Abstract
DUF1220 protein domains exhibit the most extreme human lineage–specific (HLS) copy number increase of any protein coding region in the human genome and have recently been linked to evolutionary and pathological changes in brain size (e.g., 1q21‐associated microcephaly). These findings lend support to the view that DUF1220 domain dosage is a key factor in the determination of primate (and human) brain size. Here we analyze 41 animal genomes and present the most complete account to date of the evolutionary history and genome organization of DUF1220 domains and the gene family that encodes them (NBPF). Included among the novel features identified by this analysis is a DUF1220 domain precursor in nonmammalian vertebrates, a unique predicted promoter common to all mammalian NBPF genes, six distinct clades into which DUF1220 sequences can be subdivided, and a previously unknown member of the NBPF gene family (NBPF25). Most importantly, we show that the exceptional HLS increase in DUF1220 copy number (from 102 in our last common ancestor with chimp to 272 in human; an average HLS increase of ∼28 copies every million years since the Homo/Pan split) was driven by intragenic domain hyperamplification. This increase primarily involved a 4.7 kb, tandemly repeated three DUF1220 domain unit we have named the HLS DUF1220 triplet, a motif that is a likely candidate to underlie key properties unique to the Homo sapiens brain. Interestingly, all copies of the HLS DUF1220 triplet lie within a human-specific pericentric inversion that also includes the 1q12 C‐band, a polymorphic heterochromatin expansion that is unique to the human genome. Both cytogenetic features likely played key roles in the rapid HLS DUF1220 triplet hyperamplification, which is among the most striking genomic changes specific to the human lineage.
Keywords: DUF1220, NBPF, PDE4DIP
Genome sequences encoding DUF1220 protein domains have undergone an exceptional human lineage-specific (HLS) increase in copy number that decreases generally as a function of a species\x{2019} evolutionary distance from humans (Popesco et al. 2006). The HLS DUF1220 copy number expansion was first identified as part of genome-wide survey of gene copy number change among human and great ape lineages (Fortna et al. 2004). An independent study identified the same gene family and named it neuroblastoma breakpoint family (NBPF) when a member of the family was found to be disrupted by a rearrangement in a neuroblastoma patient (Vandepoele et al. 2005). DUF1220 domains are approximately 65 amino acids in length and are encoded by sequences that show signs of positive selection and, in brain, exhibit neuron-specific expression (Popesco et al. 2006; Dumas et al. 2007). Notably, DUF1220 sequences are found in two dissimilar genomic environments: as a single, likely ancestral, domain encoded by the single- or low-copy PDE4DIP (myomegalin) gene, and separately, as tandemly repeated copies encoded by members of the NBPF gene family. It is only this latter form that underwent the dramatic copy number amplification in recent primate evolution (Vandepoele et al. 2005; Popesco et al. 2006).
DUF1220 protein domains have also generated interest because copy number variations (CNVs) in the 1q21.1 region, where most DUF1220 sequences map, have been implicated in numerous recurrent human developmental and neurogenetic diseases (Dumas and Sikela 2009). These include microcephaly and macrocephaly (Brunetti-Pierri et al. 2008; Velinov and Dolzhanskaya 2010), autism (Autism Genome Project Consortium 2007; Pinto et al. 2010), schizophrenia (International Schizophrenia Consortium 2008; Levinson et al. 2011), mental retardation (Mefford et al. 2008; Jaillard et al. 2010), congenital heart disease (Christiansen et al. 2004; Greenway et al. 2009), congenital anomalies of the kidney and urinary tract (Weber et al. 2011), and neuroblastoma (Vandepoele et al. 2008; Diskin et al. 2009). Interestingly, a recent study has implicated DUF1220 domain copy number loss in the etiology of 1q21-associated microcephaly, and provides support for the view that DUF1220 copy number may function as a key general effecter of evolutionary, pathological, and normal variation in brain size among primate (and human) lineages (Dumas et al. 2012).
These findings are also consistent with a model which proposes that 1) the strong selection pressures that drove the rapid HLS increase in DUF1220 copies favored retention of the high genomic instability of the 1q21 region, and 2) it is the resulting unstable, duplication-rich genomic architecture that is a major factor in the etiology of the numerous 1q21.1 disorders that have been reported (Dumas and Sikela 2009). Given these observations, we undertook a comprehensive reconstruction of DUF1220 evolutionary history to gain insight into both the key genomic events that underlie the rapid evolutionary expansion in DUF1220 copy number as well as how this process may be responsible for the multitude of recurrent genetic and developmental diseases associated with the 1q21.1 region.
Methods
Genome searches and nucleotide alignments
DUF1220 nucleotide and protein domains, NBPF proteins, and PDE4DIP proteins were used as BLAT/BLAST queries against 40 genomes in the UCSC (Kent et al. 2002), Ensembl (Flicek et al. 2010), or NCBI (Pruitt et al. 2009) genome databases, and proteins were searched against the NCBI protein database using HMMER3 (Finn et al. 2011). BLAT results were considered significant with an alignment of at least 50 and a score of at least 100 and BLAST results with an alignment of 50 and a score of at least e−10. All significant NBPF alignment results were additionally investigated using the Pfam (Finn et al. 2010) protein database to determine homology to DUF1220 domains at the protein level. Homology was considered significant with a score of e−5.
DUF1220 domain precursor regions were found by aligning the human PDE4DIP DUF1220 seed domain to the predicted PDE4DIP proteins from five genomes, including zebrafish, Xenopus tropicalis (frog), Anolis carolinensis (lizard), chicken, and opossum, from the UCSC genome browser using ClustalW (Larkin et al. 2007). The aligned region was then extracted from the protein, the DUF1220 precursors were realigned, and a phylogeny was created using the ClustalW program. Confirmation of the DUF1220 precursor region was performed by HMMER3 (Finn et al. 2011).
Two thousand base pairs upstream of the human NBPF4 gene was used as a BLAT query against human as well as other mammalian genomes in the UCSC and Ensembl genome browsers to determine whether a conserved NBPF promoter sequence was present within and across multiple genomes. The two thousand base pairs upstream of predicted NBPFs in eight mammalian genomes were extracted and then aligned using the VISTA global alignment tool (Frazer et al. 2004). These same regions were then searched for conserved transcription factor binding sites from the TRANSFAC database using rVISTA 2.0 (Loots and Ovcharenko 2004).
