Abstract
BACKGROUND
DNA methylation patterns differ among children and adults and play an unambiguous role in several disease processes, particularly cancers. The origin of these differences are inadequately understood, and this is a question of specific relevance to childhood and adult cancer.
METHODS
DNA methylation levels at 26,485 autosomal CpGs were assayed in 201 newborns (107 African-American and 94 Caucasian). Nonparametric analyses were performed to examine the relation between these methylation levels and maternal parity, maternal age, newborn gestational age, newborn gender,and newborn race. To identify the possible influences of confounding, stratification was additionally performed by a second and third variable. For genes containing CpGs with significant differences in DNA methylation levels between races, analyses were performed to identify highly represented gene ontological terms and functional pathways.
RESULTS
13.7% (3,623) of the autosomal CpGs exhibited significantly different levels of DNA methylation between African-Americans and Caucasians. 2% of autosomal CpGs had significantly different DNA methylation levels between male and female newborns. Cancer pathways, including four (pancreatic, prostate, bladder, and melanoma) with substantial differences in incidence between the races, were highly represented among the genes containing significant race-divergent CpGs.
CONCLUSIONS
At birth, there are significantly different DNA methylation levels between African-Americans and Caucasians at a subset of CpG dinucleotides. It is possible that some of the epigenetic precursors to cancer exist at birth and that these differences partially explain the different incidence rates of specific cancers between the races.
Keywords: DNA methylation, African-American, Caucasian, Racial difference, Stratified Kruskal-Wallis
Introduction
DNA methylation, along with other epigenetic processes, is widely recognized as an important element of cellular differentiation and gene regulation that is key to normal development. On the other hand, abnormalities in DNA methylation are a fundamental aspect of several newborn syndromes (e.g, Beckwith-Wiedemann, Silver-Russell, Prader-Willi, Angelman syndromes) and adult diseases, particularly cancer (Feinberg and Tycko, 2004). The changes to DNA methylation that are associated with these adverse outcomes are likely to be highly similar across geographic and racial groups. However, the rates of incidence of some epigenetically-influenced diseases, such as cancers, differ among racial groups, and these differences must be explained by different environmental exposures, different responses to the same exposures, or by intrinsic differences in the frequencies of DNA sequence or epigenetic variants. For example, based on national statistics, there are distinct differences in the incidences of some of the most common cancers between African-Americans and Caucasians (Edwards and others, 2005). Caucasian men and women have higher incidences of lymphoma, leukemia, and cancers of the bladder, brain, thyroid, uterus and ovary, while African-Americans have higher incidences of myeloma, and cancers of the prostate, colon, kidney, stomach, pancreas, and cervix.
In adulthood, there is evidence that various racial groups differ in terms of both their patterns of DNA methylation in healthy tissue and their patterns of change in some cancerous tissues. In a survey of global levels of DNA methylation in normal mononuclear blood cells among women in their 40s (Terry and others, 2008), statistically significant differences were observed, with African-American women having the lowest level of DNA methylation, Hispanic women the highest, and non-Hispanic Caucasians intermediate. Similarly, in a survey of LINE-1 methylation in normal colonic tissue from males and females with an average age of 57 and a history of adenoma (Figueiredo and others, 2009), African-Americans and Caucasians had similar levels of methylation, while Hispanics had slightly higher levels. A survey of DNA methylation in normal prostate tissue from prostate cancer patients revealed higher methylation in African-American men for two genes (TIMP3 and NKX2-5), while five (AR, RARβ2, SPARC, TIMP3, and NKX2-5) of six genes had higher methylation in cancer tissue in African-Americans (Kwabi-Addo and others, 2010).
There is mixed evidence for differences between African-Americans and Caucasians in DNA methylation in cancerous tissues or changes in DNA methylation during the process of oncogenesis, including cancers with distinct racial differences in incidence. Colorectal cancer has a higher incidence and mortality rate in African-Americans than Caucasians, and in a survey of 13 candidate cancer genes three (GPNMB, ICAM5, and CHD5) showed higher levels of methylation in African-Americans than Iranians (Mokarram and others, 2009). By contrast, prostate cancer has about a two-fold higher incidence in African-Americans, but among eight putative prostate cancer genes, only CD44 showed a statistically suggestive (p = 0.1 for a 1.6-fold higher rate of hypermethylation in African-Americans) difference between African-Americans and Caucasians (Woodson and others, 2004). On the other hand, increased methylation of the tumor suppressor TMS1/ASC (also called PYCARD) was observed in Caucasian prostate cancer patients relative to benign prostate hypertrophy controls (62.2% vs. 22.7%, respectively), but not in African-Americans patients (66.7% vs. 58.3% in controls; Das and others, 2006). This suggests that TMS1 methylation levels, generally associated with downregulation of the gene, are lower in non-cancerous prostate tissue of Caucasians but in cancerous tissue increase to the level observed in both healthy and cancerous tissues in African-Americans. Incidence of breast cancer is higher overall in Caucasians than African-Americans, but until 50 years of age the incidence is higher among African-Americans and they have the highest overall mortality in all age groups. Although the average later stage of diagnosis in African-American women contributes to this disparity in mortality, they also exhibit additional negative prognostic factors, such as earlier age of diagnosis, estrogen and progesterone receptor negativity (ER−/PR−), and unique mutations in some oncogenes (e.g., BRCA1 and p53). In a comparison of ER/PR positive and negative invasive ductal carcinomas, levels of methylation were similar for five putative oncogenes in all groups except African-American women less than 50 with ER-/PR- tumors, among whom there was significantly higher methylation of four genes (HIN-1, Twist, Cyclin D2, and RASSF1A, with a marginally significant [p = 0.01] increase for RAR-β) involved in apoptosis and tumor suppression. Furthermore, in a separate survey of methylation levels among 773 cancer-related genes in breast cancer samples, race was significantly associated with cluster membership after hierarchical clustering, indicating unique methylation profiles across races. Finally, Caucasians have statistically higher overall levels of DNA methylation in uninvolved bronchial epithelium and epithelial hyperplasia compared to squamous cell carcinomas (Piyathilake and others, 2003). The hypomethylation in the Caucasian squamous cell cancers brought the levels of methylation down to those observed for squamous cell carcinomas, uninvolved bronchial epithelium, and epithelial hyperplasia in African-Americans.
