Abstract
Purpose
To assess the prognostic value of EGFR molecular characteristics of head and neck squamous cell carcinoma (HNSCC).
Patients and Methods
HNSCC tumors from patients prospectively enrolled in either an Early Detection Research Network (EDRN) study and treated with surgery without an EGFR-targeted agent (N=154) or enrolled in a chemoradiation trial involving the EGFR-targeted antibody cetuximab (N=39) were evaluated for EGFR gene amplification by fluorescence in situ hybridization (FISH) and EGFR protein by immunohistochemical (IHC) staining. Fresh-frozen tumors (EDRN) were also evaluated for EGFR protein and site-specific phosphorylation at Y992 and Y1068 using reverse-phase protein array (RPPA) (n=67). Tumor (n=50) EGFR and EGFRvIII mRNA levels were quantified using real-time PCR.
Results
EGFR expression by IHC was significantly higher in the EDRN tumors with EGFR gene amplification (P<0.001), and a similar trend was noted in the cetuximab-treated cohort. In the EDRN and cetuximab-treated cohorts elevated EGFR by IHC was associated with reduced survival (p=0.019 and p=0.06, respectively). Elevated expression of total EGFR and EGFR PY1068 were independently significantly associated with reduced progression-free survival in the EDRN cohort (HR=2.75; 95% CI=1.26–6.00 and HR=3.29; 95% CI=1.34–8.14, respectively).
Conclusions
In two independent HNSCC cohorts treated with or without cetuximab, tumor EGFR levels were indicative of survival. Tumor EGFR PY1068 levels provided prognostic information independent of total EGFR.
Keywords: epidermal growth factor receptor, receptor tyrosine kinase, site-specific phosphorylation, prognosis, head and neck cancer
Introduction
In the U.S. 36,540 oral cavity and pharynx cancers and 12,720 laryngeal cancers were diagnosed in 2010 according to population-based estimates (1). The primary challenge in the successful treatment of these cancers, more than 90% of which are squamous cell carcinoma (HNSCC), is the high rate of tumor recurrence. Most HNSCC cases are diagnosed as locoregionally advanced disease (stage III or stage IV) (1). Approximately 50% of HNSCC patients treated for a locoregionally advanced primary cancer will experience disease recurrence within 5 years and most will die as a result (1).
Standard of care for locally advanced HNSCC has included multi-modality treatment involving surgery with curative intent followed by radiation therapy (RT) or chemoradiotherapy (CRT) (2). Even with combined modality treatment, five-year survival rates for HNSCC have largely remained unchanged since the 1970s (1). Aggressive multimodality treatments are associated with significant morbidities including reduced swallowing and communication. These morbidities require consideration of the necessity for aggressive treatment. Efforts to define treatment-relevant subpopulations of HNSCC patients have been ongoing, and the status of the epidermal growth factor receptor (EGFR) in HNSCC tumors has been a primary focus.
EGFR was recognized as contributing to HNSCC development and progression as early as the 1990s. Consequently, molecular characterization of EGFR in tumors, especially protein expression and gene amplification, has been a focus of several studies intending to define prognostic markers for HNSCC (3–18). High levels of EGFR mRNA and protein have been found in 92% and 40–90% of HNSCC, respectively (9, 19–20). High EGFR tumor protein by quantitative immunohistochemistry (IHC) was associated with reduced patient survival (9), and high tumor levels of EGFR by IHC have generally been found to be associated with poorer prognosis (4, 6, 9, 17–18). Increased EGFR gene copy number has been reported to be associated with reduced progression-free survival (14–17). Tumor levels of phosphorylated EGFR (pEGFR) have been evaluated for prognostic import with conflicting findings (21–22). In general, the relationships between tumor EGFR amplification, EGFR gene and protein/phosphoprotein expression have not been clearly defined, and the combined prognostic value of these tumor characteristics has not been evaluated for HNSCC.
Recently, the FDA-approved, EGFR-targeted chimeric antibody therapeutic, Erbitux (cetuximab, C225; Imclone Systems and Bristol-Myers Squibb), has been included in regimens for HNSCC treatment following a phase III clinical trial demonstrating improved survival of cetuximab plus RT compared to RT alone (23). To date, in contrast to colorectal cancer for which cetuximab has been FDA-approved but KRAS mutations are contraindicated, no molecular characteristic has been identified to be significantly associated with HNSCC response or resistance to cetuximab treatment (24–25). EGFR activating mutations, which have been associated with tumor response to EGFR tyrosine kinase inhibitors in lung cancers, have not been reported in U.S. HNSCC populations (26–30).
The present study was carried out to define the relationship between EGFR gene amplification, gene expression and protein and phosphoprotein levels in prospectively collected tumor tissues from a cohort of HNSCC patients treated with surgery with curative intent. The overall goal was to define EGFR-specific prognostic molecular characteristics and to increase our understanding of the relationships between these molecular characteristics. We further sought to determine whether primary findings could be extrapolated to an independent patient population that had received cetuximab therapy.
Materials and Methods
Study Subjects and Tissue Samples
Surgical patients who were treated with curative intent for pathologically-confirmed HNSCC of the oral cavity, oropharynx, hypopharyx, or larynx were enrolled in this Early Detection Research Network- (EDRN-) sponsored study prior to surgery (n=154) (Table 1). Patients gave written informed consent, donated tissues for study and completed an administered questionnaire about tobacco use. Fresh-frozen tumors were available for a subset of subjects for reverse-phase protein array (RPPA) (n=67) and quantitative real-time PCR (QRT-PCR) (n=50) analyses (Table 1). EDRN tumors (n=58) and paired histologically normal mucosal tissues (n=30) were arrayed in triplicate in a previously described tissue microarray (TMA) (31). Tumors were prioritized for TMA inclusion based on overlap with available fresh-frozen tissues and adequate tissue for triplicate cores; TMA-arrayed tumors were representative of the EDRN cohort with regard to patient age, sex, smoking status and tumor site (Table 1). The majority of TMA-arrayed EDRN tumor samples were also evaluated by RPPA and QRT-PCR (n=40). Arrayed paraffin-embedded tissues from a previously described cohort treated on protocol with induction docetaxel, cisplatin and cetuximab followed by concurrent radiotherapy, cisplatin and cetuximab were available for EGFR IHC analysis (32). All tissues were collected under a tissue bank protocol approved by the University of Pittsburgh Institutional Review Board.
Table 1.
