Abstract
For many infectious agents, the detection of antibodies is critical for diagnosis, monitoring and understanding vaccine responses. To facilitate the highly quantitative and simultaneous analysis of antibodies against multiple proteins from infectious agents, we have developed Luciferase Immunoprecipitation Systems (LIPS) arrays. By configuring microtiter plates with multiple antigens and testing control and infected serum samples at one time in solution, LIPS arrays provided highly reproducible antibody titers to panels of antigens with a wide dynamic range of detection. While all serum samples showed similar positive and negative immunoreactivity with internal control antigens derived from Influenza and Renilla luciferase-alone protein, respectively, antibody titers to many HCV and HIV antigens were generally 10 to over 400-fold higher in the infected versus uninfected samples. Additional screening of 18 proteins from the EBV proteome with serum samples from healthy EBV-infected individuals showed statistically significant antibody titers to 50% of the proteins tested. Antibody titers for the different EBV antigens in the healthy EBV-infected individuals were markedly heterogeneous highlighting the complexity of host humoral responses. These results suggest that LIPS arrays offer a highly discriminating platform for simultaneously profiling a wide spectrum of antibodies associated with many infectious agents.
Introduction
The study of humoral responses is an essential component for understanding and monitoring immune responses to infectious agents. Importantly, the detection of antibody responses is the primary clinical method for diagnosing many current and even past infections 1. Serology is especially critical for the diagnosis of some agents, including KSHV/HHV-8, and Borrelia burgdorfi, where nucleic acid amplification is not sensitive enough to detect the low levels of DNA in plasma. Besides determining infection status, antibody responses have the potential to yield insight into the initial time of infection and for stratifying some infection-associated conditions. Lastly for vaccine monitoring, antibody responses are used to monitor protective B-cell responses against surface proteins.
More recently, large scale antibody screening studies have been used to uncover new antigenic targets, reveal potential vaccine targets and provide an overview of immunoreactivity against the entire proteome of certain pathogens 2. These studies employ solid phase protein arrays to simultaneously evaluate antibody responses to hundreds and even thousands of recombinant antigens 2. Recombinant proteins are produced via E. coli-based in vitro transcription/translation reactions and the unpurified, recombinant proteins are immobilized on nitrocellulose membranes or slides. These solid phase arrays are then blocked with bacterial lysates, incubated with sera, and primary antibody binding is detected with fluorescently labeled secondary antibodies. Using this approach, antibody responses to the full and partial proteomes of many different pathogens including Vaccinia 3, smallpox 3, Borrelia burgdorfi 4, and Plasmodium falciparum 5 have been studied 2. Despite the impressive information generated by this approach, solid-phase arrays have limitations including high backgrounds due to contaminating E. coli proteins, a narrow dynamic range of detection, and sub-optimal detection of conformational epitopes 1.
As an alternative to solid phase formats, liquid phase assays are routinely employed to evaluate antibodies directed at conformational epitopes 6. In particular, liquid phase assays, such as radiobinding assays (RBA), are the preferred method for serological diagnosis of many autoimmune diseases because of their high sensitivity in detecting autoantibodies directed against both conformational and linear epitopes. One drawback for RBA is the need for radioactively labeled antigens, which limits the storage of the antigens and the clinical utility of the assay. As an alternative, we developed the solution-phase Luciferase Immunoprecipitation assay Systems (LIPS) which employs Renilla luciferase (Ruc)-tagged antigens for detecting antibodies to protein targets 1. In these studies, Ruc-tagged proteins have low background binding, produce highly linear enzymatic output and are stable for long periods of storage at −80°C. Not only does LIPS efficiently measure autoantibody responses, but it is also highly useful for detecting antibodies to infectious agents. From numerous studies profiling antibodies against viral, bacterial, and filarial pathogens, LIPS often has higher sensitivity and specificity, and/or a larger dynamic range than existing ELISA assays 1. For example, standard or even rapid LIPS tests for tropical diseases including Loa loa, and Onchocerciasis diagnostically out-perform existing ELISAs 7, 8. A LIPS test for Lyme disease shows high sensitivity and specificity and may be useful for disease monitoring due to the wide dynamic range of antibody detection, which spans over 10,000-fold without serum dilution 9. Unlike many existing RBAs, the highly scalable LIPS format is also practical for antibody profiling of partial and full proteomes of relatively small viruses10–13. LIPS antibody profiling can also distinguish different treatment outcomes11 and different diseases caused by the same infectious agent13, 14. Together these and other studies demonstrate the many advantages and new information that can be acquired by LIPS antibody testing.
To date, all of the described LIPS studies have been performed by sequential iterative testing of serum samples against different antigens rather than testing many individual antigens at one time 1. As an alternative, we have developed LIPS arrays to simultaneously profile antibodies to panels of antigens. We describe initial validation of the array format by antibody profiling human samples against proteins derived from the HCV, HIV and EBV proteomes.