In general, a picture is emerging of racially distinguishable patterns of DNA methylation in normal tissue and changes in methylation during oncogenesis. Our goal in this paper was to use a cohort of African-American and Caucasian newborns to determine if the differences in DNA methylation observed in adulthood are also present at birth. While the potential effects of differences between the races in maternal diet or maternal metabolism during pregnancy cannot be excluded, the presence of DNA methylation differences at birth would suggest that those differences observed later in life were not acquired only postnatally but were an initial feature of the epigenome. If so, these DNA methylation differences might affect the expression of adjacent genes and could affect genetic response to physiological and environmental influences. In particular, the presence of differences in DNA methylation among genes involved in oncogenesis at birth may form part of the explanation for later differences in the incidence and rate of mortality of cancers across races.
Material and Methods
Human subjects
Newborns (N = 216) were selected from a larger longitudinal cohort study of human development from pregnancy to age 3, the Conditions Affecting Neurocognitive Development and Learning in Early Childhood Study (CANDLE), being performed in Shelby County, Tennessee. Solicitation for inclusion in the study is via advertising and brochures in local gynecological clinics. Upon contacting study personnel by telephone, potential participants are asked screening questions for eligibility, and eligible women are requested to visit one of two research clinics administered by the study. Out of every 2.5 women screened, one is invited to participate. Of those invited, 92% are participating. Written informed consent was obtained from all mothers, and this study was approved by the institutional review boards of all the participating hospitals. Original selection criteria for this substudy were: maternal age 18–40 years, singleton pregnancy, complete data on birth weight and maternal pre-pregnancy weight, and absence of several complications, specifically sexually-transmitted disease, diabetes, oligohydramnios, preeclampsia, placental abruption, and cervical cerclage. For the purposes of examining racial differences, we further restricted attention to gestational ages of 36–42 weeks and to mother-newborn pairs whose self-declared race was only Caucasian or only African-American (not Asian, Hispanic, Native American, Pacific Islander or multiracial). After applying these additional criteria, the final sample size was N = 201 (Table 1). Three newborns were excluded based on gestational age and twelve were excluded based on racial data.
Table 1.
Characteristics of the mothers and newborns
| African-American (N = 107) |
Caucasian (N = 94) |
|||
|---|---|---|---|---|
| Median | Range | Median | Range | |
| Maternal age (years) | 24 | 18 – 39 | 29 | 18 – 39 |
| Gestational age (weeks) | 39 | 36 – 41 | 39 | 36 – 41 |
| Female (%) | 46 | 53 | ||
| Parity | 2 | 1 – 5 | 2 | 1 – 5 |
Genome-wide measurement of DNA methylation levels
Bisulfite conversion of 750ng of genomic DNA from umbilical cord blood was performed using EZ DNA Methylation reagents (Zymo Research, Orange CA, USA). Samples were then processed according to the manufacturer’s specifications and hybridized and scanned on the Humanmethylation27 BeadChip (Illumina Inc., San Diego CA, USA) in batches of 24 samples using the Illumina BeadStation. The Humanmethylation27 BeadChip bears probes for 27,578 specific CpG dinucleotides assigned to 14,495 loci. The level of methylation of each CpG is represented by a beta value, which is calculated as the level of the fluorescence for the probe specific for 5-methylcytosine divided by the fluorescence from the probes for both the methylated and unmethylated C at that position. Consequently, the beta values span the bounded range 0 – 1. The Humanmethylation27 BeadChip includes bisulfite conversion control probes targeted at a non-CpG cytosine. The level of fluorescence from the unconverted C is an indication of the efficiency of bisulfite conversion. Across all samples, the intensity of signal from this probe was below 800 fluorescence units and exhibited a similar range within both races (200–800 units). By contrast, the intensity from the probe specific for the converted C was above 2,000 in all samples, with the majority of the signals ranging from 8,000 – 30,000 in both races. The uniformly low signal from the unconverted probe and the much higher signals from the probes for the converted C indicated that bisulfite conversion was highly effective in all samples. The range in signal from the probe for converted C suggests variation due to differences in DNA concentration or efficiency of whole genome amplification and/or incorporation of fluorescent probe, while the uniformly low signal for the probe for the unconverted C suggests that comparatively few C’s remained unconverted and their signal was independent of DNA concentration or amplification/labeling efficiency.
Statistical analyses
Raw output files from array hybridization experiments were processed using GenomeStudio software (Illumina Inc.). This software reports detection p values for each CpG interrogated by the array, which are an indication of the ability to distinguish the target sequence from background. Beta values representing the proportion of methylation at each site and the corresponding detection p values were imported into Microsoft SQL Server 2005, where filtering of the data values was performed before statistical analyses. As part of our quality assurance, for each newborn, probes with detection p values ≥ 10−3 were dropped from that individual. Additionally, one probe with a median detection p value > 10−6 across newborns was dropped from all individuals. Finally, due to the unique pattern of inheritance of the X chromosome and consistent differences between males and females and between the active and inactive copy of the X, all CpG's on the X chromosome were dropped from analyses. The final data set included data for 26,485 CpG's across all of the autosomes.
Statistical analyses were performed using R version 2.10.1 (R Foundation, Vienna, Austria) and Stata version 10 (Stata Corporation, College Station, TX, USA). Several variables were considered to be potentially associated with differences in DNA methylation levels across CpG dinucleotides. Because the DNA methylation values at many CpGs are not normally distributed across newborns, univariate nonparametric analyses were performed to examine the relation between levels of DNA methylation and each of those variables. Specifically, Spearman rank correlation was calculated for maternal age, gestational age, and parity, while Wilcoxon rank-sum analysis was performed for gender and race. For those variables that exhibited some genomewide significant associations with individual CpG methylation levels (Bonferroni corrected p = 1.89 × 10−6 based on 26,485 tests per variable), Wilcoxon rank-sum and chi-square analyses were performed to determine if there were significant differences in the distribution of each variable when stratified by a second variable (e.g., significant differences in the ages of mothers between the two races) that could produce false-positive associations in the univariate analyses due to confounding.