Evolutionary analyses
The DUF1220 phylogenetic profile was inferred using the minimum evolution (ME) method (Rzhetsky and Nei 1992). The consensus tree inferred from five optimal trees is shown (Figure 2A). Branches corresponding to partitions reproduced in less than 50% trees are collapsed. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling 1965) and are in the units of the number of amino acid substitutions per site. The ME tree was searched using the close-neighbor-interchange (CNI) algorithm (Nei and Kumar 2000) at a search level of 1. The neighbor-joining algorithm (Saitou and Nei 1987) was used to generate the initial tree. All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (pairwise deletion option). There were a total of 57 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (Tamura et al. 2007).
The HLS triplets (HLS1-HLS2-HLS3) for each NBPF gene in the 1q21 were aligned using PRANK (Löytynoja and Goldman 2005). The HLS triplets were defined as the genomic sequence starting from the small exon of an HLS1 DUF1220 within an NBPF gene, across the following HLS2 and HLS3 sequences, and to the beginning of the next downstream HLS1 small exon. The phylogenetic tree of these alignments was generated using the APE (Paradis et al. 2004) package in R.
NBPF annotation, copy number organization, and ontogeny
Human NBPF genomic positions were determined by a combination of promoter start positions, the UCSC and Ref-Seq gene tracks at the Santa Cruz genome browser, corresponding DUF1220 domain positions, and the Ensembl gene prediction engine (specifically for NBPF genes predicted by promoter and DUF1220 domain position, but not predicted by the UCSC or Ref-Seq gene tracks). Chimp NBPF numbers were predicted using a combination of the number of predicted NBPF start positions when the promoter sequences were utilized as BLAT queries, the number of DUF1220 sequences sharing clades with known human and orangutan DUF1220 domains in the phylogenetic analysis, and parsimony based upon shared NBPF genes between orangutan and human genomic assemblies. The number of DUF1220 domains in the common ancestor of Pan and human was determined by subtracting the number of DUF1220 domains in chimp-specific clade expansions from the chimp genome DUF1220 count.
The organization and evolution of each NBPF gene in the human genome was evaluated based upon the phylogeny generated. A superclade was considered conserved if 50% or more of the clade was made up of nonhuman DUF1220 domains and it contained DUF1220 domains from all five primates. A superclade was classified as HLS if more than 60% of the clade was made up of human DUF1220. The superclade classification used is further supported by the fact that DUF1220 members of each clade hold a particular placement within the organization of an NBPF gene (supporting information, Table S1) and each clade is representative of one or more Pfam DUF1220 seed domains (Table S2). Determination of whether a human-specific DUF1220 domain arose either via gene duplication or domain amplification was determined by clade analysis. A DUF1220 domain that clusters with a domain from another gene was determined to have arisen via gene duplication. A DUF1220 domain that clusters with domains within the same gene was determined to have arisen via domain amplification. Timing of appearance of CON1 and CON2 domains is based on amino acid alignments of nonprimate mammalian DUF1220 domains with the CON1 and CON2 clade domains (data not shown). PDE4DIP, NBPF, and DUF1220 domain chromosomal positions in human, orangutan, macaque, marmoset, cow, dog, horse, and pig were determined using the UCSC genome browser. Chimp chromosome 1 positions were determined using the UCSC genome browser as well as inversion data mapped by Szamalek et al. (2006).
The DUF1220 domain evolutionary chronology was assembled based on timing of events found by the analysis of 41 animal genomes, the CM promoter analysis performed here, and timing of DUF1220 clades based on the phylogenetic profile. The chronology also incorporates the work from Vandepoele et al. (2009) on the acquisition of the EVI5 promoter and the altered expression work from Illarionova et al. (2007) on the incorporation of an intronic HERV(K) insertion.
Results
DUF1220 origin and copy number expansion
Using genome sequences of 41 animal species (36 mammals and 5 nonmammalian vertebrates), we have generated the most complete evolutionary history of DUF1220 domains from their first appearance to their current levels in existing lineages (Table 1). These analyses indicate that the DUF1220 protein domain first appears as part of the PDE4DIP gene at least 200 million years ago (mya) (Figure 1). Although almost all vertebrates sequenced to date have homologs of PDE4DIP, the proteins do not show levels of sequence conservation high enough to be formally classified by BLAT/BLAST as containing a DUF1220 domain until the emergence of the mammalian lineages. However, the homologous protein coding region of the PDE4DIP DUF1220 domain in nonmammalian vertebrates shows 70% similarity and 32% identity at the amino acid level as far back as bony fish (Figure S1), establishing that a DUF1220 domain precursor in PDE4DIP existed at least 450 mya. This was confirmed by an HMMER (Finn et al. 2011) search against the NCBI protein database. These findings indicate that the DUF1220 protein domain precursor likely existed as an important functional entity prior to the emergence of the mammalian order and persisted as the only DUF1220 domain form for 50–100 million years.
Table 1. Genomes searched for DUF1220 domains, NBPF genes, and PDE4DIP.
