Abstract
Selective breeding programs aiming to increase the productivity and profitability of the sheep meat industry use elite, progeny tested sires. The broad genetic traits of primary interest in the progeny of these sires include skeletal muscle yield, fat content, eating quality, and reproductive efficiency. Natural mutations in sheep that enhance muscling have been identified, while a number of genome scans have identified and confirmed quantitative trait loci (QTL) for skeletal muscle traits. The detailed phenotypic characteristics of sheep carrying these mutations or QTL affecting skeletal muscle show a number of common biological themes, particularly changes in developmental growth trajectories, alterations of whole animal morphology, and a shift toward fast twitch glycolytic fibers. The genetic, developmental, and biochemical mechanisms underpinning the actions of some of these genetic variants are described. This review critically assesses this research area, identifies gaps in knowledge, and highlights mechanistic linkages between genetic polymorphisms and skeletal muscle phenotypic changes. This knowledge may aid the discovery of new causal genetic variants and in some cases lead to the development of biochemical and immunological strategies aimed at enhancing skeletal muscle.
Keywords: sheep, skeletal muscle, gene, myostatin, Callipyge, imprinting
Introduction
Significant improvement in sheep meat production has recently been made by using selective breeding and improved animal husbandry (Fogarty, 2009; Gardner et al., 2010). The breeding programs typically select sires based on production performance of their progeny, relatives and ancestors, or combinations thereof. Genetic selection and breeding using sires with high estimated breeding values (EBVs) for muscling traits is prevalent in the sheep meat industry.
DNA marker-assisted breeding strategies in sheep are now positioned to markedly accelerate the rates of genetic gain for desirable production traits, especially those that are difficult to measure, costly, and only expressed late in life. DNA markers in linkage disequilibrium (LD) with causal genetic variants are typically used in this strategy. Most livestock production traits are complex, involving contributions from a large number of additive genetic variants each of small effect size, interactions between genes, and strong gene by environment influences. To account for the additive contributions from many genes, and perhaps also epistatic effects, there has been recent focus on the implementation of genomic selection (or whole genome selection), which is “a form of marker-assisted selection in which genetic markers covering the whole genome are used so that all quantitative trait loci (QTL) are in LD with at least one marker” (Goddard and Hayes, 2007). A DNA marker-assisted selection strategy, and whole genome selection, will likely be more accurate and more robust if it incorporates contributions from causal genetic variations rather than relying entirely on markers in LD with these variants. Moreover, the discovery of causal variations can often suggest new opportunities for biochemical and immunological modulation of muscling traits. The latter strategy has potential for rapid implementation in all livestock and its impact is likely to be highly predictive and complement the use of genetic markers. These attributes are offset by its potential cost and the necessity for reintroduction of the application in each generation.
The rate of discovery of causal genetic variants (quantitative trait nucleotides or QTN) has been slow in most mammals. This is due to (i) the inability to efficiently dissect the broad genomic regions identified as containing QTN, (ii) the typically small effect sizes of most QTN, (iii) the difficulty providing conclusive proof, which often involves mechanistic investigation, and (iv) the absence, or only recent availability, of genome sequences for many domestic animal species (Ron and Weller, 2007; Braunschweig, 2010). The bovine genome sequence has been available since 2009 and an ovine genome sequence will likely be available in 2012 (Elsik et al., 2009; International Sheep Genomics Consortium et al., 2010). Genome sequences coupled with high density SNP chips provide opportunity to markedly accelerate discovery of causal genetic polymorphisms contributing to complex traits, like muscling. Despite the above limitations, a few causal genetic variants of relatively large effect sizes impacting on muscling traits have been discovered. Moreover, novel genetic, developmental, and biochemical mechanisms associated with these genetic variants have also been revealed.
The purpose of this review is to summarize this information, identify gaps in knowledge, and speculate on biochemical and immunological strategies for enhancement of muscling. Causal genetic polymorphisms are defined as those directly implicated in contributions to trait variation. In some instances independently confirmed genetic markers associated with skeletal muscle traits are also described, although the underlying causal polymorphisms are unknown. The impacts on eating quality traits associated with enhanced muscling in sheep have been recently summarized and are not discussed in any detail herein (Warner et al., 2010; Hopkins et al., 2011; Lambe et al., 2011).
Skeletal Muscle form and Function
To understand the impacts of genetic variants affecting skeletal muscle traits in livestock, it is important to consider the normal functions and development of skeletal muscle. Skeletal muscle is superbly adapted to its functional roles. Muscles undergoing slow but continuous contractions in the post-natal environment, such as various postural muscles, are characterized by predominance of slow twitch fibers (type 1 fibers), which do not tire easily and are reliant on oxidative metabolism. They have abundant mitochondria and appear reddish in color. In contrast, muscles requiring rapid contraction generating substantial force, such as some locomotory muscles, have a greater proportion of fast twitch fibers (type IIb and IIx fibers). These muscles largely depend on glycolytic metabolism for energy generation and are whiter and have fewer mitochondria (MacIntosch et al., 2006). The number of fibers and fiber type composition are set during early to mid fetal development. During late fetal development and the rapid post-natal growth period there is extensive muscle fiber hypertrophy driven by fusion of mononuclear muscle satellite cells with multinucleated myofibers (MacIntosch et al., 2006). Satellite cells are located between the sarcolemma and basal membrane of muscle fibers, and while normally quiescent in adult muscle, upon muscle injury, they proliferate and fuse with myofibers thereby aiding repair. Muscle fibers are composed of a number of highly organized proteins, principally polymeric actin, myosin, and their ancillary proteins (Tellam, 1985; MacIntosch et al., 2006).
The transition from the ruminant fetal environment to that of the newborn is associated with major physiological changes. Skeletal muscle must rapidly adapt to meet the new demands of locomotion and postural support against gravity, whilst using different sources of energy than those available in the uterine environment. These adaptations are of great importance to newborn ruminants as they stand, walk, and run within a few hours of birth. Ovine skeletal muscle undergoes a major transition late in its development, which ensures newborn skeletal muscle is largely characterized by adult skeletal muscle fiber types (Byrne et al., 2010b). Thus, skeletal muscle is subject to a robust developmental program upon which is built responsiveness to environment cues. Mutations affecting muscling (i.e., muscle thickness relative to skeletal dimensions) typically alter the trajectory of this developmental program and change muscle fiber number, composition, and hypertrophy. Multiple ovine mutations have been discovered that enhance muscling and act through novel mechanisms impacting on this developmental program (Table 1). These mutations provide excellent models for studying muscle function and formation.
Table 1.
Characteristics of genes affecting muscling in sheep.
| Gene or QTL | Genomic location | Characteristics of QTN | Breed | Phenotypic effect (reference) |
|---|---|---|---|---|
| Myostatin | g + 6723G-A | SNP in the 3′ untranslated region of MSTN which generates an illegitimate miRNA binding site | Texel | Enhanced muscling and less fat (Clop et al., 2006; Johnson et al., 2009; Boman et al., 2010; Masri et al., 2011a,b) |
| Myostatin | g + 6723G-A | SNP in the 3′ untranslated region of MSTN which generates an illegitimate miRNA binding site | Charollais | Increased muscle depth (Hadjipavlou et al., 2008) |
| Myostatin | c.960delG | Frame-shift mutation | Norwegian White | Enhanced muscling and less fat (Boman et al., 2010) |
| Myostatin | Intron 1 | – | Baluchi | Body weight (Ansary et al., 2011) |
| Callipyge | Telomeric region of OAR18 | Intergenic SNP within an imprinted locus on the telomeric arm of OAR18 (phenotype only expressed post-natally by the paternal heterozygote) | American Dorset | Increased size of caudal muscles, leanness, shift toward type IIx and IIb fibers, increased FCE, increased toughness (Koohmaraie et al., 1995; Cockett et al., 1996, 2005; Jackson et al., 1997a,b,c; Freking et al., 1998a, 1999; Kerth et al., 1999; Siltberg and Liberles, 2002; Charlier, 2004; Vuocolo et al., 2007; White et al., 2007) |
| Carwell | 2–6 cM telomeric of CSSM18 on OAR18 | Not imprinted | Australian Poll Dorset | Increased depth of longissimus dorsi; no effect on fatness; possible change in muscle shape; shift toward IIb and IIx fibers (Nicoll et al., 1998; Greenwood et al., 2006) |
| LoinMax (rib eye muscling) | 2–6 cM telomeric of CSSM18 on OAR18 | – | Poll Dorset | Enhanced muscling in the loin (Masri et al., 2010) |
| TM-QTL | 2 cM telomeric of CSSM18 on OAR18 | Evidence for paternal expression of the muscling phenotype | British Texel | Enhanced muscling in the loin (Walling et al., 2004; Macfarlane et al., 2009, 2012; Rius-Vilarrasa et al., 2009; Matika et al., 2010) |
| Xinjiang | Telomeric region of OAR18 | – | Xinjiang | Enhanced muscling (Liu et al., 2006) |
| QTL OAR1 | 2 QTL located on OAR1 | Likely to be separate genes as QTL separated by 50 cM | Suffock and Charollais | Enhanced muscle depth or live weight (Walling et al., 2004; McRae et al., 2005; Matika et al., 2010) |
The Heritability of Muscling Traits in Production Sheep
Sheep muscling traits typically have moderate heritabilities. Various Merino and Border Leicester crosses have been examined for a variety of carcass and muscling traits (Mortimer et al., 2010). The ranges of heritabilities for muscle weight, meat yield, and carcass muscle dimensions were 0.22–0.35, 0.24–0.35, and 0.25–0.34, respectively. For United Kingdom Charollais, heritabilities for muscle depth and muscle depth corrected for live weight were 0.25 and 0.31, respectively (McRae et al., 2005), while heritabilities for muscle depth in Texel, Suffock, and Charollais sheep ranged between 0.38 and 0.54 (Matika et al., 2010). These values are similar to an average figure of 0.4, calculated for the muscling trait longissimus muscle area, in a meta-analysis of 72 studies in cattle (Utrera and Van Vleck, 2004). A number of QTL affecting muscling traits in various sheep populations have been discovered. Some of the underlying QTN have been identified, especially genetic variants with relatively large effect sizes, e.g., the myostatin (MSTN) and Callipyge mutations. Other QTL, usually of low to moderate effect sizes, have been confirmed in independent sheep populations but only localized to broad chromosomal regions containing many genes.
Myostatin
Myostatin is a potent negative regulator of muscle mass in mammals. It is a member of the TGF-β superfamily of cytokines, several of which have key roles in maintaining skeletal muscle homeostasis through regulation of growth, differentiation, and regeneration of muscle (McPherron and Lee, 1997). Natural mutations in bovine, human, ovine, caprine, and canine MSTN that either inactivate the encoded protein or suppress its quantity cause enhanced muscling (Grobet et al., 1997; McPherron et al., 1997; Clop et al., 2006; Mosher et al., 2007; Stinckens et al., 2011; Zhang et al., 2012). Genetic manipulations that inactivate MSTN in transgenic mice and fish cause a similar phenotype (McPherron and Lee, 1993; McPherron et al., 1997; Sawatari et al., 2010). MSTN gene polymorphisms are also associated with racing speed for specific lines of horses and dogs (Mosher et al., 2007; Hill et al., 2010). For the cow, there are multiple mutations in MSTN that enhance muscling (McPherron and Lee, 1997). Most major skeletal muscles are affected by these mutations, which generally increase myofibril number (hyperplasia) and to a lesser extent myofiber cross-sectional area (hypertrophy), and increase the proportion of fast twitch glycolytic fibers. There are also decreased levels of connective and intramuscular adipose tissues (Bellinge et al., 2005). The impacts of the mutations are typically larger in homozygous animals compared with heterozygous animals. In sheep, several MSTN polymorphisms associated with muscling phenotypes have been reported (Table 1). The variety of different types of mutations that inactivate MSTN, particularly in cattle, are noteworthy and correspondingly there is a range of muscling phenotypes (Joulia-Ekaza and Cabello, 2006). Thus, there is a wealth of biological information accompanying MSTN mutations that is common to many mammalian species.