Following the univariate analyses of associations with DNA methylation, those variables that exhibited any (univariate) genomewide significant relations were studied further by means of stratified Kruskal-Wallis rank sum tests (Dalgaard, 2005). In essence, such a stratified test evaluates, e.g., the effect of gender in each of the race strata by a Kruskal-Wallis test and then combines the evidence against the null hypothesis (no effect) across the strata (van Elteren, 1960). Additionally, each of the three variables exhibiting a significant relation to DNA methylation levels in univariate analyses were simultaneously stratified by the two other variables and the overall significance determined by the above described method. Race and newborn gender were used to stratify analyses into two categories. Maternal age (in years) exhibits 22 unique levels (18 – 39), such that individual strata contain zero or very few observations when it is used directly to stratify individuals. Therefore, to avoid a prolific number of strata in relation to the number of cases tertiles of maternal age (18–24, N = 77; 25–29, N = 64; 30–39, N = 60) were used for stratification.
Association between CpG methylation levels and DNA sequence variation
We found a large number of CpG sites whose methylation significantly differed between the races. However, it has been demonstrated that methylation levels at individual CpG sites can be highly associated with both local (cis) and distant (trans) sequence variation (Gibbs and others, 2010; Zhang and others, 2010). Likewise, allele frequencies among single nucleotide polymorphisms (SNPs) can differ substantially among populations with different geographic ancestries (Altshuler and others, 2010). Therefore, it is possible that the racial differences in DNA methylation we observed could be due to differences in the frequencies of SNP alleles or haplotypes between the races that influence CpG methylation levels.
For 179 of the newborns, we have also collected genomewide SNP variation data using the Genome-Wide Human SNP Arrays 5.0 and 6.0 (Affymetrix, Santa Clara CA, USA). Samples were processed according to Affymetrix's protocol and genotypes were called using the BRLMM (5.0 array) and Birdseed (6.0 array) algorithms with default parameters within the Affymetrix Genotyping Console v4.0 application. As an exploratory analysis to determine if our racial differences can be ascribed primarily to SNP allele differences, we selected the top 100 CpGs (according to their p values) associated with race and performed an association analysis of their methylation levels with SNP variation. Using the DNA methylation data as the dependent variable, we included the SNP genotypes, maternal age, gender, and the loadings on the first two principal components calculated based on the genomewide SNP data (to partially adjust for population structure) in a multiple regression model. P values were calculated by comparing the full to reduced (without SNP data) model using the SNP and Variation Suite module of the GoldenHelix v7 software (Golden Helix Inc., Bozeman MT, USA). Attention was restricted to significantly associated SNPs located within 1Mb of the CpG, referred to as local, or cis, SNPs. Even though we restricted attention to SNPs within a ±1 Mb window around the race-associated CpGs, we used a genomewide Bonferroni-corrected p value (5.5 × 10−8) to declare significance based on the 903,861 SNP genotypes called from the Affymetrix 6.0 array. We chose this stringent p value to protect against false positive associations in this analysis of a comparatively small number of individuals. Given the limitations of the sample size, these analyses must be viewed as preliminary.
Identification of enriched pathways and gene ontological terms
The Database for Annotation, Visualization, and Integrated Discovery (DAVID v6.7, Dennis and others, 2003; Huang da and others, 2009) was used to identify molecular pathways and gene ontological terms enriched among the 3,297 gene loci (tagged by 3,623 CpGs) achieving genomewide significant association with race when stratified by maternal age and gender. The nonredundant list of genes was provided to the algorithm using Entrez gene ID numbers provided in the annotation of the Illumina array. For identification of enriched ontological terms, we screened the gene list versus the GOTERM_BP_FAT database, which is a custom list of terms designed to filter out the broadest terms so that more specific terms will emerge in the results (http://david.abcc.ncifcrf.gov/forum/cgi-bin/ikonboard.cgi?act=ST;f=3;t=1336). To identify enriched pathways, we searched the gene list versus the KEGG_PATHWAY database. Default settings were used, and an uncorrected p value threshold of 0.05 was used to declare significant enrichment.
Intersection of CpG probes in genes significantly associated with race and in those characteristic of blood cell types
We used buffy coats from whole blood for this study, and these are composed of a combination of different cell types (e.g., lymphocytes, granulocytes, monocytes). Each of these types of cells, as well as their further subtypes, likely has a unique pattern of DNA methylation across genes. As a consequence, if newborns of different races have consistent, even if slight, differences in the relative proportions of these blood cell types, this could produce patterns of DNA methylation that appear to be related to race, but are in fact due to differences in cell type composition. We have no direct data on the blood cell type composition of our newborns. However, Palmer and others (2006) identified genes whose expression levels are characteristic of B-cells, T-cell subtypes, granulocytes and lymphocytes. To indirectly address the possibility that the patterns we observe are due to race-related differences in blood cell composition, we used the data of Palmer and others (2006). Our analysis is based on the assumption that the differences in the levels of expression in the blood cell type signature genes are partially due to differences in DNA methylation levels in some of those genes. Using gene symbols, we identified the overlap between the signature genes reported by Palmer and others and the genes targeted by the Illumina methylation array. After eliminating probes (N = 33) in genes present on the X chromosome (no Y-linked probes were observed and we considered only autosomes in the analyses in this article), we identified the subset of probes in the blood cell type signature genes that were also significantly related to race. We then performed chi-square analysis to determine if probes in signature genes are disproportionately represented among the significant race-related probes. If the racial differences in methylation we observed are due primarily to racial differences in blood cell type composition at birth, we would expect to find an over-representation of probes in the cell type signature genes among those showing a significant difference in methylation levels between the races.