| Genome | Coverage | PDE4DIP | Total DUF1220 | NBPF Genes | Source | Assembly | Promoter |
|---|---|---|---|---|---|---|---|
| Euarchotanglires | |||||||
| Human | >9X | 2 | 272 | 23 | UCSC | GRCh37 | CM,EVI5, HERV |
| Chimp | 4X + WGS | 3 | 125 | 19 | UCSC + Ensembl | panTro2 | CM,EVI5, HERV |
| Gorilla | 2X + Illumina | 3 | 99 | 15 | Ensemble | gorGor3 | CM,EVI5, HERV |
| Orangutan | 6X | 4 | 92 | 11 | UCSC + Ensembl | ponAbe2 | CM,EVI5 |
| Gibbon | 5.6X | 3 | 53 | 10 | Ensembl | nomLeu1 | CM,EVI5 |
| Macaque | High-BAC | 1 | 35 | 10 | UCSC + Ensembl | rheMac2 | CM,EVI5 |
| Marmoset | 6X + WGS | 1 | 31 | 11 | UCSC | calJac3 | CM,EVI5 |
| Mouse lemur | 1.93X | 1 | 2 | 1 | Ensembl | micMur1 | Seq gap |
| Bushbaby | 1.5X | 1 | 3 | 2 | Ensembl | otoGar1 | |
| Tarsier | 1.83X | 1 | 1 | 0 | Ensembl | tarSyr1 | |
| Rabbit | 7X | 1 | 8 | 3 | UCSC & Ensembl | oriCun2 | CM |
| Pika | 1.93X | 1 | 1 | 0 | Ensembl | ochPri1 | |
| Mouse | High-BAC | 1 | 1 | 0 | UCSC | mm9 | |
| Rat | High-WGS | 1 | 1 | 0 | UCSC | rn4 | |
| Guinea pig | 6.79X | 1 | 1 | 1 | UCSC & Ensembl | cavPor3 | |
| Squirrel | 1.9X | 1 | 1 | 1 | Ensembl | speTri1 | |
| Tree shrew | 2X | 1 | 4 | 3 | Ensembl | tupBel1 | CM |
| Laurasiatheria | |||||||
| Cow | 7X | 1 | 7 | 3 | UCSC & NCBI | bosTau6 | CM |
| Dolphin | 2.59X | 1 | 4 | 1 | Ensembl | turTru1 | CM |
| Pig | 4X | 1 | 3 | 1 | UCSC & Ensembl | susScr2 | CM |
| Horse | 6.79X | 1 | 8 | 3 | UCSC & NCBI | equCab2 | CM |
| Dog | 2005 WGS | 1 | 3 | 1 | UCSC | canFam2 | CM |
| Panda | 56X Illumina | 1 | 2 | 1 | UCSC | ailMel1 | CM |
| Cat | 2.8X | 1 | 3 | 2 | UCSC & Ensembl | felCat4 | CM |
| Megabat | 2.63X | 1 | 1 | 0 | Ensembl | pteVam1 | |
| Microbat | 1.7X | 1 | 1 | 0 | Ensembl | myoLuc1 | |
| Hedgehog | 1.86X | 1 | 1 | 0 | Ensembl | eriEur1 | |
| Shrew | 1.9X | 1 | 1 | 0 | Ensembl | sorAra1 | |
| Xenarthra | |||||||
| Armadillo | 2X | 1 | 1 | 0 | Ensembl | dasNov1 | |
| Sloth | 2.05X | 1 | 1 | 0 | Ensembl | choHof1 | |
| Afrotheria | |||||||
| Elephant | 7X | 1 | 1 | 2 | UCSC & Ensembl | loxAfr3 | CM |
| Hyrax | 2.19X | 1 | 1 | 0 | Ensembl | proCap1 | |
| Tenrec | 2X | 1 | 1 | 0 | Ensembl | echTel1 | |
| Metatheria | |||||||
| Opossum | 7.33X | 1 | 1 | 0 | Ensembl | monDom5 | |
| Wallaby | 2X | 1 | 1 | 0 | Ensembl | Meug1 | |
| Prototheria | |||||||
| Platypus | 6X | 1 | 1 | 0 | UCSC | ornAna1 | |
| Other vertebrate | |||||||
| Chicken | 6.6X | 0 | 0 | 0 | UCSC | galGal3 | |
| Zebra finch | 6X | 0 | 0 | 0 | UCSC | taeGut1 | |
| Lizard | 6.8X | 0 | 0 | 0 | UCSC | anoCar1 | |
| Frog | 7.65X | 0 | 0 | 0 | UCSC | xenTro2 | |
| Zebrafish | 7X | 0 | 0 | 0 | UCSC | danRer6 |
Of the 41 genomes searched, 36 were from mammals, and 5 were from nonmammalian vertebrates.
Figure 1.
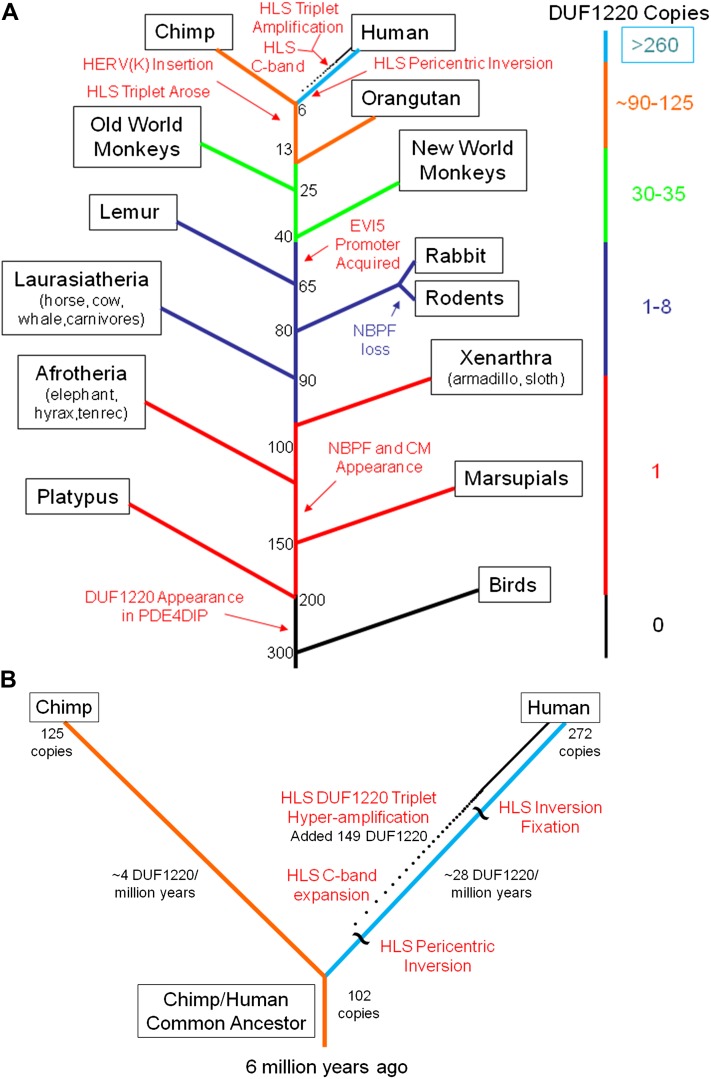
(A) Phylogenetic tree of the major events shaping the evolution of the DUF1220 domain family. Depicted are different mammalian lineages with their approximate divergence times at branch points in millions of years ago. Major events in DUF1220 evolution are identified, and approximate numbers of DUF1220 domains per genome are shown on the right. The dotted line of increasing density along the human divergence branch is indicative of the accumulation of the HLS DUF1220 triplet since the HLS pericentric inversion. (B) Expanded image of the human/chimp split in the evolutionary timeline of DUF1220 domains showing the different evolutionary fates of DUF1220 domains within each of the two lineages.