Myostatin protein processing and signaling
Myostatin is synthesized as a secreted 52 kDa precursor protein, which is proteolytically processed into the mature 12 kDa MSTN polypeptide and the 40 kDa N-terminal inhibitory propeptide (Latency Associated Peptide, LAP; Figure 1). Both the mature MSTN polypeptide (a dimer) and the latent inactive complex consisting of the MSTN polypeptide bound to LAP, circulate in blood (McPherron et al., 1997; Thies et al., 2001). Over-expression of LAP in mouse results in muscle mass enhancement. This is due to the ability of LAP to bind MSTN and inhibit its signaling activity (Li et al., 2010). Several circulating MSTN binding proteins [follistatin, decorin, small glutamine rich tetratricopeptide repeat (SGT), titan cap, Grb2-associating protein (GASP), follistatin related gene protein (FLRG)] have been identified in mouse. Over-expression of follistatin or FLRG increases skeletal muscle mass (Carnac et al., 2007). Presumably, these proteins sequester the mature MSTN polypeptide and thereby suppress its signaling capacity. Proof of this mechanism is still required for many of these proteins.
Figure 1.
Diagrammatic representations of myostatin (MSTN) gene and protein structures. The gene consists of three exons. The position of the c. *1232 G > A mutation (previously referred to as g + 6723 G-A) is shown by an asterisk. Unshaded regions in exons and mRNA represent the 5′ and 3′ untranslated regions. The lightly shaded region in the prepro-myostatin protein sequence corresponds to the signal sequence. Also shown is the site for processing of the promyostatin protein (thick arrow). The mature myostatin polypeptide forms a dimer held together by a disulfide bond.
Myostatin binds to activin type II receptors (ACVR2A and ACVR2B) and activates the SMAD signaling transduction pathway resulting in altered transcription of a number of target genes. MSTN also activates various cyclin-dependent kinase pathways. This combined intracellular signaling leads to inhibition of the G1 to S transition in the cell cycle of myoblasts, inhibition of myoblast differentiation, and inhibition of satellite cell activation and renewal (Langley et al., 2002; McCroskery et al., 2003). Thus, the primary role of MSTN is to negatively regulate myogenesis.
Down-regulation or inhibition of MSTN activity during fetal development is thought to relieve MSTN’s suppression of muscle satellite cell activation and renewal, thereby promoting myofiber formation (Joulia-Ekaza and Cabello, 2006). This results in hyperplasia and to a lesser extent hypertrophy, with the latter caused by increased fusion of satellite cells with myotubes. The myofibers typically also have greater representation of type IIb fibers (Bellinge et al., 2005; Hennebry et al., 2009). It is noteworthy that the expression of murine MSTN is normally sixfold greater in predominantly fast versus slow twitch skeletal muscles suggesting that fast twitch myofibers may be more susceptible to inactivating MSTN mutations (Allen and Loh, 2011). Moreover, murine MSTN is a positive regulator of MEF2C, a key promyogenic transcription factor that normally promotes formation of slow oxidative fibers. MSTN is also a negative regulator of MYOD, another key promyogenic transcription factor with preferential expression in fast twitch glycolytic fibers. Thus, the absence of MSTN results in decreased MEF2C mediated slow twitch fiber formation and increased MYOD mediated fast twitch fiber formation (Hennebry et al., 2009).
Recent selection for myostatin mutations
The MSTN gene, which encompasses three exons (Figure 1), and the signaling pathways through which it operates are conserved from zebrafish to humans, indicating that its functions are of ancient evolutionary origin (McPherron et al., 1997). Although mammals harboring MSTN mutations are usually viable, dystocia has been reported in some breeds of cattle (Keele and Fahrenkrug, 2001). The similar muscling phenotypes generated in widely different species carrying MSTN inactivating mutations also attest to the conservation of this muscle regulatory system. MSTN’s primary biological role may be to limit muscle size in a manner that is compatible with the size of the skeletal framework of each species.
The prevalence of mutations in the MSTN genes of cattle, sheep, goats, and to a lesser extent in racing dogs and horses, may reflect recent intensive artificial selection for muscling traits in these populations. Phylogenetic analysis measuring the ratios of non-synonymous to synonymous substitutions (Ka/Ks) indicates that the MSTN gene is subject to recent positive selection in the bovinae and caprinae–there is also some evidence for additional positive selection that predates domestication (Siltberg and Liberles, 2002; Tellgren et al., 2004; Bellinge et al., 2005). Moreover, the time to the most recent ancestor has been estimated at less than 400 years for a number of specific MSTN mutations, thereby inferring recent positive selection (O’Rourke et al., 2012). Recent, strong selection for MSTN has also been observed in three separate populations of Texel sheep (Kijas et al., 2012).
Myostatin mutations in sheep
Texel sheep are characterized by enhanced muscling compared with other sheep breeds. In an elegant exemplification of modern genetic technologies, Clop et al. (2006) identified a QTL of major effect on muscling located in a 10 cM confidence interval on OAR2 using a Romanov × Texel F2 cross. The QTL accounted for 5–25% of the phenotypic variance. Fine mapping reduced the interval, which encompassed MSTN. Sequencing of the 10.5 kbp region of MSTN for Texel and control animals identified 20 SNP, most of which were monomorphic in Texel animals and consistent with a selective sweep across the locus. None of the SNP were located in the MSTN coding sequence. Analysis of offspring from an F2 ram that inherited an intact Texel chromosome and a recombined chromosome excluded 18 SNP. One of the two remaining SNP (c. *1232 G > A, but previously denoted g + 6723 G-A) was virtually Texel specific in the A allele and located in the 3′ UTR of MSTN mRNA (Figure 1). The variant created an illegitimate motif potentially recognized by three miRNA (miR-1, miR-206, and miR-122a; Figure 2). Two of the miRNA, miR-1, and miR-206, were known to be strongly expressed in skeletal muscle. Consistent with a miRNA mediated mechanism, the total level of circulating MSTN was about one third of that in wild type sheep, although the mRNA level in skeletal muscle was unaffected. Further experiments using reporter constructs transfected into COS1 cells unambiguously proved that the A allele created an illegitimate miRNA binding site in the 3′ UTR of MSTN mRNA allowing miRNA mediated translational repression. The effect of the mutation on muscling traits has been independently confirmed in a number of sheep breeds and crosses: Charollais, New Zealand Texel, Norwegian White, Texel × Poll Dorset cross, and Texel × Welsh Mountain cross (Hadjipavlou et al., 2008; Johnson et al., 2009; Boman et al., 2010; Masri et al., 2011a,b).
Figure 2.
Alignments of the ovine miR-1 and miR-206 sequences with the region in the 3′ UTR of wild type ovine MSTN where the c. *1232 G > A mutation (underlined) occurs. Bolded nucleotides show complementarity of miR-1 and miR-206 with the MSTN 3′ UTR (center sequence). The eight nucleotide seed regions in the 5′ end of the miRNA are boxed. The G to A mutation produces an illegitimate recognition site for these miRNA and miRNA mediated translational repression of MSTN mRNA.
Additional mutations in the ovine MSTN gene affecting muscling have been described but in less detail. One frame-shift mutation (c.960delG) in Norwegian White Sheep resulted in increased muscling and less fat and these effects were stronger than in animals carrying the c. *1232 G > A mutation (Boman et al., 2009, 2010). There is also evidence for additional but as yet uncharacterized mutations possibly located in the promoter region, intron 1, intron 2, or 3′-UTR of MSTN in various sheep populations (Kijas et al., 2007; Hickford et al., 2010; Ansary et al., 2011).
Myostatin mutations have pleiotropic effects in cattle and this may also be expected in other mammalian species, including sheep. While increased muscularity and leanness are the most obvious phenotypes in ruminants, the mutation in cattle can also cause decreased sizes of several organs, fineness of limb bones, a higher incidence of underdeveloped genitalia, enlarged tongues, reduced fertility, low calf viability, increased stress susceptibility, and dystocia (Bellinge et al., 2005). These broad phenotypic effects suggest perturbation of normal system-wide development, which is most likely initiated in the embryo or early fetus, and accentuated during the period of rapid late fetal and post-natal growth. MSTN mRNA, while strongly expressed in skeletal muscle, is also expressed at lower levels in a number of adult tissues including adipose and mammary tissues. Therefore, inactivating MSTN mutations may directly affect other tissues, besides skeletal muscle (McPherron et al., 1997). In this regard, the change in body conformation and carcass “compactness” of sheep carrying the c. *1232 G > A MSTN mutation may be consistent with this concept (Boman et al., 2010; Masri et al., 2011b). Sheep carrying MSTN mutations are in widespread use in the sheep meat industry.
Biochemical and immunological strategies to enhance muscling by suppression of myostatin activity
There are a number of strategies that might be employed to enhance muscling by exploiting the genetics and biological functions of MSTN. First, because MSTN is a negative regulator of myogenesis, biochemical, and immunological intervention strategies designed to directly suppress MSTN activity may enhance muscling. Indeed, immunization of mice and pigs against MSTN induced neutralizing humoral immune responses that enhanced muscling (Tang et al., 2007; Long et al., 2009; Zhang et al., 2011). A key to success of this approach may be the generation of sustained, neutralizing IgG responses. Other comparable approaches involve the use of antibodies blocking binding of MSTN to the activin type II receptors, dominant negative forms of the receptors, or dominant negative forms of the mature MSTN polypeptide (Siriett et al., 2007). In addition, exogenous LAP, follistatin, or FLGR could be used to sequester MSTN and enhance muscling, as demonstrated in mouse models (Carnac et al., 2007). In a variation on this strategy, histone deacetylase inhibitors could be used, as these have been shown to increase follistatin expression and enhance muscle repair in mice (Minetti et al., 2006). The limiting features of these approaches are likely to be consumer acceptability, duration of efficacy, cost, and frequency and timing of administration. These interventions may also need targeting to critical windows of fetal skeletal muscle development. Second, transgenic modifications using targeted mutagenesis of MSTN, RNAi directed at MSTN mRNA or over-expression of proteins that sequester MSTN can be envisaged (Tessanne et al., 2006). The remarkable quadrupling of skeletal muscle mass in a murine transgenic mouse engineered to have a MSTN knockout and over-expression of follistatin highlights the enormous plasticity of skeletal muscle and its potential for enhancement in livestock (Lee, 2007). Thus, direct interventions in MSTN signaling and its regulatory pathways are likely to have strong potential for enhancing muscling in livestock.
Callipyge
The Callipyge mutation in sheep has provided remarkable new insights into genetics, regulation of gene expression and biology. The mutation causes skeletal muscle hypertrophy, but only in paternal heterozygous animals (NmatCpat; N, is the wild type allele and C is the allele carrying the mutation); a characteristic termed polar over-dominance (Cockett et al., 1996). The maternal heterozygote (CmatNpat) and homozygote (CmatCpat) animals show no muscling phenotypes. Specific skeletal muscles in the NmatCpat genotype are increased in size by as much as 35%, although increased meat toughness has prevented commercial exploitation of these animals (Koohmaraie et al., 1995; Jackson et al., 1997a,b,c). In affected muscles, there is a marked shift toward type IIb and IIx fibers with a corresponding decrease in type I fibers, decreases in the proportional sizes of several non-muscle organs, increased leanness, and improved feed conversion efficiency (Jackson et al., 1997a,b,c; Charlier, 2004; Vuocolo et al., 2007; White et al., 2007). Live weight is unaffected and carcasses are more compact with a shorter length and greater width at the shoulder and rump (Freking et al., 1998a).
