Abstract
Significant progress in the molecular investigation of endogenous bioelectric signals during pattern formation in growing tissues have been enabled by recently-developed techniques. Ion flows and transmembrane gradients produced by ion channels and pumps are key regulators of cell proliferation, migration, and differentiation. Now, instructive roles for bioelectrical gradients in embryogenesis, regeneration, and neoplasm are being revealed through the use of fluorescent voltage reporters and functional experiments using well-characterized channel mutants. Transmembrane voltage gradients (Vmem) determine anatomical polarity and function as master regulators during appendage regeneration and embryonic left-right patterning. A state-of-the-art recent study reveals that they can also serve as prepatterns for gene expression domains during craniofacial patterning. Continued development of novel tools and better ways to think about physical controls of cell:cell interactions will lead to mastery of the morphogenetic information stored in physiological networks. This will enable fundamental advances in basic understanding of growth and form, as well as transformative biomedical applications in regenerative medicine.
Keywords: bioelectricity, ion channels, membrane voltage, prepattern
Introduction
Embryonic patterning, regenerative repair, and suppression of cancerous disorganization all require continuous signal exchange among cells, tissues, and organ systems within the body. Alongside well-known biochemical signals exists an important and fascinating system of bioelectrical communication. These signals are mediated by endogenous ion flows, electric fields, and voltage gradients that ultimately derive from the action of ion channels and pumps. Information-bearing biophysical changes are generated, and likely received, by all cells. They are distinct from those produced by excitable nerve and muscle (occurring on time scales much slower than the more familiar action potentials - minutes to days, not milliseconds), and involve receptor mechanisms different from those mediating effects of environmental electromagnetic field exposure. Comprehensive reviews have covered the progress made on the molecular mechanisms of electric field-based guidance of cell motility and orientation in the context of wound healing [1–3], and the fields produced by epithelia [4], implicating Rho, β-adrenergic receptors, and PI(3)Kγ-dependent pathways. Here, I focus on gradients of electric potential across plasma membranes of individual cells (Vmem) in vivo, as exciting recent development of new tools has resulted in new data revealing bioelectric potentials as an important regulator of patterning information during embryogenesis and regeneration, and as a tractable control point for biomedical intervention.
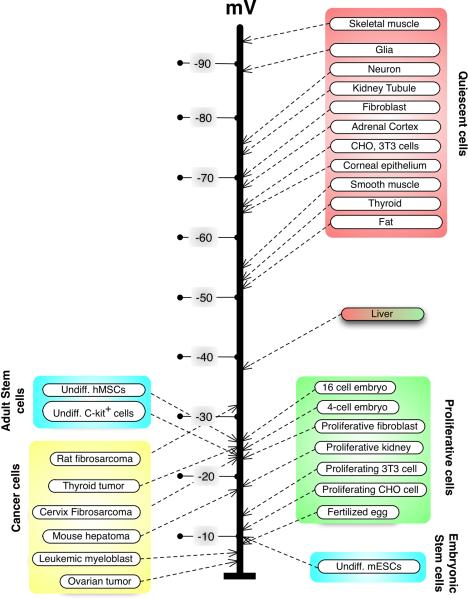
Classical studies using electrophysiology long ago demonstrated that endogenous electric fields and ion currents carry important instructive information guiding limb regeneration [5], tail development [6], cell migration and orientation through the embryo [7], oogenesis [8], and coordination of morphogenesis with histological differentiation [9] in amphibian, avian, and invertebrate model systems. Likewise, Vmem was suggested to be a key parameter mediating proliferation control, differentiation state, and neoplastic transformation [10, 11] in a wide range of cell types (Fig. 1). As molecular genetics exploded onto the scene, the continued impact of this body of work was limited by the difficulties inherent in studying a fundamentally biophysical property using methods and protocols best suited for biochemical and genetic processes. However, the recent development of state-of-the-art molecular tools has reinvigorated this field, uncovering new roles in patterning pathways. Most importantly, electric signals have now become mechanistically integrated with biochemical pathways, as studies reveal the molecular identities of the sources of ion flows in vivo as well as the downstream genetic effector cascades that ionic signaling initiates in the control of cell interactions [3, 12]. A recent study discovered a new pathway in craniofacial patterning, revealing how the activity of a proton pump produces gradients that regionalize gene expression and morphogenesis in Xenopus laevis embryos [13]. These elegant experiments are no doubt only a taste of what is to come, as the last few years have seen development of powerful approaches that reveal the importance of bioelectricity for understanding endogenous controls of growth and form, and as a potential target of biomedical interventions in regulation of cell behavior.
Figure 1.
Control of cell state by transmembrane potential. A sample of physiological measurements of various cell types (modified from [108]) reveals that quiescent, terminally differentiated cells tend to be strongly polarized, while more plastic cell types (stem cells, embryonic cells, and cancer cells) tend to be relatively depolarized. Interestingly, the liver's Vmem (abnormally low for an adult differentiated tissue) groups it with the morphogenetically labile cells, consistent with its remarkable regenerative potential. The relationship between Vmem and plasticity is a functional one; for example, mature neurons can be induced to re-enter the cell cycle by forced depolarization [109].
State-of-the-art tools for the characterization of bioelectrical signals
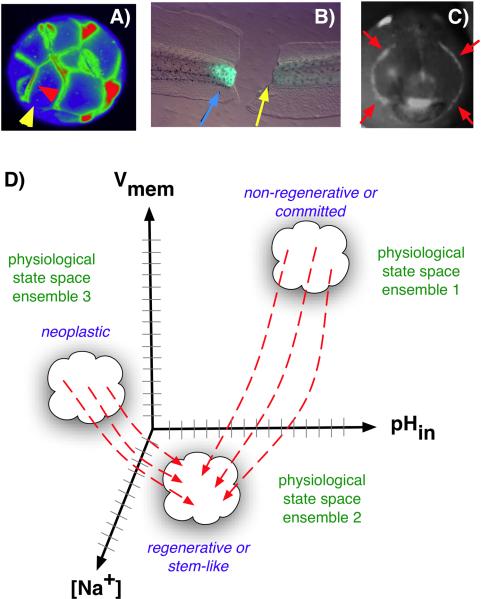
Molecular bioelectricity seeks to understand how ionic signaling participates in the orchestration of cell behaviors into the patterning needs of a host organism. Much data now indicate that specific physiological gradients underlie instructive signals in cell regulation. Thus, it is necessary to understand how patterning cues are encoded in biophysical events generated and perceived by cells and tissues during embryogenesis, regeneration, and tumor suppression. The first step to understanding the information carried by ion flows is the characterization of the spatio-temporal distributions of bioelectrical parameters in vivo and determination of how they correlate with anatomical and genetic patterning events. Much like the expression of signaling genes at key points of development (e.g. eye field specification, limb induction, etc.) reveals their involvement in driving patterning changes, correlations between alterations in ion flows/Vmem gradients are often the first entrypoint to the discovery of novel biophysically-controlled events. However, unlike mRNA and protein levels revealed by in situ hybridization and immunohistochemistry, physiological properties cannot be studied in fixed samples: reporters in the living state must be used (Fig. 2A–C). Fortunately, voltage gradients can now be visualized in 3D timelapse, using fluorescent reporters of transmembrane potential (see Box 1).
Figure 2.
Voltage gradients in vivo. A: Fluorescent voltage reporter dyes allow characterization of physiological gradients in vivo, such as this image of a 16-cell frog embryo that simultaneously reveals cells' potential levels (blue = hyperpolarized, red = depolarized) in vivo, as well as domains of distinct Vmem around a single blastomere's surface (compare the side indicated by the yellow arrowhead with the one indicated by the red arrowhead). B: Gradients of transmembrane potential demarcate important tissue domains, such as the depolarized region shortly after tail amputation in Xenopus laevis tadpoles (blue arrowhead), which will give rise to the regeneration bud, and can reveal non-regenerative conditions when the appropriate physiological state had not been achieved, or was experimentally blocked (yellow arrowhead). C: Isopotential cell fields can also demarcate subtle prepatterns existing in tissues, such as the hyperpolarized domains (red arrowheads) that presage the expression of regulatory genes such as Frizzled during frog embryo craniofacial development [13]. It is necessary to gain a quantitative understanding of the bioelectric code – to map out the linkage between physiological state with cell behavior outcomes, as a prelude to a full understanding of how 3-dimensional patterning information is stored in physiological properties of tissue. D: One hypothesis is that cell types (e.g. proliferative, or neoplastic, or undifferentiated) cluster in a multi-dimensional state space in which each axis defines the value of a physiological parameter. Additional axes (not shown) could include levels of other ions (chloride, potassium), nuclear membrane potential, cell surface charge (zeta potentials), etc. Once appropriate data are gathered, cells could be moved from their current state to a desired state by pharmacological and molecular-genetic changes shifting them along each axis, toward a different ensemble within the state space as needed for a given biomedical application.