Results
Design of the LIPS Array
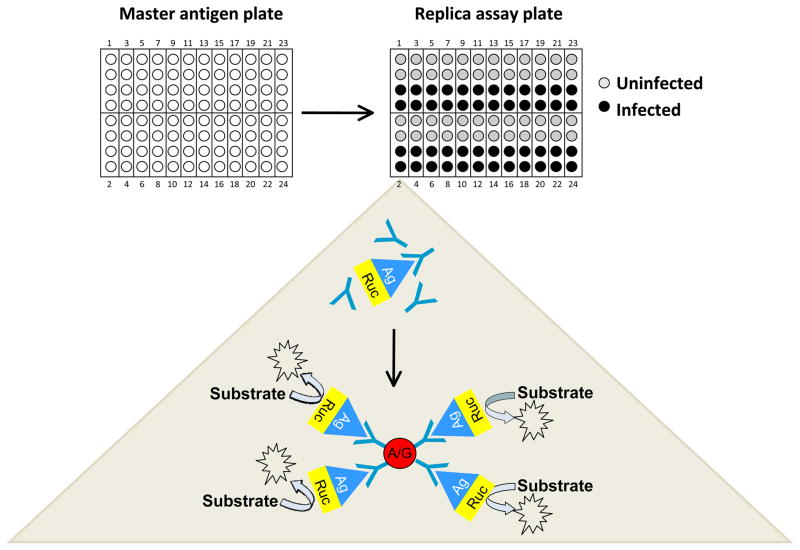
We modified the LIPS technology for simultaneously screening protein panels arranged in a 96-well microtiter plate format. For these studies, extracts of Ruc fusions with proteins from HCV, HIV, and EBV were first produced and stored frozen at −80° C until needed. These proteins were then thawed and used to produce master antigen deep-well microtiter plates containing different Ruc-antigen fusions with defined luciferase activity. For the master antigen plate, four consecutive wells of a deep well plate were used to generate an “antigen cell” for each protein tested (Fig. 1). In each cell, two wells are reserved for the test serum and two wells are for control serum or buffer. Aliquots of the luciferase-tagged antigens were then taken from the master plate and added to the replica working microtiter plates. LIPS array assays were initiated by adding duplicate samples from control and infected serum samples to the wells of each antigen cell in the replica microtiter plate (Fig. 1). In this way, control and infected serum samples can be screened in duplicate for 24 antigens simultaneously with one 96-well microtiter plate. After incubation, the serum-Ruc-antigen mixtures from each well of the microtiter plate were transferred to microtiter filter plates containing protein A/G beads. Each 96-well filter plate was then processed in the standard LIPS format for washing and measuring light units (LU) using a plate luminometer 15.
Fig. 1.
General strategy for generating LIPS arrays for testing. Antigens are thawed and used to produce a master antigen plate containing stock aliquots for the different Ruc-antigen fusions with defined luciferase activity. On the master antigen plates, four consecutive wells of a deep 96-well plate are used to generate an “antigen cell” for each protein tested (Figure 1). The master antigen plate is then used to dispense aliquots of the luciferase-tagged antigens into replica working microtiter plates. The assay is initiated by adding duplicate samples of patient and control serum to each antigen cell from the replica microtiter plate. After incubation for 1 hour at room temperature, the serum-Ruc-antigen mixtures from each well of the microtiter plate are transferred to microtiter filter plates containing protein A/G beads and then processed in the standard LIPS format for washing and measuring light units (LU) using a plate luminometer.
Simultaneous Analysis of Patient Humoral Responses against the Partial Proteome of HCV by LIPS Array
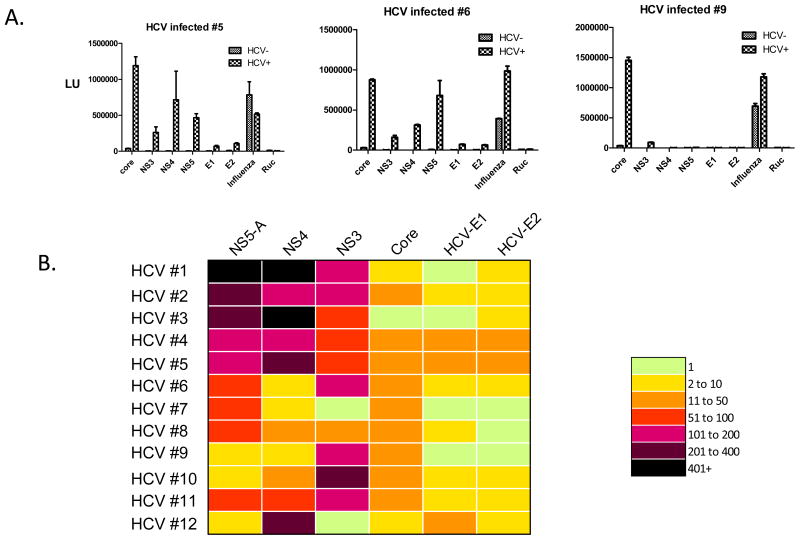
LIPS was previously used to evaluate anti-HCV antibody responses by sequential testing of antigens from the partial proteome of HCV in a cohort of pre- and post-treatment HCV patient samples 10. Here, LIPS array testing was used to analyze humoral responses against these same six proteins comprising the core, NS3, NS4, NS5A, Envelope 1, and Envelope 2 HCV proteins. Two control proteins, the highly immunoreactive HA2 from influenza 16 and the unreactive Ruc-alone protein 17 were also used. Arrays containing each of these 8 proteins were evaluated with 12 uninfected controls, 8 pre-treatment HCV patient samples and 4 post-treatment samples. Figure 2A shows representative examples of the raw data from evaluation of three of the uninfected and three HCV-infected patient samples. As expected based on the wide-spread prior exposures, the anti-HA2 influenza antibodies were present at relatively high titers in both the uninfected and HCV-infected samples, while the Ruc-alone control antigen showed little immunoreactivity (Fig. 2A). In contrast, antibodies to the six HCV antigens were statistically higher in the HCV-infected versus HCV negative samples (Mann Whitney U test; P<0.05). For example, the geometric mean titer (GMT) of anti-NS5A antibodies in 12 uninfected sera was 3,472 (95% CI, 2624–4592) LU compared to the much higher GMT of 177,600 (95% CI, 48,412–651,600) LU in the 12 HCV-infected samples. Similarly, the GMT of anti-NS4 antibodies in the uninfected samples was 1,142 (95% CI, 821–1,589) LU and was 63,790 (95% CI, 17,690–230,020) LU in the infected samples. While two of the proteins that were tested, HCV envelope 1 and HCV envelope 2, employed lower LU inputs of less than 10 million because of poor expression or solubility, significant antibody titers were still often detectable to these antigens (Fig. 2A and data not shown). It is possible that synthetically humanized antigens or protein fragments with enhanced translational efficiency and/or solubility might yield higher activity for these antigens and be even more informative.