EDRN Cohort Subject and Disease Characteristics
| All Cases | TMA Cases | RPPA Cases | QPCR Cases | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n=154 | n=58 | P* | n=67 | P* | n=50 | P* | |||||
| Gender | |||||||||||
| Male | 110 | 71.4% | 41 | 70.7% | 0.88‡ | 48 | 71.6% | 0.96‡ | 36 | 72.0% | 0.91‡ |
| Female | 44 | 28.6% | 17 | 29.3% | 19 | 28.4% | 14 | 28.0% | |||
| Ethnicity | |||||||||||
| White | 148 | 96.1% | 57 | 98.3% | 0.41∥ | 65 | 97.0% | 0.69∥ | 51 | 98.1% | 0.66∥ |
| Non-white | 6 | 4.4% | 1 | 4.4% | 2 | 4.4% | 1 | 4.4% | |||
| Age, years | |||||||||||
| Median (Range) | 58 (23–89) | 61 (23–80) | 0.42§ | 58 (23–80) | 0.76§ | 60 (23–80) | 0.44§ | ||||
| Smoking Status | |||||||||||
| Never smoker | 24 | 15.6% | 6 | 10.3% | 0.20‡ | 9 | 13.4% | 0.79‡ | 6 | 12.0% | 0.65‡ |
| Former smoker | 50 | 32.5% | 23 | 39.7% | 23 | 34.3% | 16 | 32.0% | |||
| Active Smoker | 80 | 51.9% | 29 | 50.0% | 35 | 52.2% | 28 | 56.0% | |||
| Cigarette Pack-Years | |||||||||||
| Never smoker | 24 | 15.6% | 6 | 10.3% | 0.16‡ | 9 | 13.4% | 0.42‡ | 6 | 12.0% | 0.45‡ |
| 1–59 py | 57 | 37.0% | 18 | 31.0% | 21 | 31.3% | 17 | 34.0% | |||
| ≥60 py | 58 | 37.7% | 26 | 44.8% | 28 | 41.8% | 22 | 44.0% | |||
| Unknown | 15 | 9.7% | 8 | 13.8% | 9 | 13.4% | 5 | 10.0% | |||
| Alcohol Drinker | |||||||||||
| Never drinker | 40 | 26.0% | 16 | 27.6% | 0.72‡ | 18 | 26.9% | 0.82‡ | 14 | 28.0% | 0.69‡ |
| Ever drinker | 114 | 74.8% | 42 | 74.8% | 49 | 74.8% | 36 | 74.8% | |||
| Alcohol Quantity† | |||||||||||
| Never drinker | 40 | 26.0% | 16 | 27.6% | 0.85‡ | 18 | 26.9% | 0.43‡ | 14 | 28.0% | 0.08‡ |
| 1–4 | 48 | 31.2% | 16 | 27.6% | 18 | 26.9% | 9 | 18.0% | |||
| ≥5 | 46 | 29.9% | 19 | 32.8% | 24 | 35.8% | 20 | 40.0% | |||
| Unknown | 20 | 13.0% | 7 | 12.1% | 7 | 10.4% | 7 | 14.0% | |||
| Tumor Type | |||||||||||
| Neck Metastasis | 2 | 1.3% | 2 | 3.4% | .15∥ | 2 | 3.0% | .30∥ | 1 | 2.0% | .28∥ |
| New Primary | 11 | 7.1% | 2 | 3.4% | 3 | 4.5% | 1 | 2.0% | |||
| Primary | 128 | 83.1% | 50 | 86.2% | 57 | 85.1% | 43 | 86.0% | |||
| Recurrence | 13 | 8.4% | 4 | 6.9% | 5 | 7.5% | 5 | 10.0% | |||
| Tumor Site | |||||||||||
| Oral Cavity | 70 | 45.5% | 23 | 39.7% | .32∥ | 27 | 40.3% | .23∥ | 16 | 32.0% | .06∥ |
| Oropharynx | 29 | 18.8% | 12 | 20.7% | 12 | 17.9% | 12 | 24.0% | |||
| Hypopharynx | 11 | 7.1% | 2 | 3.4% | 3 | 4.5% | 2 | 4.0% | |||
| Larynx | 40 | 26.0% | 19 | 32.8% | 22 | 32.8% | 18 | 36.0% | |||
| Neck | 4 | 2.6% | 2 | 3.4% | 3 | 4.5% | 2 | 4.0% | |||
| Disease Stage | |||||||||||
| I | 26 | 16.9% | 6 | 10.3% | .09∥ | 6 | 9.0% | .049∥ | 5 | 10.0% | .45∥ |
| II | 21 | 13.6% | 11 | 19.0% | 13 | 19.4% | 7 | 14.0% | |||
| III | 30 | 19.5% | 7 | 12.1% | 10 | 14.9% | 8 | 16.0% | |||
| IV | 61 | 39.6% | 27 | 46.6% | 30 | 44.8% | 23 | 46.0% | |||
| Recurrence/Metastasis | 15 | 9.7% | 6 | 10.3% | 7 | 10.4% | 6 | 12.0% | |||
| Unknown | 1 | 0.6% | 1 | 1.7% | 1 | 1.5% | 1 | 2.0% | |||
| Treatment | |||||||||||
| RT Only | 41 | 26.8% | 19 | 32.8% | .43∥ | 20 | 29.0% | .73∥ | 18 | 36.0% | .16∥ |
| CRT | 27 | 17.6% | 10 | 17.2% | 11 | 15.9% | 9 | 18.0% | |||
| No CRT | 79 | 51.6% | 27 | 46.6% | 35 | 50.7% | 21 | 42.0% | |||
| Unknown | 6 | 3.9% | 2 | 3.4% | 3 | 4.3% | 2 | 4.0% | |||
| Vital Status | |||||||||||
| Alive | 79 | 51.3% | 26 | 44.8% | 0.15‡ | 30 | 44.8% | 0.12‡ | 20 | 40.0% | 0.04‡ |
| Dead | 73 | 47.4% | 32 | 55.2% | 37 | 55.2% | 30 | 60.0% | |||
| Unknown | 2 | 1.3% | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | |||
| Time to Death, months | |||||||||||
| Median (Range) | 44.6 (1.5–112.3) | 42.2 (1.5–112.3) | 0.44¶ | 42.5 (1.5–112.3) | 0.21¶ | 44.2 (1.5–112.3) | 0.21¶ | ||||
| Disease Progression Status | |||||||||||
| No disease progression | 64 | 41.6% | 22 | 37.9% | 26 | 38.8% | 16 | 32.0% | |||
| 0.41‡ | 0.46‡ | 0.08‡ | |||||||||
| Disease Progression | 88 | 57.1% | 36 | 62.1% | 41 | 61.2% | 34 | 68.0% | |||
| Unknown | 2 | 1.3% | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | |||
| Progression Free Survival, months | |||||||||||
| Median (Range) | 33.8 (1.2–108.4) | 26.4 (1.2–106.9) | 0.51¶ | 26.8 (1.2–106.9) | 0.40¶ | 24.8 (1.2–106.9) | 0.28¶ | ||||
Typical number of alcohol drinks in 2 week period
Chi square test
Rank sum test
Fisher's exact test
Log rank test
Analyzed versus unanalyzed cases
Immunohistochemical (IHC) staining and quantification
IHC staining of TMA sections, deparaffinized with successive ethanol and xylene baths, was performed for P16 (P16INK4 mAb G175–405, BD Pharmingen; 1:200) following antigen retrieval. EGFR staining was performed without antigen retrieval using clone H11 antibody (EGFR M3563, Dako; 1:500), which has been previously validated and employed (33–34). Signal amplification employed a proprietary antibody-conjugated micropolymer peroxidase (ImmPRESS™, Vector). Immunoreactive cells were visualized with diaminobenzidine (DAB) chromogenic substrate (5 minutes at room temperature). Sections were counterstained with hematoxylin and lithium carbonate for morphologic detail. For EGFR, only plasma membrane and cytoplasmic staining were scored, as little to no nuclear staining was observed; for P16, combined nuclear and cytoplasmic staining was scored. A composite score was derived from the product of the percentage of stained tumor to the nearest 5% and staining intensity (0 to 3 integer scale), and scores were averaged over replicate cores to derive a final IHC Score for each tumor.
Fluorescence In Situ Hybridization (FISH)
Dual color FISH analysis was performed using a Spectrum Green-labeled chromosome 7 centromeric probe (CEN7) and a Spectrum Orange-labeled EGFR probe (Abbott Molecular) for EDRN tumor samples incorporated into a tissue microarray (TMA). Nuclei were counterstained with DAPI/antifade (Abbott Molecular) At least 60 cells were scored for each core. EGFR FISH analysis of the cetuximab-treated cohort tumor tissues has been previously described (32). Gene amplification was defined as present if the ratio of EGFR to CEN7 probe signals was greater than 2.0 in at least one core. Tissues were determined to be hyperploid if one or more cores had at had four or more copies of EGFR and CEN7 in at least 50% of cells examined. Tissues were defined as having high EGFR copy number if one of the following was present in at least one core: EGFR gene amplification, hyperploidy, or 15 or more EGFR gene copies in at least 10% of cells.
Human papillomavirus (HPV) evaluation
EDRN HPV tumor status was assessed using an HPV pan-specific DNA probe (Dako, Wide Spectrum HPV DNA Probe Cocktail, Biotinylated), which recognizes HPV subtypes 6, 11, 16, 18, 31, 33, 35, 45, 51 and 52, and bright field in situ hybridization. HPV status for the cetuximab-treated cohort has been previously described (32).
Reverse-phase protein array (RPPA)
RPPA was used to quantify EGFR proteins and phosphoproteins in fresh frozen EDRN tumors. Fresh-frozen tumor tissues were not available for analysis for the cetuximab-treated cohort. Seven 2-fold dilutions of tumor protein lysate were spotted onto nitrocellulose-coated FAST slides. Antibodies for EGFR (SC-03, Santa Cruz Biotechnology Inc.; 1:1000), EGFR PY992 (#2235, Cell Signaling Technology; 1:100) and EGFR PY1068 (#2234, Cell Signaling; 1:100) and Catalyzed Signal Amplification (CSA) System (DakoCytomation) were used for detection. The dilution series of each of the protein/phosphoprotein samples were quantified using computerized optical densities with local background adjustment (MicroVigene Software), and relative protein/phosphoprotein levels of each diluted sample were fitted and interpolated in the R Supercurve Package (MDA Bioinformatics & Computational Biology Department, http://bioinformatics.mdanderson,org/software/supercurve) (35).
