Abstract
Toxoplasma gondii, a zoonotic protozoal parasite, is well-known for its global distribution and its ability to infect virtually all warm-blooded vertebrates. Nonetheless, attempts to describe the population structure of T. gondii have been primarily limited to samples isolated from humans and domesticated animals. More recent studies, however, have made efforts to characterize T. gondii isolates from a wider range of host species and geographic locales. These findings have dramatically changed our perception of the extent of genetic diversity in T. gondii and the relative roles of sexual recombination and clonal propagation in the parasite’s lifecycle. In particular, identification of novel, disease-causing T. gondii strains in wildlife has raised concerns from both a conservation and public health perspective as to whether distinct domestic and sylvatic parasite gene pools exist. If so, overlap of these cycles may represent regions of high probability of disease emergence. Here, we attempt to answer these key questions by reviewing recent studies of T. gondii infections in wildlife, highlighting those which have advanced our understanding of the genetic diversity and population biology of this important zoonotic pathogen.
Keywords: Toxoplasma gondii, population genetics, genotype, wildlife
1. Background
1.1 Lifecycle
Toxoplasma gondii is an apicomplexan parasite and the most extensively studied of the tissue encysting coccidia, a group that comprises many species of human and veterinary medical importance, including members of the genera Sarcocystis, Neospora, Hammondia and Besnoitia. The T. gondii lifecycle involves an asexual stage in intermediate hosts and a sexual stage in a definitive host, which may be any species of domestic or wild cat (Dubey and Frenkel, 1972; Hutchison, 1965). Cats are naturally infected through the oral route and, upon ingestion, T. gondii commences the sexual stage by differentiating into male and female gametes which fuse in the intestinal epithelium to form a fertilized oocyst (Dubey and Frenkel, 1972; Ferguson et al., 1974; Ferguson, 2002; Ferguson et al., 1975a, b). Oocysts are then shed into the environment in the feces where they undergo meiosis during the process of sporulation, eventually producing eight haploid progeny that are enveloped in an environmentally stable infectious propagule (Dubey et al., 1970b; Ferguson, 2002; Frenkel et al., 1975). Over the course of one to three weeks, a cat may shed millions of oocysts, which are highly infectious orally to intermediate hosts and, to a lesser extent, other definitive hosts (Dubey, 1996a, b, 1998, 2006; Dubey and Frenkel, 1972; Dubey et al., 1996; Dubey et al., 1970a).
The intermediate host range of T. gondii is vast, inclusive of essentially all warm-blooded vertebrates, both avian and mammalian (Dubey, 2008). When intermediate hosts ingest T. gondii, the parasite crosses the intestinal epithelium and differentiates into a rapidly dividing tachyzoite form which disseminates infection throughout the host (Dubey, 1998). If the host survives this acute phase of infection, the chronic phase is initiated when tachyzoites differentiate into slowly dividing, semi-dormant bradyzoites that form tissue cysts, typically in muscle or neural cells (Dubey, 1997, 1998; Dubey and Frenkel, 1976; Dubey et al., 1998). Tissue cysts can persist for the life of the host and are orally infectious to definitive hosts through carnivory (Dubey, 2001, 2006; Frenkel et al., 1970; Su et al., 2003).
Toxoplasma gondii is not, however, an obligately heteroxenous parasite and can propagate clonally, presumably indefinitely, by cycling among intermediate hosts. This can occur vertically through transplacental transmission from mother to offspring (Dubey et al., 1997; Elbez-Rubinstein et al., 2009; Hide et al., 2007; Innes et al., 2009; Miller et al., 2008a), or, in rare cases during medical operations, horizontally through tissue transplant (Martina et al., 2010). Tissue cysts are also orally infectious to carnivorous intermediate hosts, permitting the parasite to bypass the sexual stage in the definitive host (Dubey, 2001, 2006; Khan et al., 2007; Su et al., 2003). This trait, along with its widely inclusive host range, has often been cited as unique to T. gondii among the tissue encysting coccidia (Grigg and Sundar, 2009; Khan et al., 2007; Sibley and Ajioka, 2008; Sibley et al., 2009; Su et al., 2003). However, many recent studies have shown that other tissue encysting coccidia, including Neospora caninum and Sarcocystis neurona, have much broader intermediate host ranges than originally thought, including both mammalian and avian species and predators and their prey (Cheadle et al., 2001; Costa et al., 2008; Dubey et al., 2001a; Dubey et al., 2001b; Gondim et al., 2010; Mansfield et al., 2008; Miller et al., 2009; Rejmanek et al., 2010; Wendte et al., 2010b). Until more rigorous studies are completed, the possibility cannot be excluded that these related parasites also have an ability for carnivorous oral transmission among intermediate hosts.
Infection in the definitive host stage is also not always synonymous with sexual propagation. In the feline host, T. gondii readily differentiates into the asexual tachyzoite and bradyzoite forms, eventually developing into infectious tissue cysts just as in intermediate hosts (Dubey, 1997). Moreover, initiation of the sexual pathway does not preclude clonal propagation, as T. gondii has been found to lack predetermined mating types, permitting a single genotype to form both male and female gametes that fuse to yield progeny nearly identical to the parent (Cornelissen and Overdulve, 1985; Pfefferkorn et al., 1977; Pfefferkorn and Pfefferkorn, 1980). This process, termed selfing, has been shown to have important implications for the population biology and epidemiology of T. gondii and other tissue encysting coccidia (Vaudaux et al., 2010; Wendte et al., 2010a), and will be discussed further below.
1.2 Disease
Major manifestations of T. gondii-induced disease in humans include encephalitic, ocular, and pneumatic toxoplasmosis (Montoya and Liesenfeld, 2004). Based upon seroprevalence rates, it has been assumed that the majority of infections are asymptomatic, with disease mainly linked to congenital infection or an immune-compromised state such as AIDS (Montoya and Liesenfeld, 2004). However, it is becoming increasingly appreciated that symptomatic disease can also occur in immune-competent individuals (Ajzenberg et al., 2004; Boothroyd and Grigg, 2002; De Salvador-Guillouet et al., 2006; Demar et al., 2007; Grigg et al., 2001b; Leal et al., 2007; Sacks et al., 1983). Many factors are likely to influence whether or not disease occurs including dose, parasite stage initiating infection, parasite genotype, host genotype, and various factors influencing host immune status, especially concomitant infection with other pathogens.
Descriptions of toxoplasmosis in wildlife are generally limited to post-mortem analyses except for well-monitored species such as endangered and captive or semi-captive animals. The clinical picture of toxoplasmosis in wildlife appears similar to that in humans, with lung, brain, and eye involvement prevalent in many cases, thus providing important parallels that have increased our understanding of human disease. Of particular concern are several reports of multi-organ involvement indicative of disseminated disease in rare and exotic species, perhaps signifying increased susceptibility in hosts that have only recently been exposed to this parasite through human activity. Several reports of toxoplasmosis in wildlife are summarized in Table 1 for the reader’s reference.
Table 1.
Toxoplasmosis in wildlife*
| MAMMALS | Species | Captive? | Location | Diagnostic Technique | Symptoms | Reference |
|---|---|---|---|---|---|---|
|
Alouatta belzebul (red-handed howler monkey) |
Y | Pernambuco State, Brazil |
parasite isolation, PCR | prostration, diarrhea, hyperthermia |
Pena et al. (2010) Vet Parasitol In Press |
|
|
Bettongi penicillata (woylie) |
Manjimup reserve, Australia |
PCR | neurological symptoms |
Parameswaran et al. (2010) Int J Parasitol 40: 635– 640 |
||
|
Delphinapterus leucas (beluga whale) |
Quebec, Canada | IHC | stranding, mortality | Mikaelian et al. (2000) J Comp Path 122: 73–76 |
||
|
Enhydra lutris (Northern sea otter) |
WA, USA | IHC, bioassay, PCR-RFLP | convulsions, encephalitis, mortality |
Lindsay et al. (2001) Vet Parasitol 97: 319– 327 |
||
|
Enhydra lutris nereis (Southern sea otter) |
CA, USA | IHC, serology, parasite isolation, PCR |
stranding, encephalitis, mortality |
Miller et al. (2004) Int J Parasitol 34: 275– 284 |
||
|
Enhydra lutris nereis (Southern sea otter) |
CA, USA | IHC, parasite isolation, PCR-RFLP | stranding, mortality | Cole et al. (2000) J Parasitol 86: 526–530 |
||
|
Felis margarita (sand cat) |
Y | United Arab Emirates |
IHC, serology, PCR | mortality, hepatitis, pneumonitis |
Dubey et al. (2010) Vet Parasitol 172:195– 203 |
|
|
Felis margarita (sand cat) |
Y | Qatar | IHC, serology, PCR | anorexia, weakness, mortality |
Dubey et al. (2010) Vet Parasitol 172:195- |
|
|
Lynx rufus (bobcat) |
MT, USA | IHC, serology | mortality, myocarditis, hepatitis, encephalitis |
Dubey et al. (1987) J Wildl Dis 23: 324–327 |
||
|
Macropus rufogriseus (wallaby) |
Tasmania, Australia |
parasite isolation | neurologic symptoms |
Parameswaran et al. (2010) Int J Parasitol 40: 635– 640 |
||
|
Marcropus eugenii (Tammar wallaby) |
New Zealand |
serology, parasite isolation | poor health, mortality |
Dubey and Crutchley (2008) J Parasitol 94:929–933 |
||
|
Marcropus rufogriseus (Bennett's wallaby) |
Y | Spain | IHC | apathy, depression, emaciation, mortality, pneumonia, myocarditis, cholangiohepatitis, severe gastroenteritis, uveitis |
Bermúdez et al. (2009) Vet Parasitol 160: 155–158 |
|
|
Marcropus Rufogriseus (Bennett's wallaby) |
New Zealand |
IHC, serology, parasite isolation, PCR-RFLP |
lethargy, emactiaion, incoordination, impaired vision, mortality, pneumonia, congestive heart failure |
Dubey and Crutchley (2008) J Parasitol 94:929–933 |
||
|
Marcropus Rufogriseus (Bennett's wallaby) |
Y | PA, USA | IHC, serology, parasite isolation, PCR-RFLP |
weakness, lethargy, loss of appetite, incoordination |
Dubey and Crutchley (2008) J Parasitol 94:929–933 |
|
|
Marcropus rufogriseus (Bennett's wallaby) |
Y | Argentina | IHC, serology, parasite isolation, PCR |
Mortality |
Basso et al. (2007) Vet Parasitol 144: 157–161 |
|
|
Marmota monax (woodchuck) |
NY, USA | IHC, RT-PCR | neurologic symptoms, weight loss, mortality, cerebral hemorrhage, encephalitis, myocarditis, hepatitis |
Bangari et al. (2007) J Vet Diagn Invest 19: 705–709 |
||
|
Mustela vison (American mink) |
MI, USA | IHC, PCR | neurological symptoms (lameness, ataxia, head tremors, impaired vision) |
Jones et al. (2006) J Wildl Dis 42: 865–869 |
||
|
Phoca vitulina richardsi (Pacific harbor seal) |
CA, USA | parasite isolation | meningoencephalomyelitis | Miller et al. (2001) J Parasitol 87, 816–822 |
||
|
Rupicapra pyrenaica (southern chamois) |
Pyrenees, Spain |
IHC, serology | neurological symptoms, systemic toxoplasmosis with eye and lung pathology |
Marco et al. (2009) J Vet Diagn Invest 21: 244–247 |
||
|
Sousa chinensis (Indo-Pacific Humpbacked dolphin) |
Queensland, Australia |
IHC | mortality, pneumonia, emaciation, shark attack, peritonitis, pancreatitis, ocular pathology |
Bowater et al. (2003) Aust Vet J 81: 627–632 |
||
|
Stenella coeruleoalba (striped dolphin) |
Italy | ICH, serology | mortality, encephalitis, bronchopneumonia, pulmonary atelectasis, consolidation, emphysema |
Di Guardo et al. (2010) Vet Parasitol 47: 245– 253 |
||
|
Suricata suricatta (meerkat) |
Y | Perth, Australia |
PCR | neurologic symptoms |
Parameswaran et al. (2010) Int J Parasitol 40: 635– 640 |
|
|
Tamiasciurus hudsonicus (American red squirrel) |
IN, USA | IHC, RT-PCR | weight loss, mortality, pneumonia, encephalitis |
Bangari et al. (2007) J Vet Diagn Invest 19: 705–709 |
||
|
Tursiops Truncates (bottlenose dolphin) |
Russia | IHC, serology, parasite isolation, PCR, PCR-RFLP |
disorientation, lethargy, anorexia, mortality, encephalitis |
Dubey et al. (2009) J Parasitol 95: 82–85 |
||
|
Vombatus ursinus (wombat) |
Tasmania, Australia |
Parasite isolation, PCR | neurologic symptoms |
Parameswaran et al. (2010) Int J Parasitol 40: 635– 640 |
||
|
Vombatus ursinus (wombat) |
Y | New South Wales, Australia |
IHC, serology | anorexia, mortality, dyspnoea, tachycardia, neorological and respiratory lesions |
Hartley (2006) Aust Vet J 84: 107–109 |
|
|
Vulpes cana (Blanford's fox) |
Y | United Arab Emirates |
IHC, serology | mortality | Dubey and Pas (2008) Vet Parasitol 153: 147–151 |
|
|
Vulpes lagopus (artic fox) |
Svalbard | IHC, serology | jaundice, mortality, hepatitis, pneumonia, meningitis |
Sorenson et al (2005) Res Vet Sci 78:161–167 |
||
|
Vulpes ruepelli (sand fox) |
Y | United Arab Emirates |
IHC | mortality | Pas and Dubey (2008) J Parasitol 94: 976–977 |
| BIRDS | Species | Captive? | Location | Diagnostic Technique | Symptoms | Reference |
|---|---|---|---|---|---|---|
|
Acridotheres (myna) |
Y | Mexico | electron microscopy | inappetent, shivering, mortality |
Dhillon et al. (1982) Avian Dis 26: 445–449 |
|
|
Anseranas semipalmata (magpie geese) |
Y | TX, USA | IHC, serology | mortality, pneumonia, hepatitis |
Dubey et al. (2001) J Parasitol 87:219–223 |
|
|
Caloenas nicobaria (Nicobar pigeons) |
Y | South Africa | IHC | mortality | Las and Shivaprasad (2008) J S Afr Vet Assoc 79: 149–152 |
|
|
Corvus hawaiiensis (Alala) |
HI, USA | IHC, serology, parasite isolation | emaciation, depression, mortality |
Work et al. (2000) J Wildl Dis 36: 205–212 |
||
|
Cyanoramphus spp. (kakariki) |
Y | Australia | IHC | loss of appetite, mortality | Hartley et al (2008) J Parasitol 94: 1424–1425 |
|
|
Eos bornea (red lory) |
Y | LA, USA | IHC | pneumonia, depression, mortality |
Howerth et al (1991) Avian Dis 35: 642–646 |
|
|
Eos cyanogenia (black-winged lory) |
Y | SC, USA | serology, parasite isolation, PCR- RFLP |
mortality, acute toxoplasmosis |
Dubey et al. (2004) J Parasitol 90: 1171–1174 |
|
|
Eudyptula mino (little penguin) |
Y | Tasmania, Australia |
IHC | anorectic, diarrhea, mortality | Mason et al (1991) J Parasitol 77: 328 |
|
|
Francolinus erckelii (Erckels francolin) |
HI, USA | IHC | mortlality, necrosis | Work et al (2002) J Parasitol 88: 1040–1042 |
||
|
Haliaeetus leucocephalus (bald eagle) |
unknown | IHC | respiratory distress, mortality, myocarditis |
Szabo et al. (2004) J Parasitol 90: 907–908 |
||
|
Melanerpes carolinus (red- bellied woodpecker) |
GA, USA | IHC, serology, PCR | lethargy, neurological symptoms (seizures), encephalitis |
Gerhold and Yabsley (2007) Avian Dis 51: 992–994 |
||
|
Meleagris gallopavo(wild turkey) |
WV, USA | IHC | emaciation, crusting dermatitis on head and neck, mortality, splenomegaly, multifocal necrotizing hepatitis, splenitis |
Quist et al. (1995) J Wildl Dis 31:255–258 |
||
|
Meleagris gallopavo(wild turkey) |
GA, USA | IHC, electron microscopy | mortality, pneumonia, splenomegaly |
Howerth and Rodenroth(1985) J Wildl Dis 21: 446–449 |
||
|
Nesochen sandvicensis (nene goose) |
Y | HI, USA | IHC | mortality, splenomegaly | Work et al (2002) J Parasitol 88: 1040–1042 |
|
|
Ramphastos sulfuratus (keel- billed toucan) |
Y | Costa Rica | bioassay, parasite isolation, PCR- RFLP |
Mortality | Dubey et al. (2009) J Parasitol 95: 467–468 |
|
|
Serinus canaria (Roller canary) |
Y | Victoria, Australia |
IHC | blindness, nystagmus, ataxia, head rotation, mortality |
Lindsay et al. (1995) Avian Dis 39: 204–207 |
|
|
Serinus canaria (canary) |
Y | unknown | IHC, electron microscopy | chemosis, lethargy, torticollis |
Williams et al (2001) Avian Dis 45: 262–267 |
|
|
Strix varia (barred owl) |
Quebec, Canada | IHC | car collision, anorexia, inactivity, mortality, hepatitis |
Mikaelian et al (1997) Avian Dis 41: 738–740 |
||
|
Sula Sula (red- footed booby) |
HI, USA | IHC | weak, mortality, necrosis, inflammation |
Work et al (2002) J Parasitol 88: 1040–1042 |
Search terms used were: “Toxoplasma gondii”, “mammal”, “bird”, and “wildlife” on ISI web of science and PubMed online databases.
