Abstract
Histone interactions which occur, at moderate ionic strengths, when several types of purified, renatured histones are mixed at equimolar ratios have been studied. The four histones H2A,H2B,H3 and H4 complex and form dimers. Histone H1 does not interact with the other four histone types and does not form dimers. Mixing of single histone species with preformed histone pairs as well as mixing of two different types of histone pairs, leads to exchange of histones among the pairs and formation of dimers. No trimers are formed. The dimers are in equilibrium with high-molecular weight histone structures. The results indicate that histone dimers may serve as a stable intermediate in histone assembly. Because each histone type (except H1) can interact with itself as well as with each of the other three histone types we suggest that each histone type should be considered as an interchangeable subunit of a multichain protein in which the dimer species is the most stable structure.
Full text
PDF


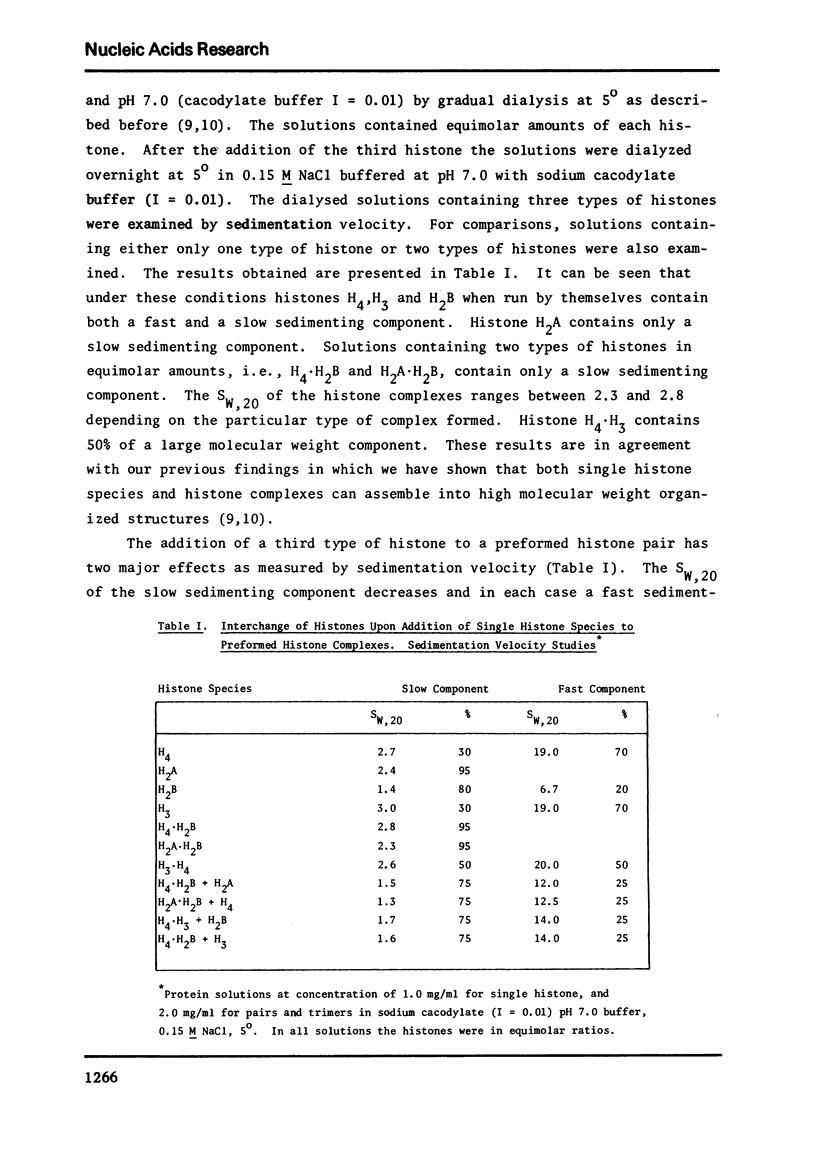

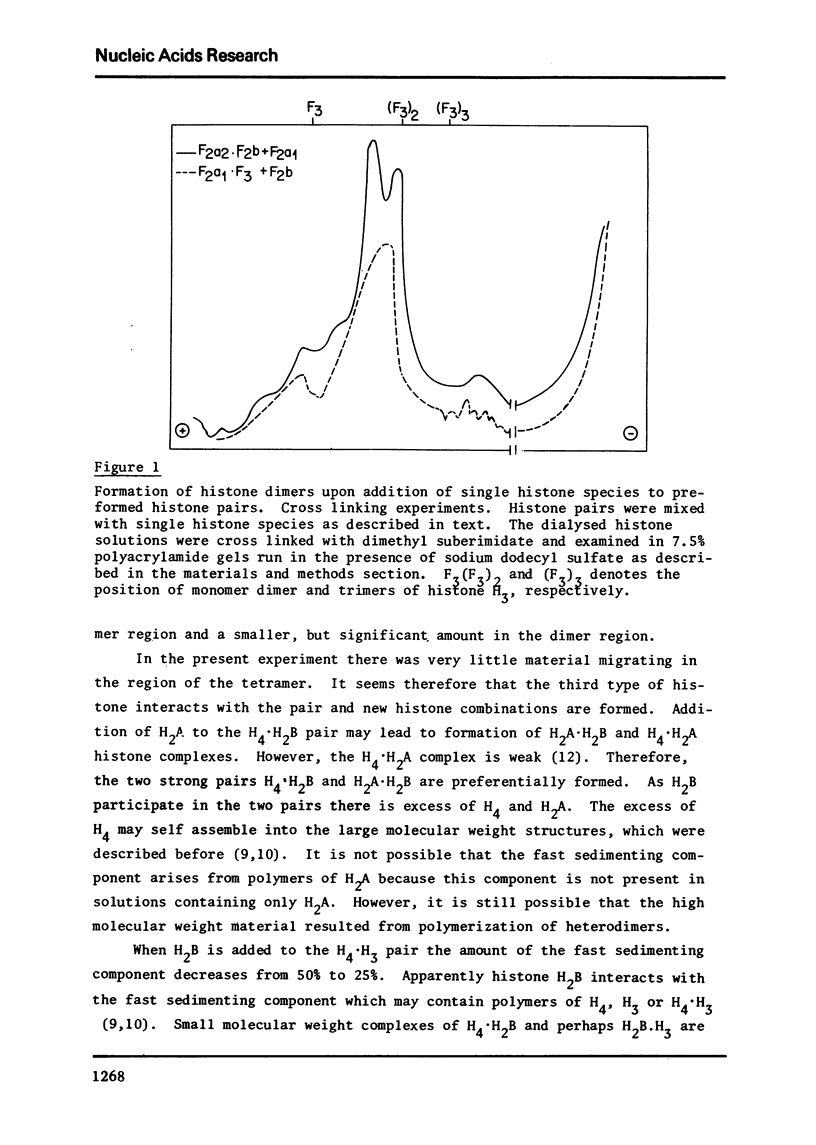

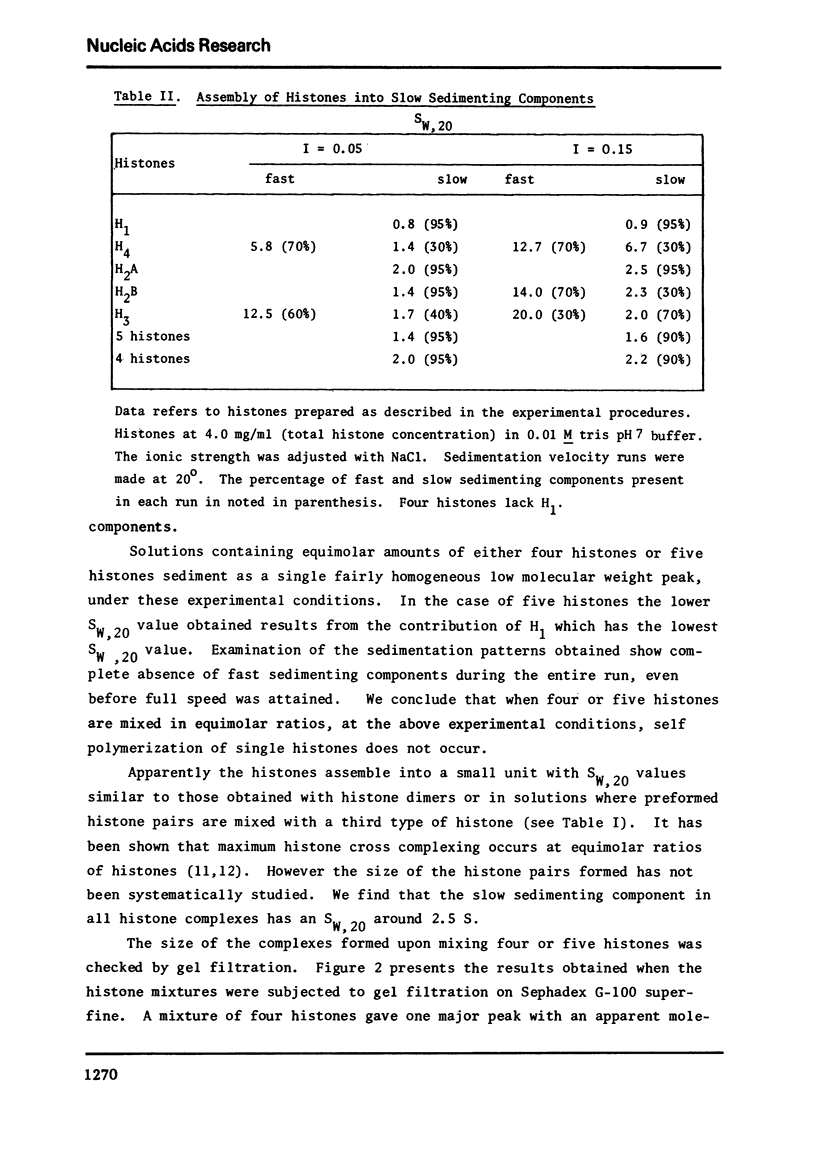
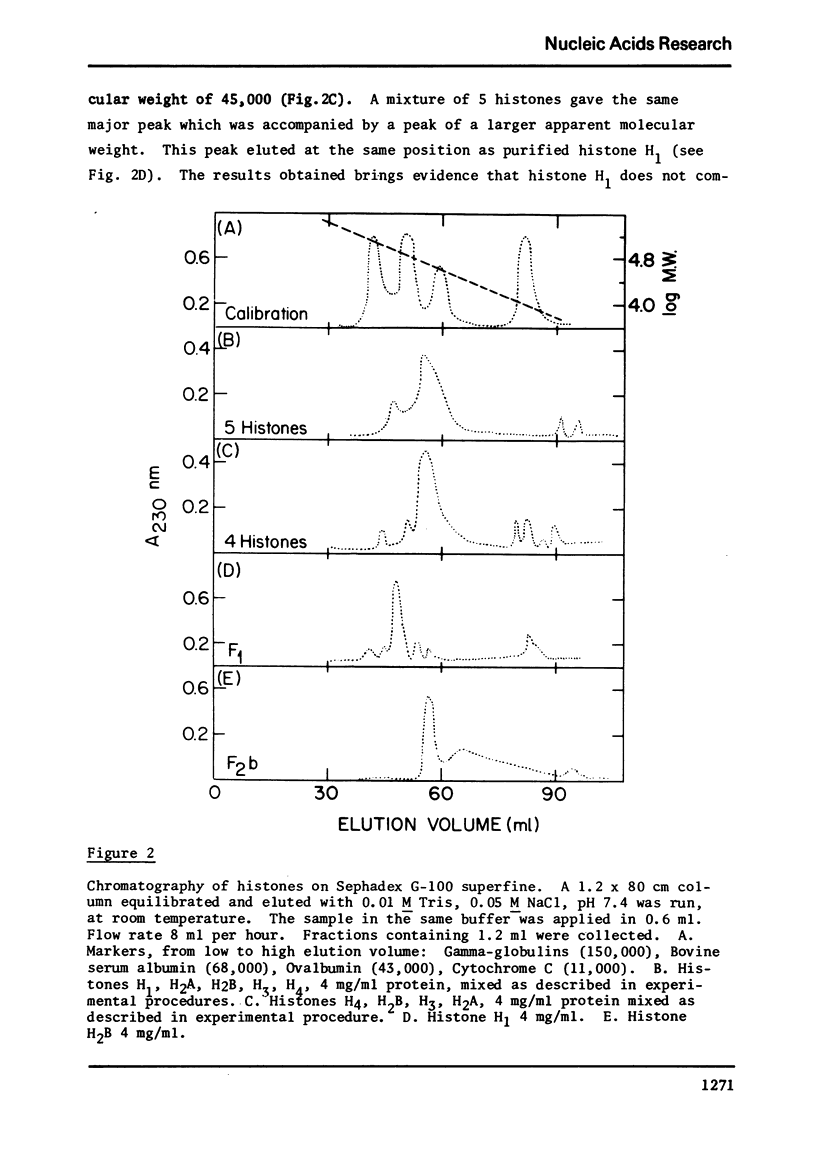
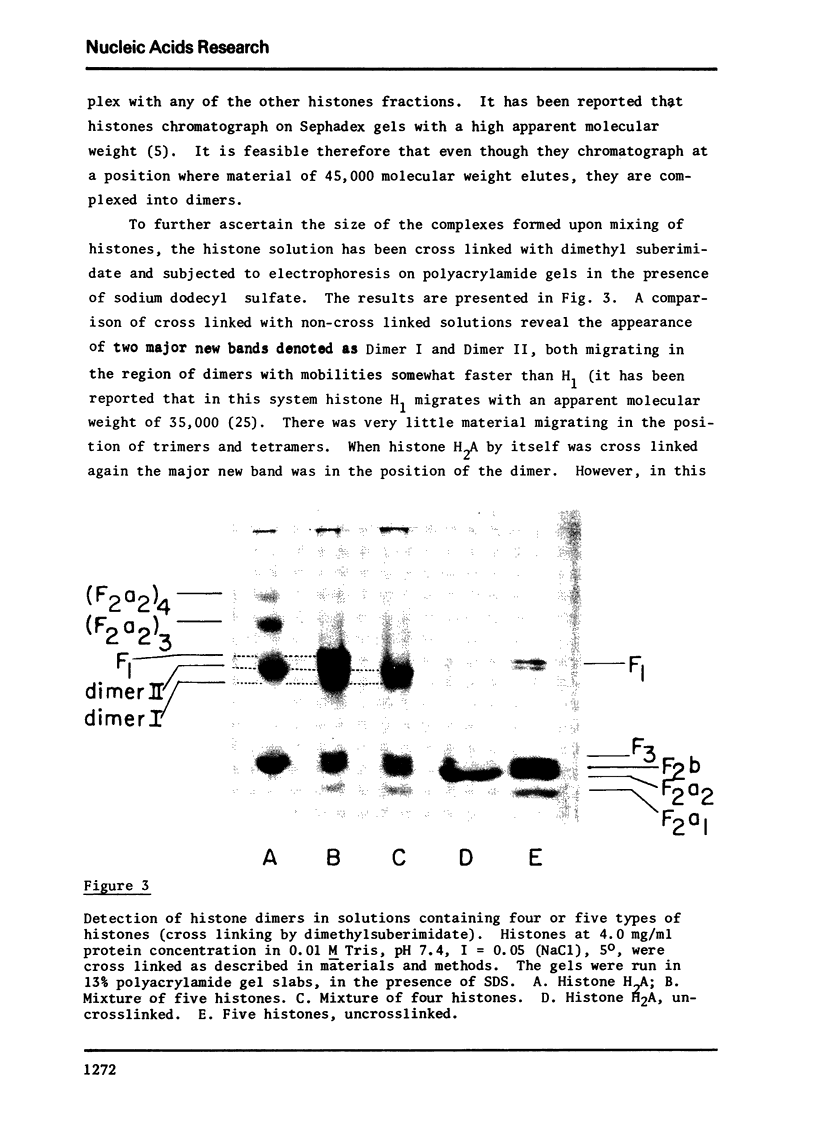
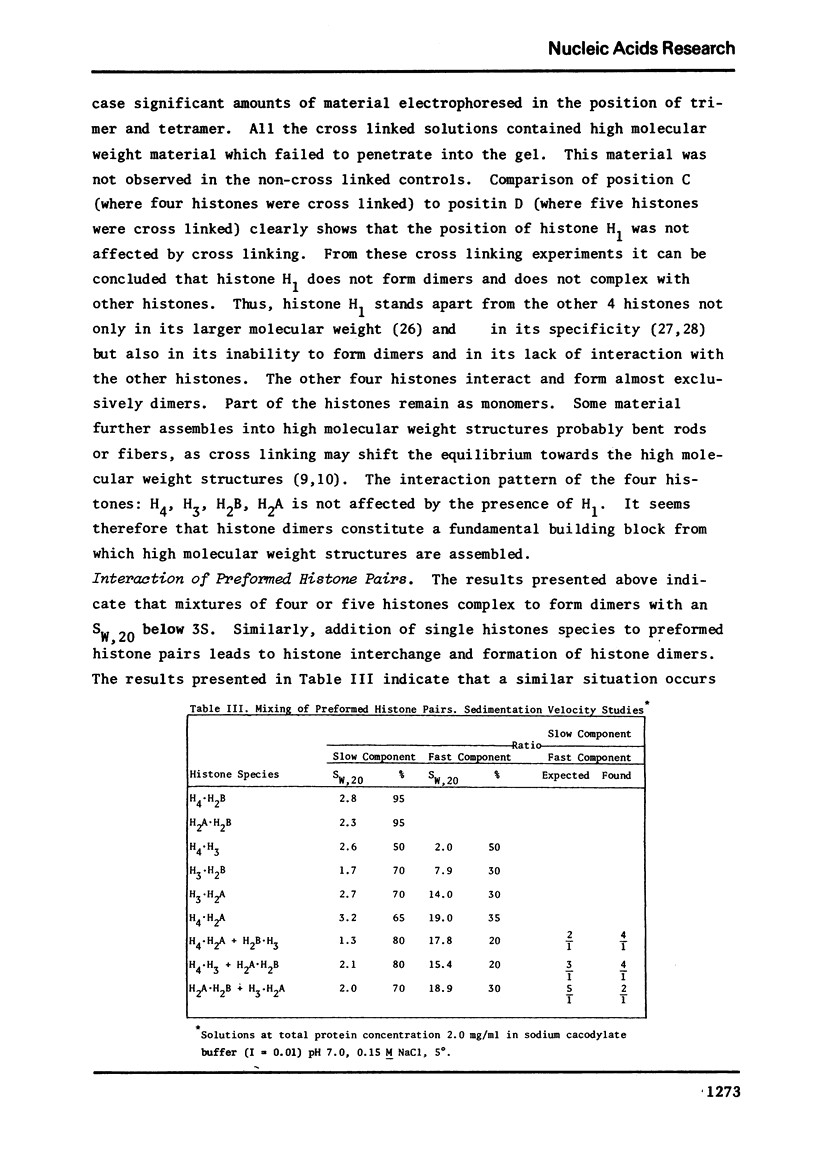


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin J. P., Boseley P. G., Bradbury E. M., Ibel K. The subunit structure of the eukaryotic chromosome. Nature. 1975 Jan 24;253(5489):245–249. doi: 10.1038/253245a0. [DOI] [PubMed] [Google Scholar]
- Bram S., Butler-Browne G., Bradbury E. M., Baldwin J., Reiss C., Ibel K. Chromatin neutron and X-ray diffraction studies and high resolution melting of DNA-histone complexes. Biochimie. 1974;56(6-7):987–994. doi: 10.1016/s0300-9084(74)80519-5. [DOI] [PubMed] [Google Scholar]
- Bustin M., Cole R. D. Species and organ specificity in very lysine-rich histones. J Biol Chem. 1968 Sep 10;243(17):4500–4505. [PubMed] [Google Scholar]