Fig. 2.
LIPS array detection of HCV antibodies. A. Shown are raw LU data for 3 representative uninfected and HCV-infected patients probed with the HCV arrays. Antibody titers in LU are plotted on the Y-axis for each antigen. Each panel includes results for one infected patient and the simultaneously run uninfected control. B. Heatmap analysis of anti-HCV profiles in 12 HCV-infected patients. The titer values, expressed as the fold greater than the uninfected controls, were color-coded from green (no fold increase) to dark purple (>400 fold increase). Each row represents the result for a single patient.
A colored heat map was produced to show the relative immunoreactivity of each of the different HCV antigens seen in the 12 HCV-infected samples compared to the uninfected controls (Fig. 2B). For this heatmap, antibody titer values for each antibody in the HCV-infected sample were calculated as a fold increase compared to the uninfected control evaluated in the same plate. The HCV core, NS3, NS4 and NS5A antigens, showed the largest fold increases. For example, antibody titers to NS5-A and NS4 antigens showed 10 to over 400-fold higher titer values in many of the HCV-infected patient samples in comparison to the uninfected samples. On the other hand, the highly divergent HCV envelope-1 and envelope-2 viral glycoproteins typically displayed lower immunoreactivity and often showed only 2–6 fold higher titers in the HCV-infected samples compared to the controls (Fig. 2B). In spite of the wide dynamic range of antibody titers detected by LIPS array, the measurements were highly reproducible. For example, examination of two different arrays probed with the same uninfected and HCV-infected serum samples showed less than an average 0.24-fold difference in antibody titers to the different antigens (data not shown). Lastly, comparison of the LIPS arrays data with the standard format showed, as expected, that the antibody titers tracked each other well (data not shown). These findings suggest that highly reproducible and robust antibody titers can be detected by the LIPS array format.
Simultaneous Antibody Profiling of the Complete HIV Proteome
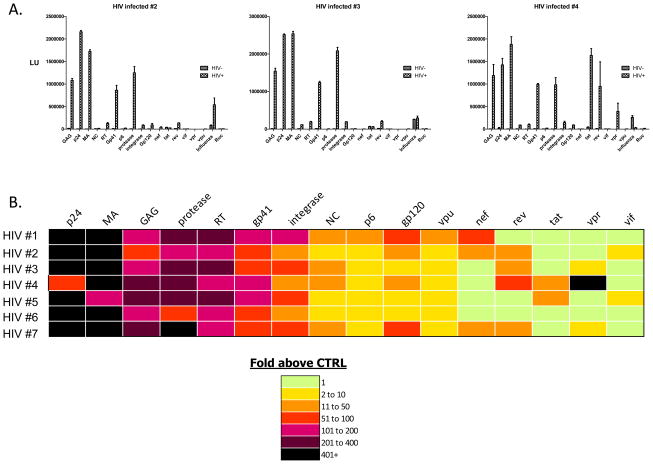
To determine if larger LIPS arrays could be employed for whole viral protein screens, uninfected and HIV-infected serum samples were tested against the complete proteome of HIV comprised of 16 different HIV proteins plus the influenza and the Ruc-alone control antigens. For testing, serum samples from seven HIV positive and seven HIV negative patients were tested in duplicate with each antigen on the array. Representative quantitative results from three different uninfected and three HIV-infected patient samples are presented in Fig. 3A. As expected, all the human serum samples were positive for anti-influenza antibodies and all serum samples were unreactive with the Ruc protein. The GMT of the 10 most informative HIV antigens for the HIV-infected and uninfected samples are shown in Table I. Some of the most highly antigenic proteins included p24, MA (matrix/p17), integrase, protease and gp41. Compared to the results seen in the HCV-infected patients, an even more dramatic strength of humoral responses was seen against the HIV proteins. For example, the GMT of anti-p24 antibodies in the seven uninfected sera was 2,229 (95% CI, 807–6,160) LU compared with the seven HIV-infected samples, which had a marked higher GMT of 2,123,000 (95% CI, 1,444,000–3,122,000) LU (Table I). Similarly, the GMT of anti-MA antibodies in the uninfected samples was 2,452 (95% CI, 1,760–3,416) LU and compared to 1,617,000 (95% CI, 633,000–4,133,000) LU in the HIV infected samples (Table I).
Fig. 3.