Unlike MSTN mutations, not all major muscles are affected by the Callipyge mutation. The muscling phenotype is first expressed 1–3 months post-birth and this occurs along a rostro-caudal gradient in the NmatCpat animal, with greatest impact on skeletal muscles innervated through lumbar and sacral roots, e.g., longissimus dorsi and semimembranosus muscles (Koohmaraie et al., 1995; Freking et al., 1998a,b, 1999; Kerth et al., 2003; Cockett et al., 2005). As a consequence of the post-natal development of the phenotype, there is no increased risk of dystocia in NmatCpat lambs. The muscling phenotype is maintained throughout adult life.
Changes in expression of imprinted genes surrounding the Callipyge mutation
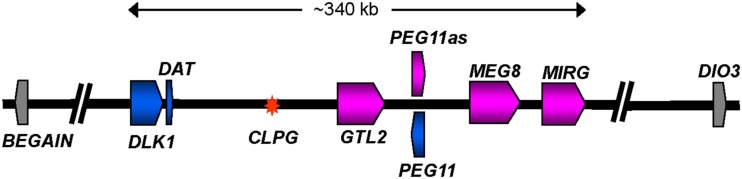
The causative point mutation (A/G) responsible for the Callipyge muscling phenotype has been located in a 12 bp conserved motif positioned near the telomeric end of OAR 18 between the protein encoding gene DLK1 (delta-like 1) and the non-protein encoding gene GTL2 (gene trap locus 2; or MEG3; Figure 3; Freking et al., 2002; Smit et al., 2003). The ram in which the mutation first arose was a germ line mosaic for the mutation (Smit et al., 2003). The broader ∼1 Mbp region defines a cluster of conserved imprinted genes (Figure 3; Schmidt et al., 2000; Charlier et al., 2001a,b; Seitz et al., 2003; Takeda et al., 2006; Byrne et al., 2010a). The expression patterns of the core group of these imprinted genes are strikingly perturbed in affected skeletal muscles from NmatCpat lambs, but are normal in unaffected muscles or the other genotypes (Bidwell et al., 2001, 2004; Charlier et al., 2001a; Murphy et al., 2005; Vuocolo et al., 2005, 2007; Fleming-Waddell et al., 2007, 2009; White et al., 2007; Caiment et al., 2010). Thus, altered gene expression at this imprinted locus is tightly linked with emergence of the phenotype. The affected genes span a core region of ∼350 kbp and include DLK1, GTL2, PEG11 (paternally expressed gene 11; also known as retrotransposon like 1 (RTL1)), PEG11AS (antisense to PEG11; also known as RTL1AS), MEG8 (maternally expressed gene 8) and MIRG (micro RNA-containing gene). While the expression of these genes is altered, their imprinted status remains unchanged. DLK1 and PEG11 are paternally expressed and both encode proteins, while the remainder are maternally expressed non-coding genes. The latter are all expressed from the same strand. GTL2 expresses a large number of splice variants of a long non-coding RNA. MIRG encodes at least 50 miRNA, while MEG8 encodes a cluster of CD snoRNA. As the muscling phenotype is only expressed in NmatCpat animals, it was reasoned that the markedly enhanced expression of one (or both) of the paternally expressed protein encoding genes, DLK1 or PEG11, was the primary driver of the muscling phenotype (Charlier, 2004). The mutation also enhances expression of the maternally expressed genes in NmatCpat animals (Vuocolo et al., 2005). However, these effects are relatively small compared with their enhanced expression in the CmatNpat and CmatCpat genotypes, both of which do not express a muscling phenotype. Hence, these genes are unlikely to be directly promoting the phenotype.
Figure 3.
Diagrammatic representation of the organization of imprinted genes located at the telomeric end of ovine chromosome 18. The diagram shows a representation of the ∼1 Mbp region from BEGAIN to DIO3. The core imprinted genes affected by the Callipyge mutation are colored while imprinted genes unaffected by the mutation are shown in gray. Affected paternally and maternally expressed genes are shaded blue and pink, respectively. The direction of transcription for each gene is indicated by an arrow. Introns are not shown. The asterisk denotes the position of the Callipyge mutation (CLPG). The precise lengths of the maternally expressed genes, which all produce non-coding RNA, are unclear. The diagram is based on that deduced by (Georges et al., 2003) supplemented with annotation for a miRNA cluster (MIRG) deduced by comparative sequence analyses with orthologous murine and human sequence regions. A larger population of miRNA scattered throughout the core region has been omitted for clarity (Caiment et al., 2010).
It has been postulated that normal post-natal down-regulation of imprinted genes acts to coordinate the post-natal growth deceleration in many tissues (Lui et al., 2008). The Callipyge mutation in NmatCpat animals prevents the normal post-natal down-regulation of DLK1 and PEG11 in skeletal muscle and recapitulates their fetal-like gene expression programs (Murphy et al., 2005; White et al., 2007). The dosage of these imprinted genes may be having major effects on the developmental program, and indeed for murine Dlk1 there is evidence for an optimum dosage that fixes the upper limit of its growth promotion (da Rocha et al., 2009). There was no effect of the Callipyge mutation on the expression ratio of the two major DLK1 splice variants in sheep skeletal muscle, even though there is a strong developmental switch in their usage (Vuocolo et al., 2005, 2007). The switch occurs during early to mid fetal development and long before the expression of the Callipyge phenotype a few weeks after birth. The splice variation produces a shorter plasma membrane-bound protein (predominant in the post-natal state) and a longer membrane-bound form (predominant in the early fetus). The latter is subject to proteolytic processing releasing a circulating form of DLK1 (Vuocolo et al., 2003).
The primary effector gene driving muscle hypertrophy in Callipyge sheep
There is significant evidence indicating that DLK1 is the effector of the muscle hypertrophy phenotype in Callipyge sheep. First, the increased expression of DLK1 as a function of genotype, development, muscle type, and muscle fiber type is strongly associated with the expression of the hypertrophy phenotype (Charlier et al., 2001a; Perkins et al., 2006; Fleming-Waddell et al., 2007; Vuocolo et al., 2007; White et al., 2007). In particular, this type 1 membrane-bound glycoprotein has increased expression in hypertrophied type IIb muscle fibers of the NmatCpat genotype (Charlier et al., 2001a; Charlier, 2004; White et al., 2007). Second, transgenic mice over-expressing Dlk1 using a myosin light chain 3F promoter were characterized by mild skeletal muscle hypertrophy (Davis et al., 2004). Third, DLK1 has been implicated in the control of proliferation and differentiation of many cell types (Wang and Sul, 2009) and is thought to be an atypical repressive ligand for the NOTCH signaling pathway. This is a key pathway negatively regulating cell commitment and differentiation (Vuocolo et al., 2007). Fourth, NmatCpat myoblasts grown in cell culture and induced to differentiate do not become hypertrophic, but they also do not express DLK1 (Lavulo et al., 2008). Fifth, consistent with the identification of DLK1 expressing mononucleated PAX7+ cells (muscle satellite cells) in fetal NmatCpat skeletal muscle, over-expression of Dlk1 in murine skeletal muscle satellite cells inhibited their proliferation and enhanced differentiation into multinucleated myotubes (White et al., 2007; Waddell et al., 2010). Sixth, genetic ablation of Dlk1 in the murine myogenic lineage caused reduced body weight, reduced skeletal muscle mass, and impaired muscle regeneration, and in cultured satellite cells ablation promoted satellite cell self-renewal (Waddell et al., 2010). Over-expression of DLK1 on the surface of the multinucleated myotubes in some post-natal NmatCpat skeletal muscles may be driving the characteristic hypertrophy of these muscle fibers. One model involves over-expression of DLK1 by nascent or regenerating myofibers which promotes the differentiation of neighboring satellite cells thereby leading to muscle hypertrophy (Waddell et al., 2010).
Despite the strong evidence for a direct role of DLK1 in initiating hypertrophy there is inconsistent information. First, a murine Dlk1 knockout resulted in obesity but no reported effect on muscling (Moon et al., 2002). Second, constitutive expression of Dlk1 in the murine myogenic cell line, C2C12, which does not express endogenous Dlk1, did not promote differentiation into multinucleated myotubes (Smas and Sul, 1993). Third, constitutive over-expression of Dlk1 using its endogenous regulatory elements in transgenic mice resulted in growth enhancement but the mice failed to survive early life and showed no reported signs of muscle hypertrophy (da Rocha et al., 2009). These contradictory results may reflect the use of different experimental species and models and, in some cases, the lack of primary focus on skeletal muscle phenotypes. Species specific differences seem an unlikely explanation since the imprinted gene locus is highly conserved in placental mammals.
The paternally expressed gene PEG11 is an alternative candidate for the effector. PEG11 is a conserved Ty3-Gypsy retrotransposon like gene with a single long open reading frame encoding a 1,333 amino acid protein with retroviral gag-pol type structure. The gene is not associated with long terminal repeats and many of its encoded retroviral-like protein domains contain mutations that collectively destroy retrotransposon activity (Byrne et al., 2010a). The conservation of PEG11 in placental mammals, and its absence from syntenic chromosomal regions of non-placental mammals, suggests that during evolution the newly retrotransposed gene was co-opted for novel functional roles within placental mammals (Lynch and Tristem, 2003). The full length PEG11 protein has been identified in nuclear fractions from Callipyge semimembranosus skeletal muscle (Byrne et al., 2010a). The biological role of PEG11 remains unclear although it has been implicated in angiogenesis during murine placentation (Sekita et al., 2008).
Like DLK1, the increased expression of PEG11 as a function of genotype, development, and muscle type is also strongly correlated with the hypertrophy phenotype (Charlier et al., 2001a; Bidwell et al., 2004; Perkins et al., 2006; Vuocolo et al., 2007; White et al., 2007). PEG11 is up-regulated about 45-fold in affected skeletal muscles from NmatCpat animals at 3 months of age. In comparison, DLK1 is up-regulated about 6- to 10-fold (Charlier et al., 2001a; Bidwell et al., 2004; Vuocolo et al., 2005, 2007; Byrne et al., 2010a). As discussed below, the maternally expressed gene PEG11AS contains a number of miRNA that cause RNA-induced silencing complex (RISC)-mediated cleavage of the paternally expressed PEG11 transcript (Davis et al., 2005). Thus, if PEG11 was the effector driving the muscling phenotype, there is an obvious mechanism that could explain the lack of phenotype in homozygote animals. The potential role of PEG11 in inducing the muscling phenotype could be directly tested by its over-expression in transgenic mouse skeletal muscle or myogenic cells. This could be achieved by direct over-expression of PEG11 or by suppression of PEG11AS, and hence suppression of miRNA targeting PEG11. It is also possible that over-expression of both PEG11 and DLK1 are required to induce muscle hypertrophy. This hypothesis could also be tested in similar ways in transgenic mice.
It is difficult to envisage how the putative effectors, PEG11 or DLK1, could be used in direct biochemical and immunological interventions to enhance muscling. Both putative effectors positively enhance muscling and hence any intervention must induce increased levels of these proteins and they must be specifically targeted to skeletal muscle to avoid potential effects in other tissues. While exogenous soluble DLK1 may be introduced into an animal, it is unclear whether it would affect muscling as DLK1 may need cell surface presentation to responsive muscle cells. The intracellular location of PEG11 precludes a similar approach. Various genetic manipulations within this imprinted locus in mice have dramatic effects on gene expression at the locus and typically result in embryonic or fetal lethality (da Rocha et al., 2009). Consequently, it is likely that any exogenous effector will need to be targeted to skeletal muscle and expressed in a specific developmental window.