Results
Characteristics of the population
There were 107 African-American and 94 Caucasian newborns (Table 1). Gestational age (median and mean = 39 weeks) and parity (median = 2 for both races, x̄ = 2.1 for African-Americans, x̄ = 1.9 for Caucasians) were similar between the two races. The median age among African-American mothers was 5 years younger than among Caucasian mothers. Although maternal ages among Caucasians are well approximated by a normal distribution in our sample (Shapiro-Wilks test, p > 0.9), this is not true for the ages among African-American mothers (p = 0.002) where we observed a high number of younger mothers (39% African-American vs. 7% Caucasian younger than 23 years). This difference is statistically significant (Wilcoxon rank-sum, p < 0.0001) and indicates that maternal age is a potentially important confounder when investigating racial differences in DNA methylation. Although there is a larger proportion of females among Caucasian newborns in this data set, the difference relative to African-American newborns is not statistically significant (Chi-square, p = 0.3). Similarly, there is no statistically significant difference in the maternal age of male versus female newborns (Wilcoxon rank-sum, p > 0.5) in either race stratum. Therefore, newborn gender appears to have relatively low likelihood of being a confounder in analyses of maternal age or race.
Univariate associations with CpG methylation
None of the individual CpGs interrogated on the microarray exhibited a relation to gestational age in the range of 36–41 weeks or with parity at genomewide significance (Supplemental Table 1). Among all newborns, 61 autosomal CpGs were significantly associated with newborn gender (Table 2). When the two races were considered separately, 41 CpGs were associated with gender in Caucasians, while only 6 were associated in African-Americans. Across both races, 38 CpGs were correlated with maternal age, but this result appeared to be due entirely to trends in African-Americans, given that 57 were correlated in African-Americans and none in Caucasians. When the newborn genders were analyzed separately, the methylation levels of none of the CpGs were associated with maternal age. Finally, 4,235 CpGs exhibited significantly different levels of methylation between the two races, with twice as many males (1,808) as females (902) showing this trend when analyzed individually.
Table 2.
Univariate analyses of association with CpG methylation levels.
| Variable | Analysis | # genomewide significant |
|---|---|---|
| Gestational age | Spearman | 0 |
| Parity | Spearman | 0 |
| Newborn gender | Wilcoxon | 61 |
| African-American only | Wilcoxon | 6 |
| Caucasian only | Wilcoxon | 41 |
| Maternal age | Spearman | 38 |
| African-American only | Spearman | 57 |
| Caucasian only | Spearman | 0 |
| Male only | Spearman | 0 |
| Female only | Spearman | 0 |
| Race | Wilcoxon | 4,235 |
| Females only | Wilcoxon | 902 |
| Males only | Wilcoxon | 1,808 |
Stratified univariate associations with CpG methylation
From univariate analyses, maternal age, newborn gender and race emerged as significant predictors of methylation levels at a subset of CpG sites. However, we wished to evaluate the possibility that associations among these variables could produce spurious false positive results or that trends apparent within unique subsets of the newborns could be obscured in analyses that ignored these distinctions. For example, both race and maternal age exhibited significant associations with individual CpG methylation levels, but the significant difference in the ages of African-American and Caucasian mothers might result in mistakenly attributing a racial difference in DNA methylation levels to an effect related to maternal age. As a way to control for confounders, we therefore analyzed the relation of each of the three variables with CpG methylation levels when stratifying by one or both of the other variables (Table 3).
Table 3.
Tests of association with individual CpG methylation levels when stratifying by additional variables.
| Main variable | # of additional stratifying variables |
Stratification variables | # genomewide significant |
|---|---|---|---|
| Maternal age | 1 | Newborn gender | 104 |
| 1 | Race | 0 | |
| Newborn gender | 1 | Maternal age | 66 |
| 1 | Race | 75 | |
| Race | 1 | Maternal age | 3,692 |
| 1 | Newborn Gender | 4,211 | |
| Maternal age | 2 | Newborn gender & Race | 0 |
| Newborn gender | 2 | Maternal age & Race | 75 |
| Race | 2 | Maternal age & Newborn gender | 3,623 |
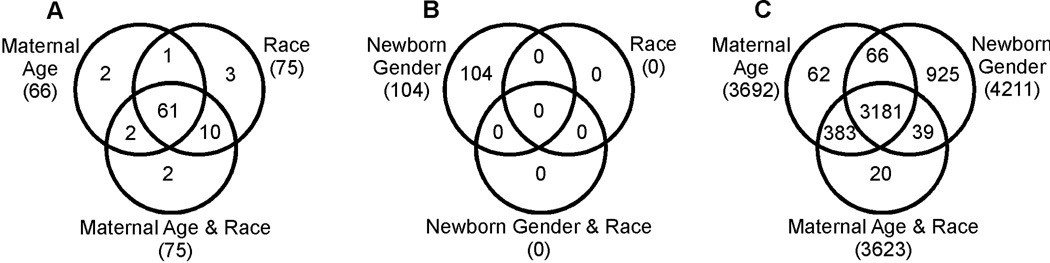
When stratifying by either maternal age or race, newborn gender was significantly associated with DNA methylation levels at 66 and 75 CpGs, respectively. When stratifying by both variables simultaneously, gender was associated with methylation levels of 75 CpGs. More importantly, 61 specific CpGs exhibited genomewide significant association of newborn gender with DNA methylation levels regardless of stratification by either one or both of the mentioned variables (Figure 1A). Therefore, it appears that newborn gender exhibits a fairly stable association with a small subset of CpGs.
Figure 1.
Venn diagrams summarizing the overlap in genomewide statistically significant CpG methylation differences when (A) newborn gender, (B) maternal age (tertiles), or (C) newborn race are further stratified by an additional one or two variables and analyzed by the Kruskal-Wallis rank sum test as described by van Elteren (1960).
When stratified by newborn gender, tertiles of maternal age are associated with methylation levels of 104 sites. Superficially, this appears in contrast to the Spearman correlation results when the genders were analyzed separately (Table 2) and no CpGs were identified for each gender group. In fact, if we apply the Kruskal-Wallis test to the gender strata separately we – in agreement with the Spearman correlation analysis results – fail to identify any significant associations of maternal age with DNA methylation levels (results not shown). Thus, the 104 significant test results might be a consequence of the increased power when simultaneously testing for association in both gender groups. However, the fact that these associations are not statistically significant when stratifying by either race or race and newborn gender jointly introduces the possibility that the statistical significance of the 104 identified CpGs might be driven by a confounding variable, such as race (Figure 1B).