In addition to transmembrane voltage potentials, individual ion species such as protons [14] and sodium [15] can readily be imaged in living tissues, and their flux detected non-invasively by extracellular self-referencing ion-selective probes [16, 17]. Physiological modeling of ion flows and their resulting voltage gradients (Fig. 3) feeds the computational integration of biophysical, genetic, and anatomical data. Knowledge of ion flows within patterning tissues reveals and explains the boundaries of physiological domains within tissues, identifies which ion concentration gradients actively contribute to voltage changes, and allows correlations of alterations in bioelectric properties with subsequent changes in cell behavior in situ. As with expression atlases for key regulatory genes, building a map of the physiological gradients is a fundamental aspect of comprehensive understanding of any complex patterning system. Thus, molecular-level detection of ion flows and their resulting voltage gradients, as well as computational integration of physiological and anatomical data (Fig. 2), reveal and explain the boundaries of physiological domains within tissues, identify which ion concentration gradients contribute to voltage changes, and allow correlations of alterations in bioelectric properties with subsequent changes in cell behavior in situ.
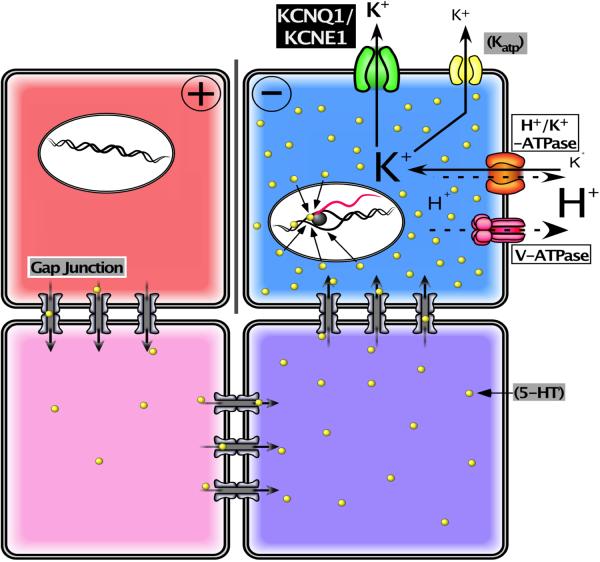
Figure 3.
A framework for modeling bioelectrical signaling. A comprehensive model synthesizing bioelectrical and genetic pathway elements must integrate physiological descriptions of the ion channels and pumps expressed in cells into a quantitative picture of the voltage gradients, their effects on movement of small signaling molecules through gap junctions and membrane transporters, and ultimately effects on second messenger pathways and transcriptional responses. Here is shown a representative system (motivated by current models of early left-right patterning of frog embryos; the schematic was modified after Fig. 7B of [110] drawn by Junji Morokuma). Differential expression and function of well-characterized channels and pumps (e.g. V-ATPase, KCNQ1, and Katp) form circuits that establish distinct Vmem levels in different cells; this process can be mathematically modeled (as has been done for kidney, lens, and inner ear tissues) based on the known physiological properties of the translocators involved. The resulting voltage gradients gate gap junction connectivity states between adjacent cells, as well as exert electromotive force that regulates the movement of small signaling molecules (e.g. 5HT serotonin). This movement can be simulated using particle-tracking or differential equations. The resulting morphogen gradients can activate transcriptional changes, setting up prepatterns of gene expression that mirror the earlier voltage gradients, regionalizing tissues and closing the link between bioelectric events and specification of anatomy.
Molecular-level techniques for functionally probing biophysical controls
Ascertaining instructive roles for bioelectric events however requires that changes in Vmem be inducible on demand in vivo, in a spatio-temporally controlled manner, to link these to cell- and tissue-level outcomes, as is routinely done in knockdown and overexpression experiments for biochemical signals. A variety of reagents and protocols (Box 1) are now available for perturbing Vmem in vivo. Loss-of-function experiments are done by inhibiting a specific channel or pump that underlies a given ion flow; for example, if a patch of cells is found to be more strongly polarized than their neighbors, pharmacological or genetic knockdown of the relevant ion translocator can be used to determine whether this regionalized Vmem state is functionally important for the cell group's subsequent developmental activities. How does one know which ion channels or pumps should be targeted to abolish an observed ion flux or voltage gradient? The endogenous targets for such experiments can be identified by inexpensive and rapid pharmacological screens (chemical genetics) [18]. The phenotypic outcomes of targeted inhibition of specific native ion translocator proteins shed light on endogenous roles of ion transport events within any given developmental or regenerative context. The converse - a gain-of-function experiment can be performed by misexpressing a well-characterized ion channel or pump in specific cells to drive a desired Vmem change (Box 1) and observe induced changes in cell behavior and large-scale patterning.
One example of the convergence of such techniques is illustrated by the discovery of a sparse but widely-distributed set of cells in the frog embryo with unique signaling properties that reveal a novel environmental factor that can confer a neoplastic-like phenotype upon stem cell derivatives [19]. An expression analysis revealed a widely-distributed, sparse population of cells expressing a glycine-gated chloride channel (GlyCl). By exposing embryos to the specific GlyCl channel activator ivermectin, and controlling the extracellular levels of chloride, the membrane potential of these specific cells could be set to any desired level (and confirmed with voltage-reporting fluorescent dyes). When depolarized, these “instructor cells” signal, at significant distance (non-cell-autonomously), to melanocytes - pigment cell derivatives of the neural crest [20]. The melanocytes then acquire three properties commonly associated with metastasis: they hyper-proliferate, change to a highly dendritic morphology, and invade tissues throughout the animal (such as blood vessels, gut, and neural tube) in a matrix metalloprotease-dependent manner. How can it be determined whether the signaling by these instructor cells is driven by voltage change per se, and not a mechanism specific to GlyCl protein or chloride levels?
The same phenotype was obtained with the native ligand of GlyCl (glycine), ruling out off-target effects of ivermectin. The phenotype could be turned on or off by modulating extracellular chloride levels, precisely as predicted by the Goldman Equation, ruling out ion-independent roles of GlyCl protein. Misexpression of mRNAs encoding sodium, potassium, or proton translocators could also phenocopy the highly specific change in melanocyte behavior induced by efflux of chloride, demonstrating that the effect is truly voltage-based and independent of any specific ion or channel protein. Moreover, the effect of GlyCl opening in the presence of low extracellular chloride could be rescued by pre-expressing a hyperpolarizing potassium channel in the exposed embryos. The downstream mechanisms of voltage control of melanocytes were found to involve regulation of serotonin efflux by depolarization in the instructor cells (which, interestingly, is also the mechanism linking voltage gradients to asymmetric gene expression during embryonic left-right patterning [21]). Together, these strategies offer a blueprint for dissecting the source and mechanism of a candidate bioelectrical mechanism. It is likely that screens targeting additional channels and pumps expressed by other, as yet unknown cell subpopulations, may reveal novel regulatory interactions, which can be likewise dissected using the available molecular physiological reagents.
Bioelectric signals control cell behavior
Large scale pattern derives from an orchestration of individual cell activities; what aspects of cell behaviors are controlled by bioelectric cues? Recent work has revealed mechanisms underlying growth cone pathfinding and cell orientation [22, 23] guided by endogenous electric fields [24–26]. A particularly thorough combination of biochemistry, transgenic mouse technology, and electric field perturbation [27] dissected the mechanisms of electrotaxis showing that mammalian wound healing requires cells to sense endogenous fields (generated by transepithelial potential) by a PTEN- and PI(3)K-γ-dependent pathway. Cell differentiation is also controlled by changes in Vmem, as has been shown in human mesenchymal stem cells [28], embryonic stem cells [29], myoblasts (in which the Kir2.1 channel plays a crucial role) [30, 31], the specification of neurotransmitter types [32], and the control of precursor differentiation [33–35] in the developing nervous system and heart. Given the known roles of Vmem in regulating migration, differentiation, and proliferation, it is not surprising that control of ion flux [36, 37] and membrane voltage [19, 20] are being increasingly implicated in cancer (Table 1a). Interestingly, electric cues can overcome competing biochemical signals; for example, depolarization trumps the induction of differentiation by insulin+dexamethasone in human mesenchymal stem cells [28], while physiological- strength electric fields override opposing chemical trophic factors, contact inhibition release, and population pressure [2].
Table 1a.
Ion transporters implicated in cancer
| Protein | Species | Reference |
|---|---|---|
| NaV1.5 sodium channel | Human | [93] |
| EAG-1 potassium channel | Human | [94] |
| KCNK9 potassium channel | Mouse | [95] |
| Ductin (proton V-ATPase component) | Mouse | [96] |
| SLC5A8 sodium/butyrate transporter | Human | [97] |
| KCNE2 potassium channel | Mouse | [98] |
Several ion transporters are now recognized as causal agents in carcinogenesis, consistent with the role of Vmem in regulating cell proliferation, migration, and differentiation. Future work remains to test the hypothesis that the patterning roles of voltage gradients are an important component of pattern disregulation as a fundamental cause of neoplasia [99, 100].