2. Toxoplasma in wildlife
2.1 Molecular genotyping and population genetics
2.1.1 History
Several techniques for detecting and characterizing T. gondii genetic material either directly from infected host tissues or from parasites isolated via bioassay in mice, cats, or tissue culture have been developed. Here, we will discuss a subset of these techniques and their relative advantages and disadvantages as pertinent to the subject matter at hand. We refer the reader to a recent extensive review of molecular genotyping of T. gondii for further details (Su et al., 2010). To adequately discuss the genetic characterization of T. gondii in wildlife, it will first be necessary to review the history of discovery and current viewpoints on the population genetic structure of this parasite.
The first picture of the T. gondii population structure came from isoenzyme and restriction fragment length polymorphism (RFLP) analyses (Darde et al., 1988, 1992; Sibley and Boothroyd, 1992). Dardé and colleagues (1992) used isoelectric focusing of six enzymes to show that 35 T. gondii isolates could be grouped into just 5 distinct zymodemes, with the majority of isolates (56%) comprising just one zymodeme. At about the same time, Sibley and Boothroyd (1992) used RFLP analysis of 27 samples at three loci to show that nine isolates that were virulent to laboratory mice comprised a distinct, identical genotype while a moderate amount of diversity was found in the remaining 21 samples. Clonality in the mouse virulent samples was confirmed with RFLP analysis of an additional seven loci (Sibley and Boothroyd, 1992). These RFLP findings were later expanded to encompass a larger sampling of 106 isolates spanning 3 continents and several host species. Using six RFLP markers, ~84%, or 89 of 106 isolates were classified into just three clonal types (Howe and Sibley, 1995). This apparently clonal population structure was especially striking since, in a few cases, samples from diverse parts of the globe were identical at all six markers (Darde et al., 1992; Howe and Sibley, 1995; Sibley and Boothroyd, 1992). These studies also prompted the conclusion that, despite presumably numerous opportunities for sexual recombination in widespread feline definitive hosts, productive sexual crosses only occur very rarely in nature (Howe and Sibley, 1995).
Closer examination of the data presented in the studies described above, however, suggest other explanations aside from a strictly clonal population structure. It is important to note that the 1992 Dardé et al. and Sibley and Boothroyd studies shared nine isolates (9/62; approx. 30% of the total in each study). Moreover, of the 53 total isolates analyzed between the two studies, 47/53 (89%) were from either the US or France, 52/53 (98%) were from humans or domestic animals, and of these, 37/53 (70%) were from humans with congenital infections, AIDS patients, and/or humans with symptomatic disease (Darde et al., 1992; Sibley and Boothroyd, 1992). Thirty-six of these 53 (68%) were again analyzed in the Howe and Sibley study (1995), which also comprised mainly human samples (68/106, 64%) from the US or France. A sample set that is highly biased towards a certain host species, towards cases of disease, or both increases the likelihood of over-sampling particular genotypes, skewing the population structure in favor of apparent clonality (Feil and Spratt, 2001). Moreover, even if parasite genotypes do not segregate according to host species or virulence, oversampling of a few genotypes does not preclude an underlying population of diverse parasite genotypes with high rates of genetic exchange, as has been described for organisms with so-called ‘epidemic’ population structures (Feil and Spratt, 2001; Maynard Smith et al., 1993). Epidemic here refers not exclusively to localized disease epidemics (though this is often the case), but instead to any circumstance where a particular genotype experiences enhanced clonal expansion relative to others in the population, at a local or global level, and is thus over-sampled (Maynard Smith et al., 1993).
It is also possible that limitations in the discriminatory power of the markers used in these early studies missed much of the genetic diversity present. When Lehmann and colleagues applied higher resolution DNA sequencing analysis of loci, they identified increased levels of polymorphism among the genotypes of sixteen isolates, including twelve previously typed by the Howe and Sibley 1995 study (Lehmann et al., 2000). This study identified more allelic diversity among strains than that identified with RFLP analyses. Genetic diversity within the three clonal archetypes was also confirmed using high resolution microsatellite typing (Ajzenberg et al., 2002a; Ajzenberg et al., 2002b; Blackston et al., 2001). Ajzenberg and colleagues (2002a) found that among 83 archetypal isolates, mostly from human clinical cases, 72 distinct genotypes could be discerned using these highly polymorphic markers, highlighting their utility for distinguishing strains within clonal groups.
The re-analysis of previously typed strains also resulted in two important observations attesting to the importance of the sexual stage for T. gondii’s population structure. First, linkage disequilibrium, while significantly present even when correcting for effects of a geographically and temporally diverse sample set (Ajzenberg et al., 2002a), was not absolute across all loci, again supporting the notion that sexual recombination is a viable process in the T. gondii lifecycle (Ajzenberg et al., 2002a; Grigg et al., 2001a; Lehmann et al., 2000). Second, independent gene tree analyses of each typing locus revealed for the majority of strains only two groupings, instead of three, and that some strains could be assigned to either one group or the other depending on the locus examined (Grigg et al., 2001a; Lehmann et al., 2000). This suggested that members of the three clonal lineages defined by Howe and Sibley (1995) were actually sibling progeny of one or a few genetic outcrosses, a finding that was later confirmed by more extensive analyses (Boyle et al., 2006). These findings revealed sexual recombination as a major force shaping the population structure of T. gondii, with important implications for the emergence of virulent strains (Grigg et al., 2001a) or strains associated with particular disease, for example ocular toxoplasmosis (Grigg et al., 2001b). It also became apparent that genetic exchange is a critical factor to account for when performing phylogenetic analyses, as it greatly increases the potential for phylogenetic non-congruency between loci, even in instances of moderate to high linkage disequilibrium, thus leading to conflicting (Blackston et al., 2001; Lehmann et al., 2000) or poorly resolved (Ajzenberg et al., 2002a) genetic clusters.
A more diverse population structure was later confirmed as typing techniques with increased resolution were applied to isolates more representative of T. gondii’s geographic distribution. Microsatellite typing of T. gondii isolates from humans and domestic animals in Brazil and French Guiana showed that many isolates comprise a genetically distinct population from that in the US and France (Ajzenberg et al., 2004; Lehmann et al., 2004). The increased levels of genetic diversity (Ajzenberg et al., 2004) and lower levels of linkage disequilibrium (Lehmann et al., 2004) detected in South American strains suggested that sexual recombination occurs more frequently in this population than in the US and France and, again, likely explains the poor phylogenetic resolution among these strains (Ajzenberg et al., 2004).
An apparently distinct and diverse gene pool in South America prompted additional studies from this area and others around the globe to gain a more comprehensive picture of genetic diversity in the T. gondii population. Su et al. (2006) developed a standardized RFLP typing scheme based on nine mostly unlinked nuclear genomic loci and one apicoplast marker to reveal a wide diversity of strains isolated from 28 domestic cats in Brazil. Importantly, this study confirmed that clonality is not the rule and re-classified many strains that were typed previously only at a single locus, SAG2, via the method developed by Howe et al. (1997) under the assumption that a single locus was sufficient to type strains in a highly clonal population. Khan et al. (2006) also typed several human clinical and domestic animal samples from Brazil using a combination of RFLP typing and DNA sequencing at a single locus to reveal additional diverse genotypes. This study employed phylogenetic analyses of the typing data and identified poor bootstrap support for several branches and conflicting groupings of strains based on RFLP and DNA sequencing data. This study did not support a truly clonal population structure, and one plausible interpretation was that the clouding of genetic histories was attributable to recombination (Khan et al., 2006). RFLP analysis of additional human and domestic animal samples by Ferreira Ade et al. (2006) also revealed an admixture of archetypal alleles across loci, potentially indicating recombinant strains.
The increasing wealth of genetic data from diverse geographic locales facilitated several comprehensive re-assessments of the global T. gondii population structure. Lehmann and colleagues (2006) analyzed 275 strains isolated from chickens, representative of North America, Central America, South America, Europe, Africa and Asia, using a combination of six microsatellite markers and one gene coding marker. Their results showed a diverse array of genotypes that could be assigned to four distinct sub-populations: two exclusive to South America, one found on all continents sampled except South America and one found world-wide (Lehmann et al., 2006). Similar results were revealed in a smaller-scale study that used eight intron loci to describe the population structure of 46 isolates from North America, Europe, and South America, many of which were previously typed in studies discussed above (Khan et al., 2007). This study also found evidence for structuring the T. gondii population into four sub-populations with the same geographic distribution as those found by Lehmann et al. (2006). Importantly, the methodology employed here permitted assignment of different parts of an individual isolate’s genotype to different populations, thus accounting for the likely event of admixture between populations in a parasite with a viable sexual cycle (Khan et al., 2007). This revealed the intriguing result that nearly all genotypes showed some degree of admixture of two or more of the four ancestral populations (Khan et al., 2007).
This study also further sub-grouped isolates into 11 haplogroups by clustering representative subsets of isolates using neighbor-joining phylogenetic analysis of concatenated sequences and by comparing the relative admixture profiles of individual genotypes. However, evidence for recombination between these groupings (Grigg and Sundar, 2009; Khan et al., 2007) indicates that concatenation was inappropriately applied as the robustness of these haplogroups breaks down if loci are examined individually. There were also large discrepancies between the similarities in admixture profiles within an individual haplogroup. For example, genotypes in haplogroups 1, 2, and 3 had essentially identical admixture profiles to other members of their respective groups, but genotypes in haplogroups 4 and 5 had admixture profiles that varied widely in the extent to which each ancestral population contributed (Khan et al., 2007). It is unclear if these groupings represent static entities that can consistently be applied in future studies, or, rather, points on a continuum that will blend together as more isolates are described. The diverse admixture distributions present among isolates, though, do reveal how the genomic position of markers chosen for genotyping studies can profoundly influence how a certain genotype is perceived to be related to ancestral parent types and extant strains.