Immunoblotting
Fresh-frozen EDRN tumors were lysed in 1% TritonX, 10 mM Tris-HCl pH 7.6, 5 mM EDTA and 50 mM NaCl containing Complete protease inhibitors (Roche, Mannheim, Germany) and PhosSTOP phosphatase inhibitor (Roche). Proteins/phosphoproteins were quantified in 40 µg of tumor protein lysates using the Odyssey Infrared Imaging System (Li-Cor Biosciences) and either goat anti-rabbit IRDye 680 or goat anti-mouse IRDye 800CW secondary antibodies (Li Cor Biosciences). Primary antibodies recognized β-actin (#4967S), Src PY416 (#2101S), STAT3 PY705 (#9131S), STAT3 (#9132S), EGFR PY992 (#2235S) or EGFR PY1068 (#2234S) (Cell Signaling Technology), Src (B-12, Santa Cruz Biotechnology) or EGFR (#610017, BD Transduction Laboratories).
Measurement of EGFR and EGFRvIII mRNA levels using Quantitative RT-PCR
cDNA was synthesized from RNA isolated from fresh-frozen EDRN tumors using random hexamers and Superscript III First-Strand kit (Invitrogen). Taqman real-time PCR quantification of EGFR, EGFRvIII and beta-glucuronidase (β-Gus) gene expression was performed in duplicate using a 7700 Sequence Detector (Applied Biosystems Inc.) with an initial 12-minute 95°C denaturation followed by 40 cycles of 95°C denaturation and 60 seconds at 60°C. EGFR primers and probe were as follows: 5’-ATACGCGGCAGGACCAAG-3’ (forward primer), 5’-GGAGCGTAATCCCAAGGATGT-3’ (reverse primer) and 5’-CATGGTCAGTTTTCTCTTGCAGTCGTC-3’ (probe). EGFRvIII primers and probe were as follows: 5’-CTCTGGAGGAAAAGAAAGGTAA-3’ (forward primer), 5’-AGGCCCTTCGCACTTCTTAC-3’ (reverse primer), 5’-TGCGTCCGAGCCTGTGGG-3’ (probe). β-Gus primers are probe were as follows: 5’-CTCATTTGGAATTTTGCCGATT-3’ (forward primer), 5’-CCGAGTGAAGATCCCCTTTTTA-3’ (reverse primer) and 5’-TGAACAGTCACCGACGAGAGTGCTGG-3’ (probe). EGFR and EGFRvIII expression were measured relative to β-Gus using the comparative CT method. EGFRvIII was scored as present or absent, EGFR mRNA was scored on a continuous scale.
Statistical analysis
Statistical analyses were performed using STATA V9 (Statacorp), SPSS V14.0 (IBM) and Prism (Graphpad). Tests were two-sided and significance was defined as p<0.05. Correlations were assessed using Spearman’s rank correlation test and associations between categorical variables were tested using Fisher’s exact tests. The intraclass correlation coefficient (ICC) was used to assess EGFR IHC staining reproducibility. Progression-free survival (PFS) was defined as time from first treatment to first subsequent upper aerodigestive cancer, metastasis, or death. Overall survival (OS) was defined as time from first treatment to death. Each EGFR molecular marker was tested for association with PFS by log rank tests, and a log rank test of trend was employed when three ordered categories were compared. Power was estimated for log rank tests using the Freedman method (36). EGFR characteristics found to be associated with survival were tested in univariate Cox proportional hazards models and, if significantly associated, tested for association with survival in multivariable models adjusted for candidate prognostic variables associated with survival in this cohort (Wald p<0.05). Candidate prognostic variables included age, sex, tumor HPV status, AJCC disease stage, smoking status, pack-year category, cancer site and adjuvant treatment as defined in Table 1. The assumption of proportional hazards was tested by evaluation of scaled Schoenfeld residuals.
Results
EGFR protein by IHC but not by RPPA was higher in EDRN tumors with amplified EGFR
We previously reported that elevated tumor EGFR protein levels in EDRN TMA-arrayed tumors tended to be associated with reduced PFS (31). We assessed tumor EGFR gene amplification status in order to characterize relationships between tumor EGFR gene amplification and EGFR protein levels.
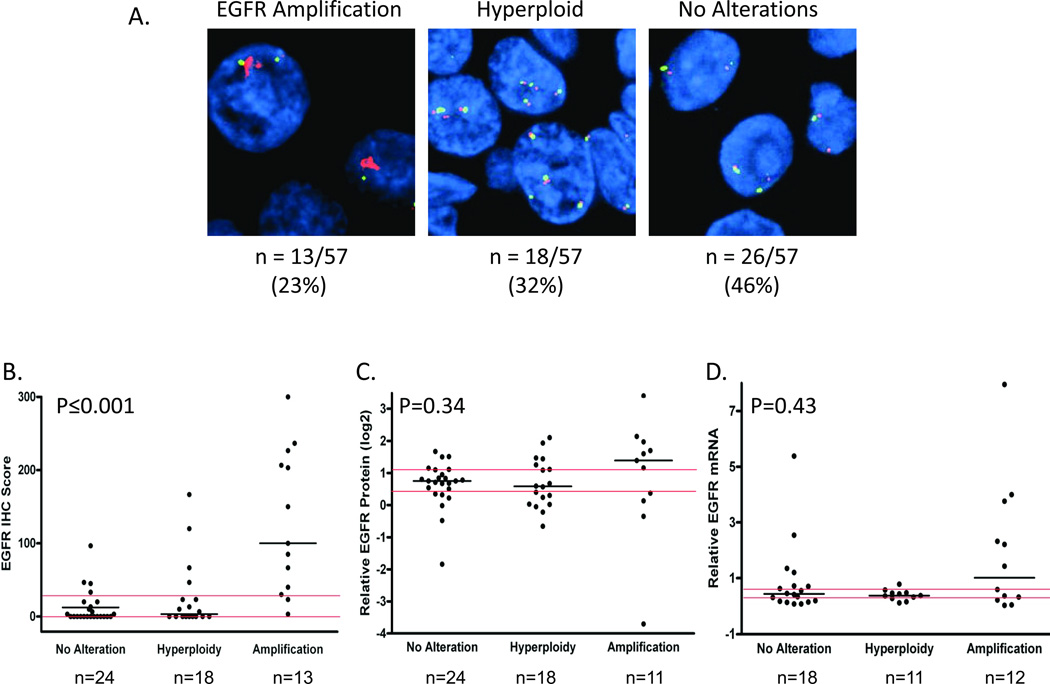
EGFR gene amplification status was categorized as follows: (1) EGFR gene amplification, (2) hyperploidy without EGFR gene amplification and (3) no hyperploidy or EGFR gene amplification. Numbers of tumors in each category are provided in Figure 1A. No EGFR gene amplification was detected in histologically normal adjacent tissues (n=30).
Figure 1. EGFR protein levels by IHC were higher in EDRN tumors with EGFR gene amplification.
(A) Representative EGFR (red) and chromosome 7 (green) FISH for tumors with EGFR gene amplification (left panel, n=13/57), hyperploidy without EGFR gene amplification (middle panel, n=18/57) and neither EGFR gene amplification nor hyperploidy (right panel, n=26/57). (B) EGFR protein levels determined by IHC staining categorized by tumor EGFR gene amplification and ploidy status. (C) EGFR protein levels determined by reverse phase protein array (RPPA) categorized by EGFR gene amplification and ploidy status. (D) Tumor EGFR mRNA level by EGFR gene amplification and ploidy status. Medians (black horizontal bars), overall tertiles (red horizontal bars) and p values (Kruskal-Wallis tests) are provided.
EGFR protein assessment by IHC demonstrated reliability with an ICC of 0.86 (95% CI=0.77–0.92) for triplicate tumor cores. EGFR protein levels by IHC were significantly higher in EDRN HNSCC tumors with EGFR gene amplification (Figure 1B). Of the 13 tumors with amplified EGFR, 10 tumors had high EGFR by IHC (IHC scores in the highest tertile: >30) and none had low levels (IHC scores equal to 0). Eighteen of 56 tumors with EGFR IHC scores had scores in the highest tertile. Of these 56% had EGFR amplification, 22% were hyperploid and 22% were neither hyperploid nor had amplified EGFR, indicating that processes in addition to EGFR amplification contributed to high tumor EGFR protein levels. Our finding of increased EGFR tumor protein by IHC with increasing EGFR copy number has been observed for lung cancer (37).
Because P16 and HPV have been identified as prognostic indicators for HNSCC, EDRN tumors incorporated into the TMA were evaluated for P16 and HPV status. Only 4 of the evaluated 48 EDRN tumors were found to be positive for HPV: 2 oropharyngeal, 1 laryngeal and 1 neck metastasis (Supplemental Table 1). All HPV-positive tumors had high P16 levels. Of the 4 HPV-positive tumors none had EGFR gene amplification nor exhibited hyperploidy and all had low EGFR protein levels by IHC (Supplemental Table 2).