In contrast to the analyses of maternal age stratified by newborn gender, when stratification was performed by race, no CpG met genomewide levels of significance, although 57 were significant when Spearman correlation was performed for African–Americans only. Even when computing the Kruskal-Wallis test for the subgroup of African-Armericans, no CpG met the criterion for significance. Recalling that Spearman’s rank correlation utilizes maternal age on its original scale, whereas the Kruskal-Wallis test is based on tertiles of maternal age leads us to the conjecture that this apparent difference in test results might be due to the loss of power generally associated with grouping of continuous variables since information is inevitably lost in the process of grouping. In any case, and somewhat speculatively, these results indicate that there might be a weak relation between maternal age and DNA methylation levels at a small subset of CpGs, but that any such relation is restricted to African-Americans. In fact, the correlation between the Spearman rho statistics calculated for the two races is weakly negative (rho = −0.047, p = 2.1 × 10−14 due to the presence of 26,485 observations), suggesting that the relation between maternal age and DNA methylation tends to be opposite in the two races. However, with the comparatively small number of individuals in the two races, the existence of statistically significant differences in trends with respect to maternal age in the two races remains speculative.
In comparison to newborn gender and maternal age, African-American versus Caucasian racial group exhibits a very strong relation to newborn umbilical cord DNA methylation levels. In the univariate analysis of race (Table 2), 4,235 CpGs were significantly related to CpG methylation. When stratified by gender in Kruskal-Wallis analysis (Table 3), a nearly identical number (4,211) were significant, suggesting that newborn gender is not a confounder in the analysis of racial effects and that the trends are similar in the two genders. When stratified by maternal age or by maternal age and newborn gender simultaneously, a slightly smaller number of sites (3,692 and 3,623, respectively) are significantly related to race, but importantly, most (3,181) CpGs remain statistically significant (Figure 1C). On the other hand, 925 CpGs only exhibit statistically significant associations of methylation levels with race if the analysis does not utilize stratification by maternal age or newborn gender. In the remainder of this paper, we will focus on the 3,623 CpG sites and their corresponding 3,297 genes that are significantly related to race after stratification by newborn gender and maternal age.
CpG-SNP associations
Of the 100 CpGs most significantly associated with race that we analyzed for association with nearby SNP variation, 7 achieved genomewide significance (p = 2.6 × 10−8 – 4.7 × 10−13; Supplementary Table 1, last column). This proportion of significant CpG – SNP associations (7%) is similar to the proportion of cis associations within 1 Mb observed among 153 human cerebellum samples (8.6%; Zhang and others, 2010) and slightly higher than that observed (4 – 5.2%) across four different sections of the brain in 150 individuals (Gibbs and others, 2010) based on the same Illumina array used in this study.
Enriched pathways and ontological terms
Pathways related to cancer (lung, pancreatic, prostate, bladder, and skin) and cellular signaling (calcium, hedgehog, cytokine and MAPK) formed two broad categories significantly enriched among the genes whose methylation levels differed between African-Americans and Caucasians (Table 4, Supplementary Table 2), while immunologic functions (antigen processing/presentation and NK cell mediated cytotoxicity) formed a minor category. Not surprisingly, given the over-representation of cancer-related pathways, ontological terms related to cellular proliferation (differentiation, proliferation, cell death, apoptosis, migration/motion, and tissue morphogenesis) were highly over-represented (Table 5, Supplementary Table 3). Additional broad categories included metabolic (phosphorous/phosphate, biosynthesis, nucleic acids, and nitrogen compounds) and endocrine (hormone response and gland development) processes.
Table 4.
KEGG pathways enriched among the loci with significant association between race and DNA methylation.
| Pathway term | Loci in Pathway |
P | Fold Enrichment |
|---|---|---|---|
| Pathways in cancer | 98 | 8.85×10−6 | 1.5 |
| Non-small cell lung cancer | 23 | 3.27×10−4 | 2.1 |
| Pancreatic cancer | 27 | 9.05×10−4 | 1.9 |
| Prostate cancer | 31 | 0.00139 | 1.8 |
| Bladder cancer | 18 | 0.00167 | 2.2 |
| Calcium signaling pathway | 51 | 0.00352 | 1.5 |
| Melanoma | 25 | 0.00387 | 1.8 |
| Hedgehog signaling pathway | 20 | 0.00922 | 1.8 |
| Cytokine-cytokine receptor interaction | 67 | 0.0181 | 1.3 |
| Small cell lung cancer | 26 | 0.0189 | 1.6 |
| Antigen processing and presentation | 25 | 0.0297 | 1.5 |
| Focal adhesion | 52 | 0.0308 | 1.3 |
| Base excision repair | 13 | 0.0322 | 1.9 |
| MAPK signaling pathway | 66 | 0.0377 | 1.2 |
| Natural killer cell mediated cytotoxicity | 36 | 0.0408 | 1.4 |
| Limonene and pinene degradation | 7 | 0.0423 | 2.5 |
Table 5.
