be compensated by averaging together 1.38 fold more data, as supported by Fig. 4. The performance level of the bootstrap metrics of the non-slice accelerated acquisition is between the performance of the 1-average and 2-average of the 3× slice accelerated acquisition. Thus, averaging together two Rsl×Rinplane=3×2 accelerated acquisitions should (and does) overcompensate for the SNR reduction of the accelerated acquisition. Thus the blipped-CAIPI simultaneous multi-slice method achieves a 3-fold acquisition time boost with an SNR cost equivalent of 1.38 acquisitions, and thus could re-create the data of the non-multi-slice acquisition with equal SNR with a time savings of 3/1.38 = 2.2 fold. We note that the g-factor related SNR reduction in Rsl×Rinplane=3×2 accelerated acquisitions can be overcome with the use of a 64-channel head array coil.
In this work, the SNR per unit time gain in the blipped-CAIPI acquisition was illustrated by assessing the uncertainty metrics of the 2 and 3 averages of the 3× slice accelerated acquisition. The work by Jones (2004) suggests that with more scan time, a larger gain in diffusion metrics robustness may be achieved by sampling more diffusion directions rather than using the extra time to acquire more averages of the original diffusion directions. Therefore, the acquisition time gained from the blipped-CAIPI sequence might be best used to acquire more diffusion directions.
The novel modified image reconstruction method, which incorporates the tailored ghost correction and the two GRAPPA kernel techniques, significantly reduced the large inter-slice ghosting artifact associated with blipped-CAIPI acquisition. This was demonstrated in Fig. 2 for the FOV/2 inter-slice image shift acquisition. With an acquisition that employs a different inter-slice shift (such as FOV/3), the inter-slice ghost artifact will appear at a spatially different location. Nonetheless, the proposed ghost correction method should still provide a good performance in removing the majority of this artifact.
The coil sensitivity profiles were used in combining coil array data to improve SNR and reduce signal bias in the magnitude images of high b-value low SNR acquisitions. Further gains in SNR from optimizing the coil combination process can be achieved by employing the coils’ noise covariance information. However, the application of slice GRAPPA and standard GRAPPA algorithms can greatly modify the noise covariance information of the reconstructed coil images (Breuer et al., 2009). Further work will be needed to correctly characterize and account for these effects.
For DSI tractography results in Fig. 8, it is possible that more stable FA and volume measurements of the labeled 18 white matter pathways could be obtained by manual labeling of the paths directly on the data, instead of using the average ROIs, which are susceptible to registration errors and are probably larger than the ROIs that a rater would draw directly on the images. However, we used the average ROIs here to avoid introducing variability due to manual labeling. In a previous study we evaluated the intra-rater and inter-rater reliability of the manual labeling procedure by performing manual labeling several times on the same data set. We found the average distance between pathways labeled by the same and different raters to be, respectively, in the order of 1 voxel and 2 voxels (Yendiki et al., 2011). In the present study we found that the distance between the pathways obtained from the 1× and 3× data sets was comparable (median distance: 2.52 mm, mean distance: 3.98 mm). Further investigation with test-retest scans is warranted to determine how the differences between the 1× and 3× results compare to the test-retest reliability of each type of scan.
The simultaneous multi-slice method does put some constraints on the number of slices. For example, the acquisition is simplified if the total number of slices is a multiple of Rsl. A more subtle effect occurs when an interleaved slice order is used. The purpose of interleaving is, of course, to avoid exciting spatially adjacent slices in rapid succession. In a standard interleaved acquisition, adjacent slices are taken TR/2 apart in time. The interleaved Rsl=3 acquisition has an additional constraint if one wishes to avoid having some spatially adjacent slices acquired in rapid succession. In the simultaneous multislice acquisition with a total of Nsl, a total of Rsl subgroups each with Nsl/Rsl slices are created. The successive excitation problem occurs between the top slice of one subgroup and the bottom slice of the subgroup above it. The problem can be avoided if the number of slices in each excitation subgroup is odd. Thus Nsl/Rsl should be an odd integer to avoid signal loss slices with imperfect slice profiles at the edge of each sub-group. If an even integer is chosen, the first slice of each subgroup will be excited right after the excitation of an adjacent slice that corresponds to the last excited slice in an adjacent slice group (from the previous TR). This leads to a signal loss from the slice crosstalk for these edge slices. This effect was observed in the Q-ball experiment where 60 axial slices were acquired with a 3× slice acceleration factor (20 slices per subgroup). In this acquisition, acquired as interleaved slices from the bottom of the head up, slice 20 (from the bottom) and slice 21 were acquired adjacent in time. Slice 20 was acquired in the last (20th) excitation of the subgroup and slice 21 was acquired as the first excitation (in the next TR period) of the second subgroup immediately after the excitation for slice 20. Therefore any overlap between the two slices would have caused slice 21 to appear darker. These slices were not selected as the slice group for the bootstrap comparison and therefore did not have an effect on the present analysis. This effect is amplified by the common practice of using a wider slice-select profile for the 180 pulse of the spin echo, in order to reduce the effect of the poorer slice profile in refocusing pulses. As mentioned above, the problem could have been avoided by using an odd number of slices in the slice subgroups thus subsequent Rsl=3 acquisitions (e.g., Nsl=63).