EDRN tumor EGFR protein as measured by RPPA analysis of fresh-frozen tumors did not differ by EGFR amplification or tumor hyperploid status in TMA arrayed tumors (Figure 1C). To validate our RPPA results, 10 fresh-frozen EDRN tumors evaluated by RPPA were assayed by immunoblot for EGFR protein. We found that tumors with high EGFR protein levels by RPPA also had significantly higher EGFR protein levels by immunoblotting than those tumors with low EGFR by RPPA (Supplemental Figure 1A). We did not find a statistically significant correlation between EGFR IHC protein levels and EGFR protein by RPPA (ρ=0.16, p=0.26).
EDRN Tumor EGFR mRNA and EGFRvIII mRNA levels did not differ by EGFR gene amplification status
In order to determine whether EGFR mRNA levels were elevated in EDRN HNSCC tumors with EGFR gene amplification, we measured tumor EGFR mRNA levels by QRT-PCR. Tumor EGFR mRNA levels did not differ by EGFR amplification or hyperploid status (Figure 1D), and tumor EGFR protein levels by IHC and EGFR mRNA levels were not correlated (ρ=0.28, p=0.12). Similarly, there was no correlation between levels of tumor EGFR protein by RPPA and tumor EGFR mRNA (ρ=0.04; p=0.81). Thus EGFR protein levels are, at least in part, likely post translationally regulated.
Transcripts encoding EGFRvIII, a variant of EGFR that lacks exons 2 through 7 (38), were detected in 9 of 49 (18%) successfully evaluated tumors. Detection of EGFRvIII expression, which was confined to HPV-negative tumors (Supplemental Table 2), did not differ with tumor EGFR gene amplification or hyperploid status (p=0.69, Fisher’s exact test). EGFRvIII was detected only in tumors with intermediate or high EGFR mRNA levels (P=0.004, Fisher’s exact test).
Analyzed tumor subsets were representative of the EDRN cohort
Tissues selected for molecular analysis depended upon incorporation into a TMA or availability of fresh-frozen tissues. Of the 58 TMA tumors, 54 were analyzed by RPPA and 42 for EGFR mRNA levels by QRT-PCR. Overall, 40 tumor samples were analyzed using all methods. Patient and tumor characteristics of analyzed tissues did not differ from tissues not analyzed by sex, age, smoking/drinking histories, tumor type or tumor site (Table 1). We noted a decrease in PFS for TMA arrayed tumors that was not statistically significant and likely reflected the need for larger tumors in order to array sufficient material. Similarly, cases analyzed by RPPA differed from non-analyzed cases with respect to disease stage, likely because advanced stage tumors provided sufficient tissue for evaluation (Table 1).
EDRN Cohort exhibited typical characteristics
In order to compare our cohort to previously described surgical cohorts, we evaluated associations between overall survival (OS) and demographic factors, tobacco use, disease stage and tumor site. Increasing age by category (<55, 55–65, or >65 years) tended to be associated with reduced OS (p=0.09; log rank test (LRT)). The ratio of men to women in our cohort was typical and approximately 2.5:1, (5, 39), and sex was not associated with OS (p=0.77; LRT). Increasing number of cigarette pack-years (PY) and active smoking status at first treatment were significantly associated with reduced OS (p=0.002 and p=0.03, respectively; LRT). Patients with higher AJCC tumor stage had shorter OS (p=0.0004; LRT) as did patients with nodal disease (p=0.02; LRT). OS differed by adjuvant chemoradiotherapy (CRT) versus radiotherapy (RT) (p=0.001; LRT) with patients who received RT only in addition to surgery having shorter OS. Eighty percent of stage IV and 40% of stage III patients received adjuvant RT or CRT. Tumor site was not significantly associated with OS (p=0.69; LRT).
We did not observe statistically significant improved survival for patients with HPV-positive/ P16 high tumors (p=0.27; LRT; Supplemental Table 1), likely reflecting the low prevalence of HPV-positive tumors in our surgical cohort (8%). Median OS for patients with HPV-negative tumors was 45.1 months while the median survival time was not reached for patients with HPV-positive tumors. Of the 4 patients with HPV-positive tumors, one patient died 42.6 months after treatment. This patient was 1 of 2 HPV-positive patients who were active smokers at the time of first treatment.
Overall, our patient cohort was typical of a surgical HNSCC cohort with the usual associations of reduced survival with higher disease stage, presence of nodal disease and heavier tobacco use.
High tumor EGFR protein by IHC was associated with reduced survival in the EDRN cohort
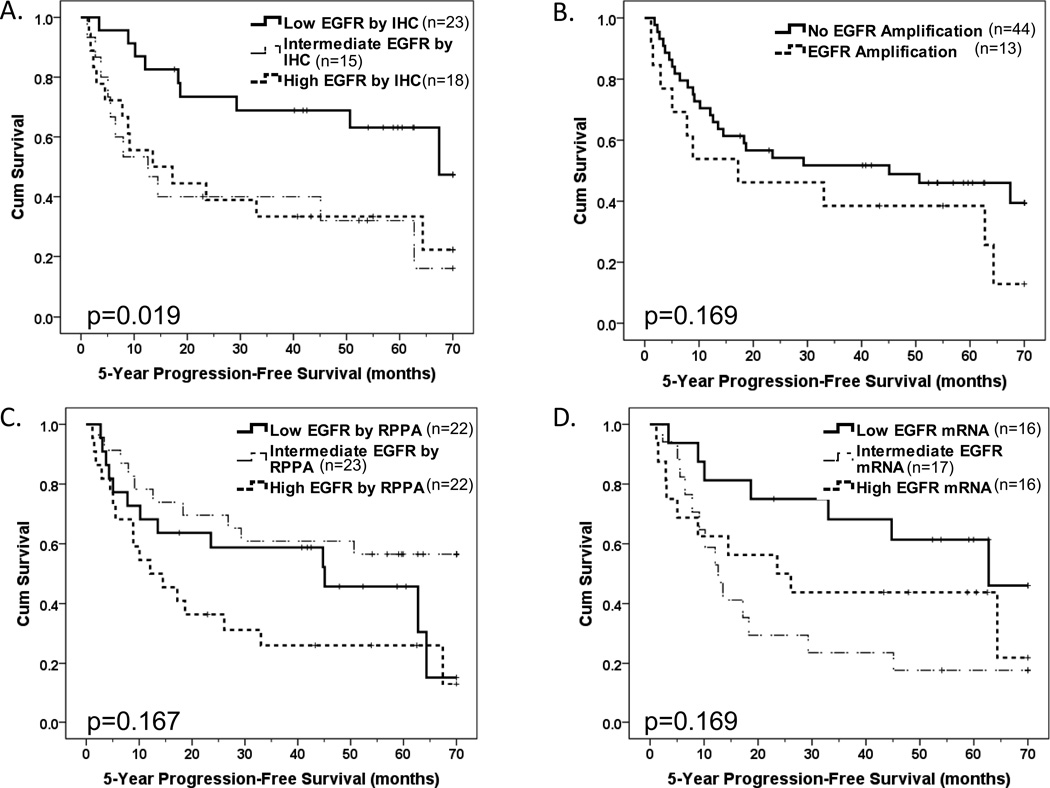
EGFR tumor protein levels by IHC were analyzed using tertiles to define high, intermediate and low EGFR expressing tumors in order to represent the EDRN tumor data in more detail (31). PFS decreased significantly with increasing tumor EGFR levels by IHC (Figure 2A). In our cohort, EGFR amplification was not a significant indicator of patient progression-free survival (PFS) (Figure 2B). High EGFR copy number, which included tumors with high polysomy and/or EGFR amplification and has been shown to be associated with reduced PFS in a similar analysis (15), tended to be associated with reduced PFS in the EDRN cohort but this did not reach statistical significance (p=0.09). We estimated our power to detect a difference in PFS by EGFR copy number to be 55%, 68% and 78% for hazards ratios of 1.8, 2.0 and 2.2, respectively, indicating that we had limited ability to detect differences of lower magnitude. In a multivariable Cox proportional hazards models adjusted for age and nodal stage, EGFR IHC levels (low versus intermediate/high), but not high EGFR copy number, was significantly independently associated with PFS (p=0.016 and p=0.685, respectively). Adjuvant therapy was found to not be statistically significantly associated with PFS (p=0.318) and, as a result, was not included in the final multivariable model. Therefore, EGFR high copy number did not provide prognostic information independent of EGFR IHC. Levels of EGFR protein by RPPA, EGFR mRNA and presence of EGFRvIII mRNA were also not significant predictors of PFS (Figures 2C, 2D, and p=0.68, respectively).