Specific geographic locales outside the US and Europe have also been the focus of recent studies. Pena et al. (2008) described 125 isolates from chickens, cats, and dogs from Brazil using 10 PCR-RFLP markers to reveal 48 genotypes, many of which were unique to this region. Importantly, this study showed that the three dominant clones identified by Howe and Sibley (1995) were present at very low frequencies or even absent (e.g. ‘Type II’) from this region, and genetic analyses revealed a highly reticulated network indicative of uncertain genetic histories consistent with recombination (Pena et al., 2008). Additional unique Brazilian genotypes have been identified in similar studies of sheep and goats from this region (da Silva et al., 2011; Ragozo et al., 2010). Also, in Africa, Mercier et al. typed 69 strains isolated from domestic animals (mostly chickens) from Gabon with 13 microsatellite loci to reveal 27 genotypes that could be structured into two populations (Mercier et al., 2010). These results are consistent with those reported for African isolates by Lehmann et al. (2006), but no previously described isolates were included to identify whether the populations described here were the same or novel.
2.1.2 Toxoplasma genotypes in wildlife
Most of the studies discussed above only sampled or analyzed strains from a small fraction (i.e. humans and domestic animals) of the host range utilized by T. gondii. It is telling, though, that a large majority of unique strains have been identified by studies of T. gondii in wildlife, advancing our understanding of the extent of genetic diversity in this species. Intriguingly, the few wildlife samples that were included in the Howe and Sibley 1995 study (2 white tailed deer, 2 bear, 1 rodent, 1 turkey, and 1 dove) were genetically distinct from the three major clones, with some of them potentially representing recombinants of the three clonotypes (Howe and Sibley, 1995). This suggested that a productive sexual stage for this parasite exists in nature and that wider sampling, more representative of T. gondii’s broad host range, would reveal much greater genetic diversity. In several genotyping reports of T. gondii in wildlife this has proved to be true.
In North America, multiple genetically diverse genotypes have been identified circulating in wild animals. From a cougar in Canada, Lehmann et al. (2000) identified a genetically highly divergent strain (from archetypal Types I, II, and III identified in Howe and Sibley (1995)) using DNA sequencing, and this result was later confirmed by RFLP typing with 10 markers (Dubey et al., 2008b). This same RFLP typing scheme, originally reported in Su et al. (2006), has been applied in other studies of terrestrial wildlife in the US and Canada to provide similar results. Eighty-one of the RFLP genotypes identified using this scheme have been classified according to a numbering system (i.e. RFLP genotype #1, #2, #3, etc.) on the public T. gondii genome database, Toxodb.org (Gajria et al., 2008). We will refer to RFLP genotypes using this classification, except for strains of the Type I, Type II, Type III, and Type X (see below) lineages (Toxodb.org designated RFLP genotypes 10, 1, 2, and 5, respectively) which we will refer to with their traditional ‘Type’ nomenclature. Strains that have an RFLP type that has not been described on Toxodb.org will be referred to with their original isolate designation.
Of 31 total samples pooled from several studies, 16 were identified as non-archetypal, including 4/15 white-tailed deer, 2/2 bears, 1/2 skunks, 5/6 raccoons, and 4/6 coyotes (Dubey et al., 2008b; Dubey et al., 2010b; Dubey et al., 2007c; Dubey et al., 2008d). Some of these distinct strains likely represent recombinants of the three main clonal types based on their allelic combinations, but this cannot be confirmed without further sequencing analysis, since this typing scheme was specifically designed to detect polymorphisms inherent to the archetypal strains (Su et al., 2006). It is increasingly evident that apparent recombinants of the three archetypal lineages detected by RFLP analysis are often later re-classified by DNA sequencing as genetically distinct admixtures bearing many non-archetypal alleles (Boyle et al., 2006; Frazao-Teixeira et al., 2011; Grigg et al., 2001a; Khan et al., 2006; Su et al., 2010).
Toxoplasma gondii has also recently been identified as an important disease agent in marine mammals in North America (see Table 1 for details). Genetic studies of marine mammal isolates have contributed greatly to our understanding of how this terrestrial pathogen has infiltrated the marine environment. Miller et al. (2004) applied a combination of RFLP analyses and DNA sequencing to show that a genetically distinct strain termed ‘Type X’ was a major cause of disease in Southern sea otters stranding along the Morro Bay area of California. These results were corroborated by application of the RFLP typing to an additional 39 sea otter isolates to independently show that ‘Type X’ was the major strain causing disease in sea otters of Morro Bay (Sundar et al., 2008). This result was contrasted with T. gondii strains isolated from sea otters near Monterey Bay, CA that were mainly infected with ‘Type II’ strains or Toxodb.org designated RFLP genotype 3, which differs from ‘Type II’ at a single locus, Apico (Sundar et al., 2008). This same ‘Type X’ genotype was later shown to be present in wild felid hosts in the adjacent terrestrial environment of California, as well as in a filter-feeding invertebrate collected near the shoreline (Miller et al., 2008b). This study provided evidence for a mechanism through which this terrestrial parasite could infiltrate the marine environment via land-to-sea run-off and bio-concentration of oocysts in prey species of sea otters and other marine mammals (Miller et al., 2008b). Toxoplasma gondii isolates from dolphins in the US (Dubey et al., 2008a) and Canada (Dubey et al., 2009a) have also been extensively PCR-RFLP genotyped, revealing two ‘Type II’ strains, one strain with RFLP genotype 3 (as designated on Toxodb.org), and one atypical genotype in four isolates analyzed. Overall, reports from wildlife in North America suggest diverse strains are circulating in this region despite apparent limited strain diversity in domestic animal and human samples (Howe and Sibley, 1995).
Wildlife T. gondii isolates from other parts of the world have been genetically characterized as well. The standardized RFLP scheme has been applied to isolates from synanthropic rodents, including a seronegative mouse and rat (Araujo et al., 2010), captive animals, including a red-handed howler monkey and a jaguarundi, and a free-ranging black eared opossum (Pena et al., 2010) from Brazil to reveal they were all infected with non-archtypal strains, two of which had not been identified previously. Also in Brazil, an extensive RFLP analysis was completed on isolates from 36 capybaras that identified 16 total genotypes, seven of which were unique to this sample set (Yai et al., 2009). Importantly, only five of the isolates could be classified as a classic archetypal strain (‘Type III’). However, without DNA sequencing, definitive classification as an archetypal strain is not possible in regions where genetic diversity is substantial. It is quite possible that these strains will harbor non-archetypal alleles and ultimately be resolved as genetically distinct admixtures. Three of the 36 samples were found to be infected with a mixture of T. gondii strains (Yai et al., 2009), which could promote the T. gondii sexual cycle in the definitive felid host. Genetic data has also been reported from the United Arab Emirates and Qatar showing two atypical and one classic ‘Type II’ genotype present in isolates from four sand cats (Dubey et al., 2010a). The unusually high prevalence of symptomatic toxoplasmosis in this felid species raises interesting questions as to the universality of felids as functional definitive hosts, though several factors could be contributing to disease in these cases (Dubey et al., 2010a).
This trend towards apparently increased genetic diversity from wildlife samples is not absolute, however, as recent studies of wildlife in France and Norway have reported high prevalence of archetypal strains, specifically the ‘Type II’ strain. From 45 French isolates, including 21 from wild boar (Richomme et al., 2009), 12 from roe deer, nine from fox, and one each from mouflon, red deer and mallard (Aubert et al., 2010), RFLP typed at three loci and microsatellite typed at six loci, only ‘Type II’ strains were identified. Similarly in the Svalbard archipelago of Norway, the majority of 55 total isolates from arctic foxes were either ‘Type II,’ ‘Type III’ or RFLP genotype 3 (Prestrud et al., 2008). It is possible that the low levels of diversity in this instance could be attributed to a founder effect, given the isolated nature of this location (Prestrud et al., 2008).
Genetic studies of T. gondii in Australia have been particularly limited, though interest in T. gondii in this area has risen due to recent reports of fatal disease in threatened marsupial fauna (Basso et al., 2007; Bermudez et al., 2009; Dubey and Crutchley, 2008; Hartley, 2006; Obendorf et al., 1996). Efforts have accordingly been made to identify parasite genotypes circulating in this area. Parameswaran et al. (2010) applied PCR and DNA sequencing to identify eight different non-archetypal and two recombinant strains from a wombat, a wallaby, two woylies, a mouse, a meerkat (from the Perth Zoo), and eight kangaroo samples. Two of the kangaroo samples and one woylie possessed only archetypal alleles at the few loci amplified indicating these animals were likely infected by ‘Type I’ or ‘Type II’ strains, and one kangaroo harbored a mixed-strain infection (Parameswaran et al., 2010). Non-archetypal or recombinant strains were also obtained from domestic animal samples (1 horse and 1 goat), along with classic ‘Type I’ strains (1 cat and 1 goat) (Parameswaran et al., 2010). That the recombinant strains were truly recombinants was supported by the high resolution of DNA sequencing in this case.
Overall, aside from studies from certain locales in Europe there appears to be a high level of diversity present in wildlife isolates, including those from areas previously reported to be genetically homogenous, such as North America. Whether this is suggestive of a distinct sylvatic cycle for T. gondii is an important topic that will be discussed further below.
2.1.3 Population structure: reassessment
Considering the extent of genetic diversity and evidence for recombination present in wildlife samples, the picture that emerges for the population structure of T. gondii is more complex than a limited number of sexual events that have potentiated the emergence of just a few dominant clones (Khan et al., 2007; Sibley and Ajioka, 2008; Sibley et al., 2009; Su et al., 2003). Certainly the dominance of a limited number of genotypes in certain sample sets and from widely geographically dispersed locales is truly remarkable, but it does not necessarily lead to classification as a clonal population (Ajzenberg et al., 2002a; Howe and Sibley, 1995; Tibayrenc and Ayala, 1991). Based on the studies described above, the T. gondii population structure is in some cases highly clonal, in others it appears to be epidemic and overall, should be classified as intermediate, with clear influences from both clonal expansion of strains and sexual recombination (Lehmann et al., 2004; Lehmann et al., 2006). This would intuitively be the case for an organism with numerous mechanisms for clonal expansion in its lifecycle and a widespread host in which sexual recombination can occur. To what degree either clonal or sexual propagation can be detected in a sample set is dependent on several factors including: 1) the number and genomic location of genetic markers used, 2) the polymorphism present in the markers, 3) the resolution of the techniques used to detect polymorphism and 4) the host and geographic location of origin of the isolates.
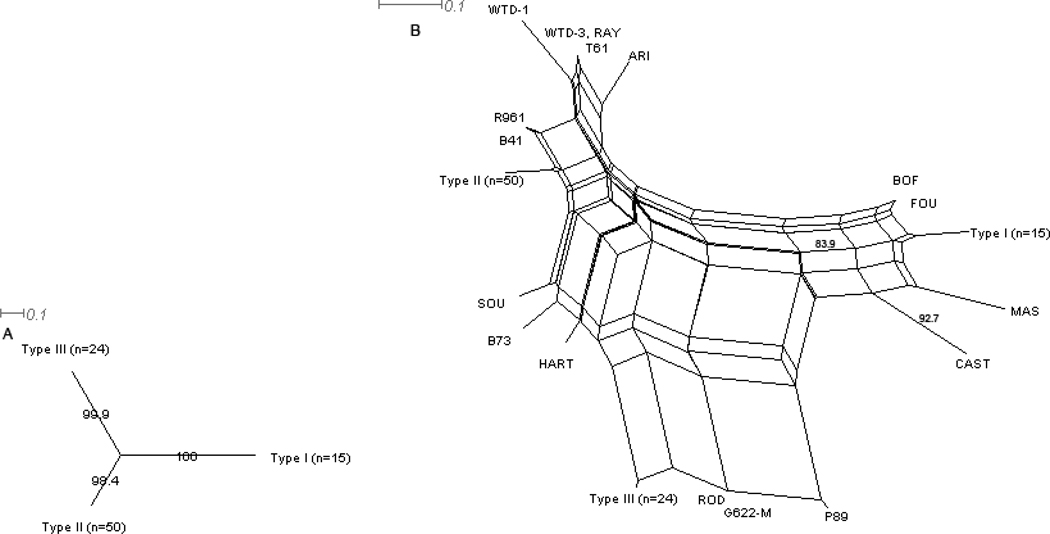
To help illustrate these important points we re-analyzed the Howe and Sibley 1995 RFLP data using the Neighbor-Net method with SplitsTree4 software (Huson and Bryant, 2006). Though this method does not create a true phylogenetic history of genotypes, it has been applied in recent T. gondii genetic studies due to its advantages in visually capturing the character conflicts in nucleotide data between genotypes that have undergone evolutionary events such as genetic exchange (recombination), gene conversion, or homoplasy and are therefore better represented by a network of nodes connected by multiple edges rather than as a bifurcating tree (Dubey et al., 2008c; Pena et al., 2008). First, we specifically input only those isolates that were members of the three clonal lineages, Types I, II, and III (essentially three data points) to illustrate the striking simplicity with which the vast majority (84%, 89/106) of the data can be described (Figure 1A). However, addition of the remaining samples (n=106 total), including those from wildlife, transforms the display into a complex network indicative of many potential pathways relating the additional strains to the clonal types and vice versa (Figure 1B).
Figure 1. SplitsTree4 analysis of Toxoplasma gondii RFLP data using the Neighbor-Net joining method.