In this work, Rsl=3× slice acceleration factor was used in the blipped-CAIPI simultaneous multi-slice acquisition scheme to accelerate diffusion acquisitions 3-fold. Further reduction in acquisition time can be achieved by increasing the slice acceleration factor and/or by combining parallel imaging simultaneous multi-slice method with the SIR technique as shown in Feinberg et al. (2010); Setsompop et al. (2010). With higher acceleration factors, SNR and SAR will need to be carefully considered. With a high slice acceleration factor, and hence short TR, the saturation effect will lower the SNR in a given acquisition, but the SNR per unit time will still improve until TR is reduced below its optimum value of 1.25 T1. On the other hand, the g-factor related SNR penalty will increases with increasing slice acceleration. In general, a reduction in SNR per acquisition (from saturation effect and g-factor) is tolerable as long as the associated acquisition time reduction overcompensates this reduction to create a net gain in SNR per unit time. However, additional consideration will need to be made in choosing the slice acceleration factor, when SNR of each acquisition is close to the noise floor (e.g., acquisition with high b values and/or spatial resolution). When diffusion weighted image (DWI) signals are close to the back-ground noise level, deviation from Gaussian noise behavior can cause signal bias that is orientationally dependent, resulting in a biased estimate of the diffusion parameters (Jones and Basser, 2004). Increasing the slice acceleration factor reduces SNR per acquisition and thereby increases this bias. Since diffusion acquisitions are very sensitive to motion, which can cause large phase changes in the image, averaging of multiple repetitions is usually performed on the magnitude image. This improves the overall SNR but does not mitigate the aforementioned signal bias issue. Therefore in choosing the slice acceleration factor, one will need to consider both the SNR per unit time and the possible signal bias that results from low SNR per acquisition.
Simultaneous multi-slice excitation can lead to an increase in RF SAR deposition. In this work, the VERSE algorithm (Conolly et al., 1988) was used to reduce the peak RF voltage and hence SAR at a cost of excitation profile degradation for off-resonance spins. This degradation was mitigated through the use of a large Time Band-Width (TBW) product value in the SLR (Pauly et al., 1991) based RF pulse design. With this approach, we were able to obtain RF excitation and refocusing pulses for Rsl=3 twice-refocused spin echo sequence (Feinberg and Jakab, 1990; Reese et al., 2003) that together provided good slice selection profiles while staying under the SAR limit. In the case of higher slice acceleration factors, slice profile fidelity will need to be traded for reductions in peak power to stay within SAR limits. The use of a single-refocused spin echo sequence would help to significantly lower SAR and mitigate this issue, but at the cost of eddy currents (Reese et al., 2003).
For the acquisition on the CONNECTOM gradient system, a single refocused spin echo sequence was employed, while the maximum allowable gradient strength was limited to 200 mT/m (rather than 300 mT/m) to limit eddy-current distortions. This was a conservative approach that account for the relatively modest b-values (bmax=10500 s/mm2) that was used. At this maximum b-value, the increase in TE from reducing the maximal gradient strength from 300 mT/m to 200 mT/m is only a few ms, while the reduction in eddy current distortion is rather significant (33%). Future work will explore the use of phase reversal based distortion correction methods (Andersson et al., 2003; Morgan et al., 2004), which could improve the mitigation of both the eddy current and local field inhomogeneity related distortions, and allow for high-quality acquisitions of ultra high b values diffusion images using the full gradient capability of the CONNECTOM system.