The differential activation of voltage-responsive transduction mechanisms on opposite sides of a cell allows bioelectric signals to regulate cell polarity. This was long ago suggested for the algae Fucus [38], and has been recently shown using high-resolution imaging and genetic techniques in yeast [39] and pollen tubes [40, 41]. This work establishes proof-of-principle for comprehensive quantitative profiling of extracellular flux and intracellular ion concentrations of protons and calcium in a genetically-tractable model system to understand the biophysics of anatomical polarity.
How do changes in membrane potential alter cell behavior?
A fundamentally physical event such as ion flow or potential change needs to be transduced into changes of gene expression. While rapid action potentials function through voltage-gated calcium channels or proteins such as CREB to shape nervous system development, several distinct mechanisms convert slow changes in resting Vmem levels into second-messenger cascades in non-excitable cells that ultimately drive transcriptional responses to biophysical cues (reviewed in detail in [42]). These include: electrophoretic pulling of small signaling molecules through gap junctional paths between cells, voltage-based regulation of membrane transporters of signaling molecules like calcium and various neurotransmitters, voltage-driven changes in conformational structure of integrin proteins, and electrophoretic separation or clustering of protein complex subunits within the plane of the cell membrane. One particularly interesting mechanism downstream of Vmem involves voltage-sensitive phosphatases that hydrolize phosphoinositides upon depolarization of membrane potential [43]. By allowing voltage changes to reversibly switch enzymatic activity of the tumor suppressor PTEN, these modular proteins illustrate another way in which electrical activity can be transduced into an important and well-studied biochemical signal.
Bioelectric cues are increasingly being found to be an important regulator of cell behavior, controlling cell number (proliferation and apoptosis), position (migration and orientation), and identity (differentiation trajectory). Recent confluence of genetic, biophysical, and molecular-physiological analysis has highlighted a continuous path from the upstream sources of ion flows, to the physiological gradients that are produced by cells, and ultimately to the machinery by which cells sense changes in their Vmem: powerful secondary messenger pathways establishing changes in gene expression in cells as a result of potential changes induced intrinsically or via their neighbors.
Gradients of transmembrane potential serve as instructive patterning cues
Perhaps the most interesting role for bioelectric signals, however, is during complex pattern regulation. One of the remarkable findings over the last 10 years has been that these bioelectrical cues are distinct from the metabolic gradients proposed by Child [44], since it is readily possible to dissociate the housekeeping functions of bioelectric gradients from more subtle, instructive roles. How can a fundamental cellular parameter be used to carry patterning information, when significant changes in Vmem might be expected to disrupt cell viability and metabolism? Much as engineers use modulation to transmit information on top of a carrier wave, Vmem gradients control tissue- and organ-level patterning in addition to providing baseline polarization needed for cell health. Indeed, it has now been repeatedly observed that artificial perturbation of Vmem usually results not in toxicity, death, or uninterpretable dysmorphias, but in specific, coherent changes of large-scale patterning [19, 45, 46].
Ion flows and the resulting Vmem changes are components of long-range conversations that orchestrate cellular activities during embryonic development, regeneration, and morphostatic tumor suppression. Bioelectric gradients mediate patterning decisions via a number of mechanisms. They can regulate the transport of diffusible signaling molecules in and out of cells (as occurs for the electrophoretic transport of maternal serotonin among early embryonic blastomeres during left-right patterning [21]), or cause the release of diffusible secondary messengers from specific cells [19]. Vmem changes in adjacent cells can propagate over long distances via conventional gap junctional paths [47] or through the more exotic nanotubes – narrow cytoplasmic structures with gap junction at their base that can conduct electrical signals between cells as a kind of nanowire [48]. These cell-autonomous and long-range signaling mechanisms allow bioelectrical gradients to carry patterning information in several different modes.
First, transmembrane potential levels can regulate differentiation of cells, and appears to specify tissue identity at the level of cell groups, such as the recent discovery that hyperpolarization to a specific Vmem range is not only responsible for regionalization of the native eye field (forming a feedback loop with Pax6 to define eye precursor cells) but is also able to induce complete ectopic eyes in locations far from the anterior neurectoderm [49]. Anterior neural tissues were previously thought to be the only ones capable of an eye fate in vertebrate embryos. These data underscore the importance of biophysical gradients in determining organ field boundaries and reveal that the fate restrictions defined by competence to respond to biochemical signals may not be absolute. Artificial control of Vmem level was able to turn many ventral and caudal tissues towards an eye fate; this does not occur with the canonical “master eye gene” Pax6, illustrating that inclusion of biophysical regulators might significantly expand our understanding of competency maps and the possible lineage relationships between different cell types.
Second, specific Vmem changes control large-scale axial (anatomical) polarity, in systems such as head-tail decisions for blastemas during planarian regeneration [45], left-right axis orientation in embryogenesis [50], and base-tip polarity in pollen tube outgrowth [40, 41]). Third, changes in voltage and ion content can provide master-regulator-like triggers (Fig. 2B) that initiate complete, highly orchestrated, self-limiting downstream patterning cascades in tissues. An example of the latter is the induction of complete tadpole tail regeneration – the construction of a complex appendage containing spinal cord, muscle, vasculature, and other tissues, kickstarted by a simple repolarization of the tissues under the wound epithelium [15, 46]. Lastly, Vmem patterns across underlying tissues carry positional information used to guide migratory cells [7] and directly establish molecular regionalization boundaries.
Bioelectrical pre-patterns underlie molecular and anatomical regionalization
Over 50 years ago it was observed that spatial patterns of bioelectric parameters (e.g. voltage difference between specific locations) quantitatively predicted anatomical features developing at much later timepoints, and thus might control morphogenesis as a kind of subtle scaffold or information-bearing prepattern [51, 52]. However, only recently has it become possible to probe the instructive nature of such physiological gradients with molecular resolution. A landmark paper [13] characterized, in real-time, the bioelectric properties of a highly dynamic morphogenetic event: the formation of the amphibian face. Using voltage-reporter dyes and timelapse microscopy, a non-invasive map was made of the many dynamic changes occurring at this time. A single frame of the movie† is shown in Figure 2C, revealing a rich regionalization of voltage gradient that demarcates interior of the neural tube, the future mouth, and thin bilateral crescents on the edge of the face (red arrowheads) that mark the position of the first pharyngeal pouch. Several of these bioelectrically-unique regions match the expression patterns of key genes that regulate differentiation and migration of tissues in the face. Using a combination of loss- and gain-of-function mutant constructs to perturb pH and transmembrane potential in the embryonic face in vivo, it was shown that these gradients are natively driven by differences in the activity of the V-ATPase proton pump; the resulting hyperpolarization occurs in three distinct waves and pharmacological experiments allowed a dissection of the timing of action of each component. Artificially perturbing the pattern of the voltage domains results in changes in the expression of important patterning genes such as Sox9, Slug, Pax8, Mitf, Frizzled3, and Otx2 and in subsequent characteristic defects in the morphology of craniofacial structures. This rich spatio-temporal profiling of native physiology, combined with detailed characterization of anatomical and molecular-genetic perturbation of the boundaries of the hyperpolarization domains, is a superb example of physiology serving as a subtle prepattern for regions of gene expression, much as transcriptional domains act as prepatterns for subsequent anatomy. Future efforts will involve developing methods for very precise spatio-temporal control of Vmem, using the derived model to repair craniofacial defects induced by genetic and mechanical means, as well as screening and subsequent functional characterization of the other embryonic voltage patterns observed but not dissected in this study.
Current gaps in knowledge
We are just beginning to scratch the surface of the bioelectric code - the mapping between voltage properties and patterning outcomes, akin to the genetic, epigenetic, and perhaps other codes remaining to be discovered. One testable, quantitative hypothesis (Fig. 2D) is that cells sharing important properties (e.g. proliferative, or neoplastic, or undifferentiated) cluster in a multi-dimensional state space in which each axis defines the value of a physiological parameter. Additional axes of such a state space could include levels of other ions (chloride, potassium), nuclear membrane potential, cell surface charge (zeta potentials), and other biophysical properties. If true, such a quantitative model would mean that cells could be moved toward a different ensemble within the state space by selecting an appropriate genetic or pharmacological reagent that will serve to move cells from their current state into a desired state. While largely focused on cell-autonomous responses (neglecting long-range and tissue-level control mechanisms), construction of such a model would go far towards predictive control of cell behavior by biophysical modulation.