Representative LIPS arrays profiling anti-HIV antibodies against the whole proteome of HIV. A. Shown are raw LU data for 3 representative uninfected and HIV-infected patients probed with the HIV arrays. Antibody titers in LU are plotted on the Y-axis for each antigen. B. Heatmap analysis of anti-HIV profiles in 7 HIV-infected patients. The titer values expressed as the fold greater than the uninfected controls were color-coded from green to dark purple. Each row represents the result for a single patient.
Table 1.
Antibody titer characteristics for informative HIV antigens
| Antigen | HIV- GMT (95% CI) | HIV+ GMT (95% CI) | P value |
|---|---|---|---|
| P24 | 2229 (807 – 6160) | 2,123,000 (1,444,000 – 3,122,000) | 0.0006 |
| MA | 2452 (1760 – 3416) | 1,617,000 (633,000 – 4,133,000) | 0.0006 |
| Protease | 4338 (2357 – 7983) | 1,288,000 (404,000 – 4,105,000) | 0.0006 |
| Gp41 | 12,300 (9018 – 16,780) | 1,155,000 (863,700 – 1,545,000) | 0.0006 |
| Integrase | 3166 (2434 – 4189) | 176,300 (90,690 – 342,800) | 0.0006 |
| Gp120 | 3491(2741 – 4446) | 45,960 (11,630 – 181,600) | 0.0006 |
| Reverse transcriptase | 908 (717 – 1149) | 154,400 (101,400 – 235,200) | 0.0002 |
| Full GAG | 14,250 (1906 – 106,600) | 1,143,000 (779,800 – 1,675,000) | 0.018 |
| NC | 4363 (2011 – 9463) | 42,320 (10,190 – 175,800) | 0.018 |
| P6 | 2062 (919 – 4623) | 10,680 (3,425 – 33,280) | 0.018 |
The relative fold increase in antibody titers to the 16 different HIV antigens with the HIV-infected patients compared to the corresponding uninfected control samples is shown in the heatmap in Fig. 3B. This heatmap highlights the major antigenic components for each infected serum sample and shows the marked heterogeneity in HIV patient antibody responses to this panel. For example, it is evident that the most prominent antibody responses are against HIV p24 and MA (Fig. 3B). Of note, the full-length GAG protein, containing the unprocessed MA, p24, NC and p6 as a single protein, showed higher background binding in the uninfected serum samples and showed less of a titer difference than the individual MA and p24 proteins in distinguishing the two populations (Table I). Furthermore, the reverse transcriptase, protease and integrase were robustly positive (typically 50–400 fold above controls) in all the HIV-infected samples. Additional LIPS analyses of much larger numbers of control uninfected and HIV-infected samples have shown that the anti-reverse transcriptase antibodies have the same high level of sensitivity and specificity as measuring anti-p24 antibodies for HIV diagnosis (data not shown). While relatively high anti-gp41 antibody titers were found in all the HIV samples, antibodies to the hypervariable gp120 glycoprotein were much lower and were only found highly reactive in a few samples. Several of the HIV accessory proteins such as TAT, NEF, VPU, and VPR were positive in only a few patients and VIF showed minimal immunoreactivity (Fig. 3B). Together these results suggest that LIPS arrays can be used for simultaneously profiling patient antibodies against complete proteomes of small viruses such as HIV.
LIPS Array Analysis of Patient Humoral Responses against the Partial Proteome of EBV
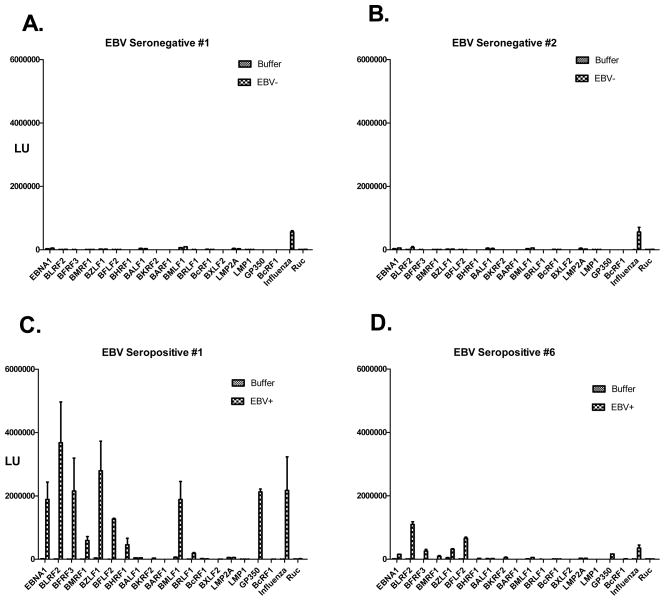
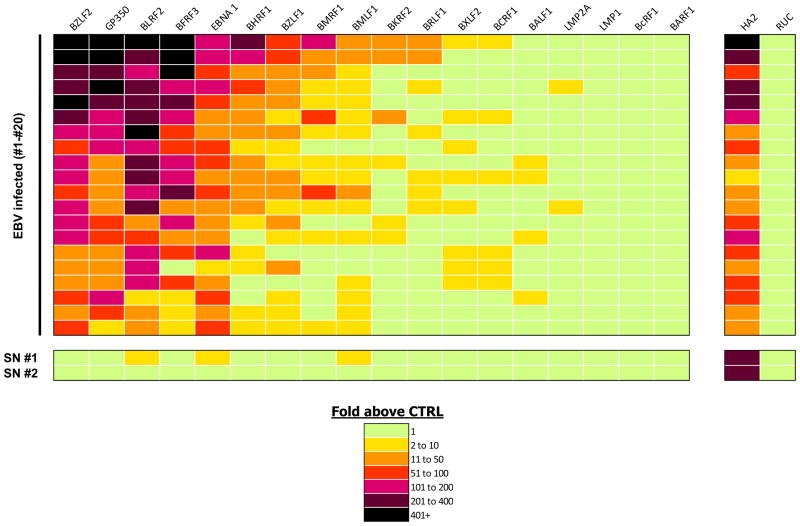
EBV is a herpes virus with a relatively small proteome consisting of approximately 85 proteins, which shows a high seroprevalence of infection in the general population of approximately 95% 18. While EBV infection is usually asymptomatic, it is linked to a large number of conditions including mononucleosis, nasopharyngeal carcinomas and Burkitt lymphoma 18. To begin to understand the normal spectrum of humoral responses to EBV, LIPS arrays were used to evaluate antibody responses in healthy individuals. For these studies, 18 different EBV antigens were tested including several known antigenic EBV targets. Due to the low frequency of EBV-negative individuals in the population, we first tested two known EBV negative serum samples and compared the results