In this work, we implemented the slice-GRAPPA kernel application as a k-space convolution in Siemen’s ICE reconstruction software environment and standard PC based reconstruction hardware. With our implementation, the reconstruction time is approximately 2× the acquisition time for the acquisitions used in this work. With a more efficient algorithm for the kernel application, such as the “split-domain” approach (Brau et al., 2008), we expect significant improvement in the reconstruction speed.
In this section, a bootstrap based comparison between sensitivity combined and SoS coil combinations for HARDI diffusion data set is provided using data from experiment 1 (Q-ball imaging with 64 directions, b=3000 s/mm2 and no parallel imaging acceleration). Figure S1A shows the diffusion-weighted images and the 95% angular confidence interval of the second ODF maxima derived from boot-strap analysis. As expected, a strong elevation of the mean noise level is seen in low signal regions for the SoS coil combination. This is mitigated in the sensitivity based coil combination method. The reduction in signal bias and the improvement in SNR from sensitivity combined reconstruction results in a lower angular uncertainty (Figure S1A right and Figure S1B histograms). The sensitivity-based coil combination produces an over 2-fold increase in the number of voxels with a 95% angular confidence interval of less than 15°. Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/j.neuroimage.2012.06.033.
Supplementary Material
Acknowledgment
Grant support: NIH: NIBIB K99EB012107, NIBIB R01EB006847, NIMH R01MH652456, NCRR P41RR14075, and the NIH Blueprint for Neuroscience Research U01MH093765 the Human Connectome project. NSF: PHY-0855161.
Footnotes
Uncited reference
Setsompop et al., 2011
References
- Andersson JL, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage. 2003;20:870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J. Magn. Reson. B. 1994;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- Blaimer M, Breuer FA, Seiberlich N, Mueller MF, Heidemann RM, Jellus V, Wiggins G, Wald LL, Griswold MA, Jakob PM. Accelerated volumetric MRI with a SENSE/GRAPPA combination. J. Magn. Reson. Imaging. 2006;24:444–450. doi: 10.1002/jmri.20632. [DOI] [PubMed] [Google Scholar]
- Bodammer N, Kaufmann J, Kanowski M, Tempelmann C. Eddy current correction in diffusion-weighted imaging using pairs of images acquired with opposite diffusion gradient polarity. Magn. Reson. Med. 2004;51:188–193. doi: 10.1002/mrm.10690. [DOI] [PubMed] [Google Scholar]
- Brau AC, Beatty PJ, Skare S, Bammer R. Comparison of reconstruction accuracy and efficiency among autocalibrating data-driven parallel imaging methods. Magn. Reson. Med. 2008;59:382–395. doi: 10.1002/mrm.21481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer FA, Blaimer M, Heidemann RM, Mueller MF, Griswold MA, Jakob PM. Controlled aliasing in parallel imaging results in higher acceleration (CAIPIRINHA) for multi-slice imaging. Magn. Reson. Med. 2005;53:684–691. doi: 10.1002/mrm.20401. [DOI] [PubMed] [Google Scholar]
- Breuer FA, Kannengiesser SA, Blaimer M, Seiberlich N, Jakob PM, Griswold MA. General formulation for quantitative G-factor calculation in GRAPPA reconstructions. Magn. Reson. Med. 2009;62:739–746. doi: 10.1002/mrm.22066. [DOI] [PubMed] [Google Scholar]
- Cohen-Adad J, Descoteaux M, Wald LL. Quality assessment of high angular resolution diffusion imaging data using bootstrap on Q-ball reconstruction. J. Magn. Reson. Imaging. 2011;33:1194–1208. doi: 10.1002/jmri.22535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conolly SM, Nishimura DG, Macovski A, Glover GH. Variable-rate selective excitation. J. Magn. Reson. 1988;78:440–458. [Google Scholar]