Gene ontological terms enriched among loci with significant association between race and DNA methylation.*
| Ontological Term | Loci | P | Fold Enrichment |
|---|---|---|---|
| Cell motion | 131 | 2.72×10−7 | 1.5 |
| Positive regulation of developmental process | 85 | 5.02×10−7 | 1.7 |
| Positive regulation of cell differentiation | 73 | 6.27×10−7 | 1.8 |
| Regulation of cell proliferation | 196 | 9.12×10−7 | 1.4 |
| Negative regulation of cell proliferation | 102 | 2.03×10−6 | 1.6 |
| Regulation of cell death | 199 | 3.08×10−6 | 1.3 |
| Regulation of programmed cell death | 198 | 3.62×10−6 | 1.3 |
| Regulation of apoptosis | 196 | 4.13×10−6 | 1.3 |
| Negative regulation of cell death | 99 | 1.02×10−5 | 1.5 |
| Cell migration | 80 | 1.05×10−5 | 1.6 |
| Response to steroid hormone stimulus | 60 | 1.37×10−5 | 1.7 |
| Negative regulation of apoptosis | 97 | 1.48×10−5 | 1.5 |
| Gland development | 46 | 1.50×10−5 | 1.9 |
| Negative regulation of programmed cell death | 98 | 1.57×10−5 | 1.5 |
| Positive regulation of cell communication | 91 | 1.89×10−5 | 1.5 |
| Positive regulation of phosphorylation | 36 | 1.96×10−5 | 2.0 |
| Positive regulation of cellular biosynthetic process | 167 | 2.29×10−5 | 1.3 |
| Positive regulation of biosynthetic process | 169 | 2.37×10−5 | 1.3 |
| Positive regulation of macromolecule metabolic process | 202 | 2.82×10−5 | 1.3 |
| Positive regulation of nucleic acid metabolic process | 153 | 3.82×10−5 | 1.4 |
| Positive regulation of nitrogen compound metabolic process | 157 | 4.04×10−5 | 1.3 |
| Response to hormone stimulus | 98 | 4.06×10−5 | 1.5 |
| Positive regulation of phosphorus metabolic process | 36 | 4.11×10−5 | 2.0 |
| Positive regulation of phosphate metabolic process | 36 | 4.11×10−5 | 2.0 |
| Cell motility | 84 | 6.15×10−5 | 1.5 |
| Localization of cell | 84 | 6.15×10−5 | 1.5 |
| Positive regulation of locomotion | 35 | 6.45×10−5 | 2.0 |
| Tissue morphogenesis | 55 | 6.46×10−5 | 1.7 |
| Response to endogenous stimulus | 105 | 7.66×10−5 | 1.4 |
| Cell-cell signaling | 146 | 8.61×10−5 | 1.3 |
Only p values ≤ 10−4 shown. Supplementary Table 3 provides the complete output of DAVID.
Relation between race and newborn DNA methylation in blood cell type signature genes
Each type of somatic cell has a unique pattern of DNA methylation across genes that presumably plays a functional role in the regulation of genes required for the proper functioning of that cell. Buffy coat from whole blood is composed of a mixture of different cell types (i.e., granulocytes, monocytes, lymphocytes). Wu and others (2011) have demonstrated that whole blood buffy coat, mononuclear cells and granulocytes differ in their global levels of DNA methylation, with granulocytes being most distinctive. Similarly, Rakyan and other (2010) and Teschendorff and others (2010) have inferred that a subset of the genes whose DNA methylation appears to change with age are the consequence of shifts in the overall composition of white blood cells as one ages. If African-American and Caucasian newborns consistently differ in their relative proportions of individual blood cell types, then those two races would exhibit statistically significant differences in their levels of methylation at the subset of genes whose methylation levels differ among blood cell types. Additionally, the biological state of individual blood cells (e.g., activated versus nonactivated granulocytes) affects their gene expression, and presumably DNA methylation, profiles (Subrahmanyam and others, 2001; Zhang and others, 2004). If African-American and Caucasian newborns generally differ in the biological status of their white blood cells, such as baseline inflammation levels, this could produce differences in their levels of DNA methylation at a subset of loci.
Of 852 loci with official gene symbols whose expression levels are signatures of blood cell types (Palmer and others, 2006), we found 658 that were assayed by the Illumina Humanmethylation27 array. These 658 genes are represented by 1,273 CpG probes on the Illumina array, of which 33 were present on the X chromosome and were ignored, leaving a set of 1,240 probes (Supplementary Table 4). Of those 1,240 probes, 178 (14.4%) were among the 3,623 (out of 26,485 total probes, or 13.7%) significantly associated with race. Based on chi-square analysis, the difference in these proportions is not significant (chi-square, 1 d.f. = 0.43, p = 0.56). This indicates that DNA methylation probes in genes whose expression levels are signatures of blood cell types are not over-represented. In fact, probes in these cell type signature genes whose levels of methylation significantly differ between the races are found in approximately the proportion we would expect by chance.
Discussion
There is extensive evidence for progressive changes in DNA methylation with age postnatally (Boks and others, 2009; Christensen and others, 2009; Rakyan and others, 2010; Teschendorff and others, 2010), but it is unknown if these changes are at all reflected in the next generation, particularly given the nearly complete demethylation and remethylation of the maternal and paternal genomes after fertilization. If the age-related changes in DNA methylation observed in somatic tissues are reflected in the germline, this would be expected to result in shifts in the distribution of DNA methylation at individual CpGs in the newborns of older parents. The evidence for such an effect in this cohort is equivocal. Stratified Kruskal-Wallis analyses found no significant association with maternal age when stratification by race was performed. However, maternal age is a continuous variable and the stratified analyses divided the maternal ages into three broad strata and did not fully utilize the maternal age data. When treated as a continuous variable, maternal age was significantly associated with newborn DNA methylation at 57 CpGs only among African-Americans. The validity of this result needs to be verified in an independent multiethnic population.
In this cohort, there is evidence for differences between the genders in the levels of DNA methylation of up to 75 CpGs assigned to 69 autosomal loci. These 75 sites included 9 (C6orf68, TLE1, GLUD1, ALX4, DPPA3, NUPL1, FLJ20582, LRRC2, and FLJ43276) of the 11 autosomal sites found to significantly associate with gender in a previous study of saliva DNA in 197 individuals using the same Illumina array (Liu and others, 2010). It appears clear that a small minority of CpGs across the genome exhibit significant differences in methylation between the genders, and this should be taken into consideration in studies of the regulation and expression of these genes.
A surprisingly large proportion (13.7%) of autosomal CpGs analyzed in this study exhibited significant differences in their levels of methylation between African-Americans and Caucasians. Based on analyses of association with SNPs within 1 Mb, about 7% of these CpGs are associated with nearby SNPs, and it is possible that racial differences in the frequencies of these SNP alleles may explain some of the race-related DNA methylation differences. However, the proportion of CpGs associated with nearby SNPs in this racially mixed cohort is similar to that observed (4 – 8.6%) in two previous Caucasian cohorts (Gibbs and others, 2010; Zhang and others, 2010), suggesting that differences in SNP allele frequencies between populations of European and African ancestry are not the predominant cause of the DNA methylation differences we observed. At this time, the underlying reason for racial differences in DNA methylation patterns at birth is unclear, although differences in maternal or fetal metabolism of substrates in the one carbon pathway, transfer of one carbon sources across the placenta, genomic signals for methylation, or diet are possible explanations.