to buffer alone without serum. Assessment with the positive control antigen showed that anti-influenza antibodies were present at relatively high titers in the two EBV seronegative serum samples and essentially undetectable in the buffer control (Fig. 4A and B). Furthermore, relatively little immunoreactivity was found against the panel of 18 different EBV proteins in the two EBV-seronegative samples and the LU measurements were similar to those seen in the buffer blanks (Fig. 4A and B). These results suggest that the background binding titers of the EBV antigens in seronegative samples and buffer alone are essentially identical. Using the EBV arrays, 20 EBV seropositive serum samples from healthy individuals and buffer alone were next tested in parallel. Examples of the immunoreactivity of two EBV seropositive subjects with a large number of the Ruc-EBV fusion proteins are shown in Fig. 4C and D. From testing the EBV seropositives samples, 9 of the 18 EBV proteins showed statistically elevated immunoreactivity in at least a few serum samples (Table 2). EBV proteins showing the highest antibody titers included BZLF2 (gp42 envelope glycoprotein), gp350 (major viral glycoprotein) and BLRF2 (p23 capsid protein). Non-structural proteins including the BHRF1 (an inhibitor of apoptosis, and BZLF1 (a transcriptional activator) also showed consistently elevated antibody titers in many of the EBV-seropositive samples. While antibodies to the latent origin binding protein EBNA1 were immunoreactive with every EBV seropositive serum sample, the titers were lower than expected. Further testing revealed that the anti-EBNA1 antibodies in the serum samples were saturated and required dilution of the serum of approximately 1:20 to obtain values in the linear range (data not shown).
Fig. 4.
Representative LIPS arrays profiling of anti-EBV antibodies in 2 seronegative and 2 seropositive serum samples. Two seronegative samples (A and B) and 2 representative seropositive EBV samples (C and D) were profiled against the EBV array. Antibody levels in LU are plotted on the Y-axis.
Table 2.
Antibody titer characteristics for informative EBV antigens
| Antigen | Buffer GMT (95% CI) | EBV+ GMT (95% CI) | P value |
|---|---|---|---|
| BZLF2 | 2781 (2373 – 3260) | 225,300 (106,200 – 477,800) | <0.0001 |
| GP350 | 1849 (1648 – 2075) | 92,970 (39,750 – 217,500) | <0.0001 |
| BLRF2 | 5875 (4735 – 7291) | 566,500 (278,300 – 1,153,000) | <0.0001 |
| BFRF3 | 2150 (1835 – 2520) | 93,460 ( 36,530 – 239,200) | <0.0001 |
| EBNA 1 | 17,510 (15,190 – 20,170) | 572,100 (350,300 – 934,300) | <0.0001 |
| BHRF1 | 1840 (1622 – 2087) | 16,190 (7485 – 35,030) | <0.0001 |
| BZLF1 | 26,370 (22310 – 31,160) | 192,000 (97,430 – 378,300) | <0.0001 |
| BMRF1 | 2211 (1827 – 2676) | 7693 (3566 – 16,600) | 0.0019 |
| BMLF1 | 29,600 (22,240 – 39,410) | 86,350 (49,570 – 150,400) | 0.0022 |
The heatmap in Figure 5 illustrates the relative fold increase in antibody titers to these different proteins with the 20 different EBV seropositive and 2 EBV seronegative samples compared to the buffer blanks. Evident from the heatmap is the marked heterogeneity of antibody responses present in healthy EBV-infected individuals, highlighting the complexity of host humoral responses to the virus. Five of the EBV antigens, BZLF2, GP350, BLRF2, BFRF3 and EBNA1 showed high signals with essentially all 20 EBV-infected serum samples (Fig. 5). An additional four EBV proteins including BHRF1, BZLF1, BMRF1, and BMLF1 showed high antibody titers in only a fraction of the EBV-positive serum samples (Fig. 5). In contrast, several integral membrane proteins including LMP-1 and LMP-2 and several potential soluble proteins including, BARF1, BcRF1 and BALF1 showed relatively little or no immunoreactivity in the samples (Fig. 5). Comparison of the antibody titers to the different EBV antigenic proteins revealed that they did not correlate with one another (data not shown). These results highlight the complexity of anti-EBV host humoral responses in healthy individuals.