Investigation of 36 mammalian genomes [one Prototheria (platypus), two Metatheria (marsupials), and 33 Eutheria (placental mammals)] reveals that the first DUF1220 domain outside of PDE4DIP appeared ∼100–150 mya when the domain likely underwent a duplicative transposition into a new genomic environment. This event generated a new DUF1220 domain copy separate from the PDE4DIP gene and created a new gene family (NBPF) that exclusively encodes DUF1220 protein domains. This timeframe is based on homologs of NBPF genes found in three of the four sister superorders of placental mammals: Afrotheria (elephant), Laurasiatheria (hoofed mammals and carnivores), and Euarchotanglires (supraprimates). A complete list of the DUF1220 domains found among all 36 mammals investigated is given in Table 1.
It should be noted that the Rodentia order has no NBPF gene homologs in the mouse, rat, Guinea pig, or squirrel genomes and only carries the single DUF1220 domain that is found in the PDE4DIP gene. The most parsimonious explanation for this absence is that NBPF homologs were lost in a common ancestor of the rodents. This conclusion is based on two observations: (1) that there is a significant number of species that contain NBPF homologs that diverged prior to the emergence of the supraprimate superorder, and (2) that their sister order Lagomorpha (rabbit) has NBPF homologs.
In nonprimate mammals, copy number expansion of the NBPF type of DUF1220 domain was minimal (1–7 NBPF DUF1220 domains per genome), with the majority of DUF1220 domains located within 0.5 Mb of the PDE4DIP gene and thus consistent with a local duplication event. This is in sharp contrast to the dramatic changes that occurred in the primate order, which show a pattern of increasing DUF1220 copy number as a function of increasing phyletic proximity to human. This progressive expansion can be seen from lemur (2–3 copies) to new and old world monkeys (30–35 copies) to apes (90–125 copies) and finally to human (272 copies in the hg19 build) (Figure 1 and Table 1). The primate NBPF genes also show elevated DUF1220 copy number per gene (2–52 DUF1220 domains) as compared with the nonprimate mammalian NBPFs (1–4 DUF1220 domains), reflecting an enhanced level of intragenic domain amplification.
Organization of DUF1220-encoding genes
Analysis of the HLS increase in DUF1220 domains is challenging due to the lack of a definitive NBPF gene annotation. Current DUF1220/NBPF annotations often lack defined gene start and stop points, and they frequently list individual NBPF genes at multiple locations on chromosome 1. Therefore, a detailed comparison of NBPF genes within and between species was undertaken to more precisely define NBPF genes and improve current annotation of the gene family.
Identification of the CM promoter:
Comparison of NBPF genes across eight species, including 2000 bp upstream of their predicted start positions, indicated that the promoter regions of human NBPFs 4–7 showed remarkable similarity to the promoter regions of many of the NBPFs in both primates and nonprimates (Figure S2). Alignment of these regions revealed that the 900 bp upstream of NBPF genes was particularly conserved, with the most divergent promoter regions (human and cow) having greater than 50% identity. When this region is searched vs. a transcription factor binding site database (TRANSFAC), two conserved binding motifs were detected: an OCT4 binding site, implicated in control of pleuripotent stem cell embryogenesis (Wang and Dai 2010), and an IRF1 binding site, implicated in induction of interferon, tumor suppression, cell-cycle control, and apoptosis (Romeo et al. 2002). We have designated this 900 bp conserved region as the CM promoter due to its conservation among all mammalian NBPF genes.
Specific criteria for defining NBPF genes:
When the CM promoter is used as a BLAT/BLAST query against the 41 mammalian genomes investigated, only three regions appear: (1) upstream regions of nonprimate NBPF genes, (2) upstream regions of the majority of primate NBPF genes, and (3) in the fourth intron upstream of the first DUF1220 domain in NBPF genes, where it is not in the promoter position. In the primate NBPF genes with an intronic CM promoter, the genes have acquired the previously documented EVI5 promoter (Vandepoele et al. 2009). Interestingly, this shows that the CM promoter region is distinct from the EVI5 promoter and may act as an alternative start site in primate NBPF genes that have acquired the EVI5 promoter. This analysis indicates that all NBPF genes have either only the predicted CM promoter or both the EVI5 and CM promoters. Based on these data, we propose that the presence of the CM promoter immediately upstream of one or more predicted DUF1220 domains is criterion that represents the sine qua non for defining NBPF genes. This new definition allowed us to generate the most comprehensive annotation so far reported of the 23 NBPF genes in the human genome (Table 2). This list includes all previously documented NBPF genes as well as an undocumented gene predicted to encode 16 DUF1220 domains, which we have named NBPF25.
Table 2. Predicted NBPF genes in the human genome.
| Name | Position | No. of DUF1220 | No. of DUF1220 in Triplet | Promoter | NBPF Type |
|---|---|---|---|---|---|
| NBPF1 | 16,890,412–16,939,982 | 7 | 0 | EVI5/CM | Primate |
| NBPF2p | 21,749,601–21,754,300 | 3 | 0 | None | Pseudogene |
| NBPF3 | 21,766,631–21,811,392 | 5 | 0 | EVI5/CM | Primate |
| NBPF4 | 108,765,963–108,786,703 | 4 | 0 | CM | Nonprimate |
| NBPF5p | 108,918,460–108,953,432 | 2 | 0 | CM | Pseudogene |
| NBPF6 | 108,992,904–109,013259 | 4 | 0 | CM | Nonprimate |
| NBPF7 | 120,377,389–120,387,779 | 2 | 0 | CM | Nonprimate |
| NBPF8 | 144,146,807–144,224,481 | 44 | 42 | Gap | HLS |
| NBPF9 | 144,593,696–144,828,810 | 7 | 0 | EVI5/CM | Primate |
| NBPF10 | 145,293,371–145,368,682 | 40 | 36 | CM | HLS |
| NBPF11 | 146,032,543–146,068,252 | 7 | 0 | EVI5/CM | Primate |
| NBPF25 | 146,214,651–146,253,110 | 16 | 11 | Gap | HLS |
| NBPF12 | 146,374,056–146,467,744 | 34 | 29 | EVI5/CM | HLS |
| NBPF13 | 146,581,444–146,596,147 | 5 | 0 | CM | Primate |
| NBPF24 | 147,574,324–147,624,601 | 7 | 0 | EVI5/CM | Primate |
| NBPF14 | 148,003,643–148,026,039 | 10 | 7 | Gap | HLS |
| NBPF20 | 148,251,113–148,346,929 | 52 | 48 | CM (? gap EVI5) | HLS |
| NBPF15 | 148,556,090–148,596,267 | 6 | 0 | EVI5/CM | Primate |
| NBPF16 | 148,739,442–148,758,311 | 6 | 0 | CM (? gap EVI5) | Primate |
| NBPF23 | 149,089,947–149,154,938 | 6 | 0 | EVI5/CM | Primate |
| NBPF18p | 151,991,138–152,015,250 | 0 | 0 | CM | Pseudogene |
| NBPF21p | Chr3: 36,657,498–36,678,949 | 1 | 0 | None | Pseudogene |
| NBPF22p | Chr5: 85,578,262–85,593,362 | 2 | 0 | CM | Pseudogene |
This table was created using the new comprehensive annotation method described here. Listed are the NBPF name, genomic position, number of encoded DUF1220 domains, number of DUF1220 domains intragenically amplified in the HLS triplet clades, and predicted promoters. Genes containing amplified HLS DUF1220 triplets are in bold.