However, a tremendous amount of physiomic profiling data must be gathered – large numbers of cells in various states, from different tissues and model species, need to be analyzed quantitatively. The maintenance and mining of such deep physiological datasets will require extensions of the databases and algorithms currently used to integrate genetic profiling and developmental atlases. Moreover, open questions remain about the exact nature of the key bioelectric properties that determine growth and form. Is the plasma membrane gradient the primary factor, or do intracellular organelles' gradients, such as the nuclear envelope potential [53, 54], play a role in regulating cell behavior during complex pattern formation? Decades of electrophysiology drove the notion of a single Vmem for a cell; however, this is an artifact of using a single electrode for measurement, and cells can contain huge numbers of domains as small as 2μm in their membranes [55, 56] with distinct voltage gradients. Is information for neighboring cells (and for intracellular processes) encoded and processed in this enormous manifold present on each cell's surface [57]? Molecular tools for individually manipulating such domains within single cells, and computational approaches (such as cellular automata theory [58]) to truly understand the complex dynamics of voltage gradients on the “grid” surface of cells will be crucial to understand the mechanisms supporting distinct voltages within a cell's membrane and testing their importance for the orchestration of cell behavior in vivo.
Order can be generated at the level of physiology, not protein/mRNA levels
A crucial shift is occurring in the field's thinking about how physiological properties underlie patterning, somewhat paralleling the lessons learned about redundancy and robustness in chemical networks. Bioelectrical states are an important source of non-genetic heterogeneity [59]. As with calcium fertilization waves or action potentials traveling down axons, spatially-patterned heterogeneities (of Vmem and gap junction-mediated connectivity) can be generated across a tissue of identical cells without changes in mRNA or protein levels. Cells expressing exactly the same voltage-regulated gap junctional proteins and ion channels/pumps can be in very different physiological state depending on which ion translocators are open or closed. This virtually guarantees that profiling at the mRNA or protein level gives a very incomplete picture of the information stored in collections of “pure” [60] populations of isolated cells or cell groups expressing the same genes.
Thus, profiling must include bioelectrical parameters. Acquiring the necessary physiomic data will be facilitated by the development of voltage reporters with robust cell membrane localization, better dynamic range, ratiometric properties, and a lack of effects on native Vmem [61, 62] together with quantitative calibration protocols. These datasets can be mined for new hypotheses about the relationships between biophysical properties and morphogenetic outcomes. Conversely, direct discovery of new roles for ionic signals will be augmented by future screens that overcome the problem of ion channel compensation and redundancy. In addition to traditional single gene knockouts, which rarely reveal Vmem roles due to the many channels and pumps that contribute to resting potential, screens involving knock-ins of dominant negatives, and the use of pharmacological screens [18, 63], should be applied to tractable model systems to identify new bioelectrical targets for patterning roles.
An exciting area for future discoveries involves the self-organizing capabilities of fields of cells that are able to generate and respond to transmembrane potential changes. The physics of ion flows and the post-translational gating of channels and pumps means that differences in cells' properties (spatio-temporal order) can arise at the level of physiology, from the interplay of gap junctions and ion channels that not only regulate ion flow but are themselves gated by voltage and pH. Positive and negative feedback loops (for example, implemented by voltage-sensitive potassium channels) can drive complex physiological dynamics via the amplification of very small initial state differences into stable cell states that differ across an anatomically and genetically homogenous cell sheet as cells regulate their own Vmem and spread this information to their neighbors. For example, self-generated (Turing-Child) wave-fronts and patterns of multicellular compartments (such as those driving somitogenesis and skin pigmentation patterns) can be formed from bioelectrical circuits, as they can from coupled chemical reaction-diffusion systems.
As cells' transmembrane potentials vary in space and time, driven by the regulation of ion translocators, cell groups will occupy stable or oscillating Vmem ranges – attractors in physiological state space. Such attractors necessarily store and reveal information about the cell's history and predicted future behavior. As in neuronal networks, systems of ion channels can implement memory or multi-state switches by allowing cells to be placed into two distinct Vmem values [64–66]. For the purpose of making targeted changes in cell behavior in biomedical contexts, and for the bioengineering of hybrid synthetic biology devices, it will be crucial to be able to predict and control the real-time behavior of electrical dynamics in cell sheets and 3D organs. Quantitative data on Vmem and the use of dynamical systems theory will be needed to fully understand the self-organizing properties of such networks. Interestingly, a successful precedent exists for understanding and exploiting systems in which information is encoded as persistent (standing) voltage gradients: the flip-flop circuit - a simple kind of computer memory, stores one bit of state information within a persistent voltage gradient, which can be reversibly altered (written) or measured (read) by external gradients applied to the circuit. Electrically coupled cells whose voltage is a function of dynamically-regulated channels can readily conduct dynamic, persistent ion flows in one of several discrete states, thus implementing a module that may allow non-excitable cells to support computational behavior normally associated with neural networks.
What is the right way to think about the genetic basis for bioelectrical signals?
The genes encoding ion channels are paramount, because they produce the gradients that determine downstream cell behaviors (and are thus an important foundation for mechanistically understanding the source of any bioelectrical event (Fig. 4)). At the same time, the specific genes are largely irrelevant when searching for the necessary and sufficient factor causing a specific phenotype. In traditional genetic pathways, a single growth factor can be associated with a particular change of cell phenotype – a mapping of gene to phenotype that is the cornerstone of many screening and pathway construction paradigms in developmental biology. In contrast, in bioelectrical signaling it is a physiological event (e.g. a depolarization into a particular Vmem range) that is causally responsible for a given patterning outcome. Whichever channel happens to drive the physiology in a tissue endogenously, it can be replaced (by evolution, or by an experimental manipulation) by a completely heterologous ion translocator that achieves similar regulation of Vmem. Thus, the information-bearing signal in many cases is not the genetic identify of a particular channel or pump, nor its sequence or structure. A cell's Vmem can be set at a particular level by many combinations of channels and pumps; conversely, a fixed set of expressed transporters can drive a cell to many different stable Vmem levels depending on the cell's real-time history of physiological inputs that post-translationally gate channel and pump activity. The main lesson revealed by recent data is that it is the voltage change (a fundamentally biophysical property), not a specific gene product (genetic element), nor even a specific ion concentration (chemical), that “codes for” or induces a specific response in cell behavior or large-scale patterning outcome.
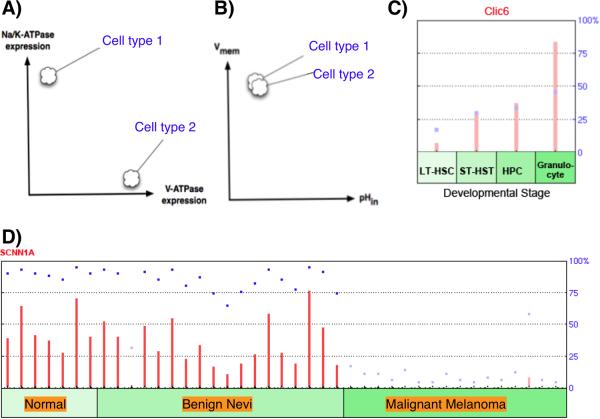
Figure 4.
Physiological profiling. A: Cells exhibiting very different expression profiles for ion transporter genes can indeed be in similar physiological states (B), based on overlapping functions for the transporters. Lack of 1:1 mapping between expression of channels/pumps and physiological state (due to compensation by other family members and post-translational gating) means that important information about cell behavior is not captured by molecular-genetic analysis without physiological profiling. Nevertheless, transcriptional data can suggest hypotheses for functional validation: the GEO database [111] can be mined for ion channel/pump profiles, such as the increased expression of the Clic6 chloride channel during HSC maturation (C), and the changes in the expression of sodium channels during progression towards melanoma in skin cells (D).
The high degree of compensation and redundancy is the bad news: single-channel knockouts are rarely informative about the true role of Vmem in patterning and its disorders, which is why the kinds of screens done to date have implicated only a few individual translocators in specific patterning phenotypes (Table 1a,b). Moreover, current profiling technologies do not capture physiological processes: any RNAi, morpholino, knockout, or microarray approach would have completely missed the earliest steps of left-right patterning, driven for 8 hours by maternal proteins and the physics of electrophoresis in frog embryos [67]. The good news however is that any convenient transporter that induces the desired changes of Vmem (by overcoming native cell conductances' activities) can be utilized during basic functional experiments or for regenerative interventions seeking to activate cell growth or pattern formation for biomedical applications [46].
Table 1b.
Genetic data identifying patterning roles for ion channels or gap junctions
| Protein | Morphogenetic role | Species | Reference |
|---|---|---|---|
| TMEM16A chloride channel | Tracheal morphogenesis | Mouse | [101] |
| Kir7.1 potassium channel | Melanosome development | Zebrafish | [102] |
| KCNH2 potassium channel | Cardiac morphology | Mouse | [103] |
| Cx41.8 gap junction | Pigmentation pattern | Zebrafish | [104] |
| Cx43 gap junction | Fin regeneration | Zebrafish | [105] |
| Cx43 gap junction | Fin size regulation | Zebrafish | [106] |
| Kir2.1 potassium channel | Craniofacial morphogenesis (Andersen-Tawil syndrome) | Mouse | [107] |
Although single gene mutation approaches are unlikely to reveal roles for Vmem(because of the high degree of compensation and redundancy among ion channel family members), a number of ion transport regulators have been identified in unbiased screens for morphogenetic mutants. A full appreciation of the involvement of bioelectric signaling in development will require screens in which Vmem is systematically altered in distinct cell types, among discrete ranges of voltage, for example by using tight physiological or optical control of genetically-misexpressed transporters as discussed above.