A model for the inheritance of the Callipyge phenotype
There are unusual genetic mechanisms associated with the inheritance of the Callipyge phenotype, as the hypertrophy phenotype is only expressed by NmatCpat animals (Cockett et al., 1994; Carpenter et al., 1996; Jackson et al., 1997a; Freking et al., 1998a,b; Bidwell et al., 2004). An elegant genetic model has been developed to explain this observation (Charlier et al., 2001a; Georges et al., 2003). The model proposes that the balance between the activities of a paternally expressed effector and a maternally expressed trans-acting repressor dictates the phenotypic outcome. The latter may exert its effects by translational suppression of the effector. In the NmatCpat genotype the paternally expressed effector gene is influenced by the mutation acting in cis, thereby causing its over-expression relative to the maternally expressed repressor. The excess influence of the effector over repressor in this genotype induces muscle hypertrophy. Conversely, for CmatNpat animals, where the animals are phenotypically normal, the mutation acts in cis to increase expression of the maternally expressed repressor, but it has no influence as the effector is not expressed from the wild type paternal allele. In the homozygote the paternally expressed effector and maternally expressed repressor are both up-regulated, but the influence of the effector does not dominate the repressor, and therefore no change in phenotype is generated.
The nature of the trans-acting maternally expressed repressor in this model is unclear although it is noted that only non-coding RNA are maternally expressed from the core of this imprinted locus (Charlier et al., 2001a). These non-coding genes are strongly up-regulated in the CmatNpat and CmatCpat genotypes, both of which do not express muscling phenotypes. In this regard, the PEG11AS transcript encodes six miRNA, which cause RISC-mediated cleavage of PEG11 (Davis et al., 2005). Hence, these miRNA are ideally placed to act as maternally expressed trans-acting repressors of paternally expressed PEG11. In this model PEG11 is the putative effector of the phenotype.
An alternative, but analogous mechanism involves some of the approximately 110 likely maternally expressed miRNA that map to the core of the imprinted domain, and in particular a subset of approximately 50 miRNA encoded by the maternally expressed non-coding gene, MIRG (Glazov et al., 2008; Caiment et al., 2010). Some MIRG miRNA are predicted to target DLK1 and many of these miRNA are up-regulated in cis with the Callipyge mutation (Hagan et al., 2009; Caiment et al., 2010). Experimental definitions of the targeting specificities for these maternally expressed miRNA are required to substantiate this hypothesis. It will also be important to define whether these miRNA act by RISC-mediated cleavage or translational repression of DLK1. A counterpoint to this mechanism is that many of the miRNA in this imprinted region show substantially increased expression in the CmatNpat and CmatCpat genotypes, which display no muscling phenotype. This result implies the effector is not expressed and there are no other mRNA targets for these miRNA in skeletal muscle cells from animals with these genotypes, despite the number of miRNA expressed from this region and their predicted broad mRNA target specificities. One possibility is that the primary role of these miRNA may involve maintenance of imprinting status at this locus rather than regulating mRNA. Intriguingly, the Callipyge mutation acting in trans, also causes a small increase in the expression of these maternally expressed miRNA, as it does for GTL2, suggesting that there is much more to learn about the complex regulatory events occurring within this imprinted gene locus (Caiment et al., 2010).
Mechanisms causing muscle hypertrophy in Callipyge sheep
Microarray analyses of affected skeletal muscles have been undertaken to gain insight into the mechanism underlying generation of the hypertrophy phenotype in Callipyge lambs (Fleming-Waddell et al., 2007; Vuocolo et al., 2007). In one model generated from these studies, increased expression of DLK1 on myofibrils suppresses the myogenic inhibitory effects of the NOTCH signaling pathway in adjacent satellite cells, possibly in conjunction with suppression of HDAC9 (histone deacetylase 9), a known negative regulator of myogenesis (Vuocolo et al., 2007). This promotes satellite cell activation and their fusion with adjacent multinucleated myotubes thereby generating larger hypertrophied myotubes. The discovery that DLK1 is expressed on the cell surface of a subset of mononuclear satellite cells and on the surface of multinuclear myofibers in NmatCpat skeletal muscle supports this possibility (White et al., 2007; Waddell et al., 2010). Another investigation suggested that activation of the AKT/mTOR signaling pathway was the primary driver of enhanced protein synthesis in muscles affected by the mutation (Fleming-Waddell et al., 2009). Both AKT/mTOR and HDAC9 are known to be important regulators of myogenesis and muscle hypertrophy in mouse models (summarized in Fleming-Waddell et al., 2007, 2009; Vuocolo et al., 2007).
Interplay between the Callipyge mutation and epigenetic marks
How does the Callipyge mutation act over considerable genomic distances to cause changes in gene expression within the core of the imprinted domain? A clue to the mechanism whereby the Callipyge mutation, acting in cis, increases expression of a number of adjacent imprinted genes can be found in changes to DNA methylation surrounding the site of the mutation. In post-natal NmatCpat samples from affected, but not unaffected, muscles, the wild type allele is strongly methylated but the Callipyge allele is hypomethylated (Murphy et al., 2006). The mutation is also associated with development of novel DNase-I hypersensitivity sites, indicating increased relaxation of the chromatin state (Takeda et al., 2006). Consistent with this permissive epigenetic state, the mutation, acting in cis, causes bidirectional long range DLK1-GTL2 intergenic transcription of non-coding RNA in the lamb (Takeda et al., 2006). The conserved 12 bp motif, in which the mutation is located, is normally likely to be a long range regulatory element, possibly an insulator or enhancer, that exerts developmental and tissue specific controls on the expression of multiple genes in the core of the imprinted region. The mutation is presumed to perturb these roles by changing binding affinity for key transcriptional co-regulators thereby preventing the normal program of post-natal down-regulation of imprinted genes in this region. The mutation also exerts influence in specific muscles toward the rear of the animal possibly through the actions of a rostro-caudal positionally graded regulator, whose pattern of expression is set early in embryonic development.
The polar over-dominant inheritance pattern characteristic of the Callipyge muscling phenotype is more widespread in mammals than widely appreciated. In pigs, the same locus is associated with growth, fatness, and body composition in a polar over-dominant manner (Kim et al., 2004; Oczkowicz, 2009). In humans, a SNP, rs1802710 situated in exon 3 of DLK1, is associated with polar over-dominant expression of child and adolescent obesity (Wermter et al., 2008). Another SNP, rs941576, located within GTL2 is associated with paternally inherited risk of type 1 diabetes (Wallace et al., 2010). Notably, Dlk1-null mice show growth retardation, accelerated obesity, and hyperlipidaemia (Moon et al., 2002). A genome scan for imprinted body weight and growth QTL in mice has identified additional unusual inheritance patterns at a variety of loci elsewhere in the genome (Wolf et al., 2008). These patterns include bipolar over-dominance (the two heterozygotes differ from each other but the two homozygotes have similar phenotypes), and polar under-dominance (one of the two heterozygotes exhibits a lower intensity of phenotype compared to the other three genotypes). Widespread allelic imbalance in gene expression and DNA methylation are likely to be the norm and not the exception (Kong et al., 2009; Meaburn et al., 2010). Some of this imbalance will be due to allelic contributions and trans-acting epistatic effects, while other contributions are likely to reflect epigenetic based parent-of-origin effects and generalized monoallelic gene silencing. Unless expressly tested, these non-Mendelian effects are likely to have been missed in current genome-wide trait association analyses (Matika et al., 2010). Re-analysis of existing datasets using a broader range of genetic models is warranted, as this may reveal new QTL with non-Mendelian behavior and simultaneously enhance sensitivity to detect standard QTL.
Confirmed Quantitative Trait Loci for Muscling in Sheep
In addition to highlighting the contributions of MSTN polymorphic variants to muscling phenotypes, several genome scans in different populations of sheep have identified QTL for muscling on the telomeric end of OAR 18. These are different from the effects of the Callipyge mutation, but located at the same general position (Cockett et al., 2005).
Carwell, LoinMAX™ and other muscling QTL located on the telomeric arm of OAR18
The Carwell (or rib eye muscling, REM) locus identified in Australian Poll Dorset rams is associated with increased muscle depth of longissimus dorsi (also named longissimus lumborum) with no effect on fatness or live weight. There is also a change in muscle shape and a shift from type IIa towards type IIb and IIx muscle fibers (Nicoll et al., 1998; Greenwood et al., 2006). The QTL is positioned 2–6 cM telomeric to the CSSM18 marker on OAR18 and overlaps with the site of the Callipyge mutation. The effect of each allele was additive and dominant, and there was no indication that the QTL was imprinted; however, this possibility was not explicitly tested. The effects of this QTL are relatively small and more variable compared with Callipyge. Recently, a QTL (LoinMAX™) for the same muscling trait was located within this genomic interval in a cross between Poll Dorset heterozygous ram carriers and British cross-bred ewes (Masri et al., 2010). In this instance, there was a small extent of enhanced muscling and no effects on carcass lean weight, fat content or body conformation scores.
In Texel and Texel–Suffolk families another confirmed muscling QTL (TM-QTL) was localized to 2 cM telomeric of the same marker (CSSM18) on OAR18 (Walling et al., 2004; Macfarlane et al., 2009, 2012; Rius-Vilarrasa et al., 2009; Matika et al., 2010). The muscling phenotype in these animals was similar to that in the Poll Dorset sheep and associated with altered muscle shape–the carcass was described as more “compact.” It was recently concluded there is evidence for imprinting of this QTL. It showed a polar over-dominance model of inheritance suggesting similarity with the Callipyge mutation, albeit the muscling effect size of the QTL was much smaller (Matika et al., 2010). In a fourth example, a polymorphism in this same region was associated with muscling traits in Xinjiang sheep (Liu et al., 2006). It is likely that discernment of finer details of the muscling phenotypes and inheritance patterns in these four sheep populations is limited by the relatively small muscling effect sizes of these QTL.
The causal genetic polymorphisms linked with these QTL are unknown although they are unlikely to be directly related to the Callipyge mutation for the following reasons. (i) Direct examination of DNA sequence at the site of the Callipyge mutation does not reveal any sequence variation in the Texel or Xinjiang animals (Liu et al., 2006; Matika et al., 2010). (ii) The absolute effects on specific muscles were much weaker for these QTL (∼2.5–7% increase in muscling compared with up to 35% in some muscles from Callipyge sheep), and effects on fat depth and muscle tenderness were also quantitatively different. The pronounced rostro-caudal muscling gradient in Callipyge animals was also not apparent in these other populations. (iii) Callipyge has a complex inheritance pattern that was not readily apparent for all of these QTL, with the exception of the Texel QTL (Matika et al., 2010). Carwell shows a simple autosomal dominant pattern of inheritance (Jopson et al., 2001), although there has been an unconfirmed suggestion of sire-dependent effects in the progeny of LoinMAX™ animals (Nicoll et al., 1998). The pathway to commercial exploitation of these QTL will critically depend on whether or not they are imprinted, as this will have a strong influence on breeding flock design. A homozygote terminal sire would likely be used. All progeny from this sire would inherit one active copy of the imprinted gene because of the paternal transmission mechanism. QTL for muscle depth and live weight have been also located on OAR1 in Suffock and Charollais populations, respectively (Walling et al., 2004; McRae et al., 2005; Matika et al., 2010). The QTL lie 50 cM apart and are therefore unlikely to be caused by polymorphisms in the same genes. The corresponding QTN have not been defined.