- Constantinides CD, Atalar E, McVeigh ER. Signal-to-noise measurements in magnitude images from NMR phased arrays. Magn. Reson. Med. 1997;38:852–857. doi: 10.1002/mrm.1910380524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descoteaux M, Angelino E, Fitzgibbons S, Deriche R. Regularized, fast, and robust analytical Q-ball imaging. Magn. Reson. Med. 2007;58:497–510. doi: 10.1002/mrm.21277. [DOI] [PubMed] [Google Scholar]
- Feinberg DA, Jakab PD. Tissue perfusion in humans studied by Fourier velocity distribution, line scan, and echo-planar imaging. Magn. Reson. Med. 1990;16:280–293. doi: 10.1002/mrm.1910160209. [DOI] [PubMed] [Google Scholar]
- Feinberg DA, Reese TG, Wedeen VJ. Simultaneous echo refocusing in EPI. Magn. Reson. Med. 2002;48:1–5. doi: 10.1002/mrm.10227. [DOI] [PubMed] [Google Scholar]
- Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Glasser MF, Miller KL, Ugurbil K, Yacoub E. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PLoS One. 2010;5:e15710. doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn. Reson. Med. 2002;47:1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Jones DK. Determining and visualizing uncertainty in estimates of fiber orientation from diffusion tensor MRI. Magn. Reson. Med. 2003;49:7–12. doi: 10.1002/mrm.10331. [DOI] [PubMed] [Google Scholar]
- Jones DK. The effect of gradient sampling schemes on measures derived from diffusion tensor MRI: a Monte Carlo study. Magn. Reson. Med. 2004;51:807–815. doi: 10.1002/mrm.20033. [DOI] [PubMed] [Google Scholar]
- Jones DK, Basser PJ. Squashing peanuts and smashing pumpkins”: how noise distorts diffusion-weighted MR data. Magn. Reson. Med. 2004;52:979–993. doi: 10.1002/mrm.20283. [DOI] [PubMed] [Google Scholar]
- Larkman DJ, Hajnal JV, Herlihy AH, Coutts GA, Young IR, Ehnholm G. Use of multicoil arrays for separation of signal from multiple slices simultaneously excited. J. Magn. Reson. Imaging. 2001;13:313–317. doi: 10.1002/1522-2586(200102)13:2<313::aid-jmri1045>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Ugurbil K. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn. Reson. Med. 2010;63:1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan PS, Bowtell RW, McIntyre DJ, Worthington BS. Correction of spatial distortion in EPI due to inhomogeneous static magnetic fields using the reversed gradient method. J. Magn. Reson. Imaging. 2004;19:499–507. doi: 10.1002/jmri.20032. [DOI] [PubMed] [Google Scholar]
- Nunes RG, Hajnal JV, Golay X, Larkman DJ. Simultaneous slice excitation and reconstruction for single shot EPI; Proceedings of the 14th Annual Meeting of ISMRM; Seattle, Washington, USA. 2006.2006. p. 293. [Google Scholar]
- Pajevic S, Basser PJ. Parametric and non-parametric statistical analysis of DT-MRI data. J. Magn. Reson. 2003;161:1–14. doi: 10.1016/s1090-7807(02)00178-7. [DOI] [PubMed] [Google Scholar]
- Paley MN, Lee KJ, Wild JM, Griffiths PD, Whitby EH. Simultaneous parallel inclined readout image technique. Magn. Reson. Imaging. 2006;24:557–562. doi: 10.1016/j.mri.2005.12.019. [DOI] [PubMed] [Google Scholar]
- Pauly J, Le Roux P, Nishimura D, Macovski A. Parameter relations for the Shinnar-Le Roux selective excitation pulse design algorithm. IEEE Trans. Med. Imaging. 1991;10:53–65. doi: 10.1109/42.75611. [DOI] [PubMed] [Google Scholar]
- Polimeni JR, Wiggins GC, Wald LL. Characterization of artifacts and noise enhancement introduced by GRAPPA reconstructions; Proceedings of the 16th Annual Meeting of ISMRM; Toronto, Canada. 2008.2008. p. 1286. [Google Scholar]
- Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn. Reson. Med. 1999;42:952–962. [PubMed] [Google Scholar]
- Reese TG, Heid O, Weisskoff RM, Wedeen VJ. Reduction of eddy-current-induced distortion in diffusion MRI using a twice-refocused spin echo. Magn. Reson. Med. 2003;49:177–182. doi: 10.1002/mrm.10308. [DOI] [PubMed] [Google Scholar]