Figure 2. High EDRN tumor EGFR protein by IHC was associated with reduced progression-free survival.
Kaplan-Meier progression-free survival plots by (A) tumor EGFR level by IHC Score tertile, (B) EGFR gene amplification status (presence versus absence), (C) EGFR protein level by RPPA by tertile and (D) EGFR mRNA level by tertile. Log rank tests with associated p values compare presence versus absence (B) or trend across low, intermediate and high tertiles (A, C & D).
An independent cetuximab-treated cohort exhibited reduced survival with increased tumor EGFR protein levels by IHC
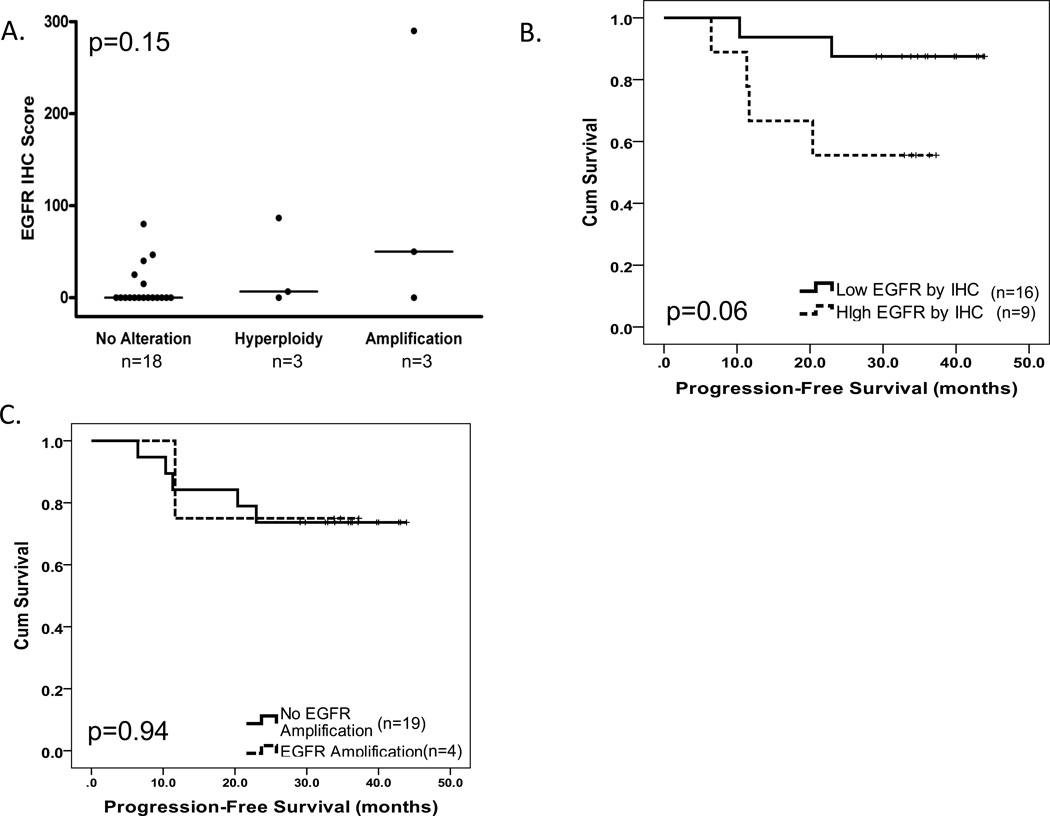
The cetuximab-treated cohort (n=39) was previously evaluated for tumor EGFR gene copy number (n=26) and HPV status (n=28); 27% had increased EGFR gene copy number defined as EGFR gene amplification or hyperploidy and 64% of tumors were HPV-positive (32). For this study, we evaluated EGFR tumor protein levels by IHC and EGFR gene amplification. Twenty-four tumors were evaluable for EGFR protein levels by IHC; large differences in EGFR levels across different tumors were observed (Supplemental Figure 2). All 24 tumors had EGFR gene amplification and HPV data available (32). We noted a distribution of EGFR IHC scores by EGFR gene amplification status that was similar to the EDRN cohort with a tendency towards increased EGFR tumor protein levels by IHC with EGFR gene amplification that did not reach statistical significance (Figure 3A). Neither EGFR protein levels nor presence of EGFR gene amplification differed by tumor HPV status (p=0.43 and p=0.19, respectively; rank sum test and Fisher’s exact test, respectively). Elevated EGFR tumor protein as defined by the median tended to be associated with reduced PFS (Figure 3B). EGFR gene amplification was not associated with PFS (Figure 3C). Neither EGFR gene copy number status nor tumor HPV status was associated with PFS in this cohort, as previously reported (32).
Figure 3. High tumor EGFR protein by IHC tended to be associated with reduced progression-free survival in the cetuximab-treated cohort.
(A) EGFR protein levels determined by IHC staining categorized by tumor EGFR gene amplification and ploidy status. Kruskal Wallis p value provided. (B) Kaplan-Meier progression-free survival plots by low versus high tumor EGFR protein by IHC level as defined by the median. (C) Kaplan-Meier plot of progression-free survival by tumor EGFR gene amplification status (presence versus absence). Log rank test-associated p values are provided for B and C.
Site-specific phosphorylation at EGFR PY1068 but not PY992 was associated with reduced survival in the EDRN cohort
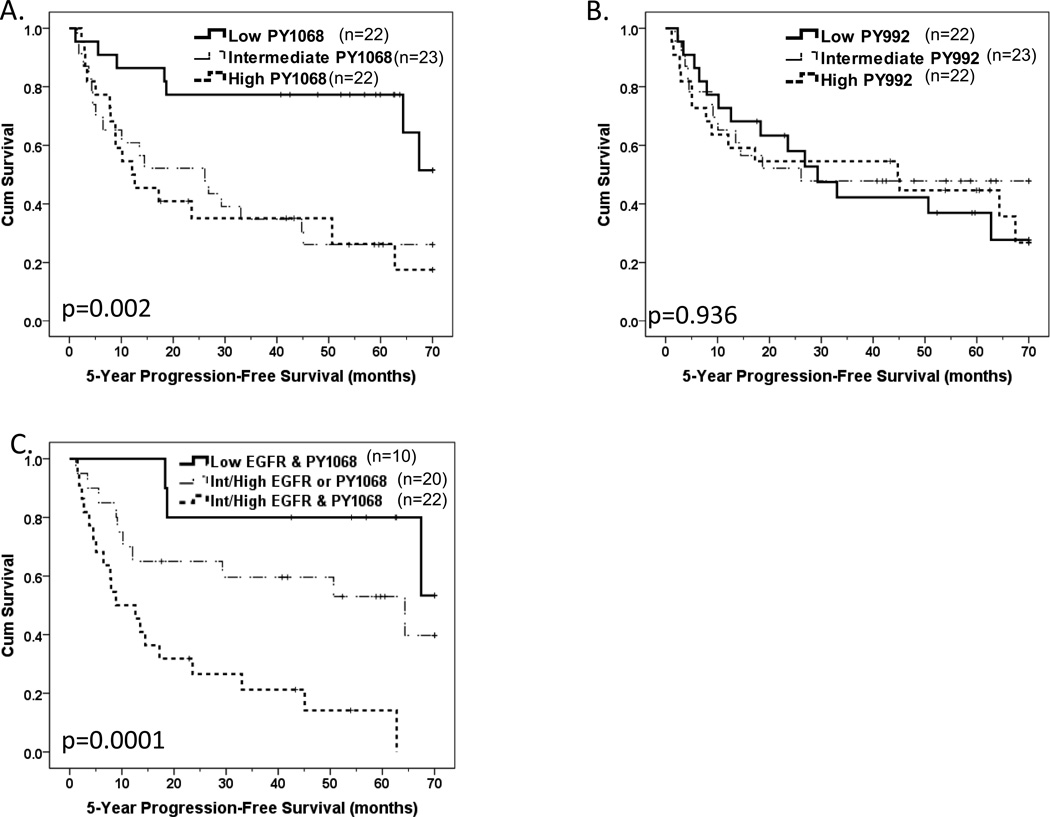
RPPA was used to measure EGFR site-specific phosphorylation at Y1068 and Y992 in fresh-frozen tumors from the EDRN cohort. We confirmed that tumors with high EGFR PY1068 or PY992 by RPPA had significantly higher levels of these EGFR phosphoproteins by immunoblotting (Supplemental Figures 1B and 1C). Intermediate or high tumor EGFR PY1068 levels were associated with significantly reduced PFS, and a statistically significant trend of decreased PFS with increasing EGFR PY1068 expression was noted across the tertiles (Figure 4A). EGFR PY992 tumor levels were not associated with PFS (Figure 4B). EGFR PY1068 and PY992 levels did not differ significantly by tumor HPV status; three of four HPV-positive EDRN tumors had low EGFR PY1068 levels while one of four HPV-positive EDRN tumors had low levels of EGFR PY992 (Supplemental Table 2).