Integration of Gene Expression with Genetic Architecture Associated with Enhanced Muscling
Muscle accretion is the result of contributions from a number of developmental and biochemical pathways. Multiple genetic variants in these pathways, acting in concert, are likely to underpin enhanced muscling traits in most production populations, particularly in the absence of genetic variants of relatively large effect sizes, e.g., the Callipyge and MSTN mutations. The impacts of these multiple genetic variants, each of small effect size, can be measured through their combined influence on these pathways. Using Poll Dorset sheep, the pathways associated with the genetics of enhanced muscling were identified by linking gene expression patterns in progeny skeletal muscle with sire based EBVs for the muscling trait, longissimus dorsi depth (Kogelman et al., 2011). In this study there was strong sire based genetic structure associated with the gene expression program in progeny skeletal muscle. Higher sire EBV was also probably associated with a shift toward fast twitch glycolytic fibers in progeny. It was concluded that sires with high muscling EBVs were characterized by progeny muscle accretion mediated by activation of pathways that include enhanced myogenesis, muscle fiber hypertrophy, and decreased protein catabolism. Low EBV status resulted from increased genetic contribution to protein degradation pathways. As may be expected, the key genetic effects dictating EBV status impacted on the balance between muscle fiber protein accretion and turnover. This type of analysis, particularly if coupled with SNP association data, has potential to accelerate the identification of additional genetic variants contributing to muscling by the discovery of pathways directly contributing to muscling traits. The analysis may also be particularly suited to dissection of complex traits. One strength of this approach is that the application of RNA-seq could allow identification of genes demonstrating allelic expression imbalance. This is a signature for a cis-acting SNP regulating gene expression (excluding imprinted genes). These SNP could be directly contributing to the trait.
Conclusion
A number of genetic variants with confirmed effects on muscling in sheep have been discovered. There are several common features associated with a high muscling phenotype in sheep irrespective of the primary genetic driver. (i) Enhanced muscling is often associated with morphology change of the animal. This suggests a common change in developmental programming that scales body shape to accommodate enhanced skeletal muscle structure. (ii) Changed developmental trajectories for skeletal muscle beginning in early life are also common and these may be linked with morphology changes. (iii) Large increases in muscularity are typically associated with a shift toward fast twitch glycolytic fibers, leanness, and poorer eating quality attributes. The reasons for these changes are unclear, although both myoblasts and preadipocytes have a common progenitor cell, suggesting a developmental link. (iv) The mutations associated with MSTN and Callipyge are typically recent and for MSTN there has been strong and positive selection in production flocks. (v) The co-occurrence in Texel sheep of a recent mutation in MSTN contributing to muscling, as well as a confirmed QTL for muscling on the telomeric end of OAR18, indicates that artificial breeding programs have enriched for genetic variants with independent and additive effects. This suggests the involvement of fundamental and independent regulatory mechanisms that presumably normally function to limit muscle growth.
The immediate future of the sheep meat industry will, see the increasing exploitation of natural genetic variation contributing to muscling. The incorporation of causal genetic variations into genomic selection strategies will enhance their accuracy and robustness, while allowing targeted selection to achieve more rapid genetic improvement. In the medium term, the discovery of developmental and biochemical pathways contributing to enhanced muscling will open new opportunities for the use of novel and acceptable biochemical and immunological interventions that may play a significant and complementary role to genetic selection in the sheep meat industry.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Allen D. L., Loh A. S. (2011). Posttranscriptional mechanisms involving microRNA-27a and b contribute to fast-specific and glucocorticoid-mediated myostatin expression in skeletal muscle. Am. J. Physiol. Cell Physiol. 300, C124–C137 10.1152/ajpcell.00142.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansary M., Tahmoorespur M., Nassiry M., Taheri A., Valeh M. (2011). Polymorphism in intron-1 of myostatin gene and its association with estimated breeding values of growth traits in Baluchi sheep. Indian J. Anim. Sci. 81, 849–852 [DOI] [PubMed] [Google Scholar]
- Bellinge R. H., Liberles D. A., Iaschi S. P., O’Brien P. A., Tay G. K. (2005). Myostatin and its implications on animal breeding: a review. Anim. Genet. 36, 1–6 10.1111/j.1365-2052.2004.01229.x [DOI] [PubMed] [Google Scholar]
- Bidwell C. A., Kramer L. N., Perkins A. C., Hadfield T. S., Moody D. E., Cockett N. E. (2004). Expression of PEG11 and PEG11AS transcripts in normal and callipyge sheep. BMC Biol. 2, 17. 10.1186/1741-7007-2-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidwell C. A., Shay T. L., Georges M., Beever J. E., Berghmans S., Cockett N. E. (2001). Differential expression of the GTL2 gene within the callipyge region of ovine chromosome 18. Anim. Genet. 32, 248–256 10.1046/j.1365-2052.2001.00776.x [DOI] [PubMed] [Google Scholar]
- Boman I. A., Klemetsdal G., Blichfeldt T., Nafstad O., Vage D. I. (2009). A frameshift mutation in the coding region of the myostatin gene (MSTN) affects carcass conformation and fatness in Norwegian White Sheep (Ovis aries). Anim. Genet. 40, 418–422 10.1111/j.1365-2052.2009.01855.x [DOI] [PubMed] [Google Scholar]
- Boman I. A., Klemetsdal G., Nafstad O., Blichfeldt T., Vage D. I. (2010). Impact of two myostatin (MSTN) mutations on weight gain and lamb carcass classification in Norwegian White Sheep (Ovis aries). Genet. Sel. Evol. 42, 4. 10.1186/1297-9686-42-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig M. H. (2010). Mutations in the bovine ABCG2 and the ovine MSTN gene added to the few quantitative trait nucleotides identified in farm animals: a mini-review. J. Appl. Genet. 51, 289–297 10.1007/BF03208858 [DOI] [PubMed] [Google Scholar]
- Byrne K., Colgrave M. L., Vuocolo T., Pearson R., Bidwell C. A., Cockett N. E., Lynn D. J., Fleming-Waddell J. N., Tellam R. L. (2010a). The imprinted retrotransposon-like gene PEG11 (RTL1) is expressed as a full-length protein in skeletal muscle from Callipyge sheep. PLoS ONE 5, e8638. 10.1371/journal.pone.0008638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne K., Vuocolo T., Gondro C., White J. D., Cockett N. E., Hadfield T., Bidwell C. A., Waddell J. N., Tellam R. L. (2010b). A gene network switch enhances the oxidative capacity of ovine skeletal muscle during late fetal development. BMC Genomics 11, 378. 10.1186/1471-2164-11-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiment F., Charlier C., Hadfield T., Cockett N., Georges M., Baurain D. (2010). Assessing the effect of the CLPG mutation on the microRNA catalog of skeletal muscle using high-throughput sequencing. Genome Res. 20, 1651–1662 10.1101/gr.108787.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnac G., Vernus B., Bonnieu A. (2007). Myostatin in the pathophysiology of skeletal muscle. Curr. Genomics 8, 415–422 10.2174/138920207783591672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter C. E., Rice O. D., Cockett N. E., Snowder G. D. (1996). Histology and composition of muscles from normal and callipyge lambs. J. Anim. Sci. 74, 388–393 [DOI] [PubMed] [Google Scholar]
- Charlier C. (2004). Towards the molecular understanding of the polar overdominance phenomenon associated with the callipyge phenotype in sheep. Bull. Mem. Acad. R. Med. Belg. 159, 490–496 [PubMed] [Google Scholar]
- Charlier C., Segers K., Karim L., Shay T., Gyapay G., Cockett N., Georges M. (2001a). The callipyge mutation enhances the expression of coregulated imprinted genes in cis without affecting their imprinting status. Nat. Genet. 27, 367–369 10.1038/86856 [DOI] [PubMed] [Google Scholar]
- Charlier C., Segers K., Wagenaar D., Karim L., Berghmans S., Jaillon O., Shay T., Weissenbach J., Cockett N., Gyapay G., Georges M. (2001b). Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 11, 850–862 10.1101/gr.172701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clop A., Marcq F., Takeda H., Pirottin D., Tordoir X., Bibe B., Bouix J., Caiment F., Elsen J. M., Eychenne F., Larzul C., Laville E., Meish F., Milenkovic D., Tobin J., Charlier C., Georges M. (2006). A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 38, 813–818 10.1038/ng1810 [DOI] [PubMed] [Google Scholar]
- Cockett N. E., Jackson S. P., Shay T. L., Farnir F., Berghmans S., Snowder G. D., Nielsen D. M., Georges M. (1996). Polar overdominance at the ovine callipyge locus. Science 273, 236–238 10.1126/science.273.5272.236 [DOI] [PubMed] [Google Scholar]
- Cockett N. E., Jackson S. P., Shay T. L., Nielsen D., Moore S. S., Steele M. R., Barendse W., Green R. D., Georges M. (1994). Chromosomal localization of the callipyge gene in sheep (Ovis aries) using bovine DNA markers. Proc. Natl. Acad. Sci. U.S.A. 91, 3019–3023 10.1073/pnas.91.8.3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockett N. E., Smit M. A., Bidwell C. A., Segers K., Hadfield T. L., Snowder G. D., Georges M., Charlier C. (2005). The callipyge mutation and other genes that affect muscle hypertrophy in sheep. Genet. Sel. Evol. 37(Suppl. 1), S65–S81 10.1186/1297-9686-37-S1-S65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha S. T., Charalambous M., Lin S. P., Gutteridge I., Ito Y., Gray D., Dean W., Ferguson-Smith A. C. (2009). Gene dosage effects of the imprinted delta-like homologue 1 (dlk1/pref1) in development: implications for the evolution of imprinting. PLoS Genet. 5, e1000392. 10.1371/journal.pgen.1000392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E., Caiment F., Tordoir X., Cavaille J., Ferguson-Smith A., Cockett N., Georges M., Charlier C. (2005). RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 15, 743–749 10.1016/j.cub.2005.02.060 [DOI] [PubMed] [Google Scholar]