Fig. 5.
LIPS array profiling the partial proteome of EBV. Heatmap analysis is shown for the anti-EBV profiles in 20 EBV seropositive and 2 EBV seronegative (SN) serum samples against 18 different antigens. As reference, antibody responses against influenza and Ruc-alone antigen are also shown. The different samples tested are listed from top to bottom. Each row represents the results for a single patient. The titer values expressed as the fold greater than the buffer controls were color-coded from green to dark purple.
Discussion
In order to gain a more complete understanding of humoral response to infectious agents we have developed the LIPS array technology to generate a broad antibody profile by simultaneously screening panels of proteins derived from infectious agents. This is the first demonstration of using liquid phase arrays to simultaneously screen antibodies to panels of antigens. The array approach is made feasible by the stability of the Ruc-antigen fusions when maintained as frozen extracts for extended periods of time. One convenient feature of the LIPS array compared to solid phase protein arrays is the ability to simultaneously test, in duplicate, control and test samples on the same array. In addition, if no uninfected serum samples are available, buffer alone produces a similar low level background binding and forms the basis of using LIPS to identify novel antigenic proteins from certain infectious agents in which there are no predetermined uninfected and infected samples. LIPS array testing with 40 proteins derived from HCV, HIV and EBV proteomes revealed that over 80% showed highly statistically significant immunoreactivity in the infected compared to the uninfected serum samples. LIPS arrays also produced highly robust antibody titers that correlated well with antibody titers using the standard LIPS format. The ability to profile multiple proteins from each of these infectious agents simultaneously enhances the reliability of diagnosis of infection status and reveals the major antigenic components for each individual.
For both HIV and HCV patients, there was marked heterogeneity in patient antibodies that likely reflects differences in HLA, genetics, length of infection, as well as differences in the infectious agents themselves. While the core antigen and certain non-structural proteins were the most antigenic for detecting anti-HCV responses, some of the most highly antigenic HIV proteins were enzymes including the reverse transcriptase, integrase and protease. All three of these HIV enzymes were as diagnostically informative as the detection of anti-p24 GAG antibodies, which is classically used to test for HIV infection. As expected, HIV patients showed high immunoreactivity with the conserved gp41 glycoprotein. In contrast, hypervariable proteins including the HCV envelope 1, HCV envelope 2 and HIV gp120 showed the poorest immunoreactivity overall, and for some infected individuals there were no detectable antibody responses to these proteins. For example, variable antibody response to gp120, likely reflects variant-specific gp120 epitopes that are missed with the current version of the HIV virus used in the LIPS array. Modification of the LIPS array to include multiple variant-specific antigens or alternatively synthetic of multi-epitope antigens could improve the performance for some of the antigens on the array.
From testing 18 selected EBV proteins, approximately 50% of the proteins were found immunoreactive with EBV-infected sera. Even minor proteins not previously known to be antigenic, such as BHRF1 involved in modulating apoptosis, were highly antigenic in some individuals. While anti-cytokine autoantibodies are known to occur in certain human diseases 19, only a few low titer antibodies were detected to BCRF1, the EBV homologue of IL-10, in these samples. The antibody responses to the different EBV target proteins correlated poorly with one another, further demonstrating the complexity of anti-EBV antibody responses in healthy, seropositive individuals. LIPS studies with another herpes virus, Kaposi-sarcoma associated herpes virus (KSHV/HHV-8) showed that only 5 of 20 selected antigens were immunoreactive in Kaposi sarcoma patients by LIPS 12. The contrast difference in antigen immunoreactivity of EBV compared to KSHV highlights the host humoral response differences to these two herpes viruses. Furthermore, other solid phase assays do not demonstrate as wide a dynamic range of detection for human herpes viruses. For example, a recent study using a protein array directed at screening humoral responses from the human herpes virus, varicella zoster virus, only observed a maximum of 20-fold titer differences between seropositive and seronegative samples 20. Since LIPS consistently demonstrates a wide dynamic range of detection, profiling the EBV proteins described here by array or the standard LIPS format may be useful for exploring various diseases including multiple sclerosis, EBV-associated lymphoproliferative diseases and EBV-associated malignancies.
One future application of the LIPS array technology will be to probe panels of properly folded surface proteins to identify neutralizing antibodies. The feasibility of this approach has already been shown by the finding that measuring antibody titers by LIPS against two EBV surface proteins, BZLF2 and gp350, correlated strongly with neutralizing activity (r=0.86) measured by the standard 6 week transformation-based assay 21. LIPS arrays for surface antigens derived from different virus isolates might be useful for serotyping the exact causative agents and for understanding the spectrum of antibodies generated against related sequences. For example, studying antibody responses to diverse envelope sequences from viruses such as HIV, HCV and influenza might yield insight into vaccine development by identifying target proteins with the broadest immunoreactivity.