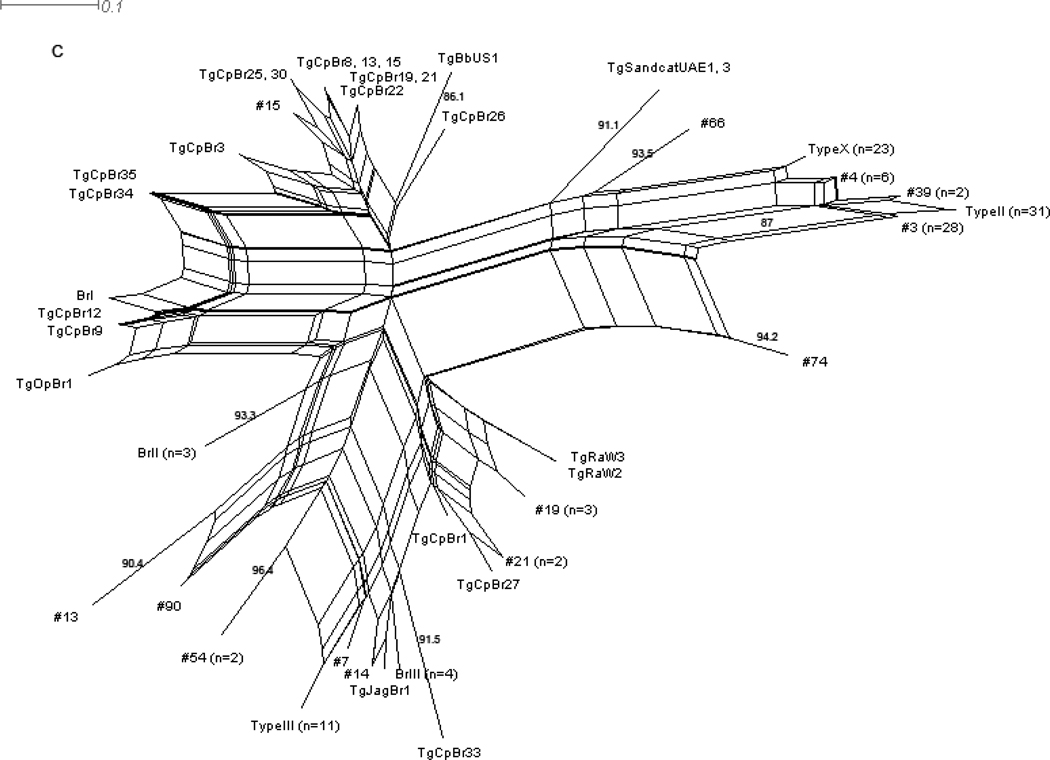
A. Input of only those strains which were classified by Howe and Sibley (1995) as one of the three main archetypal clones (Types I, II, and III) by 6 RFLP markers demonstrates the remarkable simplicity with which 84% (89/106 T. gondii isolates) of the isolates analyzed in this study can be described. B. Input of the complete data set from Howe and Sibley (1995) (n=106), including the limited number T. gondii isolates from wildlife, reveals an underlying complex population structure, despite the simplicity indicated in (A). The multiple edges connecting different nodes indicate the many possible pathways that can describe relationships between genotypes, some of which are likely indicative of recombination. C. Analysis of 147 wildlife isolates typed with 11 RFLP markers by various studies* reveals a slightly different and more complex population structure than that presented in B, highlighting the importance of including a sample set more inclusive of T. gondii’s expansive host range and utilizing typing schemes with higher resolution. Compared to B, which includes mostly samples from humans and domestic animals, Type II and III strains are still revealed as dominant clones, but Type I strains are absent. Additional dominant clones are revealed in wildlife samples as well, including Type X. High genotypic diversity related through a highly reticulated network is again consistent with a viable sexual cycle for T. gondii in nature. D. Analysis of 182 isolates from pigs in the US RFLP typed with 11 markers by Velmurugan et al. (2009) reveals a much more complex picture of the T. gondii population structure for domestic animals in the US than that originally reported in Howe and Sibley (1995), reiterating the importance of more extensive genotyping. Similar to the wildlife samples, Types II and III are dominant clones and Type I strains are absent. Additional clonal types dominate as well, including RFLP genotypes 3, 4, and 8**. *Wildlife RFLP data was obtained from Arajuo et al. (2010), Dubey et al. (2010a, b, 2009a, 2008a, b, d, 2007c), Pena et al. (In Press), Prestrud et al. (2008), Sundar et al. (2008), Yai et al. (2009). **Numbers refer to RFLP genotype designations reported on Toxodb.org. Bootstrap values (1000 replicates) are only shown for those edges with greater than or equal to 80% support. The distance measure used was uncorrected P. Note: Samples with mixed strain infections or incomplete typing data were removed from the wildlife data set.
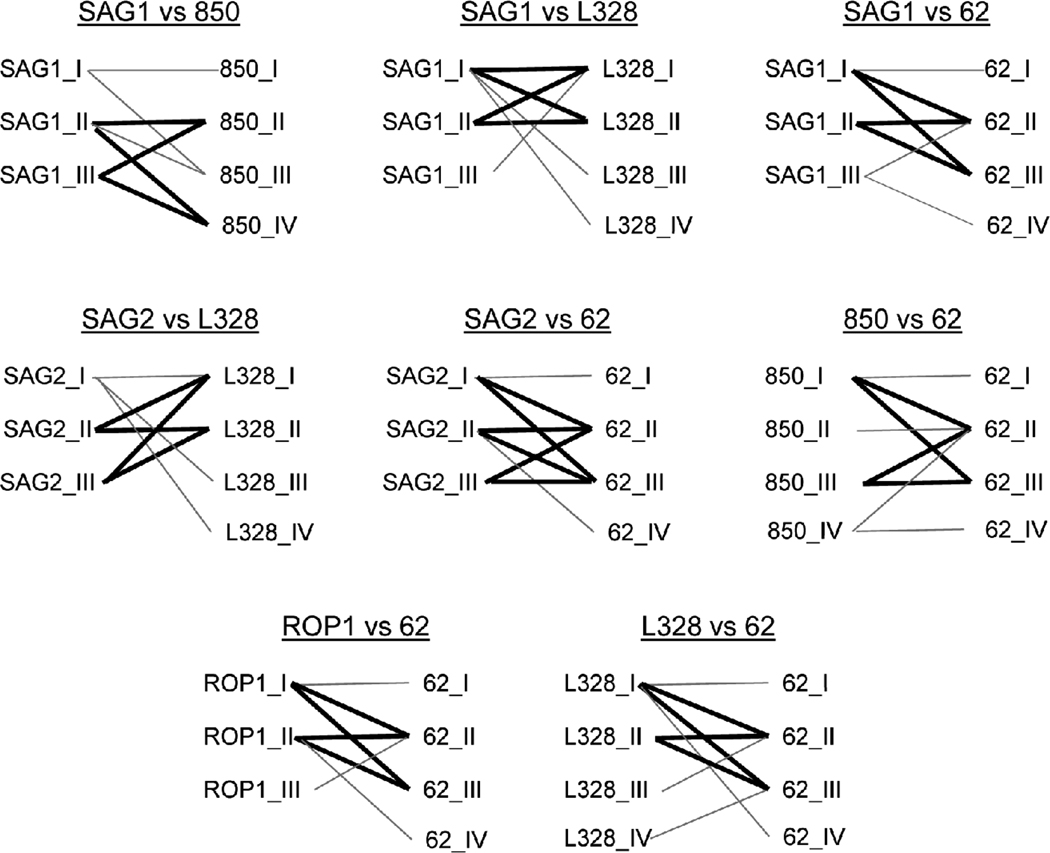
As was reported previously (Howe and Sibley, 1995), implicating sexual recombination as the mechanism for convoluting relatedness among the genotypic data, opposed to events such as homoplasy, gene conversion, or other phenomena that can create conflicts in molecular data (Morrison, 2005), is supported by comparing allele combinations across loci. In total, eight of the 15 possible locus pair comparisons have at least one example of bi-allelic combinations compatible with a recombination (or at least reassortment) event in that all four expected combinations of alleles are present in the sample set (Figure 2). These data show evidence for recombination not only among the three main clonal types, but also between exotic strains (see SAG1 vs. 850 in Figure 2). As noted above, a potential caveat to interpreting strains as recombinants of the 3 main clonal lineages with RFLP typing is that certain of these isolates have been found to harbor unique alleles at the loci in question upon further sequencing analysis (Boyle et al., 2006; Frazao-Teixeira et al., 2011; Khan et al., 2006; Su et al., 2010). However, this is not consistently the case (Parameswaran et al., 2010), and it is likely that at least a certain percentage of isolates are truly recombinants of either the archetypal clonal lines or archetypal and atypical strains. In fact, sequencing analysis of strain MAS, included in the 1995 study, confirmed that it harbored alleles that were likely the result of meiotic recombination (Grigg et al., 2001a). In contrast, strain P89, originally classified as a I/III recombinant (Howe and Sibley, 1995), was later found to likely represent one of the parent lineages that gave rise to the archetypal I, II, and III lineages (Boyle et al., 2006).
Figure 2. Bi-allelic analysis for recombination among Toxoplasma gondii isolates.
Lines connect allelic combinations present in the RFLP genotypes reported by Howe and Sibley (1995) to describe 106 T. gondii isolates. Bold lines indicate the hour glass shape expected when all four combinations of alleles that would be expected after a recombination (or reassortment) event are present in the isolates. Bi-allelic combinations that did not show evidence for recombination include: SAG1 vs SAG2, SAG1 vs ROP1, SAG2 vs 850, SAG2 vs ROP1, 850 vs ROP1, 850 vs L328, and ROP1 vs L328 (not shown).
Intriguingly, when we analyzed more recent RFLP data pooled from the various studies of wildlife isolates discussed above, a somewhat different picture emerges (Figure 1C). Among the 147 isolates analyzed, ‘Type II’ and ‘Type III’ are still dominant clones, accounting for 42/147 (29%) of the samples. However, Type I strains are absent and other clones are found to dominate, including ‘Type X’ (23/147, 16%) and RFLP genotype 3 (28/147, 19%), which together account for more samples than Types II and III. The overall genotypic diversity is also greater, with 36 distinct genotypes present in the 147 wildlife isolates (0.24 genotypes per isolate) compared to 14 in the Howe and Sibley 1995 data (0.13 genotypes per isolate), highlighting the effects a more encompassing sample set and more sensitive genotyping markers can have in changing perceptions of the genetic structure.
Until now, the consensus view of the global population structure of T. gondii based on recent studies in Europe, the Americas, Africa, and, to a limited extent, Asia (Khan et al., 2007; Lehmann et al., 2006; Mercier et al., 2010) was of a diverse array of genotypes derived from infrequent admixture among four ancestral populations and punctuated over time by recombination and clonal expansions (Sibley and Ajioka, 2008; Sibley et al., 2009). The degree to which recombination or clonal propagation has been interpreted as shaping T. gondii’s population structure has varied depending on the study and the samples analyzed, likely indicating that no study has truly analyzed a sample set fully representative of the T. gondii population. Indeed, many recent and unique strains described from both domestic animal and wildlife sources, such as those described in Brazil (da Silva et al., 2011; Frazao-Teixeira et al., 2011; Pena et al., 2008; Pena et al., 2010; Ragozo et al., 2010; Yai et al., 2009), the US (Dubey et al., 2010b), and Australia (Parameswaran et al., 2010) were not included in these previous global analyses (Khan et al., 2007; Lehmann et al., 2006). In fact, in Brazil alone at least 88 unique genotypes have been reported based on RFLP typing (da Silva et al., 2011) and only a small fraction of these have been incorporated in studies mapping the T. gondii population genetic structure. A combination of increased sample sizes that are more representative of T. gondii’s host range and of more extensive genetic data derived from each isolate will lead to further and potentially extensive revisions of the current consensus.
3. Sylvatic and domestic cycles: do they exist?
The high diversity in T. gondii genotypes isolated from wildlife samples as compared to those from domestic animals raises the question as to whether distinct gene pools exist for domestic and sylvatic hosts. The relevance of this question derives from concern that when distinct parasite gene pools overlap, most likely due to human environmental encroachment, disease outbreaks may occur in either human/domestic animal or wildlife populations due to exposure of hosts to novel parasite genotypes (Bengis et al., 2004; Cleaveland et al., 2001; Daszak et al., 2000; Thompson et al., 2009). Unfortunately, no sampling efforts or genetic analyses of T. gondii have been systematically applied in a way that would allow for rigorous comparison of domestic and sylvatic gene pools. It is possible that wildlife samples are perceived to be more genetically diverse than domestic samples because: 1. more extensive and sensitive typing has been applied to the more recently acquired wildlife isolates and/or 2. a highly clinically biased sample set has been used repeatedly in multiple studies to represent strains in domestic animals and humans.
We have compiled some recently reported data to help illustrate factors that likely will need to be accounted for when addressing this complex but important question. First, it is evident that genotype prevalence can vary both between host species and over geographic distributions. It has been noted previously that parasite genotype distributions differ between humans and domestic animals (Howe and Sibley, 1995), which is an important factor in determining which host species should be used to represent the domestic cycle. Additional evidence for this emerges when we re-analyze recently reported RFLP data for 182 isolates from pigs in the US (Velmurugan et al., 2009) using the SplitsTree4 software as described above. Comparing the network generated for the pig isolates (Figure 1D) to that for the data reported in the Howe and Sibley 1995 study (Figure 1B), in which the domestic cycle was largely represented by human isolates, reveals some important differences. “Type II’ and ‘Type III’ are also dominant clones in the pig population, but, in contrast to the human population, ‘Type I’ strains are absent, confirming what was reported originally (Howe and Sibley, 1995). It is also notable that other clones absent from the human sample set, including RFLP genotypes 3, 4 and 8, are quite prevalent in these pigs, but this difference may be attributable to the higher resolution typing applied to the pig sample set. Overall, the distribution of dominant clones in the pig data is more similar to that in the wildlife sample set (Figure 1C), but differences exist here as well, including the greater number of unique genotypes and the increased prevalence of ‘Type X’ strains (largely due to sea otter samples) in the wildlife data.