DUF1220 phylogeny
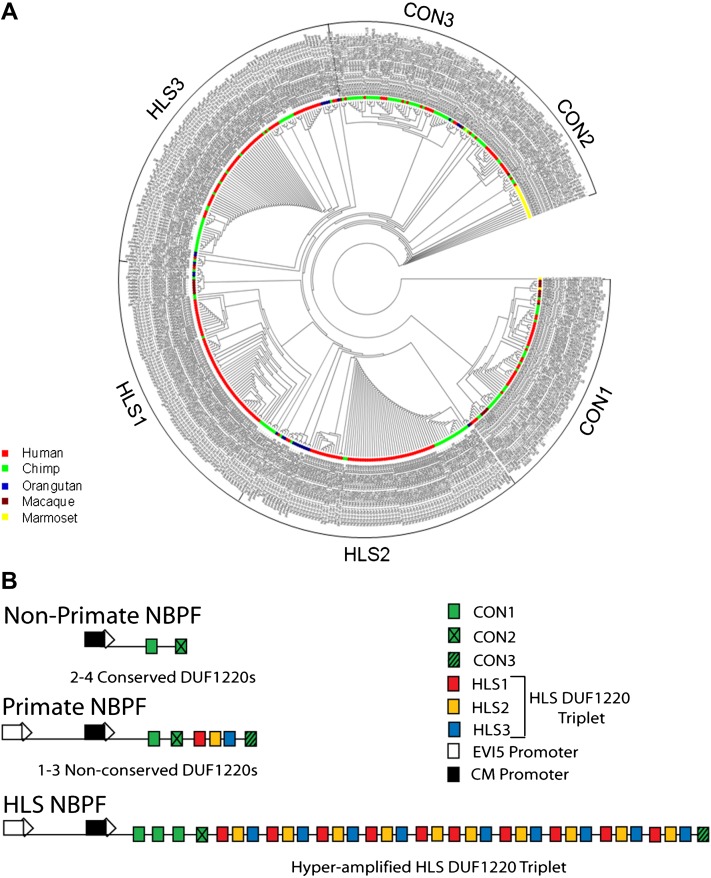
To determine the evolutionary relationship of the DUF1220 domains within defined NBPF genes, a phylogenetic profile of DUF1220 evolution among primates was created for 429 predicted DUF1220 domains in the human, chimp, orangutan, rhesus macaque, and marmoset genomes (Figure 2A). This analysis indicates that DUF1220 sequences can be subdivided into six superclades, which can be distinguished by both their evolutionary conservation within the primate order as well as their position and order within each gene (Table S1). We have classified three of the six clades as conserved (CON1–3), as DUF1220 domains from all primates investigated have members of these three clades and greater than 50% of the DUF1220 copies in these clades are also found in species other than human. Interestingly, each CON clade maintains a distinct position within each NBPF gene: CON1 clade members occur at the N-terminus of all NBPF genes, CON2 members are adjacent to CON1 in all NBPF genes (this is the C-terminal DUF1220 domain in some NBPFs), and CON3 members reside at the C-terminus of the great majority of NBPF genes (Figures 2B and 3). Amino acid alignments to nonprimate DUF1220 domains reveal that CON1 and CON2 are found in nonprimate mammals as well (data not shown). The other three clades, HLS1–3, typically fall between CON2 and CON3 in old world monkey, ape, and human NBPF genes but have been greatly expanded in copy number, specifically in the human lineage.
Figure 2.
(A) Consensus tree of evolutionary relationships of 429 DUF1220 sequences. The evolutionary history was inferred using the Minimum Evolution method with the consensus tree inferred from five optimal trees. Species covered by the tree are color coded. The different types of DUF1220 clades are bracketed. (B) Gene organization of the three distinct types of NBPF genes. Top: Nonprimate mammal NBPF organization (NBPFs 4, 6, and 7). Middle: Typical primate NBPF organization (NBPFs 1, 3, 9, 11, 13, 24, 15, 16. and 23). Bottom: HLS NBPF organization (NBPFs 8, 10, 12, 25, 14, and 20).
Figure 3.
Gene organization of all NBPF genes in the human genome. The DUF1220 domains are shown as colored boxes that correspond to the different clades shown in Figure 2A. The hyperamplified HLS DUF1220 triplets are bracketed in the six human NBPF genes in which they occur.
Comparison of this evolutionary profile of the NBPF-encoded DUF1220 domains with their gene organization indicates that, excluding four predicted pseudogenes, there are three types of functional NBPF genes in the human genome and that these exhibit widely different evolutionary histories from one another (Figure 2B and Table 2). First, the three genes that make up the group of nonprimate NBPF genes (NBPF 4, 6, and 7) exhibit a gene organization shared with nonprimate mammals, map exclusively to the human 1p-arm, and are composed entirely of conserved (CON1–3) DUF1220 domains. Second, the nine genes in the group of primate NBPF genes (NBPF 1, 3, 9, 11, 13, 24, 15, 16, and 23) show a type of gene organization that emerged only in the primate lineage and are located primarily in the 1q21.1–q21.2 region. These genes begin with N-terminal DUF1220 domains from the conserved CON1 and CON2 clades, followed by 1–5 HLS-type DUF1220 domains, and end with a C-terminal CON3 domain. Third, the six genes in the group of HLS NBPF genes (NBPF 8, 10, 12, 25, 14, and 20) are only found in the human lineage and map exclusively to the 1q21.1–q21.2 region. The most striking feature that distinguishes the HLS NBPF gene organization from the other two is that it encodes a hyperamplified number of HLS DUF1220 domains (7–48) in the middle of the gene between the CON2 and CON3 domains (Figure 2B and Table 2). Such intragenic domain hyperamplification is absent in all other primate genomes examined.