The ultimate goal of this research is to develop LIPS arrays for simultaneously screening and diagnosing a wide spectrum of infectious diseases. Analogous to nucleic acid pathogen arrays 22, LIPS arrays could be used to potentially screen for hundreds of infectious agents covering most of the human infectome in one test. The ability to detect antibodies associated with current infections, as well as past infections, offers new opportunities for understanding the link between infection and certain human diseases including pathogens associated with cancer, neuroinflammation and autoimmunity. A comprehensive array of infectious agents could also be used to explore human infection patterns, interactions between infectious agents and identifying novel infectious agents. A LIPS mixture format using a cocktail of antigens in a single well, rather than testing only one antigen per well, could be employed to simplify screening and increase the diagnostic sensitivity as shown by our studies with KSHV 12 and Onchocerciasis 7. Moreover, since LIPS assays using reaction volumes of less than 10 μl are feasible (Burbelo, unpublished), increasing the array to 384 microtiter plate format is also possible. In a 384-well array format, 96 antigens could be simultaneously evaluated with a control and test sample in duplicate, saving on reagent costs. In a 384-well format, robotic liquid handling would test 1 μl or less of serum in a total volume of approximately 30 microliters. A more extensive wash routine with smaller volumes of buffer than the standard format would be needed to maintain the low background binding. Alternatively, a microfluidic device23 in which protein A/G capture reagents are immobilized to the chip surface could be employed in the LIPS assay. These potential advances, along with the highly quantative data described here, suggest that LIPS arrays are a highly useful alternative to solid-phase protein arrays for profiling antibodies.
In summary, LIPS arrays show extraordinary potential as high-throughput screening tools to identify antibodies against infectious agents. Although our initial efforts were focused on relatively small numbers of proteins from the HCV, HIV and EBV proteomes, even larger proteomes are possible. These studies will require alternative, high efficiency cloning procedures to generate the needed Ruc-antigen fusions such as the Gateway system 24 and other recombination methods 25. Implementation of these higher throughput approaches and additional configurations of LIPS arrays will likely offer new tools for basic, translational and clinical studies to generate useful antibody profiles against a wide range of infectious agents.
Material and Methods
Patient Samples
Serum samples were obtained from patients and volunteers under institutional review board-approved protocols at the Clinical Center and National Institute of Allergy and Infectious Diseases, NIH (Bethesda, MD). All samples were collected and stored at −80° C until needed. These serum samples included well-characterized patients diagnosed with HCV, and HIV infection. The samples studied for HCV infection included 8 pre-treatment HCV patient samples and 4 post-treatment samples 11. For studying EBV humoral responses, a cohort of healthy individuals were first tested by a standard EBV ELISA for serological status and then used for LIPS antibody profiling.
Renilla Luciferase Constructs and Fusion Protein Production
The mammalian Renilla luciferase (Ruc) expression vector, pREN2 15, was used for generating the antigen constructs unless otherwise stated. The panel of antigen used for HCV 10 and HIV proteins 11 have been previously described. However, one additional HIV protein containing the unprocessed GAG, as a single protein, was also constructed in pREN2 and used for testing. The HA2 influenza protein 16 and two of the EBV proteins, BZLF2 (gp42) and gp350 have been described previously 21. Sixteen other EBV constructs were generated by PCR amplification using EBV genomic and cDNA clones. The primer adapter sequences used to clone these EBV open reading frames are available on request. DNA sequencing was used to confirm the integrity of all the DNA constructs. Plasmid DNA was then prepared from the different pREN2 expression vectors using a Qiagen Midi preparation kit. Following transfection of mammalian expression vectors into Cos1 cells, crude protein extracts were obtained as described and stored frozen at −80 ° C until needed 15.
Construction of the LIPS Arrays
For generating the LIPS arrays, Ruc-fusion protein extracts were thawed and light unit activity (LU) determined. The antigen master plates were constructed by first diluting Ruc-protein extracts in assay buffer A (50 mM Tris, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1% Triton X-100) to approximately 2×108 LU/ml. However, some Ruc fusion protein extracts with lower luciferase activity contained as little as 1×106 LU/ml. These diluted Ruc-fusion protein extracts were then placed into each of 4 wells from the master 96 deep-well polypropylene microtiter plates.
For testing, 50 microliters of diluted extract from the master plates for HCV, HIV and EBV antigens were transferred using an 8 channel micropipette into 96-well microtiter assay plates (Fig. 1). Serum samples were diluted 1:50 in buffer A and then 50 microliters of diluted serum from uninfected (or buffer) and infected serum samples were added to paired wells of each “antigen cell” on working assay plates. The general strategy for constructing the replica plates and testing is depicted in Fig. 1. After incubation for 1 hour at room temperature, the serum-Ruc-antigen mixtures from each well of the microtiter plate was transferred to microtiter filter plates containing protein A/G beads and then processed in the standard LIPS format 15. The washing steps of the retained protein A/G beads were performed on a BioMek FX work station (Beckman Coulter, Fullerton, CA) using an integrated vacuum manifold. After the final wash, LU were measured in a Berthold LB 960 Centro microplate luminometer (Berthold Technologies, Bad Wilbad, Germany) using coelenterazine substrate mix (Promega, Madison, WI).
Data Analysis
All presented LU data was determined from the average of duplicate values obtained from array testing. From over 700 titer values obtained by the LIPS arrays, the data represent the original values except for two samples for EBV antigens which showed discordant and questionable binding from duplicates. Upon retesting, these 2 EBV antigen-serum pairs were corrected to reflect no significant binding over buffer alone. Statistical analysis was performed using the GraphPad Prism software. The level of statistical significance for all tests was set at P<0.05 and P values were determined using the Mann Whitney U test. Heatmaps were also used to visualize the antibody profiles of the participants. In order to create these heatmaps, antibody titer values for each antibody in the infected sample were calculated as a fold increase compared to the uninfected control or buffer blank. For heatmap organization, the samples were rank ordered from left to right with respect to the most informative antigen for HCV, HIV or EBV diagnosis.