Conclusion
The most powerful future advances will fully integrate bioelectric signaling pathways with known genetic and biochemical cascades. Such progress will require an order-of-magnitude improvement in the sensitivity and spatial resolution of our tools for detection and control of bioelectrical properties in situ. The use of optogenetic lines of model species must be coupled with advances in physiological modeling, adapting the tools currently in use for understanding kidney and cornea [68] circuits to predictive control of developmental physiology. Increased quantitative understanding of how ion flows and physiological networks underlie changes in cell behavior will lead directly to intervention strategies, such as in vivo bioreactors [69] in which light stimuli and pharmacological cocktails [15, 70] can be delivered directly to damaged regions to provide exquisite spatio-temporal regulation of the bioelectric changes needed induce desired changes in growth and form.
A fundamental understanding of how the geometric shape of complex organs is stored in the real-time dynamics of electrically-coupled cells awaits the application of existing tools from cellular automata [58] and dynamical systems [71] theories to tractable in vivo models of stable patterning in ion flow dynamics among cell groups. The rewards will be truly vast; for example, such networks exhibit properties of hysteresis (memory) [64], suggesting that all cells (not just neurons) could support the information-processing capabilities of neural networks [72–74] and the tantalizing possibility that some of this computational capacity could be directly used to make decisions during regulative morphogenesis. This would have important implications for understanding native highly adaptive, flexible, and robust patterning processes, as well as capitalizing on electrically-mediated cellular computation for engineered tissues with novel regulatory functions. The ability to store and extract patterning information from living tissues will have transformative implications for advances in regenerative medicine, bioengineering, and synthetic biology.
Box 1.
Tools for dissecting bioelectrical signals
Voltage gradients can now be visualized continuously, in situ, using fluorescent reporters of transmembrane potential - a significant improvement on physiological impalement of single cells (far less invasive, and able to report multiple Vmem values within cell membrane domains). Reagents include cell-permeant dyes such as CC2-DMPE and DiSBAC2(3) [75–77] and genetically-encoded protein reporters [78, 79]. While many of these reagents have been optimized for rapid (neuronal) electrical signals [80], they are adaptable to the slower dynamics of patterning events because of their modular nature. A voltage-sensing domain long known to occur in voltage-sensitive ion channels was recently discovered in non-channel proteins such as Ci-VSP [81]. When its cytosolic phosphatase domain was replaced with a FRET pair, a versatile genetically-encoded voltage sensor was created that can be expressed under any promoter. Optimization produced FRET-based proteins with better plasma membrane targeting, red-shifted fluorescence (for better spectral separation from endogenous autofluorescence and better penetration of longer-wavelength light), e.g. VSFP2.3 [82]. In addition to transmembrane voltage potentials, individual ion species such as protons [14] and sodium [15] can readily be imaged in living tissues, and their flux detected noninvasively by extracellular self-referencing ion-selective probes [16, 17]. These quantitative data are analyzed by differential equations [83], particle tracking simulations [84], and novel object-oriented software tools for physiological modeling [85], to reveal the complex dynamic behavior and self-generated order of biophysical gradients.
A crucial component of characterizing bioelectrical properties is the ability to specifically perturb ionic signaling in patterning systems. Direct application of electric fields is a technique often used to study cell responses to physiological-strength electric signals in vitro; however, the complex impedance of living tissues makes it difficult to control when applied in vivo. Fortunately, a number of molecular-genetic loss- and gain-of-function strategies have become available for probing bioelectrical signals' roles in cell interactions. Highly targeted gain-of-function experiments, such as the misexpression of a K+ channel or P-type proton pump to hyperpolarize cells, can now be performed using well-characterized ion transporter proteins to induce known changes in ion content and transmembrane potential in specific cells; such plasmids, derived from the work of gut and kidney physiologists, form a rich toolkit for molecular investigations of bioelectricity. Misexpression of the P-type (single protein) proton pump was recently shown to be sufficient to induce regeneration of the tadpole tail when the native V-ATPase (a 13-subunit H+ pump complex, normally required for regenerative response) had been inhibited [46]. This illustrates an important principle: hyperpolarization produced by the pump activity was the crucial factor for initiating tail regeneration, not the specific structure or sequence of the gene product. While genetic perturbation has the highest potential for high-resolution information about bioelectric patterning control, the uncertainties of gene therapy require that pharmacological techniques also be developed for use in biomedical strategies [70]. One recent example is the design of a sodium ionophore cocktail, which induced full regeneration of the tadpole tail (a complex neuromuscular appendage including spinal cord) in a range of non-regenerative conditions after just one hour of exposure [15].
One highly promising technology for achieving tight spatio-temporal control of Vmem is the relatively young field of optogenetics [86–88]. By misexpressing optically-gated ion channels such as Channelrhodopsin, cells can be depolarized by exposure to blue light. Similarly, variants of Halorhodopsin can be expressed in cells to depolarize them upon exposure to yellow wavelengths. A large number of light-gated ion transporter mutants have now been characterized, to improve sensitivity, response times, and expression levels. Currently, these reagents are optimized for neurons, using very rapidly inactivating channels to allow experimental induction of action potentials and thus control whole animal behavior with light pulses at appropriate times. By optimizing channel and pump variants for expression in non-excitable cells, and extending the open times for the existing step-function opsins (channels that remain open for seconds, following a brief exposure to light stimulus), such proteins could be made to regulate Vmem in any cell of interest. Indeed, transgenic animals expressing optogenetic proteins have been made in tractable model systems such as zebrafish [89] and mouse [90], although they are largely restricted to neural promoters. Several labs are now working to create transgenic Xenopus laevis lines in which any cell or tissue of interest will express voltage reporter proteins or light-sensitive hyperpolarizing/depolarizing channels, allowing immediate access to bioelectrical experiments in cell, developmental, or regenerative biology. A promising new approach (an alternative to genetic misexpression of exogenous light-sensitive proteins) confers photoregulation upon existing (native) potassium channels [91, 92]. Such chemical strategies are ideal for probing endogenous channel roles, and for biomedical applications to bioelectrically control growth without the need for transgenesis. Together, biochemical, physiological, and genetic approaches are converging to provide a powerful set of methodologies for functionally probing the roles of ion flow in vivo.
Box 2.
Abbreviations + Definitions
Bioelectricity = long-term (lasting on the scale of minutes to days) ion flows, voltage gradients (transmembrane or trans-epithelial), and electric fields generated endogenously within living systems and mediating instructive information (serving as signals among cells and tissues) on top of their basic housekeeping functions.
Ion translocator = a channel, pump, co-transporter, or gap junction protein complex that allows (passively) or forces (using energy against a concentration gradient) the movement of charged molecules across a biological membrane.
Morphogenesis = the generation/unfolding of specific shapes, including not only the genetic networks that specify cell identity but all of the physical interactions required to build complex anatomical form.
Physiological Prepattern = spatial heterogeneity in Vmem across a cell field, with some groups of cells maintaining a different Vmem than adjacent cells. These are maintained by patterns of gap junctional connectivity and ion channel function (not necessarily expression), and can determine the expression domains of regulatory genes much as gene expression patterns (e.g. Hox genes) determine subsequent anatomical patterning.
Physiomics = comprehensive profiling of real-time physiological states of living cells, including high-resolution, quantitative state information on transmembrane potential (perhaps involving intracellular membranes as well as plasma membrane) and the content of major ion species within the cell.
Vmem = voltage gradient across the plasma membrane of cells, measured in milliVolts, typically −10 to −90 mV with inside negative with respect to outside. A bioelectric signal is carried by Vmem, not by a specific ion flux or by some other function of a given channel or pump, when the same downstream patterning response can be evoked by similar Vmem changes arrived at by completely different physiological means (e.g. a change of chloride or potassium flux).
Acknowledgements
This paper is dedicated to the late Lionel Jaffe, whose pioneering work on bioelectricity continues to inspire many researchers in this field. Thanks are due to Michael Romero, Richard Nuccitelli, Richard Borgens, Ken Robinson, Mustafa Djamgoz, Colin McCaig, and the members of the Levin lab for many useful discussions, to Junji Morokuma for the drawings in Fig. 3, and to Dany S. Adams, AiSun Tseng, and Laura N. Vandenberg for their helpful comments on the manuscript. This work was supported by NIH grant EY018168 and the G. Harold and Leila Y. Mathers Charitable Foundation.