Figure 4. Phosphorylation at EGFR PY1068 was associated with reduced progression-free survival in the EDRN cohort.
Kaplan-Meier progression-free survival (PFS) plots by tertiles (low, intermediate and high) of tumor levels of (A) EGFR PY1068 and (B) EGFR PY992 as assessed by RPPA. Differences in PFS phosphorylation site-specific pEGFR levels across tertiles were tested using the log rank trend test. (C) EGFR intermediate/high protein levels by IHC combined with EGFR PY1068 intermediate/high levels versus lowest tertile reference. Log rank trend test-associated p value comparing across ordered categories of (1) low in EGFR and PY1068, (2) intermediate/high EGFR or PY1068 and (3) intermediate/high in both EGFR and PY1068 is provided.
EGFR protein by RPPA was positively correlated with both PY1068 and PY992 levels (rho=0.24, p=0.053; rho=0.26, p=0.034, respectively). However, EGFR PY1068 and PY992 levels were not significantly correlated with each other (rho=0.15, p=0.24), indicating tumors with high EGFR tended to have higher phosphorylated EGFR, but phosphorylation site-specific differences were present. With limited remaining tissue we assessed levels of activated Src family kinase (SFK) and STAT3 by quantitative immunoblotting with anti-SFK PY416 or anti-PSTAT3 PY705, respectively, in 5 tumors ranked high by EGFR PY1068 and 5 tumors ranked low by EGFR PY1068 (Supplemental Figure 3). SFK PY416 levels did not differ with EGFR PY1068 levels, but tumors with high EGFR PY1068 had higher levels of STAT3 PY705 (Supplemental Figure 3) suggesting that activation of the STAT3 pathway may be an important downstream effector of EGFR PY1068 activity.
Cox proportional hazards models were used to determine whether EGFR PY1068 was associated with PFS when also considering tumor EGFR protein levels by IHC. Low versus intermediate/high levels of tumor EGFR protein by IHC and EGFR PY1068 were defined by the lowest tertile versus the intermediate and high tertiles for each marker. Intermediate/high tumor EGFR by IHC and intermediate/high tumor EGFR PY1068 were both independently associated with reduced PFS in a multivariable Cox proportional hazards model containing both (HR= 2.75; 95% CI=1.26–6.00 and HR=3.29; 95% CI=1.34–8.14, respectively), indicating that both contributed significantly as prognostic indicators. When combining EGFR by IHC and EGFR PY1068 levels into three groups, low in both, intermediate/high in one or intermediate/high in both, there was a significant trend towards reduced PFS across these three categories (Figure 4C). In univariate Cox proportional hazards models, intermediate/high in both tumor EGFR protein by IHC and EGFR PY1068 was associated with significantly reduced PFS compared to low tumor levels of both EGFR protein by IHC and EGFR PY1068 (HR=7.49; 95% CI=2.11–26.48). This association was significant even after adjusting for nodal stage and patient age (HR=5.55; 95% CI =1.42–21.79).
Discussion
Of the EGFR molecular characteristics evaluated in paraffin-embedded formalin-fixed tumors, we found that elevated tumor levels of EGFR by IHC were significantly associated with PFS in a patient cohort treated with surgery with curative intent without an EGFR-targeted therapy. Though our IHC staining method did not detect nuclear EGFR, which has been previously reported to be associated with increased local HNSCC recurrence rates (34), our results are similar to a recent study finding high predominately plasma membrane and cytoplasmic tumor EGFR levels to be associated with reduced survival (40). We found a similar decrease in PFS with elevated tumor EGFR by IHC for patients treated with chemoradiotherapy and EGFR-targeted agent cetuximab. Our two cohorts differed in treatment type and HPV-positive tumor representation, suggesting that EGFR IHC levels may be prognostic for both HPV-positive and negative tumor types.
Though we found no significant association between EGFR gene amplification and PFS in either cohort, we did note a trend towards reduced PFS with increased EGFR gene copy number in the EDRN surgical cohort that was similar to the association reported by Chung et al. (15). Our finding of lack of significant association between increased EGFR gene copy number and PFS was probably not due to a minor difference defining high EGFR copy number where we included 4 or more EGFR copies in at least 50% of cells while Chung et al. delineated 40% as the cut-point (15). Though our follow-up time was longer than the Chung et al. study, our cohort was smaller, which may decrease power in our study to detect a difference in PFS by copy number status. Acknowledging these caveats, our data demonstrated that tumor EGFR gene amplification status, which is currently assessed using an expensive and challenging assay, did not function as a stand-alone prognostic marker nor did it improve prognostic information provided by EGFR IHC, which is a relatively inexpensive assay performed routinely. Our finding that tumor EGFR levels by IHC had prognostic value while EGFR gene copy number did not is consistent with two recent studies in HNSCC that report similar findings (40–41).
Even though tumor EGFR levels by IHC were associated with PFS, EGFR levels by RPPA were not. We speculate that the differences are largely technical in nature. Recent reports have demonstrated that EGFR protein levels assessed by IHC versus immunoblotting methods can differ somewhat (33, 42), and that the magnitude of these differences varies depending upon the antibody used (33). We employed the anti-EGFR antibody clone H11 for our IHC studies. This clone H11 antibody was recently shown to have only a modest correlation between EGFR protein levels by automated quantitative analysis (AQUA) of IHC stained cell lines and EGFR protein levels quantified from immunoblots of lysates from these same cell lines (33). Of four anti-EGFR antibodies tested in a study of 642 breast cancer tumors in this same report by Anagnostou et al., EGFR levels by AQUA analysis of clone H11-stained tumors did not correlate with EGFR levels by AQUA of IHC results obtained using the other anti-EGFR antibodies tested (33). Because we employed the clone H11 antibody for IHC and a different anti-EGFR antibody for RPPA, the differences between IHC and RPPA results likely reflect inherent antibody and assay performance differences. Of interest, of the anti-EGFR antibodies tested for prognostic value in breast cancer by Anagnostou et al., only the clone H11 antibody demonstrated prognostic value (p<0.05), with shorter survival observed for patients whose tumors had high or intermediate tumor levels of EGFR as assessed by AQUA and IHC. Because of assay validation specifications, we used a different anti-EGFR antibody for our RPPA studies. Therefore, a portion of difference between IHC and RPPA results is likely attributed to differences in the antibodies performance.
In addition to inherent assay performance differences such as antibody differences and possible epitope availability differences in arrayed fresh-frozen tissue lysates compared to formalin-fixed tissues, pathologist-interpreted EGFR protein levels by IHC were confined to the cytoplasm and plasma membrane of tumor cells because nuclear EGFR was not detected using the clone H11 antibody. Subcellular architecture was lost when making lysates for RPPA. Therefore, it is possible that EGFR tumor protein in the cytoplasm, membrane and nucleus may contribute differentially to signaling.
Though the stromal compartments of tumors are appreciated as important contributors to HNSCC development and progression, it is possible that stromal tissue present in RPPA samples contributed to the lack of correlation between IHC and RPPA analyses, though we were not able to retrospectively assess this. EGFR tumor levels by IHC were confined to the tumor portion of the specimen by the evaluating pathologist. EGFR levels by RPPA would represent tumor and stromal tissue levels. Our fresh-frozen tumor tissues contained at least 70% tumor; it is possible that up to 30% of our RPPA analyzed tumor tissues were stromal tissues. Our lack of concordance between tumor EGFR levels by IHC and EGFR in protein lysates was similar to previous reports comparing tumor EGFR levels by IHC and by enzyme-linked immunosorbant assays (43–44).
We did find that high tumor levels of EGFR PY1068 but not EGFR PY992 in fresh-frozen tumor lysates contributed prognostic information alone and in addition to tumor EGFR levels by IHC, suggesting that EGFR PY1068 evaluation was not substantially hindered by subcellular localization or stromal contamination issues. Reports evaluating associations between clinical parameters and tyrosine-phosphorylated EGFR that is not site-specific have yielded mixed results (3, 21–22). Our findings suggest that there are biologically relevant differences between phosphorylation at specific sites that could impact patient survival. Our data further suggest that STAT3 signaling downstream of EGFR PY1068 may be important. We evaluated only EGFR PY992 and PY1068 because the quality of these antibodies was sufficient for RPPA. It is possible that evaluation of additional EGFR phosphorylation sites will yield more information. EGFR PY1068 had prognostic value in our predominately HPV-negative cohort. It will be important to assess its prognostic value in independent cohorts, including those with higher HPV-positive tumor representation.