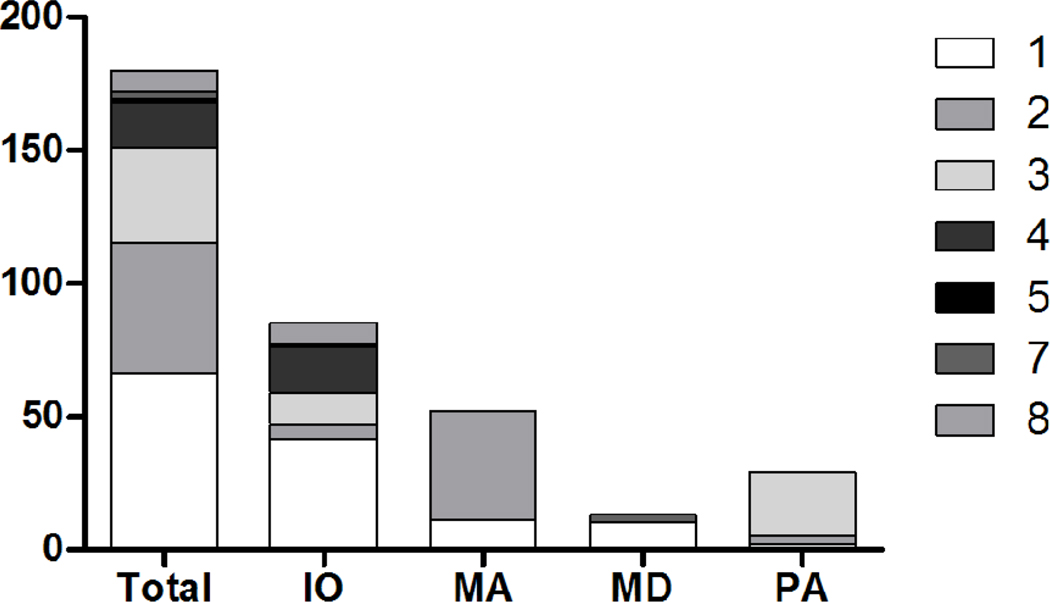
Geographic effects can also play a role in shaping our perception of which parasite genotypes are associated with which host species. This is certainly the case on a global level (e.g. North vs. South America), but has been noted on more local levels as well. For example, in Gabon, Mercier et al. (2010) provided evidence for distinct T. gondii populations between chickens acquired in different towns, which ranged from 105 to 510 kilometers distant. As mentioned above, Sundar et al. (2008) also found distinct strain compositions between the sea otters of Monterey Bay and Morro Bay, California, which are separated by approximately 200 kilometers. Similarly, when we re-examined the pig isolate data reported by Velmurugan et al. (2009) with pig genotypes classified by state of origin, different proportions of genotypes were found in different geographic sub-groupings (Figure 3).
Figure 3. RFLP genotype distribution among pig samples from the US.
Colored blocks indicate the proportion of T. gondii isolates from pigs from various states in the US that are accounted for by RFLP genotype numbers 1–5, 7, and 8 (Numbers refer to those reported on Toxodb.org). Markedly different genotype distributions from an identical host species among the 4 states illustrates the importance of geographic effects on strain prevalence. Pig RFLP data is from Velmurugan et al. (2009). Note: isolates not typed at all loci were removed from the analysis. IO=Iowa, MA=Massachusetts, MD=Maryland, PA=Pennsylvania
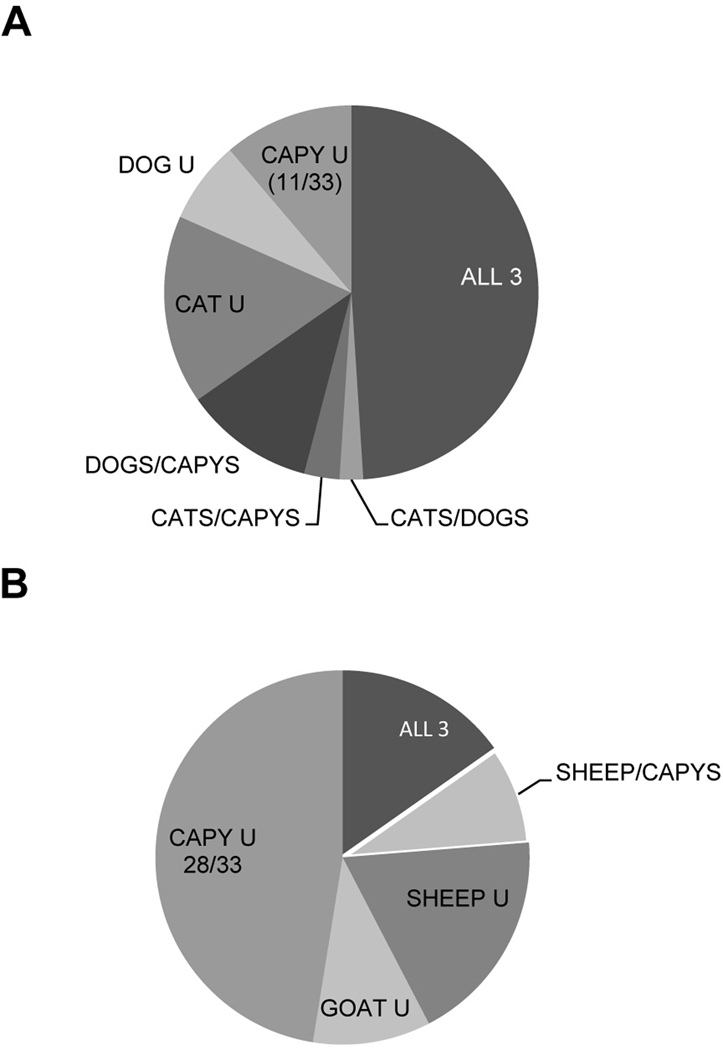
In contrast to these data, however, Lehmann et al. (2004) found no difference measured by the Fst statistic between chicken isolates from two locations over 600 kilometers apart in Brazil, perhaps indicating more expansive transmission in this region. Brazil is also one of the few locations where recent sampling efforts have identified a significant number of wildlife and domestic animal isolates with a comparable typing scheme in a localized area. In the state of Sao Paulo, a standardized RFLP typing scheme has been applied to isolates from 36 capybaras (Yai et al., 2009), 19 dogs (Dubey et al., 2007a), 44 cats (Pena et al., 2008), 16 sheep and 10 goats (Ragozo et al., 2010). Using the capybara samples as representative of wildlife, we compared the proportions of shared genotypes between wildlife and domestic carnivores (cats and dogs) (Figure 4A) and between wildlife and domestic herbivores (sheep and goats) (Figure 4B). In both cases there were several genotypes that were unique at the host species level, but comparing capybaras to domestic carnivores revealed that the genotypes for 22 out of the 33 capybara samples (mixed infections excluded) were also found in the domestic animal samples in contrast to only five isolates sharing genotypes with domestic herbivore samples (Figure 4A and B). Though it is unclear whether these results could be attributed to geographic differences within the state of Sao Paulo, they do give weight to the notion that defining the T. gondii population circulating in domestic animals could be highly dependent upon which host species are chosen to represent the domestic cycle.
Figure 4. Comparison of Toxoplasma gondii genotypes from wildlife and domestic carnivores and herbivores in Sao Paulo state, Brazil.
A. RFLP genotypes from T. gondii isolates from capybaras (representative of wildlife) and cats and dogs (representative of domestic carnivores) were analyzed for their distribution among seven categories: 1. Genotypes found in all three species (ALL3); 2. Genotypes found in cats and dogs only (CATS/DOGS); 3. Genotypes found in cats and capybaras only (CATS/CAPYS); 4. Genotypes found in dogs and capybaras only (DOGS/CAPYS); and genotypes that were unique to 5. Cats (CAT U); 6. Dogs (DOG U); and 7. Capybaras (CAPY U). Results show that many of the isolates possessed genotypes that were unique to their respective species of origin within this sample set, though nearly half of the isolates possessed a genotype found in all three species. Only 11 of the 33 total isolates from capybaras possessed a genotype that was not also found in a domestic carnivore. B. The same analysis was conducted to compare the capybara sample set to domestic herbivores, represented by sheep and goats. In contrast to the domestic carnivore comparison, nearly all the capybara isolates (28/33) possessed a genotype that was not found in domestic herbivores. All isolates were obtained from animals from Sao Paulo state, Brazil. Capybara data was from Yai et al. (2009), cat data from Pena et al. (2008), and dog data from Dubey et al. (2007a).
Overall, it is unclear to what extent distinct T. gondii gene pools exist between domestic and sylvatic cycles. It is likely that the answer to this question could vary with geographic location and host species chosen to define the respective cycles. Another potential caveat is variation in parasite genotype frequency over time. A systematic longitudinal sampling and genotyping effort across several host species over an urban to rural gradient in diverse geographic locations would do much to help address these concerns and provide more definitive conclusions.
4. Conclusions and Future Directions
The most important conclusion that can be drawn from recent genotyping efforts of T. gondii isolates from wildlife is that the so-called ‘exotic’ or ‘atypical’ strains are not insignificant anomalies in the population structure of this parasite, but rather important members of the gene pool that provide a much better representation of the vast host range utilized by this parasite. There are clearly genotypes that dominate in the T. gondii population in addition to the originally identified Types I, II, and III, even in areas previously thought to contain only these original genotypes, such as North America. Future efforts to define the genetic structure of this parasite should incorporate these newly found complexities while realizing that this will likely lead to refinement of many pre-conceived notions of simplicity in Toxoplasma population genetics.
It will also be important to gather empirical and experimental evidence for the inferences drawn from these new genetic studies of T. gondii. This is especially true for the debates over the extent to which clonal expansion and sexual recombination occur in nature and over the mechanistic basis for how certain clones come to dominate in the population structure. As discussed above, there are many potential caveats in extrapolating from a clonal sample set to conclude that a population is clonal in nature (Feil and Spratt, 2001; Maynard Smith et al., 1993). Likewise, the extent of recombination can be over-estimated depending upon the sampling techniques used (Awadalla, 2003; Prugnolle and De Meeus, 2010). Given the vast and widespread population of definitive felid hosts (approx. 90 million in the US alone (Dabritz and Conrad, 2010)), it is very likely that a frequent and productive sexual cycle exists for this parasite, despite a predominance of certain clones. Support for or against this possibility could be gained by attempting to determine the extent of co-infections in future sampling efforts, especially those which represent prey species of felids, because a simultaneous infection of two or more T. gondii strains is necessary for a productive cross to occur. In this regard, it is worth noting that the number of studies that have reported at least some mixed strain infections in a wide variety of hosts, including cats, has increased greatly since more sensitive genotyping techniques have been applied (Al-Kappany et al., 2010; Aspinall et al., 2003; Boughattas et al., 2010; Dubey et al., 2007b; Dubey et al., 2009b; Dubey et al., 2006a; Dubey et al., 2006b; Dubey et al., 2006c; Dubey et al., 2005; Elbez-Rubinstein et al., 2009; Lindstrom et al., 2008; Mercier et al., 2010; Parameswaran et al., 2010; Ragozo et al., 2010; Sundar et al., 2008; Yai et al., 2009). This is in spite of the fact that the majority of studies still obtain isolates through bioassay, a technique that has been shown to preferentially isolate certain strains over others (Lindstrom et al., 2008), and only test tissues from a single organ, even though evidence exists that different strains localize to different organs (Dubey, 1997; Saeij et al., 2005). Techniques now exist that provide the sensitivity needed to allow extensive parasite genotyping directly from multiple host tissues to adequately address this question (Opsteegh et al., 2010).
Another possibility would be to directly test oocyst samples for multiple genotypes using these more sensitive typing techniques. It would also be of great benefit to experimentally verify whether a mixed strain infection in a single intermediate host is necessary for outcrossing in the felid host, or if consumption of multiple, singly-infected prey over the course of the one to three week patent period of oocyst shedding would suffice. For studies of T. gondii infections in wildlife, it is also highly relevant to determine sources of infection and examine whether differences exist between domestic and wild felid species in their fecundity and ability to promote sexual outcrossing and/or self-mating for T. gondii. Clearly there is still much information to be gathered that could shape our perception of the likelihood and frequency of productive sexual crosses in nature.
Regardless of the ambiguities surrounding the extent of sexual recombination in the T. gondii population, it is undeniable that certain clones have expanded and persist in both domestic and wildlife populations over time. How and why this has occurred has been the focus of several studies over the past decade due to concerns that the mechanistic basis of clonal expansion may represent a threat for the emergence of virulent genotypes. Of the many mechanisms that exist in the T. gondii lifecycle for clonal propagation, it was originally proposed that the three archetypal strains, Types I, II, and III (Howe and Sibley, 1995), were unique among T. gondii strains in that they had recently acquired the ability for enhanced oral transmission of tissue cysts among intermediate hosts, thus allowing for clonal transmission that bypassed the sexual stage in the definitive host (Su et al., 2003). This hypothesis was based on limited laboratory studies and showed that tissue cysts from a small sample (3 isolates) of non-archetypal strains had greatly reduced oral transmission among laboratory mice compared to Types I, II, and III (Su et al., 2003). However, with additional sampling, it has since been shown that virtually all isolates of T. gondii, even rare genotypes, are capable of oral transmission among intermediate hosts (Khan et al., 2007), suggesting this trait may not be solely responsible for the global dominance of certain genotypes.
A case has also been made suggesting that vertical transmission, either transplacentally or through ingestion of milk, may play an important role in maintaining clonal dominance of certain strains in nature (Johnson, 1997). This hypothesis was advanced based upon knowledge of the presumably important role transplacental transmission plays for infection with the related parasite, Neospora caninum, in domestic cattle and dogs (Johnson, 1997). Numerous studies have demonstrated that T. gondii is capable of vertical transmission in a variety of hosts, including humans, but few have examined the possibility that strict vertical transmission is maintained over several generations in natural host populations. High prevalence of Toxoplasma in certain hosts that are geographically isolated from definitive felid hosts, such as the arctic fox populations mentioned above (Prestrud et al., 2008), are intriguing scenarios to speculate whether vertical transmission is important, but evidence has yet to be gathered to support this hypothesis. Much remains to be done to delineate the relative roles of vertical versus oral, carnivorous transmission in maintaining clonality in nature.