- Bustin M. Conservative amino-acid replacement in the tyrosine region of the lysine-rich histones. Eur J Biochem. 1972 Sep 18;29(2):263–267. doi: 10.1111/j.1432-1033.1972.tb01983.x. [DOI] [PubMed] [Google Scholar]
- Bustin M., Stollar B. D. Immunochemical specificity in lysine-rich histone subfractions. J Biol Chem. 1972 Sep 25;247(18):5716–5721. [PubMed] [Google Scholar]
- Böhm E. L., Strickland W. N., Strickland M., Thwaits B. H., van der Westhuizen D. R., von Holt C. Purification of the five main calf thymus histone fractions by gel exclusion chromatography. FEBS Lett. 1973 Aug 15;34(2):217–221. doi: 10.1016/0014-5793(73)80797-5. [DOI] [PubMed] [Google Scholar]
- D'Anna J. A., Jr, Isenberg I. A histone cross-complexing pattern. Biochemistry. 1974 Nov 19;13(24):4992–4997. doi: 10.1021/bi00721a019. [DOI] [PubMed] [Google Scholar]
- D'Anna J. A., Jr, Isenberg I. Interactions of histone LAK (f2a2) with histones KAS (f2b) and GRK (f2a1). Biochemistry. 1974 May 7;13(10):2098–2104. doi: 10.1021/bi00707a016. [DOI] [PubMed] [Google Scholar]
- Davies G. E., Stark G. R. Use of dimethyl suberimidate, a cross-linking reagent, in studying the subunit structure of oligomeric proteins. Proc Natl Acad Sci U S A. 1970 Jul;66(3):651–656. doi: 10.1073/pnas.66.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldblatt D., Bustin M. Exposure of histone antigenic determinants in chromatin. Biochemistry. 1975 Apr 22;14(8):1689–1695. doi: 10.1021/bi00679a022. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Matsutera E., Oba Y. A theoretical consideration of the abnormal behavior of histones on sodium dodecylsulfate gel electrophoresis. Biochim Biophys Acta. 1974 Mar 14;342(1):185–194. doi: 10.1016/0005-2795(74)90120-2. [DOI] [PubMed] [Google Scholar]