HLS DUF1220 triplet
This phylogenetic profile also shows that 73% (199/272) of the human DUF1220 domains cluster to just the three HLS clades (HLS1–3) (Figure 2A). When the genomic positions of the DUF1220 domains that fall into these clades are compared with the predicted NBPF gene positions, 173 of the 199 HLS1–3 DUF1220 domains cluster to only 6 of the predicted 23 NBPF genes in the genome (Figure 3). Most strikingly, when the order of the HLS domains within each of these 6 HLS NBPF genes is examined, a tandemly repeated 4.7 kb triplet pattern, comprised of one member from each of the three HLS clades, emerges which we have named the HLS DUF1220 triplet. The triplet is made up of six exons and six introns, with 90% of the 4.7 kb being intronic sequence. Each triplet shows high similarity to one another, with members of the same HLS clade exhibiting 96–100% nucleotide sequence identify. Although HLS1 and HLS3 clades show the greatest interclade divergence among the HLS clades, they still maintain high sequence similarity to one another (85–90%).
It is noteworthy that although the HLS DUF1220 triplet is present in the chimp and gorilla genomes, indicating that the triplet arose prior to the split of human and gorilla, it did not undergo the pronounced expansion seen in the human lineage. In both of these genomes, the triplet is found only in the unexpanded primate NBPF form mentioned in the previous section. The largest predicted chimp and gorilla NBPFs have five of the HLS clade DUF1220s in a single complete gene, indicating that there has not been a triplet duplication within the chimp genome. This lack of triplet expansion is further supported by copy number estimates of DUF1220 domains based on sequence read depth (Sudmant et al. 2010), which estimates that copy number in chimp of the HLS DUF1220 triplet-containing gene NBPF14 is a fifth of that in human.
Interestingly, the DUF1220 domains within the HLS DUF1220 triplet clades are more closely related to other HLS triplet domains within the same gene than they are to the triplets in a different gene, indicating they arose predominantly via intragenic domain amplification rather than gene duplication (Figure 4). Remarkably, the intragenic hyperamplification of the HLS DUF1220 triplet added ∼149 DUF1220 copies to the human genome, whereas only ∼21 copies were added via gene duplication. Thus, in total, the HLS DUF1220 triplet domain hyperamplification accounts for 82% of the 182 HLS clade domain copies in the human 1q21 region and 88% (149/170) of the DUF1220 domains added to the human genome since our last common ancestor with the Pan genus (an average HLS gain of ∼28 DUF1220 copies every million years since our divergence with Pan).
Figure 4.
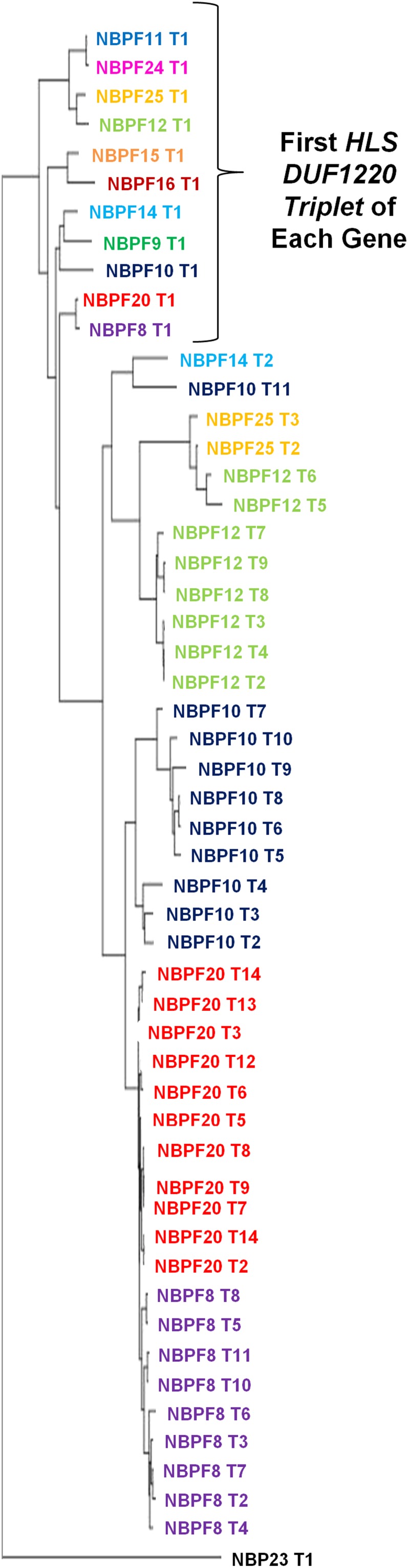
Evolutionary relationship of HLS DUF1220 triplets. Phylogenetic sequence-based alignments of the HLS DUF1220 triplets reveal that the first triplets in each NBPF gene are most similar to one another. Each successive triplet is then most similar to the triplets contained within each NBPF gene, indicating that the hyperamplification of the HLS DUF1220 triplet occurred as the result of intragenic domain amplification.
Discussion
The data presented here represent the most complete chronology of the evolutionary history of DUF1220 protein domains so far reported and include the identification of several key events related to the unusually rapid copy number amplification of these sequences. These include identification of a DUF1220 precursor dating as far back as ∼450 mya; the emergence of the first NBPF gene roughly ∼100–150 mya, which was likely due to the local duplication of a DUF1220-encoding segment of the PDE4DIP gene and its acquisition of the novel CM promoter; accelerated evolution of the DUF1220 domains in the primate lineage, including both gene and domain copy number increases; and the appearance of new DUF1220 clades.
The most striking finding, however, was the accelerated rise in copy number of the new DUF1220 domains in the human lineage due to intragenic domain hyperamplification involving the HLS DUF1220 triplet. To deduce a plausible explanation for this copy number increase in the human lineage, the HLS DUF1220 triplet sequences and their genomic locations were examined. Although the presence of transposable element (TE) insertions in intronic regions has been documented in other cases of domain amplification (Björklund et al. 2010), we found no evidence of insertions in any of the HLS triplets and hypothesize that the triplets may have something inherent to their sequence that makes them prone to amplification.