Given the much greater infective potential inherent to the definitive host stage (a single infected cat can shed hundreds of millions of oocysts into the environment (Dubey, 2001)), we recently tested whether self-mating during this stage was a viable mechanism for the expansion of a single genotype in nature. We extensively genotyped oocyst samples recovered from a reservoir linked to a waterborne T. gondii outbreak in humans in Brazil and demonstrated that they were an identical genetic clone (Wendte et al., 2010a). Combining these results with serologic typing evidence from infected people (Vaudaux et al., 2010) confirmed that the outbreak, which was attributed to ingestion of oocyst contaminated water, was indeed clonal and apparently the result of a selfing event in a felid definitive host (Wendte et al., 2010a). Since well over a hundred people were affected by this outbreak, this result demonstrated the major role selfing in the definitive host can play as a potential mechanism for clonal expansion of a disease-producing genotype in nature as compared to vertical transmission or oral transmission via carnivory (Wendte et al., 2010a). Importantly, self-mating was also the cause of a devastating clonal outbreak of the related parasite Sarcocystis neurona that caused a point-source mass mortality event in a threatened Southern sea otters due to oocyst/sporocyst contamination of waterways; a result that expands the explanatory scope of selfing as a mechanism of clonality to other tissue cyst coccidia (Wendte et al., 2010a).
The genetic basis for the expansion of certain clones has also been proposed in population genetic studies. The archetypal clones have been associated with certain alleles for the rhoptry kinase protein, ROP18, associated with virulence in the mouse model (Khan et al., 2009) and a monomorphic chromosome Ia (Khan et al., 2007), leading to the conclusion that these loci may contribute to the success of these clonotypes in nature. Fortunately, the identification of a genetically distinct, dominant clone circulating in wildlife, ‘Type X’ (see Figure 1C), should allow future comparative genomic analyses to further refine potential candidate genes accounting for the success of certain genotypes to carry forward in experimental studies.
Notably, while this manuscript was under review, a study was published further characterizing many ‘Type X’ and apparently ‘Type X-like’ strains at five intron and 3 antigen loci with a variety of phylogenetic and population analyses techniques (Khan et al., 2011). This study grouped ‘Type X’ and many closely related strains into a new ‘haplogroup 12’ according to the previously published methodology of Khan et al. (2007). Similar to our discussion above of the 2007 study, close examination of the data reveals that consistent delineations between certain haplogroups break down to varying degrees depending on the analysis technique used (Khan et al., 2011). In fact, inconsistent delineations between ‘haplogroup 12’ (‘Type X’) and ‘haplogroup 2’ (‘Type II’) due to a bi-allelic inheritance pattern at many of the loci analyzed led to the conclusion that ‘haplogroup 12’ (‘Type X’) strains are the result of a cross between ‘Type II’ and a distinct ancestral type (Khan et al., 2011). This intriguing conclusion again speaks to the importance of the sexual cycle in the T. gondii population structure as a mechanism for the emergence of new strains that can go on to dominate clonally. It is also telling that what were once considered minor (and sometimes insignificant) genetic differences by RFLP and other typing schemes among ‘Type X’ genotypes compared against ‘Type II’ genotypes were indicative of major genomic level diversity. As the field moves forward with much more extensive, whole-genome level analyses, it is likely that several strains once thought to be identical clones will be found to be comprised of multiple, diverse genetic backgrounds.
In many ways, genotyping studies of T. gondii in wildlife have caused researchers to re-consider established viewpoints of the population genetic structure and relative roles of the various lifecycle stages in shaping the population biology of this important zoonotic pathogen. Yet much work remains to be done to uncover the extent and implications of the parasite genetic diversity circulating in wild animal populations and the degree to which sylvatic and domestic cycles are synonymous or distinct. Future studies addressing these issues will be highly relevant to efforts aimed at minimizing disease in both wild and domestic populations.
Acknowledgements
Thanks to all members of the Grigg lab for helpful discussions. This work was supported by the Intramural Research program of the NIH and NIAID, grant #AI001018 and The Morris Animal Foundation wildlife training fellowship grant #D10ZO-416 (JMW). MEG is a scholar of the Canadian Institute for Advanced Research (CIFAR) Program for Integrated Microbial Biodiversity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there is no conflict of interest.
References
- Ajzenberg D, Banuls AL, Su C, Dumetre A, Demar M, Carme B, Darde ML. Genetic diversity, clonality and sexuality in Toxoplasma gondii. Int J Parasitol. 2004;34:1185–1196. doi: 10.1016/j.ijpara.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Ajzenberg D, Banuls AL, Tibayrenc M, Darde ML. Microsatellite analysis of Toxoplasma gondii shows considerable polymorphism structured into two main clonal groups. Int J Parasitol. 2002a;32:27–38. doi: 10.1016/s0020-7519(01)00301-0. [DOI] [PubMed] [Google Scholar]
- Ajzenberg D, Cogne N, Paris L, Bessieres MH, Thulliez P, Filisetti D, Pelloux H, Marty P, Darde ML. Genotype of 86 Toxoplasma gondii isolates associated with human congenital toxoplasmosis, and correlation with clinical findings. J Infect Dis. 2002b;186:684–689. doi: 10.1086/342663. [DOI] [PubMed] [Google Scholar]
- Al-Kappany YM, Rajendran C, Abu-Elwafa SA, Hilali M, Su C, Dubey JP. Genetic Diversity of Toxoplasma gondii Isolates in Egyptian Feral Cats Reveals New Genotypes. J Parasitol. 2010;96:1112–1114. doi: 10.1645/GE-2608.1. [DOI] [PubMed] [Google Scholar]
- Araujo JB, da Silva AV, Rosa RC, Mattei RJ, da Silva RC, Richini-Pereira VB, Langoni H. Isolation and multilocus genotyping of Toxoplasma gondii in seronegative rodents in Brazil. Vet Parasitol. 2010;174:328–331. doi: 10.1016/j.vetpar.2010.08.039. [DOI] [PubMed] [Google Scholar]
- Aspinall TV, Guy EC, Roberts KE, Joynson DH, Hyde JE, Sims PF. Molecular evidence for multiple Toxoplasma gondii infections in individual patients in England and Wales: public health implications. Int J Parasitol. 2003;33:97–103. doi: 10.1016/s0020-7519(02)00230-8. [DOI] [PubMed] [Google Scholar]
- Aubert D, Ajzenberg D, Richomme C, Gilot-Fromont E, Terrier ME, de Gevigney C, Game Y, Maillard D, Gibert P, Darde ML, Villena I. Molecular and biological characteristics of Toxoplasma gondii isolates from wildlife in France. Vet Parasitol. 2010;171:346–349. doi: 10.1016/j.vetpar.2010.03.033. [DOI] [PubMed] [Google Scholar]
- Awadalla P. The evolutionary genomics of pathogen recombination. Nat Rev Genet. 2003;4:50–60. doi: 10.1038/nrg964. [DOI] [PubMed] [Google Scholar]
- Basso W, Venturini MC, More G, Quiroga A, Bacigalupe D, Unzaga JM, Larsen A, Laplace R, Venturini L. Toxoplasmosis in captive Bennett's wallabies (Macropus rufogriseus) in Argentina. Vet Parasitol. 2007;144:157–161. doi: 10.1016/j.vetpar.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Bengis RG, Leighton FA, Fischer JR, Artois M, Morner T, Tate CM. The role of wildlife in emerging and re-emerging zoonoses. Rev Sci Tech. 2004;23:497–511. [PubMed] [Google Scholar]
- Bermudez R, Failde LD, Losada AP, Nieto JM, Quiroga MI. Toxoplasmosis in Bennett's wallabies (Macropus rufogriseus) in Spain. Vet Parasitol. 2009;160:155–158. doi: 10.1016/j.vetpar.2008.10.082. [DOI] [PubMed] [Google Scholar]
- Blackston CR, Dubey JP, Dotson E, Su C, Thulliez P, Sibley D, Lehmann T. High-resolution typing of Toxoplasma gondii using microsatellite loci. J Parasitol. 2001;87:1472–1475. doi: 10.1645/0022-3395(2001)087[1472:HRTOTG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Boothroyd JC, Grigg ME. Population biology of Toxoplasma gondii and its relevance to human infection: do different strains cause different disease? Curr Opin Microbiol. 2002;5:438–442. doi: 10.1016/s1369-5274(02)00349-1. [DOI] [PubMed] [Google Scholar]
- Boughattas S, Ben-Abdallah R, Siala E, Souissi O, Aoun K, Bouratbine A. Direct genotypic characterization of Toxoplasma gondii strains associated with congenital toxoplasmosis in Tunisia (North Africa) Am J Trop Med Hyg. 2010;82:1041–1046. doi: 10.4269/ajtmh.2010.09-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle JP, Rajasekar B, Saeij JP, Ajioka JW, Berriman M, Paulsen I, Roos DS, Sibley LD, White MW, Boothroyd JC. Just one cross appears capable of dramatically altering the population biology of a eukaryotic pathogen like Toxoplasma gondii. Proc Natl Acad Sci U S A. 2006;103:10514–10519. doi: 10.1073/pnas.0510319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle MA, Tanhauser SM, Dame JB, Sellon DC, Hines M, Ginn PE, MacKay RJ, Greiner EC. The nine-banded armadillo (Dasypus novemcinctus) is an intermediate host for Sarcocystis neurona. Int J Parasitol. 2001;31:330–335. doi: 10.1016/s0020-7519(01)00177-1. [DOI] [PubMed] [Google Scholar]
- Cleaveland S, Laurenson MK, Taylor LH. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:991–999. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen AW, Overdulve JP. Sex determination and sex differentiation in coccidia: gametogony and oocyst production after monoclonal infection of cats with free-living and intermediate host stages of Isospora (Toxoplasma) gondii. Parasitology. 1985;90(Pt 1):35–44. doi: 10.1017/s003118200004899x. [DOI] [PubMed] [Google Scholar]
- Costa KS, Santos SL, Uzeda RS, Pinheiro AM, Almeida MA, Araujo FR, McAllister MM, Gondim LF. Chickens (Gallus domesticus) are natural intermediate hosts of Neospora caninum. Int J Parasitol. 2008;38:157–159. doi: 10.1016/j.ijpara.2007.10.008. [DOI] [PubMed] [Google Scholar]
- da Silva RC, Langoni H, Su C, da Silva AV. Genotypic characterization of Toxoplasma gondii in sheep from Brazilian slaughterhouses: new atypical genotypes and the clonal type II strain identified. Vet Parasitol. 2011;175:173–177. doi: 10.1016/j.vetpar.2010.09.021. [DOI] [PubMed] [Google Scholar]
- Dabritz HA, Conrad PA. Cats and Toxoplasma: implications for public health. Zoonoses Public Health. 2010;57:34–52. doi: 10.1111/j.1863-2378.2009.01273.x. [DOI] [PubMed] [Google Scholar]
- Darde ML, Bouteille B, Pestre-Alexandre M. Isoenzymic characterization of seven strains of Toxoplasma gondii by isoelectrofocusing in polyacrylamide gels. Am J Trop Med Hyg. 1988;39:551–558. doi: 10.4269/ajtmh.1988.39.551. [DOI] [PubMed] [Google Scholar]
- Darde ML, Bouteille B, Pestre-Alexandre M. Isoenzyme analysis of 35 Toxoplasma gondii isolates and the biological and epidemiological implications. J Parasitol. 1992;78:786–794. [PubMed] [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD. Emerging infectious diseases of wildlife--threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- De Salvador-Guillouet F, Ajzenberg D, Chaillou-Opitz S, Saint-Paul MC, Dunais B, Dellamonica P, Marty P. Severe pneumonia during primary infection with an atypical strain of Toxoplasma gondii in an immunocompetent young man. J Infect. 2006;53:e47–e50. doi: 10.1016/j.jinf.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Demar M, Ajzenberg D, Maubon D, Djossou F, Panchoe D, Punwasi W, Valery N, Peneau C, Daigre JL, Aznar C, Cottrelle B, Terzan L, Darde ML, Carme B. Fatal outbreak of human toxoplasmosis along the Maroni River: epidemiological, clinical, and parasitological aspects. Clin Infect Dis. 2007;45:e88–e95. doi: 10.1086/521246. [DOI] [PubMed] [Google Scholar]
- Dubey JP. Infectivity and pathogenicity of Toxoplasma gondii oocysts for cats. J Parasitol. 1996a;82:957–961. [PubMed] [Google Scholar]
- Dubey JP. Pathogenicity and infectivity of Toxoplasma gondii oocysts for rats. J Parasitol. 1996b;82:951–956. [PubMed] [Google Scholar]
- Dubey JP. Tissue cyst tropism in Toxoplasma gondii: a comparison of tissue cyst formation in organs of cats, and rodents fed oocysts. Parasitology. 1997;115(Pt 1):15–20. doi: 10.1017/s0031182097008949. [DOI] [PubMed] [Google Scholar]
- Dubey JP. Advances in the life cycle of Toxoplasma gondii. Int J Parasitol. 1998;28:1019–1024. doi: 10.1016/s0020-7519(98)00023-x. [DOI] [PubMed] [Google Scholar]