- Davis E., Jensen C. H., Schroder H. D., Farnir F., Shay-Hadfield T., Kliem A., Cockett N., Georges M., Charlier C. (2004). Ectopic expression of DLK1 protein in skeletal muscle of padumnal heterozygotes causes the callipyge phenotype. Curr. Biol. 14, 1858–1862 10.1016/j.cub.2004.09.079 [DOI] [PubMed] [Google Scholar]
- Elsik C. G., Tellam R. L., Worley K. C., Gibbs R. A., Muzny D. M., Weinstock G. M., Adelson D. L., Eichler E. E., Elnitski L., Guigo R., Hamernik D. L., Kappes S. M., Lewin H. A., Lynn D. J., Nicholas F. W., Reymond A., Rijnkels M., Skow L. C., Zdobnov E. M., Schook L., Womack J., Alioto T., Antonarakis S. E., Astashyn A., Chapple C. E., Chen H. C., Chrast J., Camara F., Ermolaeva O., Henrichsen C. N., Hlavina W., Kapustin Y., Kiryutin B., Kitts P., Kokocinski F., Landrum M., Maglott D., Pruitt K., Sapojnikov V., Searle S. M., Solovyev V., Souvorov A., Ucla C., Wyss C., Anzola J. M., Gerlach D., Elhaik E., Graur D., Reese J. T., Edgar R. C., McEwan J. C., Payne G. M., Raison J. M., Junier T., Kriventseva E. V., Eyras E., Plass M., Donthu R., Larkin D. M., Reecy J., Yang M. Q., Chen L., Cheng Z., Chitko-McKown C. G., Liu G. E., Matukumalli L. K., Song J., Zhu B., Bradley D. G., Brinkman F. S., Lau L. P., Whiteside M. D., Walker A., Wheeler T. T., Casey T., German J. B., Lemay D. G., Maqbool N. J., Molenaar A. J., Seo S., Stothard P., Baldwin C. L., Baxter R., Brinkmeyer-Langford C. L., Brown W. C., Childers C. P., Connelley T., Ellis S. A., Fritz K., Glass E. J., Herzig C. T., Iivanainen A., Lahmers K. K., Bennett A. K., Dickens C. M., Gilbert J. G., Hagen D. E., Salih H., Aerts J., Caetano A. R., Dalrymple B., Garcia J. F., Gill C. A., Hiendleder S. G., Memili E., Spurlock D., Williams J. L., Alexander L., Brownstein M. J., Guan L., Holt R. A., Jones S. J., Marra M. A., Moore R., Moore S. S., Roberts A., Taniguchi M., Waterman R. C., Chacko J., Chandrabose M. M., Cree A., Dao M. D., Dinh H. H., Gabisi R. A., Hines S., Hume J., Jhangiani S. N., Joshi V., Kovar C. L., Lewis L. R., Liu Y. S., Lopez J., Morgan M. B., Nguyen N. B., Okwuonu G. O., Ruiz S. J., Santibanez J., Wright R. A., Buhay C., Ding Y., Dugan-Rocha S., Herdandez J., Holder M., Sabo A., Egan A., Goodell J., Wilczek-Boney K., Fowler G. R., Hitchens M. E., Lozado R. J., Moen C., Steffen D., Warren J. T., Zhang J., Chiu R., Schein J. E., Durbin K. J., Havlak P., Jiang H., Liu Y., Qin X., Ren Y., Shen Y., Song H., Bell S. N., Davis C., Johnson A. J., Lee S., Nazareth L. V., Patel B. M., Pu L. L., Vattathil S., Williams R. L., Jr., Curry S., Hamilton C., Sodergren E., Wheeler D. A., Barris W., Bennett G. L., Eggen A., Green R. D., Harhay G. P., Hobbs M., Jann O., Keele J. W., Kent M. P., Lien S., McKay S. D., McWilliam S., Ratnakumar A., Schnabel R. D., Smith T., Snelling W. M., Sonstegard T. S., Stone R. T., Sugimoto Y., Takasuga A., Taylor J. F., Van Tassell C. P., Macneil M. D., Abatepaulo A. R., Abbey C. A., Ahola V., Almeida I. G., Amadio A. F., Anatriello E., Bahadue S. M., Biase F. H., Boldt C. R., Carroll J. A., Carvalho W. A., Cervelatti E. P., Chacko E., Chapin J. E., Cheng Y., Choi J., Colley A. J., de Campos T. A., De Donato M., Santos I. K., de Oliveira C. J., Deobald H., Devinoy E., Donohue K. E., Dovc P., Eberlein A., Fitzsimmons C. J., Franzin A. M., Garcia G. R., Genini S., Gladney C. J., Grant J. R., Greaser M. L., Green J. A., Hadsell D. L., Hakimov H. A., Halgren R., Harrow J. L., Hart E. A., Hastings N., Hernandez M., Hu Z. L., Ingham A., Iso-Touru T., Jamis C., Jensen K., Kapetis D., Kerr T., Khalil S. S., Khatib H., Kolbehdari D., Kumar C. G., Kumar D., Leach R., Lee J. C., Li C., Logan K. M., Malinverni R., Marques E., Martin W. F., Martins N. F., Maruyama S. R., Mazza R., McLean K. L., Medrano J. F., Moreno B. T., More D. D., Muntean C. T., Nandakumar H. P., Nogueira M. F., Olsaker I., Pant S. D., Panzitta F., Pastor R. C., Poli M. A., Poslusny N., Rachagani S., Ranganathan S., Razpet A., Riggs P. K., Rincon G., Rodriguez-Osorio N., Rodriguez-Zas S. L., Romero N. E., Rosenwald A., Sando L., Schmutz S. M., Shen L., Sherman L., Southey B. R., Lutzow Y. S., Sweedler J. V., Tammen I., Telugu B. P., Urbanski J. M., Utsunomiya Y. T., Verschoor C. P., Waardenberg A. J., Wang Z., Ward R., Weikard R., Welsh T. H., Jr., White S. N., Wilming L. G., Wunderlich K. R., Yang J., Zhao F. Q. (2009). The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science 324, 522–528 10.1126/science.1169588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming-Waddell J. N., Olbricht G. R., Taxis T. M., White J. D., Vuocolo T., Craig B. A., Tellam R. L., Neary M. K., Cockett N. E., Bidwell C. A. (2009). Effect of DLK1 and RTL1 but not MEG3 or MEG8 on muscle gene expression in Callipyge lambs. PLoS ONE 4, e7399. 10.1371/journal.pone.0007399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming-Waddell J. N., Wilson L. M., Olbricht G. R., Vuocolo T., Byrne K., Craig B. A., Tellam R. L., Cockett N. E., Bidwell C. A. (2007). Analysis of gene expression during the onset of muscle hypertrophy in callipyge lambs. Anim. Genet. 38, 28–36 10.1111/j.1365-2052.2006.01562.x [DOI] [PubMed] [Google Scholar]
- Fogarty N. M. (2009). “Meat sheep breeding–where we are at and future challenges,” in Proceedings of the Association for the Advancement of Animal Breeding and Genetics, Barossa Valley, 414 [Google Scholar]
- Freking B. A., Keele J. W., Nielsen M. K., Leymaster K. A. (1998a). Evaluation of the ovine callipyge locus: II. Genotypic effects on growth, slaughter, and carcass traits. J. Anim. Sci. 76, 2549–2559 [DOI] [PubMed] [Google Scholar]
- Freking B. A., Keele J. W., Beattie C. W., Kappes S. M., Smith T. P., Sonstegard T. S., Nielsen M. K., Leymaster K. A. (1998b). Evaluation of the ovine callipyge locus: I. Relative chromosomal position and gene action. J. Anim. Sci. 76, 2062–2071 [DOI] [PubMed] [Google Scholar]
- Freking B. A., Keele J. W., Shackelford S. D., Wheeler T. L., Koohmaraie M., Nielsen M. K., Leymaster K. A. (1999). Evaluation of the ovine callipyge locus: III. Genotypic effects on meat quality traits. J. Anim. Sci. 77, 2336–2344 [DOI] [PubMed] [Google Scholar]
- Freking B. A., Murphy S. K., Wylie A. A., Rhodes S. J., Keele J. W., Leymaster K. A., Jirtle R. L., Smith T. P. (2002). Identification of the single base change causing the callipyge muscle hypertrophy phenotype, the only known example of polar overdominance in mammals. Genome Res. 12, 1496–1506 10.1101/gr.571002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner G. E., Williams A., Siddell J., Ball A. J., Mortimer S., Jacob R. H., Pearce K. L., Edwards J. E. H., Rowe J. B., Pethick D. W. (2010). Using Australian sheep breeding values to increase lean meat yield percentage. Anim. Prod. Sci. 50, 1098–1106 10.1071/AN10144 [DOI] [Google Scholar]
- Georges M., Charlier C., Cockett N. (2003). The callipyge locus: evidence for the trans interaction of reciprocally imprinted genes. Trends Genet. 19, 248–252 10.1016/S0168-9525(03)00082-9 [DOI] [PubMed] [Google Scholar]
- Glazov E. A., McWilliam S., Barris W. C., Dalrymple B. P. (2008). Origin, evolution, and biological role of miRNA cluster in DLK-DIO3 genomic region in placental mammals. Mol. Biol. Evol. 25, 939–948 10.1093/molbev/msn045 [DOI] [PubMed] [Google Scholar]
- Goddard M. E., Hayes B. J. (2007). Genomic selection. J. Anim. Breed. Genet. 124, 323–330 10.1111/j.1439-0388.2007.00702.x [DOI] [PubMed] [Google Scholar]
- Greenwood P. L., Davis J. J., Gaunt F. M., Ferrier G. R. (2006). Influences on the loin and cellular characteristics of the m. longissimus lumborum of Australian Poll Dorset-sired lambs. Aust. J. Agric. Res. 57, 1–12 10.1071/AR05205 [DOI] [Google Scholar]
- Grobet L., Martin L. J., Poncelet D., Pirottin D., Brouwers B., Riquet J., Schoeberlein A., Dunner S., Menissier F., Massabanda J., Fries R., Hanset R., Georges M. (1997). A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 17, 71–74 10.1038/ng0997-71 [DOI] [PubMed] [Google Scholar]
- Hadjipavlou G., Matika O., Clop A., Bishop S. C. (2008). Two single nucleotide polymorphisms in the myostatin (GDF8) gene have significant association with muscle depth of commercial Charollais sheep. Anim. Genet. 39, 346–353 10.1111/j.1365-2052.2008.01734.x [DOI] [PubMed] [Google Scholar]
- Hagan J. P., O’Neill B. L., Stewart C. L., Kozlov S. V., Croce C. M. (2009). At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS ONE 4, e4352. 10.1371/journal.pone.0004352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennebry A., Berry C., Siriett V., O’Callaghan P., Chau L., Watson T., Sharma M., Kambadur R. (2009). Myostatin regulates fiber-type composition of skeletal muscle by regulating MEF2 and MyoD gene expression. Am. J. Physiol. Cell Physiol. 296, C525–C534 10.1152/ajpcell.00259.2007 [DOI] [PubMed] [Google Scholar]
- Hickford J. G., Forrest R. H., Zhou H., Fang Q., Han J., Frampton C. M., Horrell A. L. (2010). Polymorphisms in the ovine myostatin gene (MSTN) and their association with growth and carcass traits in New Zealand Romney sheep. Anim. Genet. 41, 64–72 10.1111/j.1365-2052.2010.02050.x [DOI] [PubMed] [Google Scholar]
- Hill E. W., Gu J., Eivers S. S., Fonseca R. G., McGivney B. A., Govindarajan P., Orr N., Katz L. M., MacHugh D. E. (2010). A sequence polymorphism in MSTN predicts sprinting ability and racing stamina in thoroughbred horses. PLoS ONE 5, e8645. 10.1371/journal.pone.0008645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins D. L., Fogarty N. M., Mortimer S. I. (2011). Genetic related effects on sheep meat quality. Small Rumin. Res. 101, 160–172 10.1016/j.smallrumres.2011.09.036 [DOI] [Google Scholar]
- International Sheep Genomics Consortium, Archibald A. L., Cockett N. E., Dalrymple B. P., Faraut T., Kijas J. W., Maddox J. F., McEwan J. C., Hutton Oddy V., Raadsma H. W., Wade C., Wang J., Wang W., Xun X. (2010). The sheep genome reference sequence: a work in progress. Anim. Genet. 41, 449–453 10.1111/j.1365-2052.2010.02100.x [DOI] [PubMed] [Google Scholar]