The assessment of phosphoproteins in tissues is generally recognized as requiring special care because of the labile nature of the phoshorylation. We collected and processed tissues under a protocol that involved coordination of personnel and immediate transport of collected tissues to the pathology lab for examination, cataloging and freezing in order to minimize time to freezing. Beginning in 2003, our tissue bank recorded time from surgical resection to freezing, and typical banking time was within 40 minutes of resection. For evaluations of EGFR PY1068 tumor levels, time to freezing logistics will need to be considered before general use.
EGFR transcription/translational regulatory mechanisms in HNSCC are likely complex and only just beginning to be defined. Our observation of association between EGFR gene amplification and elevated EGFR protein by IHC but lack of association of EGFR mRNA levels with either characteristic suggests complex regulation and contradicts somewhat a previously published report of elevated EGFR mRNA in EGFR FISH positive lung cancer (45). Similar to a previous report by Sok et al., we found EGFRvIII expression to be associated with EGFR wild-type expression (38). EGFRvIII expression, which has been associated with resistance to EGFR-targeted therapies (38), was not prognostic in our surgical cohort that was not treated with an EGFR-targeted therapy. However, our detection of EGFRvIII in approximately 20% of tumors that were EGFR-targeting agent naive suggests that EGFRvIII expression may have relevance for HNSCC de novo resistance to EGFR-targeted therapies.
Strengths of our study include the prospective enrollment of our subjects and limited heterogeneity of treatments within each of the two cohorts evaluated. For the EDRN cohort, evaluations extended beyond paraffin-embedded tumor tissues to include fresh-frozen tissues. This study presents a multifaceted evaluation of HNSCC that includes the assessment of molecular characteristics that currently require fresh-frozen specimens. A limitation of this study was our inability to assess all patient tumors for all parameters. This limitation reflects the challenges of obtaining sufficient fresh-frozen material for study. Importantly, there are few specimen selection biases, and the evidence indicates that studied specimens are representative of the general cohort.
In conclusion, we report EGFR tumor levels by IHC assessment, which is performed routinely, are informative regarding patient prognosis. Because independent cohorts treated with and without an EGFR targeting agent had similar survival profiles by tumor EGFR IHC status, EGFR IHC likely provides prognostic rather than predictive value. We noted a trend towards reduced survival with high EGFR gene copy number; however, EGFR gene copy status did not provide prognostic information alone or in combination with tumor EGFR IHC data. We do not anticipate that EGFR FISH analysis will provide prognostic information above that acquired by tumor EGFR IHC analysis in the general HNSCC patient population. Of interest, we report that EGFR site-specific phosphorylation at PY1068 provided prognostic information that was independent of tumor EGFR levels by IHC. These data suggest a further exploration of EGFR site-specific phosphorylation events and EGFR PY1068-specific downstream signaling could provide additional insights into HNSCC progression.
Supplementary Material
Translational Significance.
EGFR has been recognized as contributing to HNSCC development and progression, but the prognostic value of many specific EGFR molecular characteristics have not been evaluated alone or in combination. We assessed EGFR molecular characteristics including tumor EGFR gene amplification, EGFR gene expression, EGFRvIII gene expression, EGFR protein and phosphoprotein for a patient cohort treated with surgery with curative intent. We identified tumor EGFR protein and EGFR PY1068 levels to be independent prognostic indicators and determined that EGFR gene amplification was not an indicator of prognosis in this surgical cohort. We validated the utility of EGFR tumor protein expression but not EGFR gene amplification as an indicator of prognosis in an independent cohort treated with the anti-EGFR antibody therapeutic, cetuximab, thereby defining elevated EGFR tumor protein level as a prognostic rather than predictive indicator. Defining EGFR molecular characteristics relevant to patient prognosis is an important step towards deciding treatment while considering the morbidities associated with aggressive multimodal therapies. In addition to identifying important prognostic indicators, the work presented here defines relationships between different tumor EGFR molecular characteristics thus providing insights regarding the aberrant regulation of EGFR.
Acknowledgments
We thank UPMC tissue bank personnel, especially M. Fichera, for their tissue collection, processing and banking efforts. We thank V. Lui and Q. Zhang for assistance processing tumors for RPPA studies.
Support: P50CA097190, R01CA098372, U01CA84968 and the American Cancer Society (JRG) University of Pittsburgh Cancer Institute Cancer Epidemiology Small Grants Program and K07CA137140 (AME) 1F31DE020223 - 01A1 (SW) and Kleberg Center for Molecular Markers at MDACC, Cancer Center Support Grant CA16672 at MDACC
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest
References
- 1.Altekruse SFKC, Krapcho M, Neyman N, Aminou R, Waldron W, Ruhl J, Howlader N, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Cronin K, Chen HS, Feuer EJ, Stinchcomb DG, Edwards BK, editors. SEER Cancer Statistics Review, 1975–2007. based on November 2009 SEER data submission. Bethesda, MD: National Cancer Institute; 2010. [Google Scholar]
- 2.Conley BA. Treatment of advanced head and neck cancer: what lessons have we learned? J Clin Oncol. 2006;24:1023–1025. doi: 10.1200/JCO.2005.05.0682. [DOI] [PubMed] [Google Scholar]
- 3.Harris SL, Thorne LB, Seaman WT, Neil Hayes D, Couch ME, Kimple RJ. Association of p16(INK4a) overexpression with improved outcomes in young patients with squamous cell cancers of the oral tongue. Head Neck. 2010;33:1622–1627. doi: 10.1002/hed.21650. [DOI] [PubMed] [Google Scholar]
- 4.Muller S, Su L, Tighiouart M, Saba N, Zhang H, Shin DM, et al. Distinctive E-cadherin and epidermal growth factor receptor expression in metastatic and nonmetastatic head and neck squamous cell carcinoma: predictive and prognostic correlation. Cancer. 2008;113:97–107. doi: 10.1002/cncr.23557. [DOI] [PubMed] [Google Scholar]
- 5.Chen B, van den Brekel MW, Buschers W, Balm AJ, van Velthuysen ML. Validation of tissue array technology in head and neck squamous cell carcinoma. Head Neck. 2003;25:922–930. doi: 10.1002/hed.10308. [DOI] [PubMed] [Google Scholar]
- 6.Ang KK, Berkey BA, Tu X, Zhang HZ, Katz R, Hammond EH, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Research. 2002;62:7350–7356. [PubMed] [Google Scholar]
- 7.Irish JC, Bernstein A. Oncogenes in head and neck cancer. Laryngoscope. 1993;103:42–52. doi: 10.1288/00005537-199301000-00009. [DOI] [PubMed] [Google Scholar]
- 8.Maurizi M, Almadori G, Ferrandina G, Distefano M, Romanini ME, Cadoni G, et al. Prognostic significance of epidermal growth factor receptor in laryngeal squamous cell carcinoma. Br J Cancer. 1996;74:1253–1257. doi: 10.1038/bjc.1996.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grandis JR, Melhem MF, Gooding WE, Day R, Holst VA, Wagener MM, et al. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst. 1998;90:824–832. doi: 10.1093/jnci/90.11.824. [DOI] [PubMed] [Google Scholar]
- 10.Ishitoya J, Toriyama M, Oguchi N, Kitamura K, Ohshima M, Asano K, et al. Gene amplification and overexpression of EGF receptor in squamous cell carcinomas of the head and neck. Br J Cancer. 1989;59:559–562. doi: 10.1038/bjc.1989.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryott M, Wangsa D, Heselmeyer-Haddad K, Lindholm J, Elmberger G, Auer G, et al. EGFR protein overexpression and gene copy number increases in oral tongue squamous cell carcinoma. Eur J Cancer. 2009;45:1700–1708. doi: 10.1016/j.ejca.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freier K, Joos S, Flechtenmacher C, Devens F, Benner A, Bosch FX, et al. Tissue microarray analysis reveals site-specific prevalence of oncogene amplifications in head and neck squamous cell carcinoma. Cancer Res. 2003;63:1179–1182. [PubMed] [Google Scholar]