- Dubey JP. Oocyst shedding by cats fed isolated bradyzoites and comparison of infectivity of bradyzoites of the VEG strain Toxoplasma gondii to cats and mice. J Parasitol. 2001;87:215–219. doi: 10.1645/0022-3395(2001)087[0215:OSBCFI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Dubey JP. Comparative infectivity of oocysts and bradyzoites of Toxoplasma gondii for intermediate (mice) and definitive (cats) hosts. Vet Parasitol. 2006;140:69–75. doi: 10.1016/j.vetpar.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Dubey JP. The history of Toxoplasma gondii--the first 100 years. J Eukaryot Microbiol. 2008;55:467–475. doi: 10.1111/j.1550-7408.2008.00345.x. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Crutchley C. Toxoplasmosis in wallabies (Macropus rufogriseus and Macropus eugenii): blindness, treatment with atovaquone, and isolation of Toxoplasma gondii. J Parasitol. 2008;94:929–933. doi: 10.1645/GE-1448.1. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Fair PA, Sundar N, Velmurugan G, Kwok OC, McFee WE, Majumdar D, Su C. Isolation of Toxoplasma gondii from bottlenose dolphins (Tursiops truncatus) J Parasitol. 2008a;94:821–823. doi: 10.1645/GE-1444.1. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Frenkel JK. Cyst-induced toxoplasmosis in cats. J Protozool. 1972;19:155–177. doi: 10.1111/j.1550-7408.1972.tb03431.x. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Frenkel JK. Feline toxoplasmosis from acutely infected mice and the development of Toxoplasma cysts. J Protozool. 1976;23:537–546. doi: 10.1111/j.1550-7408.1976.tb03836.x. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Gennari SM, Sundar N, Vianna MC, Bandini LM, Yai LE, Kwok CH, Suf C. Diverse and atypical genotypes identified in Toxoplasma gondii from dogs in Sao Paulo, Brazil. J Parasitol. 2007a;93:60–64. doi: 10.1645/GE-972R.1. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Lindsay DS, Saville WJ, Reed SM, Granstrom DE, Speer CA. A review of Sarcocystis neurona and equine protozoal myeloencephalitis (EPM) Vet Parasitol. 2001a;95:89–131. doi: 10.1016/s0304-4017(00)00384-8. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Lindsay DS, Speer CA. Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin Microbiol Rev. 1998;11:267–299. doi: 10.1128/cmr.11.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey JP, Lopez-Torres HY, Sundar N, Velmurugan GV, Ajzenberg D, Kwok OC, Hill R, Darde ML, Su C. Mouse-virulent Toxoplasma gondii isolated from feral cats on Mona Island, Puerto Rico. J Parasitol. 2007b;93:1365–1369. doi: 10.1645/GE-1409.1. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Lunney JK, Shen SK, Kwok OC, Ashford DA, Thulliez P. Infectivity of low numbers of Toxoplasma gondii oocysts to pigs. J Parasitol. 1996;82:438–443. [PubMed] [Google Scholar]
- Dubey JP, Mergl J, Gehring E, Sundar N, Velmurugan GV, Kwok OC, Grigg ME, Su C, Martineau D. Toxoplasmosis in captive dolphins (Tursiops truncatus) and walrus (Odobenus rosmarus) J Parasitol. 2009a;95:82–85. doi: 10.1645/GE-1764.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey JP, Miller NL, Frenkel JK. Characterization of the new fecal form of Toxoplasma gondii. J Parasitol. 1970a;56:447–456. [PubMed] [Google Scholar]
- Dubey JP, Miller NL, Frenkel JK. The Toxoplasma gondii oocyst from cat feces. J Exp Med. 1970b;132:636–662. doi: 10.1084/jem.132.4.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey JP, Moura L, Majumdar D, Sundar N, Velmurugan GV, Kwok OC, Kelly P, Krecek RC, Su C. Isolation and characterization of viable Toxoplasma gondii isolates revealed possible high frequency of mixed infection in feral cats ( Felis domesticus) from St Kitts, West Indies. Parasitology. 2009b;136:589–594. doi: 10.1017/S0031182009006015. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Pas A, Rajendran C, Kwok OC, Ferreira LR, Martins J, Hebel C, Hammer S, Su C. Toxoplasmosis in Sand cats (Felis margarita) and other animals in the Breeding Centre for Endangered Arabian Wildlife in the United Arab Emirates and Al Wabra Wildlife Preservation, the State of Qatar. Vet Parasitol. 2010a;172:195–203. doi: 10.1016/j.vetpar.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey JP, Quirk T, Pittt JA, Sundar N, Velmurugan GV, Kwok OC, Leclair D, Hill R, Su C. Isolation and genetic characterization of Toxoplasma gondii from raccoons (Procyon lotor), cats (Felis domesticus), striped skunk (Mephitis mephitis), black bear (Ursus americanus), and cougar (Puma concolor) from Canada. J Parasitol. 2008b;94:42–45. doi: 10.1645/GE-1349.1. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Rajendran C, Ferreira LR, Kwok OC, Sinnett D, Majumdar D, Su C. A new atypical highly mouse virulent Toxoplasma gondii genotype isolated from a wild black bear in Alaska. J Parasitol. 2010b;96:713–716. doi: 10.1645/GE-2429.1. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Saville WJ, Stanek JF, Lindsay DS, Rosenthal BM, Oglesbee MJ, Rosypal AC, Njoku CJ, Stich RW, Kwok OC, Shen SK, Hamir AN, Reed SM. Sarcocystis neurona infections in raccoons (Procyon lotor): evidence for natural infection with sarcocysts, transmission of infection to opossums (Didelphis virginiana), and experimental induction of neurologic disease in raccoons. Vet Parasitol. 2001b;100:117–129. doi: 10.1016/s0304-4017(01)00500-3. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Shen SK, Kwok OC, Thulliez P. Toxoplasmosis in rats (Rattus norvegicus): congenital transmission to first and second generation offspring and isolation of Toxoplasma gondii from seronegative rats. Parasitology. 1997;115(Pt 1):9–14. doi: 10.1017/s0031182097008950. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Su C, Cortes JA, Sundar N, Gomez-Marin JE, Polo LJ, Zambrano L, Mora LE, Lora F, Jimenez J, Kwok OC, Shen SK, Zhang X, Nieto A, Thulliez P. Prevalence of Toxoplasma gondii in cats from Colombia, South America and genetic characterization of T. gondii isolates. Vet Parasitol. 2006a;141:42–47. doi: 10.1016/j.vetpar.2006.04.037. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Sundar N, Hill D, Velmurugan GV, Bandini LA, Kwok OC, Majumdar D, Su C. High prevalence and abundant atypical genotypes of Toxoplasma gondii isolated from lambs destined for human consumption in the USA. Int J Parasitol. 2008c;38:999–1006. doi: 10.1016/j.ijpara.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Sundar N, Nolden CA, Samuel MD, Velmurugan GV, Bandini LA, Kwok OC, Bodenstein B, Su C. Characterization of Toxoplasma gondii from raccoons (Procyon lotor), coyotes (Canis latrans), and striped skunks (Mephitis mephitis) in Wisconsin identified several atypical genotypes. J Parasitol. 2007c;93:1524–1527. doi: 10.1645/GE-1245.1. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Sundar N, Pineda N, Kyvsgaard NC, Luna LA, Rimbaud E, Oliveira JB, Kwok OC, Qi Y, Su C. Biologic and genetic characteristics of Toxoplasma gondii isolates in free-range chickens from Nicaragua, Central America. Vet Parasitol. 2006b;142:47–53. doi: 10.1016/j.vetpar.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Velmurugan GV, Ulrich V, Gill J, Carstensen M, Sundar N, Kwok OC, Thulliez P, Majumdar D, Su C. Transplacental toxoplasmosis in naturally-infected white-tailed deer: Isolation and genetic characterisation of Toxoplasma gondii from foetuses of different gestational ages. Int J Parasitol. 2008d;38:1057–1063. doi: 10.1016/j.ijpara.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Vianna MC, Sousa S, Canada N, Meireles S, Correia da Costa JM, Marcet PL, Lehmann T, Darde ML, Thulliez P. Characterization of Toxoplasma gondii isolates in free-range chickens from Portugal. J Parasitol. 2006c;92:184–186. doi: 10.1645/GE-652R.1. [DOI] [PubMed] [Google Scholar]
- Dubey JR, Bhaiyat MI, de Allie C, Macpherson CN, Sharma RN, Sreekumar C, Vianna MC, Shen SK, Kwok OC, Miska KB, Hill DE, Lehmann T. Isolation, tissue distribution, and molecular characterization of Toxoplasma gondii from chickens in Grenada, West Indies. J Parasitol. 2005;91:557–560. doi: 10.1645/GE-463R. [DOI] [PubMed] [Google Scholar]
- Elbez-Rubinstein A, Ajzenberg D, Darde ML, Cohen R, Dumetre A, Yera H, Gondon E, Janaud JC, Thulliez P. Congenital toxoplasmosis and reinfection during pregnancy: case report, strain characterization, experimental model of reinfection, and review. J Infect Dis. 2009;199:280–285. doi: 10.1086/595793. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Spratt BG. Recombination and the population structures of bacterial pathogens. Annu Rev Microbiol. 2001;55:561–590. doi: 10.1146/annurev.micro.55.1.561. [DOI] [PubMed] [Google Scholar]
- Ferguson DJ, Hutchiso Wm, Dunachie JF, Siim JC. Ultrastructural Study of Early Stages of Asexual Multiplication and Microgametogony of Toxoplasma-Gondii in Small-Intestine of Cat. Acta Pathologica Et Microbiologica Scandinavica Section B-Microbiology B. 1974;82:167–181. doi: 10.1111/j.1699-0463.1974.tb02309.x. [DOI] [PubMed] [Google Scholar]
- Ferguson DJP. Toxoplasma gondii and sex: essential or optional extra? Trends in Parasitology. 2002;18:355–359. [PubMed] [Google Scholar]
- Ferguson DJP, Hutchison WM, Siim JC. Ultrastructural Development of Macrogamete and Formation of Oocyst Wall of Toxoplasma-Gondii. Acta Pathologica Et Microbiologica Scandinavica Section B-Microbiology. 1975a;83:491–505. doi: 10.1111/j.1699-0463.1975.tb00130.x. [DOI] [PubMed] [Google Scholar]
- Ferguson DJP, Hutchison WM, Siim JC. Ultrastructural Study of Development of Macrogamete and Oocyst of Toxoplasma-Gondii. Journal of Protozoology. 1975b;22:A51–A51. [Google Scholar]
- Ferreira Ade M, Vitor RW, Gazzinelli RT, Melo MN. Genetic analysis of natural recombinant Brazilian Toxoplasma gondii strains by multilocus PCR-RFLP. Infect Genet Evol. 2006;6:22–31. doi: 10.1016/j.meegid.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Frazao-Teixeira E, Sundar N, Dubey JP, Grigg ME, de Oliveira FC. Multi-locus DNA sequencing of Toxoplasma gondii isolated from Brazilian pigs identifies genetically divergent strains. Vet Parasitol. 2011;175:33–39. doi: 10.1016/j.vetpar.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel JK, Dubey JP, Miller NL. Toxoplasma gondii in cats: fecal stages identified as coccidian oocysts. Science. 1970;167:893–896. doi: 10.1126/science.167.3919.893. [DOI] [PubMed] [Google Scholar]
- Frenkel JK, Ruiz A, Chinchilla M. Soil survival of toxoplasma oocysts in Kansas and Costa Rica. Am J Trop Med Hyg. 1975;24:439–443. doi: 10.4269/ajtmh.1975.24.439. [DOI] [PubMed] [Google Scholar]
- Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, Heiges M, Iodice J, Kissinger JC, Mackey AJ, Pinney DF, Roos DS, Stoeckert CJ, Jr, Wang H, Brunk BP. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:D553–D556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondim LS, Abe-Sandes K, Uzeda RS, Silva MS, Santos SL, Mota RA, Vilela SM, Gondim LF. Toxoplasma gondii and Neospora caninum in sparrows (Passer domesticus) in the Northeast of Brazil. Vet Parasitol. 2010;168:121–124. doi: 10.1016/j.vetpar.2009.09.055. [DOI] [PubMed] [Google Scholar]
- Grigg ME, Bonnefoy S, Hehl AB, Suzuki Y, Boothroyd JC. Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science. 2001a;294:161–165. doi: 10.1126/science.1061888. [DOI] [PubMed] [Google Scholar]
- Grigg ME, Ganatra J, Boothroyd JC, Margolis TP. Unusual abundance of atypical strains associated with human ocular toxoplasmosis. J Infect Dis. 2001b;184:633–639. doi: 10.1086/322800. [DOI] [PubMed] [Google Scholar]
- Grigg ME, Sundar N. Sexual recombination punctuated by outbreaks and clonal expansions predicts Toxoplasma gondii population genetics. Int J Parasitol. 2009;39:925–933. doi: 10.1016/j.ijpara.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley MP. Toxoplasma gondii infection in two common wombats (Vombatus ursinus) Aust Vet J. 2006;84:107–109. doi: 10.1111/j.1751-0813.2006.tb12242.x. [DOI] [PubMed] [Google Scholar]
- Hide G, Gerwash O, Morley EK, Williams RH, Hughes JM, Thomasson D, Elmahaishi MS, Elmahaishi KH, Terry RS, Smith JE. Does vertical transmission contribute to the prevalence of toxoplasmosis? Parassitologia. 2007;49:223–226. [PubMed] [Google Scholar]
- Howe DK, Honore S, Derouin F, Sibley LD. Determination of genotypes of Toxoplasma gondii strains isolated from patients with toxoplasmosis. J Clin Microbiol. 1997;35:1411–1414. doi: 10.1128/jcm.35.6.1411-1414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis. 1995;172:1561–1566. doi: 10.1093/infdis/172.6.1561. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Hutchison WM. Experimental transmission of Toxoplasma gondii. Nature. 1965;206:961–962. doi: 10.1038/206961a0. [DOI] [PubMed] [Google Scholar]
- Innes EA, Bartley PM, Buxton D, Katzer F. Ovine toxoplasmosis. Parasitology. 2009;136:1887–1894. doi: 10.1017/S0031182009991636. [DOI] [PubMed] [Google Scholar]
- Johnson AM. Speculation on possible life cycles for the clonal lineages in the genus toxoplasma. Parasitol Today. 1997;13:393–397. doi: 10.1016/s0169-4758(97)01129-0. [DOI] [PubMed] [Google Scholar]