Among the CpGs that exhibit significantly different levels of methylation between African-Americans and Caucasians, the predominant pattern is for lower levels of methylation among African-Americans. Of the 3,623 CpGs reaching genomewide significance, 2,475 (68%) have average levels of methylation 1 – 27% lower among African-Americans, while the remainder exhibit either no (1%) or an increased (31%) level of methylation ranging from 1 to 21% (Supplementary Table 5). As summarized earlier, there have been few previous studies of differences in methylation levels among races in healthy tissue. However, our results suggest that slight racial differences exist at birth and are consistent with those of Terry and others (2008) who found lower global levels of DNA methylation among healthy middle-aged African-American women relative to Caucasians.
It is striking that cancers dominate the highly represented KEGG pathways (Table 4), although this can be partially attributed to the fact that the Illumina array is enriched for genes related to oncogenesis. Four (pancreatic, prostate, bladder, and melanoma) of the five types of cancers whose pathways were highly represented also exhibit substantial differences in their incidence rates between African-Americans and Caucasians (Edwards and others, 2005). A proportion of these differences in rates of incidence undoubtedly can be ascribed to differences in the frequencies of SNP and DNA repeat alleles (Ashktorab and others, 2003; Hunt and others, 2002; Pernick and others, 2003), behavioral/socioeconomic factors (Siahpush and others, 2010; Surgeon General, 1998), or skin pigmentation in the case of melanoma. However, as summarized in the introduction, distinct differences in levels of DNA methylation or extents of change in DNA methylation in cancer tissues have been observed between African-Americans and Caucasians among a subset of candidate genes involved in oncogenesis. The 98 genes identified in the top-ranked KEGG category, pathways in cancer, are represented by 116 CpGs with significantly different levels of methylation between the races. Similarly to the race-related CpGs as a whole, 88 (76%) have 1 – 9% lower levels of methylation among the African-American newborns, while the remainder have 1 – 9% lower levels among Caucasians, suggesting an overall trend towards lower levels of methylation among the African-Americans. Among the cancer-related genes, there were one to six CpGs per gene exhibiting significant differences between the races, including six (four hyper- and two hypomethylated among African-Americans) in the retinoblastoma 1 (RB1) gene. Although at age 15 there is no significant difference in the rate of retinoblastoma between African-Americans and Caucasians (Pendergrass and Davis, 1980), there does appear to be about a 2.5 fold higher rate of this cancer among African-American children up to age 3 (Jensen and Miller, 1971). Although our observations are based on a small sample size and there is not yet any follow-up data on cancer incidence, these patterns suggest the possibility that the epigenetic precursors to cancer exist at the time of birth.
In summary, we have found evidence of significant differences in levels of DNA methylation between African-American and Caucasian newborns at 13.7% of the autosomal CpG dinucleotides interrogated. At about 2% of autosomal CpGs there are also significant differences in DNA methylation levels between the genders. Although stringent Bonferroni-adjusted significance thresholds were employed and adjustment was made for possible confounders, there are additional possible factors that should be considered. This study was based upon results from a single population and a single method of measuring DNA methylation, and replication in another population and with an independent method of measuring methylation should be performed before these results are confidently accepted. Additionally, the underlying functional basis for the observed differences remains unknown. There is the possibility that consistent differences between the two races in umbilical cord blood cell type composition, maternal diet (e.g., availability of methyl donors, folate supplementation), or functional SNP allele frequencies could explain the majority of the differences. We do not have direct measurements of maternal nutrient composition in most of the mothers and have not tested for their influence. However, we do have indirect evidence that blood cell type composition and local SNP patterns appear to explain only a minority of the DNA methylation differences. We observed the same small number of SNP-DNA methylation associations as did previous studies, and CpG probes in the genes whose expression levels are signatures of blood cell types occur among our significant race-related results no more frequently than would be expected by chance.
Supplementary Material
Acknowledgements
We gratefully acknowledge the laboratory expertise of Jeanette Peeples and Joycelynn Butler, the analytical and data management skills of Priyanka Jani and Yanhua Qu, the participant recruitment and sample collection by CANDLE staff, and particularly the mothers who consented to participate.