Footnotes
References
- 1.Nuccitelli R. A role for endogenous electric fields in wound healing. Curr Top Dev Biol. 2003;58:1–26. doi: 10.1016/s0070-2153(03)58001-2. [DOI] [PubMed] [Google Scholar]
- 2.Zhao M. Electrical fields in wound healing-An overriding signal that directs cell migration. Semin Cell Dev Biol. 2009;20:674–82. doi: 10.1016/j.semcdb.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 3.McCaig CD, Song B, Rajnicek AM. Electrical dimensions in cell science. J Cell Sci. 2009;122:4267–76. doi: 10.1242/jcs.023564. [DOI] [PubMed] [Google Scholar]
- 4.Robinson K, Messerli M. Electric embryos: the embryonic epithelium as a generator of developmental information. In: McCaig C, editor. Nerve Growth and Guidance. Portland Press; Portland: 1996. pp. 131–41. [Google Scholar]
- 5.Jenkins LS, Duerstock BS, Borgens RB. Reduction of the current of injury leaving the amputation inhibits limb regeneration in the red spotted newt. Dev Biol. 1996;178:251–62. doi: 10.1006/dbio.1996.0216. [DOI] [PubMed] [Google Scholar]
- 6.Hotary KB, Robinson KR. Evidence of a role for endogenous electrical fields in chick embryo development. Development. 1992;114:985–96. doi: 10.1242/dev.114.4.985. [DOI] [PubMed] [Google Scholar]
- 7.Shi R, Borgens RB. Three-dimensional gradients of voltage during development of the nervous system as invisible coordinates for the establishment of embryonic pattern. Dev Dyn. 1995;202:101–14. doi: 10.1002/aja.1002020202. [DOI] [PubMed] [Google Scholar]
- 8.Woodruff R, Telfer W. Electrophoresis of proteins in intercellular bridges. Nature. 1980;286:84–6. doi: 10.1038/286084a0. [DOI] [PubMed] [Google Scholar]
- 9.Borgens RB, Shi R. Uncoupling histogenesis from morphogenesis in the vertebrate embryo by collapse of the transneural tube potential. Dev Dynam. 1995;203:456–67. doi: 10.1002/aja.1002030408. [DOI] [PubMed] [Google Scholar]
- 10.Blackiston DJ, McLaughlin KA, Levin M. Bioelectric controls of cell proliferation: ion channels, membrane voltage and the cell cycle. Cell Cycle. 2009;8:3519–28. doi: 10.4161/cc.8.21.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cone CD, Cone CM. Induction of mitosis in mature neurons in central nervous system by sustained depolarization. Science. 1976;192:155–8. doi: 10.1126/science.56781. [DOI] [PubMed] [Google Scholar]
- 12.Levin M. Bioelectric mechanisms in regeneration: Unique aspects and future perspectives. Semin Cell Dev Biol. 2009;20:543–56. doi: 10.1016/j.semcdb.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandenberg LN, Morrie RD, Adams DS. V-ATPase-dependent ectodermal voltage and pH regionalization are required for craniofacial morphogenesis. Dev Dyn. 2011;240:1889–904. doi: 10.1002/dvdy.22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tantama M, Hung YP, Yellen G. Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. J Am Chem Soc. 2011;133:10034–7. doi: 10.1021/ja202902d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tseng AS, Beane WS, Lemire JM, Masi A, et al. Induction of vertebrate regeneration by a transient sodium current. J Neurosci. 2010;30:13192–200. doi: 10.1523/JNEUROSCI.3315-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith PJS, Sanger RS, Messerli MA. Principles, Development and Applications of Self-Referencing Electrochemical Microelectrodes to the Determination of Fluxes at Cell Membranes. In: Michael AC, editor. Methods and New Frontiers in Neuroscience. CRC Press; 2006. [PubMed] [Google Scholar]
- 17.Reid B, Nuccitelli R, Zhao M. Non-invasive measurement of bioelectric currents with a vibrating probe. Nat Protoc. 2007;2:661–9. doi: 10.1038/nprot.2007.91. [DOI] [PubMed] [Google Scholar]
- 18.Adams DS, Levin M. Inverse drug screens: a rapid and inexpensive method for implicating molecular targets. Genesis. 2006;44:530–40. doi: 10.1002/dvg.20246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackiston D, Adams DS, Lemire JM, Lobikin M, et al. Transmembrane potential of GlyCl-expressing instructor cells induces a neoplastic-like conversion of melanocytes via a serotonergic pathway. Dis Model Mech. 2011;4:67–85. doi: 10.1242/dmm.005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morokuma J, Blackiston D, Adams DS, Seebohm G, et al. Modulation of potassium channel function confers a hyperproliferative invasive phenotype on embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:16608–13. doi: 10.1073/pnas.0808328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukumoto T, Kema IP, Levin M. Serotonin signaling is a very early step in patterning of the left-right axis in chick and frog embryos. Curr Biol. 2005;15:794–803. doi: 10.1016/j.cub.2005.03.044. [DOI] [PubMed] [Google Scholar]
- 22.Yao L, McCaig CD, Zhao M. Electrical signals polarize neuronal organelles, direct neuron migration, and orient cell division. Hippocampus. 2009;19:855–68. doi: 10.1002/hipo.20569. [DOI] [PubMed] [Google Scholar]
- 23.Pan L, Borgens RB. Perpendicular organization of sympathetic neurons within a required physiological voltage. Exp Neurol. 2010;222:161–4. doi: 10.1016/j.expneurol.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Nishiyama M, von Schimmelmann MJ, Togashi K, Findley WM, et al. Membrane potential shifts caused by diffusible guidance signals direct growth-cone turning. Nat Neurosci. 2008;11:762–71. doi: 10.1038/nn.2130. [DOI] [PubMed] [Google Scholar]
- 25.Ozkucur N, Perike S, Sharma P, Funk RH. Persistent directional cell migration requires ion transport proteins as direction sensors and membrane potential differences in order to maintain directedness. BMC Cell Biol. 2011;12:4. doi: 10.1186/1471-2121-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao L, Pu J, Zhao M. GSK-3beta is essential for physiological electric field-directed Golgi polarization and optimal electrotaxis. Cell Mol Life Sci. 2011;68:3081–93. doi: 10.1007/s00018-010-0608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao M, Song B, Pu J, Wada T, et al. Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-gamma and PTEN. Nature. 2006;442:457–60. doi: 10.1038/nature04925. [DOI] [PubMed] [Google Scholar]
- 28.Sundelacruz S, Levin M, Kaplan DL. Membrane potential controls adipogenic and osteogenic differentiation of mesenchymal stem cells. PLoS One. 2008;3:e3737. doi: 10.1371/journal.pone.0003737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng SY, Chin CH, Lau YT, Luo J, et al. Role of voltage-gated potassium channels in the fate determination of embryonic stem cells. J Cell Physiol. 2010;224:165–77. doi: 10.1002/jcp.22113. [DOI] [PubMed] [Google Scholar]
- 30.Hinard V, Belin D, Konig S, Bader CR, et al. Initiation of human myoblast differentiation via dephosphorylation of Kir2.1 K+ channels at tyrosine 242. Development. 2008;135:859–67. doi: 10.1242/dev.011387. [DOI] [PubMed] [Google Scholar]
- 31.Li F, Yin J, Yue T, Liu L, et al. The chloride intracellular channel 5 (CLIC5) involved in C2C12 myoblasts proliferation and differentiation. Cell Biol Int. 2010;34:379–84. doi: 10.1042/CBI20090334. [DOI] [PubMed] [Google Scholar]
- 32.Root CM, Velazquez-Ulloa NA, Monsalve GC, Minakova E, et al. Embryonically expressed GABA and glutamate drive electrical activity regulating neurotransmitter specification. J Neurosci. 2008;28:4777–84. doi: 10.1523/JNEUROSCI.4873-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasuda T, Adams DJ. Physiological roles of ion channels in adult neural stem cells and their progeny. J Neurochem. 2010;114:946–59. doi: 10.1111/j.1471-4159.2010.06822.x. [DOI] [PubMed] [Google Scholar]
- 34.Lange C, Prenninger S, Knuckles P, Taylor V, et al. The H(+) vacuolar ATPase maintains neural stem cells in the developing mouse cortex. Stem Cells Dev. 2011;20:843–50. doi: 10.1089/scd.2010.0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Vliet P, de Boer TP, van der Heyden MA, El Tamer MK, et al. Hyperpolarization induces differentiation in human cardiomyocyte progenitor cells. Stem Cell Rev. 2010;6:178–85. doi: 10.1007/s12015-010-9142-5. [DOI] [PubMed] [Google Scholar]
- 36.Park JY, Helm JF, Zheng W, Ly QP, et al. Silencing of the candidate tumor suppressor gene solute carrier family 5 member 8 (SLC5A8) in human pancreatic cancer. Pancreas. 2008;36:e32–9. doi: 10.1097/MPA.0b013e3181630ffe. [DOI] [PubMed] [Google Scholar]