- 13.Koynova DK, Tsenova VS, Jankova RS, Gurov PB, Toncheva DI. Tissue microarray analysis of EGFR and HER2 oncogene copy number alterations in squamous cell carcinoma of the larynx. J Cancer Res Clin Oncol. 2005;131:199–203. doi: 10.1007/s00432-004-0627-y. [DOI] [PubMed] [Google Scholar]
- 14.Morrison LE, Jacobson KK, Friedman M, Schroeder JW, Coon JS. Aberrant EGFR and chromosome 7 associate with outcome in laryngeal cancer. Laryngoscope. 2005;115:1212–1218. doi: 10.1097/01.MLG.0000163755.21035.8F. [DOI] [PubMed] [Google Scholar]
- 15.Chung CH, Ely K, McGavran L, Varella-Garcia M, Parker J, Parker N, et al. Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol. 2006;24:4170–4176. doi: 10.1200/JCO.2006.07.2587. [DOI] [PubMed] [Google Scholar]
- 16.Temam S, Kawaguchi H, El-Naggar AK, Jelinek J, Tang H, Liu DD, et al. Epidermal growth factor receptor copy number alterations correlate with poor clinical outcome in patients with head and neck squamous cancer. J Clin Oncol. 2007;25:2164–2170. doi: 10.1200/JCO.2006.06.6605. [DOI] [PubMed] [Google Scholar]
- 17.Sheu JJ, Hua CH, Wan L, Lin YJ, Lai MT, Tseng HC, et al. Functional genomic analysis identified epidermal growth factor receptor activation as the most common genetic event in oral squamous cell carcinoma. Cancer Res. 2009;69:2568–2576. doi: 10.1158/0008-5472.CAN-08-3199. [DOI] [PubMed] [Google Scholar]
- 18.Xia W, Lau YK, Zhang HZ, Xiao FY, Johnston DA, Liu AR, et al. Combination of EGFR, HER-2/neu, and HER-3 is a stronger predictor for the outcome of oral squamous cell carcinoma than any individual family members. Clin Cancer Res. 1999;5:4164–4174. [PubMed] [Google Scholar]
- 19.Lee JW, Soung YH, Kim SY, Nam HK, Park WS, Nam SW, et al. Somatic mutations of EGFR gene in squamous cell carcinoma of the head and neck. Clin Cancer Res. 2005;11:2879–2882. doi: 10.1158/1078-0432.CCR-04-2029. [DOI] [PubMed] [Google Scholar]
- 20.Soulieres D, Senzer NN, Vokes EE, Hidalgo M, Agarwala SS, Siu LL. Multicenter phase II study of erlotinib, an oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with recurrent or metastatic squamous cell cancer of the head and neck. J Clin Oncol. 2004;22:77–85. doi: 10.1200/JCO.2004.06.075. [DOI] [PubMed] [Google Scholar]
- 21.Hama T, Yuza Y, Saito Y, J Ou, Kondo S, Okabe M, et al. Prognostic significance of epidermal growth factor receptor phosphorylation and mutation in head and neck squamous cell carcinoma. Oncologist. 2009;14:900–908. doi: 10.1634/theoncologist.2009-0058. [DOI] [PubMed] [Google Scholar]
- 22.Keller J, Shroyer KR, Batajoo SK, Zhao HL, Dong LM, Hayman MJ, et al. Combination of phosphorylated and truncated EGFR correlates with higher tumor and nodal stage in head and neck cancer. Cancer Invest. 2010;28:1054–1062. doi: 10.3109/07357907.2010.512602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 24.Burtness BGM, Flood W, Mattar B, Forastiere AA Group ECO. Phase III randomized trial of cisplatin plus placebo compared with cisplatin plus cetuximab in metastatic/recurrent head and neck cancer: an Eastern Cooperative Oncology Group study. Journal of Clinical Oncology. 2005;23:8646–8654. doi: 10.1200/JCO.2005.02.4646. [DOI] [PubMed] [Google Scholar]
- 25.Licitra L, Mesia R, Rivera F, Remenar E, Hitt R, Erfan J, et al. Evaluation of EGFR gene copy number as a predictive biomarker for the efficacy of cetuximab in combination with chemotherapy in the first-line treatment of recurrent and/or metastatic squamous cell carcinoma of the head and neck: EXTREME study. Annals of Oncology. 2011;22:1078–1087. doi: 10.1093/annonc/mdq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 27.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 28.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science (New York, N Y ) 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science (New York, N Y ) 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egloff AM, Rothstein ME, Seethala R, Siegfried JM, Grandis JR, Stabile LP. Cross-talk between estrogen receptor and epidermal growth factor receptor in head and neck squamous cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:6529–6540. doi: 10.1158/1078-0432.CCR-09-0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Argiris A, Heron DE, Smith RP, Kim S, Gibson MK, Lai SY, et al. Induction docetaxel, cisplatin, and cetuximab followed by concurrent radiotherapy, cisplatin, and cetuximab and maintenance cetuximab in patients with locally advanced head and neck cancer. J Clin Oncol. 2010;28:5294–5300. doi: 10.1200/JCO.2010.30.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anagnostou VK, Welsh AW, Giltnane JM, Siddiqui S, Liceaga C, Gustavson M, et al. Analytic variability in immunohistochemistry biomarker studies. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19:982–991. doi: 10.1158/1055-9965.EPI-10-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Psyrri A, Yu Z, Weinberger PM, Sasaki C, Haffty B, Camp R, et al. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:5856–5862. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- 35.Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB. Non-parametric quantification of protein lysate arrays. Bioinformatics. 2007;23:1986–1994. doi: 10.1093/bioinformatics/btm283. [DOI] [PubMed] [Google Scholar]
- 36.Freedman LS. Tables of the number of patients required in clinical trials using the logrank test. Statistics in Medicine. 1982;1:121–129. doi: 10.1002/sim.4780010204. [DOI] [PubMed] [Google Scholar]
- 37.Dziadziuszko R, Holm B, Skov BG, Osterlind K, Sellers MV, Franklin WA, et al. Epidermal growth factor receptor gene copy number and protein level are not associated with outcome of non-small-cell lung cancer patients treated with chemotherapy. Ann Oncol. 2007;18:447–452. doi: 10.1093/annonc/mdl407. [DOI] [PubMed] [Google Scholar]
- 38.Sok JC, Coppelli FM, Thomas SM, Lango MN, Xi S, Hunt JL, et al. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res. 2006;12:5064–5073. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- 39.Gillison ML, D'Souza G, Westra W, Sugar E, Xiao W, Begum S, et al. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100:407–420. doi: 10.1093/jnci/djn025. [DOI] [PubMed] [Google Scholar]
- 40.Pectasides E, Rampias T, Kountourakis P, Sasaki C, Kowalski D, Fountzilas G, et al. Comparative prognostic value of epidermal growth factor quantitative protein expression compared with FISH for head and neck squamous cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:2947–2954. doi: 10.1158/1078-0432.CCR-10-2040. [DOI] [PubMed] [Google Scholar]
- 41.Huang SF, Cheng SD, Chien HT, Liao CT, chen IH, Wang HM, et al. Relationship between epidermal growth factor receptor gene copy number and protein expression in oral cavity squamous cell carcinoma. Oral Oncol. 2011 doi: 10.1016/j.oraloncology.2011.06.511. [DOI] [PubMed] [Google Scholar]
- 42.Thariat J, Etienne-Grimaldi MC, Grall D, Bensadoun RJ, Cayre A, Penault-Llorca F, et al. Epidermal Growth Factor Receptor protein detection in head and neck cancer patients: a many faceted picture. Clin Can Res. 2012 doi: 10.1158/1078-0432.CCR-11-2339. [DOI] [PubMed] [Google Scholar]
- 43.Spindler K-LG, Lindebjerg J, Nielsen JN, Olsen DA, Bisgard C, Brandslund I, et al. Epidermal growth factor receptor analyses in colorectal cancer: a comparison of methods. International Journal of Oncology. 2006;29:1159–1165. [PubMed] [Google Scholar]
- 44.Nannini M, Pantaleo MA, Paterini P, Piazzi G, Ceccarelli C, La Rovere S, et al. Molecular detection of epidermal growth factor receptor in colorectal cancer: does it still make sense? Colorectal Dis. 2011;13:542–548. doi: 10.1111/j.1463-1318.2010.02212.x. [DOI] [PubMed] [Google Scholar]
- 45.Dziadziuszko R, Witta SE, Cappuzzo F, Park S, Tanaka K, Danenberg PV, et al. Epidermal growth factor receptor messenger RNA expression, gene dosage, and gefitinib sensitivity in non-small cell lung cancer. Clin Cancer Res. 2006;12:3078–3084. doi: 10.1158/1078-0432.CCR-06-0106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.