Support: This work was funded by grants from the National Institutes of Health (R01-HD060713) and the University of Tennessee Health Science Center Clinical and Translational Science Institute to RMA and from The Urban Child Institute to FAT. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
References
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Gonzaga-Jauregui C, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashktorab H, Smoot DT, Carethers JM, Rahmanian M, Kittles R, Vosganian G, Doura M, Nidhiry E, Naab T, Momen B, Shakhani S, Giardiello FM. High incidence of microsatellite instability in colorectal cancer from African Americans. Clin Cancer Res. 2003;9(3):1112–1117. [PubMed] [Google Scholar]
- Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, Kahn RS, Ophoff RA. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One. 2009;4(8):e6767. doi: 10.1371/journal.pone.0006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, Sugarbaker DJ, Yeh RF, Wiencke JK, Kelsey KT. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard P. [R] Kruskal-Wallis stratified rank sum test. 2005 [Google Scholar]
- Das PM, Ramachandran K, Vanwert J, Ferdinand L, Gopisetty G, Reis IM, Singal R. Methylation mediated silencing of TMS1/ASC gene in prostate cancer. Mol Cancer. 2006;5:28. doi: 10.1186/1476-4598-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- Edwards BK, Brown ML, Wingo PA, Howe HL, Ward E, Ries LA, Schrag D, Jamison PM, Jemal A, Wu XC, Friedman C, Harlan L, Warren J, Anderson RN, Pickle LW. Annual report to the nation on the status of cancer, 1975–2002, featuring population-based trends in cancer treatment. J Natl Cancer Inst. 2005;97(19):1407–1427. doi: 10.1093/jnci/dji289. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Figueiredo JC, Grau MV, Wallace K, Levine AJ, Shen L, Hamdan R, Chen X, Bresalier RS, McKeown-Eyssen G, Haile RW, Baron JA, Issa JP. Global DNA hypomethylation (LINE-1) in the normal colon and lifestyle characteristics and dietary and genetic factors. Cancer Epidemiol Biomarkers Prev. 2009;18(4):1041–1049. doi: 10.1158/1055-9965.EPI-08-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, Arepalli S, Dillman A, Rafferty IP, Troncoso J, Johnson R, Zielke HR, Ferrucci L, Longo DL, Cookson MR, Singleton AB. Abundant quantitative trait Loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6(5):e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hunt JD, Strimas A, Martin JE, Eyer M, Haddican M, Luckett BG, Ruiz B, Axelrad TW, Backes WL, Fontham ET. Differences in KRAS mutation spectrum in lung cancer cases between African Americans and Caucasians after occupational or environmental exposure to known carcinogens. Cancer Epidemiol Biomarkers Prev. 2002;11(11):1405–1412. [PubMed] [Google Scholar]
- Jensen RD, Miller RW. Retinoblastoma: epidemiologic characteristics. N Engl J Med. 1971;285(6):307–311. doi: 10.1056/NEJM197108052850602. [DOI] [PubMed] [Google Scholar]
- Kwabi-Addo B, Wang S, Chung W, Jelinek J, Patierno SR, Wang BD, Andrawis R, Lee NH, Apprey V, Issa JP, Ittmann M. Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clin Cancer Res. 2010;16(14):3539–3547. doi: 10.1158/1078-0432.CCR-09-3342. [DOI] [PubMed] [Google Scholar]
- Liu J, Morgan M, Hutchison K, Calhoun VD. A study of the influence of sex on genome wide methylation. PLoS One. 2010;5(4):e10028. doi: 10.1371/journal.pone.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokarram P, Kumar K, Brim H, Naghibalhossaini F, Saberi-firoozi M, Nouraie M, Green R, Lee E, Smoot DT, Ashktorab H. Distinct high-profile methylated genes in colorectal cancer. PLoS One. 2009;4(9):e7012. doi: 10.1371/journal.pone.0007012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Diehn M, Alizadeh AA, Brown PO. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics. 2006;7:115. doi: 10.1186/1471-2164-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergrass TW, Davis S. Incidence of retinoblastoma in the United States. Arch Ophthalmol. 1980;98(7):1204–1210. doi: 10.1001/archopht.1980.01020040056003. [DOI] [PubMed] [Google Scholar]
- Pernick NL, Sarkar FH, Philip PA, Arlauskas P, Shields AF, Vaitkevicius VK, Dugan MC, Adsay NV. Clinicopathologic analysis of pancreatic adenocarcinoma in African Americans and Caucasians. Pancreas. 2003;26(1):28–32. doi: 10.1097/00006676-200301000-00006. [DOI] [PubMed] [Google Scholar]
- Piyathilake CJ, Henao O, Frost AR, Macaluso M, Bell WC, Johanning GL, Heimburger DC, Niveleau A, Grizzle WE. Race- and age-dependent alterations in global methylation of DNA in squamous cell carcinoma of the lung (United States) Cancer Causes Control. 2003;14(1):37–42. doi: 10.1023/a:1022573630082. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, Leslie RD, Deloukas P, Spector TD. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20(4):434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siahpush M, Singh GK, Jones PR, Timsina LR. Racial/ethnic and socioeconomic variations in duration of smoking: results from 2003, 2006 and 2007 Tobacco Use Supplement of the Current Population Survey. J Public Health (Oxf) 2010;32(2):210–218. doi: 10.1093/pubmed/fdp104. [DOI] [PubMed] [Google Scholar]
- Subrahmanyam YV, Yamaga S, Prashar Y, Lee HH, Hoe NP, Kluger Y, Gerstein M, Goguen JD, Newburger PE, Weissman SM. RNA expression patterns change dramatically in human neutrophils exposed to bacteria. Blood. 2001;97(8):2457–2468. doi: 10.1182/blood.v97.8.2457. [DOI] [PubMed] [Google Scholar]
- Surgeon General. Tobacco use among U.S. racial/ethnic minority groups--African Americans, American Indians and Alaska Natives, Asian Americans and Pacific Islanders, Hispanics. A Report of the Surgeon General. Executive summary. MMWR Recomm Rep. 1998;47(RR-18):v–xv. 1–16. [PubMed] [Google Scholar]
- Terry MB, Ferris JS, Pilsner R, Flom JD, Tehranifar P, Santella RM, Gamble MV, Susser E. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol Biomarkers Prev. 2008;17(9):2306–2310. doi: 10.1158/1055-9965.EPI-08-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, Savage DA, Mueller-Holzner E, Marth C, Kocjan G, Gayther SA, Jones A, Beck S, Wagner W, Laird PW, Jacobs IJ, Widschwendter M. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20(4):440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elteren PH. On the combination of independent two-sample tests of Wilcoxon. Bulletin of the International Statistical Institute. 1960;37:351–361. [Google Scholar]
- Woodson K, Hanson J, Tangrea J. A survey of gene-specific methylation in human prostate cancer among black and white men. Cancer Lett. 2004;205(2):181–188. doi: 10.1016/j.canlet.2003.11.027. [DOI] [PubMed] [Google Scholar]
- Wu HC, Delgado-Cruzata L, Flom JD, Kappil M, Ferris JS, Liao Y, Santella RM, Terry MB. Global methylation profiles in DNA from different blood cell types. Epigenetics. 2011;6(1) doi: 10.4161/epi.6.1.13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, Craig DW, Redman M, Gershon ES, Liu C. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86(3):411–419. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kluger Y, Nakayama Y, Poddar R, Whitney C, DeTora A, Weissman SM, Newburger PE. Gene expression in mature neutrophils: early responses to inflammatory stimuli. J Leukoc Biol. 2004;75(2):358–372. doi: 10.1189/jlb.0903412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.