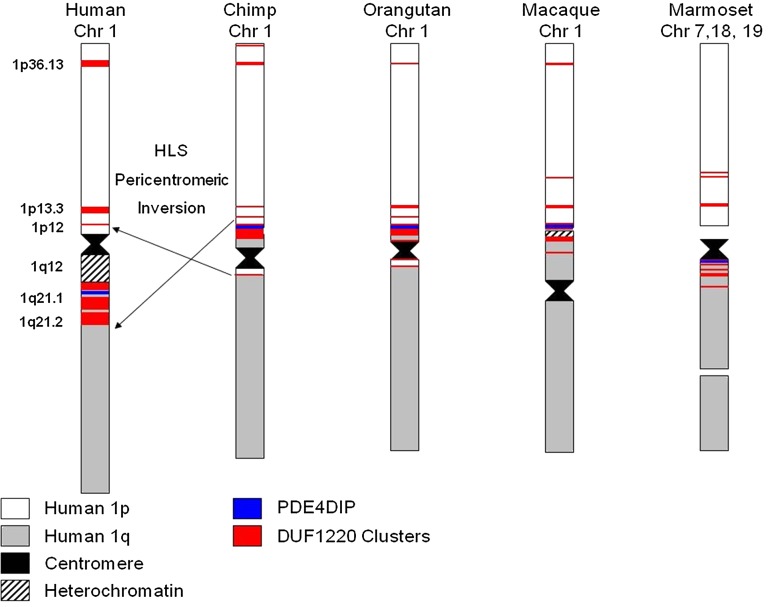
Mapping of the chromosomal locations of DUF1220 domains using the UCSC browser and data from Szamalek et al. (2006) (Figure 5) revealed that all copies of the HLS DUF1220 triplet lie in a unique human-specific genomic environment. Interestingly, although this analysis showed that all five primates tested have their largest cluster of DUF1220 domains at a homologous chromosomal position to human 1q21.1–q21.2, in human this entire region underwent an HLS pericentric inversion (1p11.2–q21.2). All HLS DUF1220 triplets map within this HLS inversion to a 7 Mb segment from 142.6 Mb to 149.7 Mb (Feb 2009 assembly, hg19). This interval along with the flanking 1q12 region was involved in the inversion, and both regions underwent dramatic human-specific genomic expansions (Figure 1B). 1q12, which has long been documented as a cytogenetically visible C-band, is a highly polymorphic, repeat-rich heterochromatic chromosomal band unique to humans that may have expanded after the inversion took place (Yunis and Prakash 1982). Both the 1q12 expansion and the 1p11.2–q21.2 inversion represent major alterations in the genome architecture of the pericentric region of human chromosome 1, and as such, may have played key roles in the rapid HLS DUF1220 triplet expansion.
Figure 5.
DUF1220 localization and cytogenetic architecture from five primate species. An ideogram depicting chromosome 1 of human, chimp, orangutan, macaque, and marmoset is shown, with arm color designations in relation to the human chromosomes 1p (white) and 1q (gray) gene order. Centromere position is indicated by a black block, and heterochromatin bands are indicated by black hash marks. The position of the PDE4DIP gene is indicated by a blue line, and clusters of DUF1220 copies are shown in red.
A plausible explanation for these genomic changes focuses on the fact that when the inversion occurred early in the human lineage both inverted and noninverted chromosomes were present. As a result, in matings involving inverted/noninverted chromosomes, proper meiotic pairing and interchromosomal recombination of this region of chromosome 1 may have been temporarily impaired. In turn, this may have augmented allele fixation and led to several rounds of intrachromosomal nonallelic homologous recombination (NAHR), as has been observed for the Y chromosome (Rozen et al. 2003). The reduced interchromosomal allelic recombination may have favored enhanced rates of intrachromosomal NAHR that, combined with positive selection, may have triggered the fixation of chromosomes with intragenic DUF1220 gene and/or domain amplification(s). In this manner, the merging of these forces could have allowed DUF1220 domain copy number to hyperamplify, expanding from ∼102 DUF1220 copies in our last common ancestor with the Pan lineage to 272 in modern human.
Most importantly, these studies have laid the groundwork for future studies into DUF1220 function. Discovery of the six DUF1220 clades and their distinct evolutionary histories and gene positions indicates that they may have different functional roles within an NBPF protein. Future studies should focus on what roles the different clades play, especially the NBPF genes that contain the HLS DUF1220 triplet. In addition, selection for the amplification of the HLS DUF1220 triplet may indicate that the triplet itself confers a functional advantage, such as the super repeat seen in nebulin proteins, which has a higher binding affinity than a single nebulin repeat alone (Björklund et al. 2010).
In summary, the human-specific hyperamplification of the HLS DUF1220 triplet described here represents the primary mechanism responsible for the unprecedented DUF1220 copy number expansion in the human lineage. This rapid burst in copy number is likely to be highly adaptive within the human lineage, and it may have contributed to, and/or been facilitated by, the duplication-promoting genome architecture that characterizes the 1q21 region. As this genomic feature is only found in the human lineage, it is likely that only the human species bears the high recurrent disease burden that has been associated with the 1q21 region (Dumas and Sikela 2009).
See Table 3 for a glossary of terms used in this article.
Table 3. Glossary.
| DUF1220 Terminology | Description |
|---|---|
| CM promoter | Conserved mammalian promoter |
| CON1 | Conserved clade; 1st DUF1220 domain in NBPF genes |
| CON2 | Conserved clade; 2nd DUF1220 domain in NBPF genes |
| CON3 | Conserved clade; last DUF1220 domain in NBPF genes |
| HLS1 | Human Lineage-Specific expansion clade; 1st member HLS DUF1220 triplet |
| HLS2 | Human Lineage-Specific expansion clade; 2nd member HLS DUF1220 triplet |
| HLS3 | Human Lineage-Specific expansion clade; 3rd member HLS DUF1220 triplet |
Supplementary Material
Acknowledgments
We would like to thank Mark Johnson, Franz van Roy, and Karl Vandepoele for their helpful discussions. This work was supported by a grant from the National Institute for Mental Health (R01 MH-081203-1). M.S.O. was supported in part by a postdoctoral training grant from the National Institute of Alcohol Abuse and Alcoholism (5T32 AA-007464-32), and C.M.D. by National Institutes of Health Computational Bioscience Program training grant (5T15 LM-009451-05).