- 37.House CD, Vaske CJ, Schwartz AM, Obias V, et al. Voltage-gated Na+ channel SCN5A is a key regulator of a gene transcriptional network that controls colon cancer invasion. Cancer Res. 2010;70:6957–67. doi: 10.1158/0008-5472.CAN-10-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaffe LF. Electrical currents through the developing fucus egg. Proc Natl Acad Sci USA. 1966;56:1102–9. doi: 10.1073/pnas.56.4.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minc N, Chang F. Electrical control of cell polarization in the fission yeast Schizosaccharomyces pombe. Curr Biol. 2010;20:710–6. doi: 10.1016/j.cub.2010.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michard E, Alves F, Feijo JA. The role of ion fluxes in polarized cell growth and morphogenesis: the pollen tube as an experimental paradigm. Int J Dev Biol. 2009;53:1609–22. doi: 10.1387/ijdb.072296em. [DOI] [PubMed] [Google Scholar]
- 41.Certal AC, Almeida RB, Carvalho LM, Wong E, et al. Exclusion of a proton ATPase from the apical membrane is associated with cell polarity and tip growth in Nicotiana tabacum pollen tubes. Plant Cell. 2008;20:614–34. doi: 10.1105/tpc.106.047423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levin M. Large-scale biophysics: ion flows and regeneration. Trends Cell Biol. 2007;17:262–71. doi: 10.1016/j.tcb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 43.Lacroix J, Halaszovich CR, Schreiber DN, Leitner MG, et al. Controlling the activity of a phosphatase and tensin homolog (PTEN) by membrane potential. J Biol Chem. 2011;286:17945–53. doi: 10.1074/jbc.M110.201749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Child CM. Patterns and problems of development. ix. The University of Chicago press; Chicago, Ill.: 1941. p. 811. [Google Scholar]
- 45.Beane WS, Morokuma J, Adams DS, Levin M. A chemical genetics approach reveals H,K-ATPase-mediated membrane voltage is required for planarian head regeneration. Chem Biol. 2011;18:77–89. doi: 10.1016/j.chembiol.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams DS, Masi A, Levin M. H+ pump-dependent changes in membrane voltage are an early mechanism necessary and sufficient to induce Xenopus tail regeneration. Development. 2007;134:1323–35. doi: 10.1242/dev.02812. [DOI] [PubMed] [Google Scholar]
- 47.Esser AT, Smith KC, Weaver JC, Levin M. Mathematical model of morphogen electrophoresis through gap junctions. Dev Dyn. 2006;235:2144–59. doi: 10.1002/dvdy.20870. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Veruki ML, Bukoreshtliev NV, Hartveit E, et al. Animal cells connected by nanotubes can be electrically coupled through interposed gap-junction channels. Proc Natl Acad Sci USA. 2010;107:17194–9. doi: 10.1073/pnas.1006785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pai VP, Aw S, Shomrat T, Lemire JM, et al. Transmembrane voltage potential conrols embryonic eye patterning in Xenopus laevis. Development. 2012 doi: 10.1242/dev.073759. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aw S, Levin M. Is left-right asymmetry a form of planar cell polarity? Development. 2009;136:355–66. doi: 10.1242/dev.015974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burr HS, Sinnott EW. Electrical correlates of form in cucurbit fruits. Am J Bot. 1944;31:249–53. [Google Scholar]
- 52.Burr HS, Hovland CI. Bio-electric correlates of development in amblystoma. Yale J Biol Med. 1937;9:540–9. [PMC free article] [PubMed] [Google Scholar]
- 53.Yamashita M. Fluctuations in nuclear envelope's potential mediate synchronization of early neural activity. Biochem Biophys Res Commun. 2011;406:107–11. doi: 10.1016/j.bbrc.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Tyner KM, Kopelman R, Philbert MA. “Nanosized voltmeter” enables cellular-wide electric field mapping. Biophys J. 2007;93:1163–74. doi: 10.1529/biophysj.106.092452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Connell KM, Rolig AS, Whitesell JD, Tamkun MM. Kv2.1 potassium channels are retained within dynamic cell surface microdomains that are defined by a perimeter fence. J Neurosci. 2006;26:9609–18. doi: 10.1523/JNEUROSCI.1825-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Connell KM, Tamkun MM. Targeting of voltage-gated potassium channel isoforms to distinct cell surface microdomains. J Cell Sci. 2005;118:2155–66. doi: 10.1242/jcs.02348. [DOI] [PubMed] [Google Scholar]
- 57.Wallace R. Neural membrane microdomains as computational systems: Toward molecular modeling in the study of neural disease. Biosystems. 2007;87:20–30. doi: 10.1016/j.biosystems.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 58.Bonchev D, Thomas S, Apte A, Kier LB. Cellular automata modelling of biomolecular networks dynamics. SAR QSAR Environ Res. 2010;21:77–102. doi: 10.1080/10629360903568580. [DOI] [PubMed] [Google Scholar]
- 59.Brock A, Chang H, Huang S. Non-genetic heterogeneity - a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet. 2009;10:336–42. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- 60.Yu K, Ruan DY, Ge SY. Three electrophysiological phenotypes of cultured human umbilical vein endothelial cells. Gen Physiol Biophys. 2002;21:315–26. [PubMed] [Google Scholar]
- 61.Akemann W, Lundby A, Mutoh H, Knopfel T. Effect of voltage sensitive fluorescent proteins on neuronal excitability. Biophys J. 2009;96:3959–76. doi: 10.1016/j.bpj.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mennerick S, Chisari M, Shu HJ, Taylor A, et al. Diverse voltage-sensitive dyes modulate GABAA receptor function. J Neurosci. 2010;30:2871–9. doi: 10.1523/JNEUROSCI.5607-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams DS, Levin M. Strategies and techniques for investigation of biophysical signals in patterning. In: Whitman M, Sater AK, editors. Analysis of Growth Factor Signaling in Embryos. Taylor and Francis Books; 2006. pp. 177–262. [Google Scholar]
- 64.Gallaher J, Bier M, van Heukelom JS. First order phase transition and hysteresis in a cell's maintenance of the membrane potential-An essential role for the inward potassium rectifiers. Biosystems. 2010;101:149–55. doi: 10.1016/j.biosystems.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 65.Sachdeva G, Kalyanasundaram K, Krishnan J, Chakravarthy VS. Bistable dynamics of cardiac cell models coupled by dynamic gap junctions linked to cardiac memory. Biol Cybern. 2010;102:109–21. doi: 10.1007/s00422-009-0352-3. [DOI] [PubMed] [Google Scholar]
- 66.Xiong W, Ferrell JE., Jr. A positive-feedback-based bistable `memory module' that governs a cell fate decision. Nature. 2003;426:460–5. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 67.Levin M. Is the early left-right axis like a plant, a kidney, or a neuron? The integration of physiological signals in embryonic asymmetry. Birth Defects Res C Embryo Today. 2006;78:191–223. doi: 10.1002/bdrc.20078. [DOI] [PubMed] [Google Scholar]
- 68.Fischbarg J, Diecke FP. A mathematical model of electrolyte and fluid transport across corneal endothelium. J Membr Biol. 2005;203:41–56. doi: 10.1007/s00232-004-0730-7. [DOI] [PubMed] [Google Scholar]
- 69.Hechavarria D, Dewilde A, Braunhut S, Levin M, et al. BioDome regenerative sleeve for biochemical and biophysical stimulation of tissue regeneration. Med Eng Phys. 2010;32:1065–73. doi: 10.1016/j.medengphy.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reid B, Graue-Hernandez EO, Mannis MJ, Zhao M. Modulating endogenous electric currents in human corneal wounds--a novel approach of bioelectric stimulation without electrodes. Cornea. 2011;30:338–43. doi: 10.1097/ICO.0b013e3181f7f2de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Skinner JE, Molnar M, Vybiral T, Mitra M. Application of chaos theory to biology and medicine. Integr Physiol Behav Sci. 1992;27:39–53. doi: 10.1007/BF02691091. [DOI] [PubMed] [Google Scholar]
- 72.Turner CH, Robling AG, Duncan RL, Burr DB. Do bone cells behave like a neuronal network? Calcif Tissue Int. 2002;70:435–42. doi: 10.1007/s00223-001-1024-z. [DOI] [PubMed] [Google Scholar]
- 73.Caporale N, Kolstad KD, Lee T, Tochitsky I, et al. LiGluR restores visual responses in rodent models of inherited blindness. Mol Ther. 2011;19:1212–9. doi: 10.1038/mt.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roberts A, Stirling CA. Properties and propagation of a cardiac-like impulse in skin of young tadpoles. Zeitschrift Fur Vergleichende Physiologie. 1971;71:295. [Google Scholar]