- Hyde J. E., Walker I. O. A model for chromatin sub-structure incorporating symmetry considerations of histone oligomers. Nucleic Acids Res. 1975 Mar;2(3):405–421. doi: 10.1093/nar/2.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde J. E., Walker I. O. Covalent cross-linking of histones in chromatin. FEBS Lett. 1975 Feb 1;50(2):150–154. doi: 10.1016/0014-5793(75)80477-7. [DOI] [PubMed] [Google Scholar]
- Kelley R. I. Isolation of a histone IIb1-IIb2 complex. Biochem Biophys Res Commun. 1973 Oct 15;54(4):1588–1594. doi: 10.1016/0006-291x(73)91168-6. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li H. J. A model for chromatin structure. Nucleic Acids Res. 1975 Aug;2(8):1275–1289. doi: 10.1093/nar/2.8.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Wilkins M. H. A super-coil model for nucleohistone. J Mol Biol. 1972 Jul 14;68(1):115–124. doi: 10.1016/0022-2836(72)90267-7. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Allfrey V. G. A method for the purification of histone fraction F3 by affinity chromatography. Arch Biochem Biophys. 1973 Jan;154(1):185–191. doi: 10.1016/0003-9861(73)90047-7. [DOI] [PubMed] [Google Scholar]
- Skidmore C., Walker I. O., Pardon J. F., Richards B. M. The structure of partially histone depleted nucleohistone. FEBS Lett. 1973 May 15;32(1):175–178. doi: 10.1016/0014-5793(73)80765-3. [DOI] [PubMed] [Google Scholar]
- Sperling R., Bustin M. Dynamic equilibrium in histone assembly: self-assembly of single histones and histone pairs. Biochemistry. 1975 Jul 29;14(15):3322–3331. doi: 10.1021/bi00686a006. [DOI] [PubMed] [Google Scholar]
- Teller D. C., Kinkade J. M., Jr, Cole R. D. The molecular weight of lysine-rich histone. Biochem Biophys Res Commun. 1965 Sep 22;20(6):739–744. doi: 10.1016/0006-291x(65)90079-3. [DOI] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]
- Weintraub H., Palter K., Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975 Sep;6(1):85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]