- Khan A, Dubey JP, Su C, Ajioka JW, Rosenthal BM, Sibley LD. Genetic analyses of atypical Toxoplasma gondii strains reveal a fourth clonal lineage in North America. Int J Parasitol. 2011 doi: 10.1016/j.ijpara.2011.01.005. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Fux B, Su C, Dubey JP, Darde ML, Ajioka JW, Rosenthal BM, Sibley LD. Recent transcontinental sweep of Toxoplasma gondii driven by a single monomorphic chromosome. Proc Natl Acad Sci U S A. 2007;104:14872–14877. doi: 10.1073/pnas.0702356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Jordan C, Muccioli C, Vallochi AL, Rizzo LV, Belfort R, Jr, Vitor RW, Silveira C, Sibley LD. Genetic divergence of Toxoplasma gondii strains associated with ocular toxoplasmosis, Brazil. Emerg Infect Dis. 2006;12:942–949. doi: 10.3201/eid1206.060025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Taylor S, Ajioka JW, Rosenthal BM, Sibley LD. Selection at a single locus leads to widespread expansion of Toxoplasma gondii lineages that are virulent in mice. PLoS Genet. 2009;5:e1000404. doi: 10.1371/journal.pgen.1000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal FE, Cavazzana CL, de Andrade HF, Jr, Galisteo AJ, Jr, de Mendonca JS, Kallas EG. Toxoplasma gondii pneumonia in immunocompetent subjects: case report and review. Clin Infect Dis. 2007;44:e62–e66. doi: 10.1086/511871. [DOI] [PubMed] [Google Scholar]
- Lehmann T, Blackston CR, Parmley SF, Remington JS, Dubey JP. Strain typing of Toxoplasma gondii: comparison of antigen-coding and housekeeping genes. J Parasitol. 2000;86:960–971. doi: 10.1645/0022-3395(2000)086[0960:STOTGC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Lehmann T, Graham DH, Dahl ER, Bahia-Oliveira LM, Gennari SM, Dubey JP. Variation in the structure of Toxoplasma gondii and the roles of selfing, drift, and epistatic selection in maintaining linkage disequilibria. Infect Genet Evol. 2004;4:107–114. doi: 10.1016/j.meegid.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Lehmann T, Marcet PL, Graham DH, Dahl ER, Dubey JP. Globalization and the population structure of Toxoplasma gondii. Proc Natl Acad Sci U S A. 2006;103:11423–11428. doi: 10.1073/pnas.0601438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom I, Sundar N, Lindh J, Kironde F, Kabasa JD, Kwok OC, Dubey JP, Smith JE. Isolation and genotyping of Toxoplasma gondii from Ugandan chickens reveals frequent multiple infections. Parasitology. 2008;135:39–45. doi: 10.1017/S0031182007003654. [DOI] [PubMed] [Google Scholar]
- Mansfield LS, Mehler S, Nelson K, Elsheikha HM, Murphy AJ, Knust B, Tanhauser SM, Gearhart PM, Rossano MG, Bowman DD, Schott HC, Patterson JS. Brown-headed cowbirds (Molothrus ater) harbor Sarcocystis neurona and act as intermediate hosts. Vet Parasitol. 2008;153:24–43. doi: 10.1016/j.vetpar.2007.12.016. [DOI] [PubMed] [Google Scholar]
- Martina MN, Cervera C, Esforzado N, Linares L, Torregrosa V, Sanclemente G, Hoyo I, Cofan F, Oppenheimer F, Miro JM, Campistol JM, Moreno A. Toxoplasma gondii primary infection in renal transplant recipients. Two case reports and literature review. Transpl Int. 2010 doi: 10.1111/j.1432-2277.2010.01173.x. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J, Smith NH, O'Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci U S A. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier A, Devillard S, Ngoubangoye B, Bonnabau H, Banuls AL, Durand P, Salle B, Ajzenberg D, Darde ML. Additional haplogroups of Toxoplasma gondii out of Africa: population structure and mouse-virulence of strains from Gabon. PLoS Negl Trop Dis. 2010;4:e876. doi: 10.1371/journal.pntd.0000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Conrad P, James ER, Packham A, Toy-Choutka S, Murray MJ, Jessup D, Grigg M. Transplacental toxoplasmosis in a wild southern sea otter (Enhydra lutris nereis) Vet Parasitol. 2008a;153:12–18. doi: 10.1016/j.vetpar.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Miller MA, Barr BC, Nordhausen R, James ER, Magargal SL, Murray M, Conrad PA, Toy-Choutka S, Jessup DA, Grigg ME. Ultrastructural and molecular confirmation of the development of Sarcocystis neurona tissue cysts in the central nervous system of southern sea otters (Enhydra lutris nereis) Int J Parasitol. 2009;39:1363–1372. doi: 10.1016/j.ijpara.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Grigg ME, Kreuder C, James ER, Melli AC, Crosbie PR, Jessup DA, Boothroyd JC, Brownstein D, Conrad PA. An unusual genotype of Toxoplasma gondii is common in California sea otters (Enhydra lutris nereis) and is a cause of mortality. Int J Parasitol. 2004;34:275–284. doi: 10.1016/j.ijpara.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Miller MA, Miller WA, Conrad PA, James ER, Melli AC, Leutenegger CM, Dabritz HA, Packham AE, Paradies D, Harris M, Ames J, Jessup DA, Worcester K, Grigg ME. Type X Toxoplasma gondii in a wild mussel and terrestrial carnivores from coastal California: new linkages between terrestrial mammals, runoff and toxoplasmosis of sea otters. Int J Parasitol. 2008b;38:1319–1328. doi: 10.1016/j.ijpara.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Montoya JG, Liesenfeld O. Toxoplasmosis. Lancet. 2004;363:1965–1976. doi: 10.1016/S0140-6736(04)16412-X. [DOI] [PubMed] [Google Scholar]
- Morrison DA. Networks in phylogenetic analysis: new tools for population biology. Int J Parasitol. 2005;35:567–582. doi: 10.1016/j.ijpara.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Obendorf DL, Statham P, Driessen M. Detection of agglutinating antibodies to Toxoplasma gondii in sera from free-ranging eastern barred bandicoots (Perameles gunnii) J Wildl Dis. 1996;32:623–626. doi: 10.7589/0090-3558-32.4.623. [DOI] [PubMed] [Google Scholar]
- Opsteegh M, Langelaar M, Sprong H, den Hartog L, De Craeye S, Bokken G, Ajzenberg D, Kijlstra A, van der Giessen J. Direct detection and genotyping of Toxoplasma gondii in meat samples using magnetic capture and PCR. Int J Food Microbiol. 2010;139:193–201. doi: 10.1016/j.ijfoodmicro.2010.02.027. [DOI] [PubMed] [Google Scholar]
- Parameswaran N, Thompson RC, Sundar N, Pan S, Johnson M, Smith NC, Grigg ME. Non-archetypal Type II-like and atypical strains of Toxoplasma gondii infecting marsupials of Australia. Int J Parasitol. 2010;40:635–640. doi: 10.1016/j.ijpara.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena HF, Gennari SM, Dubey JP, Su C. Population structure and mouse-virulence of Toxoplasma gondii in Brazil. Int J Parasitol. 2008;38:561–569. doi: 10.1016/j.ijpara.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Pena HF, Marvulo MF, Horta MC, Silva MA, Silva JC, Siqueira DB, Lima PA, Vitaliano SN, Gennari SM. Isolation and genetic characterisation of Toxoplasma gondii from a red-handed howler monkey (Alouatta belzebul), a jaguarundi (Puma yagouaroundi), and a black-eared opossum (Didelphis aurita) from Brazil. Vet Parasitol. 2010 doi: 10.1016/j.vetpar.2010.10.015. In Press. [DOI] [PubMed] [Google Scholar]
- Pfefferkorn ER, Pfefferkorn LC, Colby ED. Development of gametes and oocysts in cats fed cysts derived from cloned trophozoites of Toxoplasma gondii. J Parasitol. 1977;63:158–159. [PubMed] [Google Scholar]
- Pfefferkorn LC, Pfefferkorn ER. Toxoplasma gondii: genetic recombination between drug resistant mutants. Exp Parasitol. 1980;50:305–316. doi: 10.1016/0014-4894(80)90034-x. [DOI] [PubMed] [Google Scholar]
- Prestrud KW, Asbakk K, Mork T, Fuglei E, Tryland M, Su C. Direct high-resolution genotyping of Toxoplasma gondii in arctic foxes (Vulpes lagopus) in the remote arctic Svalbard archipelago reveals widespread clonal Type II lineage. Vet Parasitol. 2008;158:121–128. doi: 10.1016/j.vetpar.2008.08.020. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, De Meeus T. Apparent high recombination rates in clonal parasitic organisms due to inappropriate sampling design. Heredity. 2010;104:135–140. doi: 10.1038/hdy.2009.128. [DOI] [PubMed] [Google Scholar]
- Ragozo AM, Pena HF, Yai LE, Su C, Gennari SM. Genetic diversity among Toxoplasma gondii isolates of small ruminants from Brazil: novel genotypes revealed. Vet Parasitol. 2010;170:307–312. doi: 10.1016/j.vetpar.2010.02.024. [DOI] [PubMed] [Google Scholar]
- Rejmanek D, Miller MA, Grigg ME, Crosbie PR, Conrad PA. Molecular characterization of Sarcocystis neurona strains from opossums (Didelphis virginiana) and intermediate hosts from Central California. Vet Parasitol. 2010;170:20–29. doi: 10.1016/j.vetpar.2009.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richomme C, Aubert D, Gilot-Fromont E, Ajzenberg D, Mercier A, Ducrot C, Ferte H, Delorme D, Villena I. Genetic characterization of Toxoplasma gondii from wild boar (Sus scrofa) in France. Vet Parasitol. 2009;164:296–300. doi: 10.1016/j.vetpar.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Sacks JJ, Delgado DG, Lobel HO, Parker RL. Toxoplasmosis infection associated with eating undercooked venison. Am J Epidemiol. 1983;118:832–838. doi: 10.1093/oxfordjournals.aje.a113701. [DOI] [PubMed] [Google Scholar]
- Saeij JP, Boyle JP, Grigg ME, Arrizabalaga G, Boothroyd JC. Bioluminescence imaging of Toxoplasma gondii infection in living mice reveals dramatic differences between strains. Infect Immun. 2005;73:695–702. doi: 10.1128/IAI.73.2.695-702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley LD, Ajioka JW. Population structure of Toxoplasma gondii: clonal expansion driven by infrequent recombination and selective sweeps. Annu Rev Microbiol. 2008;62:329–351. doi: 10.1146/annurev.micro.62.081307.162925. [DOI] [PubMed] [Google Scholar]
- Sibley LD, Boothroyd JC. Virulent strains of Toxoplasma gondii comprise a single clonal lineage. Nature. 1992;359:82–85. doi: 10.1038/359082a0. [DOI] [PubMed] [Google Scholar]
- Sibley LD, Khan A, Ajioka JW, Rosenthal BM. Genetic diversity of Toxoplasma gondii in animals and humans. Philos Trans R Soc Lond B Biol Sci. 2009;364:2749–2761. doi: 10.1098/rstb.2009.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Evans D, Cole RH, Kissinger JC, Ajioka JW, Sibley LD. Recent expansion of Toxoplasma through enhanced oral transmission. Science. 2003;299:414–416. doi: 10.1126/science.1078035. [DOI] [PubMed] [Google Scholar]
- Su C, Shwab EK, Zhou P, Zhu XQ, Dubey JP. Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology. 2010;137:1–11. doi: 10.1017/S0031182009991065. [DOI] [PubMed] [Google Scholar]
- Su C, Zhang X, Dubey JP. Genotyping of Toxoplasma gondii by multilocus PCR-RFLP markers: a high resolution and simple method for identification of parasites. Int J Parasitol. 2006;36:841–848. doi: 10.1016/j.ijpara.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Sundar N, Cole RA, Thomas NJ, Majumdar D, Dubey JP, Su C. Genetic diversity among sea otter isolates of Toxoplasma gondii. Vet Parasitol. 2008;151:125–132. doi: 10.1016/j.vetpar.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Thompson RC, Kutz SJ, Smith A. Parasite zoonoses and wildlife: emerging issues. Int J Environ Res Public Health. 2009;6:678–693. doi: 10.3390/ijerph6020678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M, Ayala FJ. Towards a population genetics of microorganisms: The clonal theory of parasitic protozoa. Parasitol Today. 1991;7:228–232. doi: 10.1016/0169-4758(91)90234-f. [DOI] [PubMed] [Google Scholar]
- Vaudaux JD, Muccioli C, James ER, Silveira C, Magargal SL, Jung C, Dubey JP, Jones JL, Doymaz MZ, Bruckner DA, Belfort R, Jr, Holland GN, Grigg ME. Identification of an atypical strain of Toxoplasma gondii as the cause of a waterborne outbreak of toxoplasmosis in Santa Isabel do Ivai, Brazil. J Infect Dis. 2010;202:1226–1233. doi: 10.1086/656397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmurugan GV, Su C, Dubey JP. Isolate designation and characterization of Toxoplasma gondii isolates from pigs in the United States. J Parasitol. 2009;95:95–99. doi: 10.1645/GE-1746.1. [DOI] [PubMed] [Google Scholar]
- Wendte JM, Miller MA, Lambourn DM, Magargal SL, Jessup DA, Grigg ME. Self-mating in the definitive host potentiates clonal outbreaks of the Apicomplexan parasites Sarcocystis neurona and Toxoplasma gondii. PLoS Genet. 2010a;6:e1001261. doi: 10.1371/journal.pgen.1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendte JM, Miller MA, Nandra AK, Peat SM, Crosbie PR, Conrad PA, Grigg ME. Limited genetic diversity among Sarcocystis neurona strains infecting southern sea otters precludes distinction between marine and terrestrial isolates. Vet Parasitol. 2010b;169:37–44. doi: 10.1016/j.vetpar.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yai LE, Ragozo AM, Soares RM, Pena HF, Su C, Gennari SM. Genetic diversity among capybara (Hydrochaeris hydrochaeris) isolates of Toxoplasma gondii from Brazil. Vet Parasitol. 2009;162:332–337. doi: 10.1016/j.vetpar.2009.03.007. [DOI] [PubMed] [Google Scholar]