Footnotes
Communicating editor: A. H. Paterson
Literature Cited
- Autism Genome Project Consortium, 2007. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 39: 319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund J., Light S., Sagit R., Elofsson A., 2010. Nebulin: a study of protein repeat evolution. J. Mol. Biol. 402: 38–51 [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N., Berg J. S., Scaglia F., Belmont J., Bacino C. A., et al. , 2008. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat. Genet. 40: 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen J., Dyck J. D., Elyas B. G., Lilley M., Bamforth J. S., et al. , 2004. Chromosome 1q21.1 contiguous gene deletion is associated with congenital heart disease. Circ. Res. 94: 1401–140215192033 [Google Scholar]
- Diskin S. J., Hou C., Glessner J. T., Attiyeh E. F., Laudenslager M., et al. , 2009. Copy number variation at 1q21.1 associated with neuroblastoma. Nature 459: 987–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas L., Kim Y. H., Karimpour-Fard A., Cox M., Hopkins J., et al. , 2007. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 17: 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas L., Sikela J., 2009. DUF1220 domains, cognitive disease, and human brain evolution. Cold Spring Harb. Symp. Quant. Biol. 74: 375–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas L., O'Bleness M. S., Davis J. M., Dickens C. M., Anderson N., et al. , 2012. DUF1220 domain copy number implicated in human brain size pathology and evolution. Am. J. Hum. Genet. 91: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Mistry J., Tate J., Coggill P., Heger A., et al. , 2010. The Pfam protein families database. Nucleic Acids Res. 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Clements J., Eddy S. R., 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39: W29–W37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P., Aken B. L., Ballester B., Beal K., Bragin E., et al. , 2010. Ensembl’s 10th year. Nucleic Acids Res. 38: D557–D562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortna A., Kim Y., MacLaren E., Marshall K., Hahn G., et al. , 2004. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2: E207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K., Pachter L., Poliakov A., Rubin E., Dubchak I., 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32: W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway S. C., Pereira A. C., Lin J. C., DePalma S. R., Israel S. J., et al. , 2009. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat. Genet. 41: 931–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illarionova A. E., Vinogradova T. V., Sverdlov E. D., 2007. Only those genes of the KIAA1245 gene subfamily that contain HERV(K) LTRs in their introns are transcriptionally active. Virology 358: 39–47 [DOI] [PubMed] [Google Scholar]
- International Schizophrenia Consortium, 2008. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455: 178–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillard S., Drunat S., Bendavid C., Aboura A., Etcheverry A., et al. , 2010. Identification of gene copy number variations in patients with mental retardation using array-CGH: novel syndromes in a large French series. Eur. J. Med. Genet. 53: 66–75 [DOI] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., et al. , 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., et al. , 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Levinson D. F., Duan J., Oh S., Wang K., Sanders A. R., et al. , 2011. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am. J. Psychiatry 168: 302–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots G., Ovcharenko I., 2004. rVista 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32: W217–W221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A., Goldman N., 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci. USA 102: 10557–10562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T., Eichler E. E., 2009. The evolution of human segmental duplications and the core duplicon hypothesis. Cold Spring Harb. Symp. Quant. Biol. 74: 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford H. C., Sharp A. J., Baker C., Itsara A., Jiang Z., et al. , 2008. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N. Engl. J. Med. 359: 1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Kumar S., 2000. Molecular Evolution and Phylogenetics, Oxford University Press, New York [Google Scholar]
- Paradis E., Claude J., Strimmer K., 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290 [DOI] [PubMed] [Google Scholar]
- Pinto D., Pagnamenta A. T., Klei L., Anney R., Merico D., et al. , 2010. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466: 368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popesco M. C., Maclaren E. J., Hopkins J., Dumas L., Cox M., et al. , 2006. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science 313: 1304–1307 [DOI] [PubMed] [Google Scholar]
- Pruitt K., Tatusova T., Klimke W., Maglott D., 2009. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 37: D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo G., Fiorucci G., Chiantore M. V., Percario Z. A., Vannucchi S., et al. , 2002. IRF-1 as a negative regulator of cell proliferation. J. Interferon Cytokine Res. 22: 39–47 [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., Marszalek J. D., Minx P. J., Cordum H. S., et al. , 2003. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423: 873–876 [DOI] [PubMed] [Google Scholar]
- Rzhetsky A., Nei M., 1992. A simple method for estimating and testing minimum evolution trees. Mol. Biol. Evol. 9: 945–967 [Google Scholar]
- Saitou N., Nei M., 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sudmant P. H., Kitzman J. O., Antonacci F., Alkan C., Malig M., et al. , 2010. Diversity of human copy number variations and multicopy genes. Science 330: 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamalek J., Goidts V., Cooper D., Hameister H., Kherer-Sawatzki H., 2006. Characterization of the human lineage-specific pericentric inversion that distinguishes human chromosome 1 from the homologous chromosomes of the great apes. Hum. Genet. 120: 126–138 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S., 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Vandepoele K., Van Roy N., Staes K., Speleman F., van Roy F., 2005. A novel gene family NBPF: intricate structure generated by gene duplications during primate evolution. Mol. Biol. Evol. 22: 2265–2274 [DOI] [PubMed] [Google Scholar]
- Vandepoele K., Andries V., van Roy N., Staes K., Vandesompele J., et al. , 2008. A constitutional translocation t(1;17)(p36.2;q11.2) in a neuroblastoma patient disrupts the human NBPF1 and ACCN1 genes. PLoS ONE 3: e2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Andries V., van Roy F., 2009. The NBPF1 promoter has been recruited from the unrelated EVI5 gene before simian radiation. Mol. Biol. Evol. 26: 1321–1332 [DOI] [PubMed] [Google Scholar]
- Velinov M., Dolzhanskaya N., 2010. Clavicular pseudoarthrosis, anomalous coronary artery and extra crease of the fifth finger--previously unreported features in individuals with class II 1q21.1 microdeletions. Eur. J. Med. Genet. 53: 213–216 [DOI] [PubMed] [Google Scholar]
- Wang X., Dai J., 2010. Concise review: isoforms of OCT4 contribute to the confusing diversity in stem cell biology. Stem Cells 28: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S., Landwehr C., Renkert M., Hoischen A., Wühl E., et al. , 2011. Mapping candidate regions and genes for congenital anomalies of the kidneys and urinary tract (CAKUT) by array-based comparative genomic hybridization. Nephrol. Dial. Transplant. 26: 136–143 [DOI] [PubMed] [Google Scholar]
- Yunis J., Prakash O., 1982. The origin of man: a chromosomal pictorial legacy. Science 215: 1525–1530 [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L., 1965. Evolutionary divergence and convergence in proteins, pp. 97–166 in Evolving Genes and Proteins, edited by V. Bryson, and H. J. Vogel Academic Press, New York [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.