- 75.Adams DS, Robinson KR, Fukumoto T, Yuan S, et al. Early, H+-V-ATPase-dependent proton flux is necessary for consistent left-right patterning of non-mammalian vertebrates. Development. 2006;133:1657–71. doi: 10.1242/dev.02341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oviedo NJ, Nicolas CL, Adams DS, Levin M. Live imaging of planarian membrane potential using DiBAC4(3) CSH Protoc. 2008 doi: 10.1101/pdb.prot5055. 2008: pdb prot5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ozkucur N, Epperlein HH, Funk RH. Ion imaging during axolotl tail regeneration in vivo. Dev Dyn. 2010;239:2048–57. doi: 10.1002/dvdy.22323. [DOI] [PubMed] [Google Scholar]
- 78.Tsutsui H, Karasawa S, Okamura Y, Miyawaki A. Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat Methods. 2008;5:683–5. doi: 10.1038/nmeth.1235. [DOI] [PubMed] [Google Scholar]
- 79.Mutoh H, Perron A, Akemann W, Iwamoto Y, et al. Optogenetic monitoring of membrane potentials. Exp Physiol. 2011;96:13–8. doi: 10.1113/expphysiol.2010.053942. [DOI] [PubMed] [Google Scholar]
- 80.Akemann W, Mutoh H, Perron A, Rossier J, et al. Imaging brain electric signals with genetically targeted voltage-sensitive fluorescent proteins. Nat Methods. 2010;7:643–9. doi: 10.1038/nmeth.1479. [DOI] [PubMed] [Google Scholar]
- 81.Murata Y, Iwasaki H, Sasaki M, Inaba K, et al. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–43. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 82.Mutoh H, Perron A, Dimitrov D, Iwamoto Y, et al. Spectrally-resolved response properties of the three most advanced FRET based fluorescent protein voltage probes. PLoS One. 2009;4:e4555. doi: 10.1371/journal.pone.0004555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schiffmann Y. Turing-Child field underlies spatial periodicity in Drosophila and planarians. Prog Biophys Mol Biol. 2011;105:258–69. doi: 10.1016/j.pbiomolbio.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Y, Levin M. Particle tracking model of electrophoretic morphogen movement reveals stochastic dynamics of embryonic gradient. Dev Dyn. 2009;238:1923–35. doi: 10.1002/dvdy.22016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garny A, Nickerson DP, Cooper J, Weber dos Santos R, et al. CellML and associated tools and techniques. Philos Transact A Math Phys Eng Sci. 2008;366:3017–43. doi: 10.1098/rsta.2008.0094. [DOI] [PubMed] [Google Scholar]
- 86.Schultheis C, Liewald JF, Bamberg E, Nagel G, et al. Optogenetic long-term manipulation of behavior and animal development. PLoS One. 2011;6:e18766. doi: 10.1371/journal.pone.0018766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arrenberg AB, Stainier DY, Baier H, Huisken J. Optogenetic control of cardiac function. Science. 2010;330:971–4. doi: 10.1126/science.1195929. [DOI] [PubMed] [Google Scholar]
- 88.Wyart C, Del Bene F, Warp E, Scott EK, et al. Optogenetic dissection of a behavioural module in the vertebrate spinal cord. Nature. 2009;461:407–10. doi: 10.1038/nature08323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arrenberg AB, Del Bene F, Baier H. Optical control of zebrafish behavior with halorhodopsin. Proc Natl Acad Sci USA. 2009;106:17968–73. doi: 10.1073/pnas.0906252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tomita H, Sugano E, Fukazawa Y, Isago H, et al. Visual properties of transgenic rats harboring the channelrhodopsin-2 gene regulated by the thy-1.2 promoter. PLoS One. 2009;4:e7679. doi: 10.1371/journal.pone.0007679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fortin DL, Dunn TW, Kramer RH. Engineering light-regulated ion channels. Cold Spring Harb Protoc. 2011;2011:579–85. doi: 10.1101/pdb.top112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fortin DL, Banghart MR, Dunn TW, Borges K, et al. Photochemical control of endogenous ion channels and cellular excitability. Nat Methods. 2008;5:331–8. doi: 10.1038/nmeth.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Onkal R, Djamgoz MB. Molecular pharmacology of voltage-gated sodium channel expression in metastatic disease: clinical potential of neonatal Nav1.5 in breast cancer. Eur J Pharmacol. 2009;625:206–19. doi: 10.1016/j.ejphar.2009.08.040. [DOI] [PubMed] [Google Scholar]
- 94.Pardo LA, del Camino D, Sanchez A, Alves F, et al. Oncogenic potential of EAG K(+) channels. Embo J. 1999;18:5540–7. doi: 10.1093/emboj/18.20.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pei L, Wiser O, Slavin A, Mu D, et al. Oncogenic potential of TASK3 (Kcnk9) depends on K+ channel function. Proc Natl Acad Sci USA. 2003;100:7803–7. doi: 10.1073/pnas.1232448100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saito T, Schlegel R, Andresson T, Yuge L, et al. Induction of cell transformation by mutated 16K vacuolar H+-ATPase (ductin) is accompanied by down-regulation of gap junctional intercellular communication and translocation of connexin 43 in NIH3T3 cells. Oncogene. 1998;17:1673–80. doi: 10.1038/sj.onc.1202092. [DOI] [PubMed] [Google Scholar]
- 97.Gupta N, Martin PM, Prasad PD, Ganapathy V. SLC5A8 (SMCT1)-mediated transport of butyrate forms the basis for the tumor suppressive function of the transporter. Life Sci. 2006;78:2419–25. doi: 10.1016/j.lfs.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 98.Roepke TK, Purtell K, King EC, La Perle KM, et al. Targeted deletion of Kcne2 causes gastritis cystica profunda and gastric neoplasia. PLoS One. 2010;5:e11451. doi: 10.1371/journal.pone.0011451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dinicola S, D'Anselmi F, Pasqualato A, Proietti S, et al. A systems biology approach to cancer: fractals, attractors, and nonlinear dynamics. OMICS. 2011;15:93–104. doi: 10.1089/omi.2010.0091. [DOI] [PubMed] [Google Scholar]
- 100.Potter JD. Morphogens, morphostats, microarchitecture and malignancy. Nat Rev Cancer. 2007;7:464–74. doi: 10.1038/nrc2146. [DOI] [PubMed] [Google Scholar]
- 101.Rock JR, Futtner CR, Harfe BD. The transmembrane protein TMEM16A is required for normal development of the murine trachea. Dev Biol. 2008;321:141–9. doi: 10.1016/j.ydbio.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 102.Iwashita M, Watanabe M, Ishii M, Chen T, et al. Pigment pattern in jaguar/obelix zebrafish is caused by a Kir7.1 mutation: Implications for the regulation of melanosome movement. PLoS Genet. 2006;2:e197. doi: 10.1371/journal.pgen.0020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Teng GQ, Zhao X, Lees-Miller JP, Quinn FR, et al. Homozygous missense N629D hERG (KCNH2) potassium channel mutation causes developmental defects in the right ventricle and its outflow tract and embryonic lethality. Circ Res. 2008;103:1483–91. doi: 10.1161/CIRCRESAHA.108.177055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Watanabe M, Iwashita M, Ishii M, Kurachi Y, et al. Spot pattern of leopard Danio is caused by mutation in the zebrafish connexin41.8 gene. EMBO Rep. 2006;7:893–7. doi: 10.1038/sj.embor.7400757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoptak-Solga AD, Nielsen S, Jain I, Thummel R, et al. Connexin43 (GJA1) is required in the population of dividing cells during fin regeneration. Dev Biol. 2008;317:541–8. doi: 10.1016/j.ydbio.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Iovine MK, Higgins EP, Hindes A, Coblitz B, et al. Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev Biol. 2005;278:208–19. doi: 10.1016/j.ydbio.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 107.Bendahhou S, Donaldson MR, Plaster NM, Tristani-Firouzi M, et al. Defective potassium channel Kir2.1 trafficking underlies Andersen-Tawil syndrome. J Biol Chem. 2003;278:51779–85. doi: 10.1074/jbc.M310278200. [DOI] [PubMed] [Google Scholar]
- 108.Binggeli R, Weinstein R. Membrane potentials and sodium channels: hypotheses for growth regulation and cancer formation based on changes in sodium channels and gap junctions. J Theor Biol. 1986;123:377–401. doi: 10.1016/s0022-5193(86)80209-0. [DOI] [PubMed] [Google Scholar]
- 109.Stillwell EF, Cone CM, Cone CD. Stimulation of DNA synthesis in CNS neurones by sustained depolarisation. Nat New Biol. 1973;246:110–1. doi: 10.1038/newbio246110a0. [DOI] [PubMed] [Google Scholar]
- 110.Morokuma J, Blackiston D, Levin M. KCNQ1 and KCNE1 K+ channel components are involved in early left-right patterning in Xenopus laevis embryos. Cell Physiol Biochem. 2008;21:357–72. doi: 10.1159/000129628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Barrett T, Troup DB, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets - 10 years on. Nucleic Acids Res. 2011;39:D1005–10. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]