- Jackson S. P., Green R. D., Miller M. F. (1997a). Phenotypic characterization of rambouillet sheep expressing the callipyge gene: I. Inheritance of the condition and production characteristics. J. Anim. Sci. 75, 14–18 [DOI] [PubMed] [Google Scholar]
- Jackson S. P., Miller M. F., Green R. D. (1997b). Phenotypic characterization of Rambouillet sheep expressing the callipyge gene: II. Carcass characteristics and retail yield. J. Anim. Sci. 75, 125–132 [DOI] [PubMed] [Google Scholar]
- Jackson S. P., Miller M. F., Green R. D. (1997c). Phenotypic characterization of rambouillet sheep expression the callipyge gene: III. Muscle weights and muscle weight distribution. J. Anim. Sci. 75, 133–138 [DOI] [PubMed] [Google Scholar]
- Johnson P. L., Dodds K. G., Bain W. E., Greer G. J., McLean N. J., McLaren R. J., Galloway S. M., van Stijn T. C., McEwan J. C. (2009). Investigations into the GDF8 g+6723G-A polymorphism in New Zealand Texel sheep. J. Anim. Sci. 87, 1856–1864 10.2527/jas.2008-1519 [DOI] [PubMed] [Google Scholar]
- Jopson N. B., Nicoll G. B., Stevenson-Barry J. M., Duncan S., Greer G. J., Bain W. E., Gerard E. M., Glass B. C., Broad T. E., McEwan J. C. (2001). “Mode of inheritance and effects on meat quality of the rib-eye muscling (REM) QTL in sheep,” in The Association for the Advancement of Animal Breeding and Genetics (Queenstown: ), 111–114 [Google Scholar]
- Joulia-Ekaza D., Cabello G. (2006). Myostatin regulation of muscle development: molecular basis, natural mutations, physiopathological aspects. Exp. Cell Res. 312, 2401–2414 10.1016/j.yexcr.2006.07.010 [DOI] [PubMed] [Google Scholar]
- Keele J. W., Fahrenkrug S. C. (2001). Optimum mating systems for the myostatin locus in cattle. J. Anim. Sci. 79, 2016–2022 [DOI] [PubMed] [Google Scholar]
- Kerth C. R., Cain T. L., Jackson S. P., Ramsey C. B., Miller M. F. (1999). Electrical stimulation effects on tenderness of five muscles from Hampshire x Rambouillet crossbred lambs with the callipyge phenotype. J. Animal Sci. 77, 2951–2955 [DOI] [PubMed] [Google Scholar]
- Kerth C. R., Jackson S. P., Ramsey C. B., Miller M. F. (2003). Characterization and consumer acceptance of three muscles from Hampshire x Rambouillet cross sheep expressing the callipyge phenotype. J. Anim. Sci. 81, 2213–2218 [DOI] [PubMed] [Google Scholar]
- Kijas J. W., Lenstra J. A., Hayes B., Boitard S., Porto Neto L. R., San Cristobal M., Servin B., McCulloch R., Whan V., Gietzen K., Paiva S., Barendse W., Ciani E., Raadsma H., McEwan J., Dalrymple B. (2012). Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 10, e1001258. 10.1371/journal.pbio.1001258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijas J. W., McCulloch R., Edwards J. E., Oddy V. H., Lee S. H., van der Werf J. (2007). Evidence for multiple alleles effecting muscling and fatness at the ovine GDF8 locus. BMC Genet. 8, 80. 10.1186/1471-2156-8-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. S., Kim J. J., Dekkers J. C., Rothschild M. F. (2004). Polar overdominant inheritance of a DLK1 polymorphism is associated with growth and fatness in pigs. Mamm. Genome 15, 552–559 10.1007/s00335-004-2383-3 [DOI] [PubMed] [Google Scholar]
- Kogelman L. J., Byrne K., Vuocolo T., Watson-Haigh N. S., Kadarmideen H. N., Kijas J. W., Oddy H. V., Gardner G. E., Gondro C., Tellam R. L. (2011). Genetic architecture of gene expression in ovine skeletal muscle. BMC Genomics 12, 607. 10.1186/1471-2164-12-607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Steinthorsdottir V., Masson G., Thorleifsson G., Sulem P., Besenbacher S., Jonasdottir A., Sigurdsson A., Kristinsson K. T., Frigge M. L., Gylfason A., Olason P. I., Gudjonsson S. A., Sverrisson S., Stacey S. N., Sigurgeirsson B., Benediktsdottir K. R., Sigurdsson H., Jonsson T., Benediktsson R., Olafsson J. H., Johannsson O. T., Hreidarsson A. B., Sigurdsson G., Ferguson-Smith A. C., Gudbjartsson D. F., Thorsteinsdottir U., Stefansson K. (2009). Parental origin of sequence variants associated with complex diseases. Nature 462, 868–874 10.1038/nature08625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koohmaraie M., Shackelford S. D., Wheeler T. L., Lonergan S. M., Doumit M. E. (1995). A muscle hypertrophy condition in lamb (callipyge): characterization of effects on muscle growth and meat quality traits. J. Anim. Sci. 73, 3596–3607 [DOI] [PubMed] [Google Scholar]
- Lambe N. R., Richardson R. I., Macfarlane J. M., Nevison I., Haresign W., Matika O., Bunger L. (2011). Genotypic effects of the Texel Muscling QTL (TM-QTL) on meat quality in purebred Texel lambs. Meat Sci. 89, 125–132 10.1016/j.meatsci.2011.03.014 [DOI] [PubMed] [Google Scholar]
- Langley B., Thomas M., Bishop A., Sharma M., Gilmour S., Kambadur R. (2002). Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. J. Biol. Chem. 277, 49831–49840 10.1074/jbc.M204291200 [DOI] [PubMed] [Google Scholar]
- Lavulo L. T., Uaesoontrachoon K., Mirams M., White J. D., Cockett N. E., Mackie E. J., Pagel C. N. (2008). Myoblasts isolated from hypertrophy-responsive callipyge muscles show altered growth rates and increased resistance to serum deprivation-induced apoptosis. Cells Tissues Organs (Print) 187, 141–151 10.1159/000110080 [DOI] [PubMed] [Google Scholar]
- Lee S. J. (2007). Quadrupling muscle mass in mice by targeting TGF-beta signaling pathways. PLoS ONE 2, e789. 10.1371/journal.pone.0000789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhao B., Kim Y. S., Hu C. Y., Yang J. (2010). Administration of a mutated myostatin propeptide to neonatal mice significantly enhances skeletal muscle growth. Mol. Reprod. Dev. 77, 76–82 10.1002/mrd.21239 [DOI] [PubMed] [Google Scholar]
- Liu G. Q., Dai R., Ren H. X., Wang X. H., Liu S. R., Sun Y. L., Yang L. G. (2006). Polymorphism analysis of genes associated with hindquarters muscular development on chromosome 18 in Xinjiang meat sheep. Yi Chuan 28, 815–820 [PubMed] [Google Scholar]
- Long D. B., Zhang K. Y., Chen D. W., Ding X. M., Yu B. (2009). Effects of active immunization against myostatin on carcass quality and expression of the myostatin gene in pigs. Anim. Sci. J. 80, 585–590 10.1111/j.1740-0929.2009.00666.x [DOI] [PubMed] [Google Scholar]
- Lui J. C., Finkielstain G. P., Barnes K. M., Baron J. (2008). An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R189–R196 10.1152/ajpregu.00182.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch C., Tristem M. (2003). A co-opted gypsy-type LTR-retrotransposon is conserved in the genomes of humans, sheep, mice, and rats. Curr. Biol. 13, 1518–1523 10.1016/S0960-9822(03)00618-3 [DOI] [PubMed] [Google Scholar]
- Macfarlane J. M., Lambe N. R., Bishop S. C., Matika O., Rius-Vilarrasa E., McLean K. A., Haresign W., Wolf B. T., McLaren R. J., Bunger L. (2009). Effects of the Texel muscling quantitative trait locus on carcass traits in crossbred lambs. Animal 3, 189–199 10.1017/S175173110800356X [DOI] [PubMed] [Google Scholar]
- Macfarlane J. M., Lambe N. R., Haresign W., Bunger L. (2012). The effect of the Texel muscling QTL on live and carcass weight in Texel lambs. Small Rumin. Res. 105, 117–121 10.1016/j.smallrumres.2012.02.011 [DOI] [Google Scholar]
- MacIntosch B. R., Gardiner P. F., McComas A. J. (2006). Skeletal Muscle Form and Function. Champaign: Human Kinetics [Google Scholar]
- Masri A. Y., Lambe N. R., Macfarlane J. M., Brotherstone S., Haresign W., Bunger L. (2011a). Evaluating the effects of a single copy of a mutation in the myostatin gene (c.*1232G>A) on carcass traits in crossbred lambs. Meat Sci. 87, 412–418 10.1016/j.meatsci.2010.11.019 [DOI] [PubMed] [Google Scholar]
- Masri A. Y., Lambe N. R., Macfarlane J. M., Brotherstone S., Haresign W., Bunger L. (2011b). Evaluating the effects of the c.*1232G>A mutation and TM-QTL in Texel x Welsh Mountain lambs using ultrasound and video image analyses. Small Rumin. Res. 99, 99–109 10.1016/j.smallrumres.2011.03.047 [DOI] [Google Scholar]
- Masri A. Y., Lambe N. R., Macfarlane J. M., Brotherstone S., Haresign W., Rius-Vilarrasa E., Bunger L. (2010). The effects of a loin muscling quantitative trait locus (LoinMAX (TM)) on carcass and VIA-based traits in crossbred lambs. Animal 4, 407–416 10.1017/S175173110999125X [DOI] [PubMed] [Google Scholar]
- Matika O., Sechi S., Pong-Wong R., Houston R. D., Clop A., Woolliams J. A., Bishop S. C. (2010). Characterization of OAR1 and OAR18 QTL associated with muscle depth in British commercial terminal sire sheep. Anim. Genet. 42, 172–180 10.1111/j.1365-2052.2010.02121.x [DOI] [PubMed] [Google Scholar]
- McCroskery S., Thomas M., Maxwell L., Sharma M., Kambadur R. (2003). Myostatin negatively regulates satellite cell activation and self-renewal. J. Cell Biol. 162, 1135–1147 10.1083/jcb.200207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron A. C., Lawler A. M., Lee S. J. (1997). Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387, 83–90 10.1038/387083a0 [DOI] [PubMed] [Google Scholar]
- McPherron A. C., Lee S. J. (1993). GDF-3 and GDF-9: two new members of the transforming growth factor-beta superfamily containing a novel pattern of cysteines. J. Biol. Chem. 268, 3444–3449 [PubMed] [Google Scholar]
- McPherron A. C., Lee S. J. (1997). Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. U.S.A. 94, 12457–12461 10.1073/pnas.94.23.12457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae A. F., Bishop S. C., Walling G. A., Wilson A. D., Visscher P. M. (2005). Mapping of multiple quantitative trait loci for growth and carcass traits in a complex commercial sheep pedigree. Anim. Sci. 80, 135–141 10.1079/ASC41040135 [DOI] [PubMed] [Google Scholar]
- Meaburn E. L., Schalkwyk L. C., Mill J. (2010). Allele-specific methylation in the human genome: implications for genetic studies of complex disease. Epigenetics 5, 578–582 10.4161/epi.5.7.12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minetti G. C., Colussi C., Adami R., Serra C., Mozzetta C., Parente V., Fortuni S., Straino S., Sampaolesi M., Di Padova M., Illi B., Gallinari P., Steinkuhler C., Capogrossi M. C., Sartorelli V., Bottinelli R., Gaetano C., Puri P. L. (2006). Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat. Med. 12, 1147–1150 10.1038/nm1479 [DOI] [PubMed] [Google Scholar]