- Reese TG, Benner T, Wang R, Feinberg DA, Wedeen VJ. Halving imaging time of whole brain diffusion spectrum imaging and diffusion tractography using simultaneous image refocusing in EPI. J. Magn. Reson. Imaging. 2009;29:517–522. doi: 10.1002/jmri.21497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson PM, Grant AK, Madhuranthakam AJ, Lattanzi R, Sodickson DK, McKenzie CA. Comprehensive quantification of signal-to-noise ratio and g-factor for image-based and k-space-based parallel imaging reconstructions. Magn. Reson. Med. 2008;60:895–907. doi: 10.1002/mrm.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K, Gagoski BA, Polimeni J, Witzel TW, V J, Wald LL. Blipped CAIPIRHINA for simultaneous multi-slice EPI with reduced g-factor penalty; Proceedings of the 18th Annual Meeting of ISMRM; Stockholm, Sweden. 2010.2010. p. 551. [Google Scholar]
- Setsompop K, Gagoski BA, Polimeni J, Witzel T, Wedeen VJ, Wald LL. Blipped-Controlled Aliasing in Parallel Imaging (blipped-CAIPI) for simultaneous multi-slice EPI with reduced g-factor penalty. Magn. Reson. Med. 2011 doi: 10.1002/mrm.23097. early view. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K, Gagoski BA, Polimeni J, Witzel TW, V J, Wald LL. Blipped-Controlled Aliasing in Parallel Imaging (blipped-CAIPI) for simultaneous multi-slice EPI with reduced g-factor penalty. Magn. Reson. Med. 2012;67:1210–1224. doi: 10.1002/mrm.23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodickson DK. Tailored SMASH image reconstructions for robust in vivo parallel MR imaging. Magn. Reson. Med. 2000;44:243–251. doi: 10.1002/1522-2594(200008)44:2<243::aid-mrm11>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Sodickson DK, Manning WJ. Simultaneous acquisition of spatial harmonics (SMASH): fast imaging with radiofrequency coil arrays. Magn. Reson. Med. 1997;38:591–603. doi: 10.1002/mrm.1910380414. [DOI] [PubMed] [Google Scholar]
- Stejskal EO, Tanner JE. Spin diffusion measurements: spin echoes in the presence of a time-dependent field gradient. J. Chem. Phys. 1965;42:288–292. [Google Scholar]
- Tournier JD, Calamante F, Gadian DG, Connelly A. Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. Neuroimage. 2004;23:1176–1185. doi: 10.1016/j.neuroimage.2004.07.037. [DOI] [PubMed] [Google Scholar]
- Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Diffusion MRI of complex neural architecture. Neuron. 2003;40:885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- Wakana S, Caprihan A, Panzenboeck MM, Fallon JH, Perry M, Gollub RL, Hua K, Zhang J, Jiang H, Dubey P, Blitz A, van Zijl P, Mori S. Reproducibility of quantitative tractography methods applied to cerebral white matter. Neuroimage. 2007;36:630–644. doi: 10.1016/j.neuroimage.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Benner T, Sorensen AG, Wedeen VJ. Diffusion toolkit: a software package for diffusion imaging data processing and tractography; Proceedings of the 15th Annual Meeting of ISMRM; Berlin, Germany. 2007.2007. p. 3720. [Google Scholar]
- Weaver JB. Simultaneous multislice acquisition of MR images. Magn. Reson. Med. 1988;8:275–284. doi: 10.1002/mrm.1910080305. [DOI] [PubMed] [Google Scholar]
- Wedeen VJ, Hagmann P, Tseng WY, Reese TG, Weisskoff RM. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn. Reson. Med. 2005;54:1377–1386. doi: 10.1002/mrm.20642. [DOI] [PubMed] [Google Scholar]
- Yendiki A, Panneck P, Srinivasan P, Stevens A, Zollei L, Augustinack J, Wang R, Salat D, Ehrlich S, Behrens T, Jbabdi S, Gollub R, Fischl B. Automated probabilistic reconstruction of white-matter pathways in health and disease using an atlas of the underlying anatomy. Front Neuroinform. 2011;5:23. doi: 10.3389/fninf.2011.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.