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Dental and Craniofacial Research, the National Institute of Allergy and Infectious Diseases, the Clinical Center, NIH, and in part, by a Bench to Bedside award from the NIH Clinical Research Center.
References
- 1.Burbelo PD, Ching KH, Bush ER, Han BL, Iadarola MJ. Expert Rev. Vaccines. 2010;9:567–578. doi: 10.1586/erv.10.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vigil A, Davies DH, Felgner PL. Future Microbiol. 2010;5:241–251. doi: 10.2217/fmb.09.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies DH, Molina DM, Wrammert J, Miller J, Hirst S, Mu Y, Pablo J, Unal B, Nakajima-Sasaki R, Liang X, Crotty S, Karem KL, Damon IK, Ahmed R, Villarreal L, Felgner PL. Proteomics. 2007;7:1678–1686. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- 4.Barbour AG, Jasinskas A, Kayala MA, Davies DH, Steere AC, Baldi P, Felgner PL. Infect Immun. 2008;76:3374–3389. doi: 10.1128/IAI.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doolan DL, Mu Y, Unal B, Sundaresh S, Hirst S, Valdez C, Randall A, Molina D, Liang X, Freilich DA, Oloo JA, Blair PL, Aguiar JC, Baldi P, Davies DH, Felgner PL. Proteomics. 2008;8:4680–4694. doi: 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu E, Eisenbarth GS. Clin Immunol. 2007;125:120–126. doi: 10.1016/j.clim.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burbelo PD, Leahy HP, Iadarola MJ, Nutman TB. PLoS Negl Trop Dis. 2009;3:e438. doi: 10.1371/journal.pntd.0000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burbelo PD, Ramanathan R, Klion AD, Iadarola MJ, Nutman TB. J Clin Microbiol. 2008;46:2298–2304. doi: 10.1128/JCM.00490-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burbelo PD, Issa AT, Ching KH, Cohen JI, Iadarola MJ, Marques A. Clin Vaccine Immunol. 2010;17:904–909. doi: 10.1128/CVI.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burbelo PD, Ching KH, Mattson TL, Light JS, Bishop LR, Kovacs JA. Biochem Biophys Res Commun. 2007;352:889–895. doi: 10.1016/j.bbrc.2006.11.140. [DOI] [PubMed] [Google Scholar]
- 11.Burbelo PD, Kovacs JA, Ching KH, Issa AT, Iadarola MJ, Murphy AA, Schlaak JF, Masur H, Polis MA, Kottilil S. J Infect Dis. 2010;202:894–898. doi: 10.1086/655780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burbelo PD, Leahy HP, Groot S, Bishop LR, Miley W, Iadarola MJ, Whitby D, Kovacs JA. Clin Vaccine Immunol. 2009;16:621–627. doi: 10.1128/CVI.00474-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burbelo PD, Meoli E, Leahy HP, Graham J, Yao K, Oh U, Janik JE, Mahieux R, Kashanchi F, Iadarola MJ, Jacobson S. Retrovirology. 2008;5:96. doi: 10.1186/1742-4690-5-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burbelo PD, Issa AT, Ching KH, Wyvill KM, Little RF, Iadarola MJ, Kovacs JA, Yarchoan R. J Infect Dis. 2010;201:1919–1922. doi: 10.1086/652869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burbelo PD, Ching KH, Klimavicz CM, Iadarola MJ. J Vis Exp. 2009:32. doi: 10.3791/1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beigel JH, Voell J, Huang CY, Burbelo PD, Lane HC. J Infect Dis. 2009;200:501–509. doi: 10.1086/599992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burbelo PD, Goldman R, Mattson TL. BMC Biotechnol. 2005;5:22. doi: 10.1186/1472-6750-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutok JL, Wang F. Annu Rev Pathol. 2006;1:375–404. doi: 10.1146/annurev.pathol.1.110304.100209. [DOI] [PubMed] [Google Scholar]
- 19.de Lemos Rieper C, Galle P, Hansen MB. Cytokine Growth Factor Rev. 2009;20:61–75. doi: 10.1016/j.cytogfr.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Ceroni A, Sibani S, Baiker A, Pothineni VR, Bailer SM, LaBaer J, Haas J, Campbell CJ. Mol Biosyst. 2010;6:1604–1610. doi: 10.1039/c003798b. [DOI] [PubMed] [Google Scholar]
- 21.Sashihara J, Burbelo PD, Savoldo B, Pierson TC, Cohen JI. Virology. 2009;391:249–256. doi: 10.1016/j.virol.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. Proc Natl Acad Sci U S A. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng AH, Uddayasankar U, Wheeler AR. Anal Bioanal Che. 2010;397:991–1007. doi: 10.1007/s00216-010-3678-8. [DOI] [PubMed] [Google Scholar]
- 24.Matsuyama A, Yoshida M. Methods Mol Biol. 2009;577:11–24. doi: 10.1007/978-1-60761-232-2_2. [DOI] [PubMed] [Google Scholar]
- 25.Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, Romero KM, Nguyen TT, Kalantari-Dehaghi M, Crotty S, Baldi P, Villarreal LP, Felgner PL. Proc Natl Acad Sci U S A. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]