- Moon Y. S., Smas C. M., Lee K., Villena J. A., Kim K. H., Yun E. J., Sul H. S. (2002). Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol. Cell. Biol. 22, 5585–5592 10.1128/MCB.22.15.5585-5592.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer S. I., van der Werf J. H. J., Jacob R. H., Pethick D. W., Pearce K. L., Warner R. D., Geesink G. H., Edwards J. E. H., Gardner G. E., Ponnampalam E. N., Kitessa S. M., Ball A. J., Hopkins D. L. (2010). Preliminary estimates of genetic parameters for carcass and meat quality traits in Australian sheep. Anim. Prod. Sci. 50, 1135–1144 10.1071/AN10126 [DOI] [Google Scholar]
- Mosher D. S., Quignon P., Bustamante C. D., Sutter N. B., Mellersh C. S., Parker H. G., Ostrander E. A. (2007). A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 3, e79. 10.1371/journal.pgen.0030079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S. K., Freking B. A., Smith T. P., Leymaster K., Nolan C. M., Wylie A. A., Evans H. K., Jirtle R. L. (2005). Abnormal postnatal maintenance of elevated DLK1 transcript levels in callipyge sheep. Mamm. Genome 16, 171–183 10.1007/s00335-004-2421-1 [DOI] [PubMed] [Google Scholar]
- Murphy S. K., Nolan C. M., Huang Z., Kucera K. S., Freking B. A., Smith T. P., Leymaster K. A., Weidman J. R., Jirtle R. L. (2006). Callipyge mutation affects gene expression in cis: a potential role for chromatin structure. Genome Res. 16, 340–346 10.1101/gr.4389306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll G. B., Burkin H. R., Broad T. E., Jopson N. B., Greer G. J., Bain W. E., Wright C. S., Dodds K. G., Fennessy P. F., McEwan J. C. (1998). “Genetic linkage of microsatellite markers to the Carwell locus for rib-eye muscling in sheep,” in Proceedings of the 6th World Congress on Genetics Applied to Livestock Production (Armidale, NSW: University of New England; ), 529–532 [Google Scholar]
- Oczkowicz M. (2009). Polar overdominance–a putative molecular mechanism and the new examples in mammals. J. Anim. Feed Sci. 18, 17–27 [Google Scholar]
- O’Rourke B. A., Greenwood P. L., Authur P. F., Goddard M. E. (2012). Inferring the recent ancestry of myostatin alleles affecting muscle mass in cattle. Anim. Genet. [Epub ahead of print]. 10.1111/j.1365-2052.2012.02354.x [DOI] [PubMed] [Google Scholar]
- Perkins A. C., Kramer L. N., Spurlock D. M., Hadfield T. S., Cockett N. E., Bidwell C. A. (2006). Postnatal changes in the expression of genes located in the callipyge region in sheep skeletal muscle. Anim. Genet. 37, 535–542 10.1111/j.1365-2052.2006.01518.x [DOI] [PubMed] [Google Scholar]
- Rius-Vilarrasa E., Roehe R., Macfarlane J. M., Lambe N. R., Matthews K. R., Haresign W., Matika O., Bunger L. (2009). Effects of a quantitative trait locus for increased muscularity on carcass traits measured by subjective conformation and fat class scores and video image analysis in crossbred lambs. Animal 3, 1532–1543 10.1017/S1751731109990413 [DOI] [PubMed] [Google Scholar]
- Ron M., Weller J. I. (2007). From QTL to QTN identification in livestock–winning by points rather than knock-out: a review. Anim. Genet. 38, 429–439 10.1111/j.1365-2052.2007.01640.x [DOI] [PubMed] [Google Scholar]
- Sawatari E., Seki R., Adachi T., Hashimoto H., Uji S., Wakamatsu Y., Nakata T., Kinoshita M. (2010). Overexpression of the dominant-negative form of myostatin results in doubling of muscle-fiber number in transgenic medaka (Oryzias latipes). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 155, 183–9 10.1016/j.cbpa.2009.10.030 [DOI] [PubMed] [Google Scholar]
- Schmidt J. V., Matteson P. G., Jones B. K., Guan X. J., Tilghman S. M. (2000). The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 14, 1997–2002 [PMC free article] [PubMed] [Google Scholar]
- Seitz H., Youngson N., Lin S. P., Dalbert S., Paulsen M., Bachellerie J. P., Ferguson-Smith A. C., Cavaille J. (2003). Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 34, 261–262 10.1038/ng1171 [DOI] [PubMed] [Google Scholar]
- Sekita Y., Wagatsuma H., Nakamura K., Ono R., Kagami M., Wakisaka N., Hino T., Suzuki-Migishima R., Kohda T., Ogura A., Ogata T., Yokoyama M., Kaneko-Ishino T., Ishino F. (2008). Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 40, 243–248 10.1038/ng.2007.51 [DOI] [PubMed] [Google Scholar]
- Siltberg J., Liberles D. A. (2002). A simple covarion-based approach to analyse nucleotide substitution rates. J. Evol. Biol. 15, 588–594 10.1046/j.1420-9101.2002.00416.x [DOI] [Google Scholar]
- Siriett V., Salerno M. S., Berry C., Nicholas G., Bower R., Kambadur R., Sharma M. (2007). Antagonism of myostatin enhances muscle regeneration during sarcpenia. Mol. Ther. 15, 1463–1470 10.1038/sj.mt.6300182 [DOI] [PubMed] [Google Scholar]
- Smas C. M., Sul H. S. (1993). Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 73, 725–734 10.1016/0092-8674(93)90252-L [DOI] [PubMed] [Google Scholar]
- Smit M., Segers K., Carrascosa L. G., Shay T., Baraldi F., Gyapay G., Snowder G., Georges M., Cockett N., Charlier C. (2003). Mosaicism of Solid Gold supports the causality of a noncoding A-to-G transition in the determinism of the callipyge phenotype. Genetics 163, 453–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinckens A., Georges M., Buys N. (2011). Mutations in the myostatin gene leading to hypermuscularity in mammals: indications for a similar mechanism in fish? Anim. Genet. 42, 229–234 10.1111/j.1365-2052.2010.02144.x [DOI] [PubMed] [Google Scholar]
- Takeda H., Caiment F., Smit M., Hiard S., Tordoir X., Cockett N., Georges M., Charlier C. (2006). The callipyge mutation enhances bidirectional long-range DLK1-GTL2 intergenic transcription in cis. Proc. Natl. Acad. Sci. U.S.A. 103, 8119–8124 10.1073/pnas.0602844103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Yan Z., Wan Y., Han W., Zhang Y. (2007). Myostatin DNA vaccine increases skeletal muscle mass and endurance in mice. Muscle Nerve 36, 342–348 10.1002/mus.20791 [DOI] [PubMed] [Google Scholar]
- Tellam R. (1985). Mechanism of CaCl2-induced actin polymerization. Biochemistry 24, 4455–4460 10.1021/bi00337a029 [DOI] [PubMed] [Google Scholar]
- Tellgren A., Berglund A. C., Savolainen P., Janis C. M., Liberles D. A. (2004). Myostatin rapid sequence evolution in ruminants predates domestication. Mol. Phylogenet. Evol. 33, 782–790 10.1016/j.ympev.2004.07.004 [DOI] [PubMed] [Google Scholar]
- Tessanne K., Golding M. C., Long C. R., Peoples M. D., Hannon G., Westhusin M. E. (2006). Production of transgenic calves expressing an shRNA targeting myostatin. Mol. Reprod. Dev. 79, 176–185 10.1002/mrd.22007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies R. S., Chen T., Davies M. V., Tomkinson K. N., Pearson A. A., Shakey Q. A., Wolfman N. M. (2001). GDF-8 propeptide binds to GDF-8 and antagonizes biological activity by inhibiting GDF-8 receptor binding. Growth Factors 18, 251–259 10.3109/08977190109029114 [DOI] [PubMed] [Google Scholar]
- Utrera A. R., Van Vleck L. D. (2004). Heritability estimates for carcass traits of cattle: a review. Genet. Mol. Res. 3, 380–394 [PubMed] [Google Scholar]
- Vuocolo T., Byrne K., White J., McWilliam S., Reverter A., Cockett N. E., Tellam R. L. (2007). Identification of a gene network contributing to hypertrophy in callipyge skeletal muscle. Physiol. Genomics 28, 253–272 [DOI] [PubMed] [Google Scholar]
- Vuocolo T., Cockett N. E., Tellam R. L. (2005). Expression of imprinted genes surrounding the callipyge mutation in ovine skeletal muscle. Aust. J. Exp. Agric. 45, 879–892 10.1071/EA05049 [DOI] [Google Scholar]
- Vuocolo T., Pearson R., Campbell P., Tellam R. L. (2003). Differential expression of Dlk-1 in bovine adipose tissue depots. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 134, 315–333 10.1016/S1096-4959(02)00265-8 [DOI] [PubMed] [Google Scholar]
- Waddell J. N., Zhang P., Wen Y., Gupta S. K., Yevtodiyenko A., Schmidt J. V., Bidwell C. A., Kumar A., Kuang S. (2010). Dlk1 is necessary for proper skeletal muscle development and regeneration. PLoS ONE 5, e15055. 10.1371/journal.pone.0015055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace C., Smyth D. J., Maisuria-Armer M., Walker N. M., Todd J. A., Clayton D. G. (2010). The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat. Genet. 42, 68–71 10.1038/ng.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling G. A., Visscher P. M., Wilson A. D., McTeir B. L., Simm G., Bishop S. C. (2004). Mapping of quantitative trait loci for growth and carcass traits in commercial sheep populations. J. Anim. Sci. 82, 2234–2245 [DOI] [PubMed] [Google Scholar]
- Wang Y., Sul H. S. (2009). Pref-1 regulates mesenchymal cell commitment and differentiation through Sox9. Cell Metab. 9, 287–302 10.1016/j.cmet.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner R. D., Greenwood P. L., Pethick D. W., Ferguson D. M. (2010). Genetic and environmental effects on meat quality. Meat Sci. 86, 171–183 10.1016/j.meatsci.2010.04.042 [DOI] [PubMed] [Google Scholar]
- Wermter A. K., Scherag A., Meyre D., Reichwald K., Durand E., Nguyen T. T., Koberwitz K., Lichtner P., Meitinger T., Schafer H., Hinney A., Froguel P., Hebebrand J., Bronner G. (2008). Preferential reciprocal transfer of paternal/maternal DLK1 alleles to obese children: first evidence of polar overdominance in humans. Eur. J. Hum. Genet. 16, 1126–1134 10.1038/ejhg.2008.64 [DOI] [PubMed] [Google Scholar]
- White J. D., Vuocolo T., McDonagh M., Grounds M. D., Harper G. S., Cockett N. E., Tellam R. (2007). Analysis of the callipyge phenotype through skeletal muscle development; association of Dlk1 with muscle precursor cells. Differentiation 76, 283–298 10.1111/j.1432-0436.2007.00208.x [DOI] [PubMed] [Google Scholar]
- Wolf J. B., Cheverud J. M., Roseman C., Hager R. (2008). Genome-wide analysis reveals a complex pattern of genomic imprinting in mice. PLoS Genet. 4, e1000091. 10.1371/journal.pgen.1000091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Liu Y., Xu D., Wen Q., Li X., Zhang W., Yang L. (2012). Polymorphisms of myostatin gene (MSTN) in four goat breeds and their effects on Boer goat growth performance. Mol. Biol. Rep. 39, 3081–3087 10.1007/s11033-011-0737-y [DOI] [PubMed] [Google Scholar]
- Zhang T., Yang H., Wang R., Xu K., Xin Y., Ren G., Zhou G., Zhang C., Wang L., Zhang Z. (2011). Oral administration of myostatin-specific whole recombinant yeast Saccharomyces cerevisiae vaccine increases body weight and muscle composition in mice. Vaccine 29, 8412–8416 10.1016/j.vaccine.2011.09.050 [DOI] [PubMed] [Google Scholar]