Abstract
Peyer's patches consist of domains of specialized intestinal epithelium overlying gut-associated lymphoid tissue (GALT). Luminal antigens reach the GALT by translocation through epithelial gatekeeper cells, the so-called M cells. We recently demonstrated that all epithelial cells required for the digestive functions of the intestine are generated from Lgr5-expressing stem cells. Here, we show that M cells also derive from these crypt-based Lgr5 stem cells. The Ets family transcription factor SpiB, known to control effector functions of bone marrow-derived immune cells, is specifically expressed in M cells. In SpiB−/− mice, M cells are entirely absent, which occurs in a cell-autonomous fashion. It has been shown that Tnfsf11 (RankL) can induce M cell development in vivo. We show that in intestinal organoid (“minigut”) cultures, stimulation with RankL induces SpiB expression within 24 h and expression of other M cell markers subsequently. We conclude that RankL-induced expression of SpiB is essential for Lgr5 stem cell-derived epithelial precursors to develop into M cells.
INTRODUCTION
The self-renewing epithelium of the small intestine is organized into crypts and villi (4). In the mouse, the epithelium is renewed every 5 days. Stem cells are located near crypt bottoms (2). Two stem cell types have been described: cycling crypt base columnar cells, first described by Leblond and marked by Lgr5 (3), and quiescent stem cells, located at position +4 marked among others by Bmi1 (30), Hopx (26), and Lrig1 (29). These stem cells generate highly proliferative, transit-amplifying daughter cells that subsequently differentiate into enterocytes, goblet cells, enteroendocrine cells, tuft cells (8), and Paneth cells (2). The first four cell types migrate onto the villus, where they execute their physiological functions in nutrient digestion and uptake. They are lost by apoptosis upon reaching the top of the villus. Paneth cells reside at the bottom of the crypts, intermingled with the stem cells. The intestinal epithelium absorbs nutrients yet serves as a natural barrier to constrain the complex bacterial flora within the lumen (12). Peyer's patches (PPs) are secondary immune organs located in the mucosa of the gut (21). They harbor follicles containing B lymphocytes and intrafollicular T lymphocyte areas. Over each follicle lies a dome region that contains mature B and T lymphocytes, macrophages, and dendritic cells. The outer rim of the dome region consists of a network of reticular cells that express Tnfsf11, also known as RankL, which is in direct contact with a specialized intestinal epithelial lining (14). This follicle-associated epithelium (FAE) typically lacks goblet, Paneth, and enteroendocrine cells. Instead, it consists of enterocytes, the predominant cell type throughout the small intestine, and a unique sixth intestinal cell type, the M cell. RankL interaction with its receptors on this epithelial lining induces the M cell phenotype (14). An important aspect of this relates to the expression of GP2, a receptor involved in recognition and transport of substances derived from potential pathogens within the lumen to the gut-associated lymphoid tissue (GALT), in order to stimulate mucosal and systemic immune reactions (11, 15, 19).
Although several indirect observations suggest that FAE derives from crypt-borne stem cells (18), this has never been definitively established. In particular, the origin of the unique M cells within the FAE has remained unknown. Here, we identify the Lgr5 stem cell as the origin of M cells. We further define the SpiB transcription factor as a critical factor in the determination of the fate of these cells. We finally show that RankL also induces the M cell phenotype in cultured intestinal organoids, also referred to as enteroids (24). Not only does RankL induce the specific M cell markers UEA-1, annexin V, and GP2, it also strongly upregulates SpiB expression.
MATERIALS AND METHODS
Mice.
Generation of the LGR-5–enhanced green fluorescent protein (EGFP)–Ires–CreERT2 allele has been described previously (3), as have SpiB targeted mice and their genotyping (25). Rosa26RlacZ reporter mice were obtained from Jackson Labs. Wild-type bone marrow cells for transplantation were obtained from F1 animals, derived from a 129SV × C57BL/6 (Jackson Labs) cross.
Lgr5 lineage tracing.
Rosa26-LacZ (23) reporter mice were crossed with mice carrying an Lgr5-activatable CreERT (3). Mice >8 weeks old were injected intraperitoneally with 200 μl tamoxifen in sunflower oil at 10 mg ml−1. Three days later and 6 months later, Peyer's patch-containing regions of the small intestine were isolated, fixed with 4% formalin, and embedded in paraffin. Analysis for LacZ activity was as previously described (2).
Preparation of intestinal sections for in situ hybridization and immunohistochemistry.
Intestinal sections were prepared for in situ hybridization and immunohistochemistry as previously described (17). A T3 RNA polymerase in vitro-transcribed digoxigenin (DIG)-labeled probe for SpiB was prepared from IMAGE cDNA clone 382307. PU.1 was detected immunohistochemically (antibody from Santa Cruz).
Whole-mount in situ hybridization on mouse embryos.
Whole-mount in situ hybridization was essentially performed as previously described (13). In vitro DIG-labeled antisense RNA probes for SpiB and VCAM were prepared using IMAGE clones 382307 and 4196681 as templates, respectively.
High-resolution immunofluorescence staining semithin sections of Peyer's patches.
Cryosectioning was performed as previously described (20). The FAE area was visualized using rat anti-LAMP-1 antibody (CD107a; BD Pharmingen) in combination with goat anti-rat antibody–Alexa Fluor 488 (Invitrogen). For the combination of GP2 with Lamp1 we used rat anti GP2 (MBL) combined with goat anti-rat antibody–Texas Red X (Invitrogen T-6392), and rabbit anti-Lamp1 (ab24170; Abcam) followed by goat anti-rabbit antibody–Alexa Fluor 488 (Invitrogen). For M cell detection, biotin-conjugated UEA-1 lectin (Sigma) was used, followed by rabbit antibiotin (Rockland) and goat anti-rabbit antibody–Texas Red X (Invitrogen). Rabbit anti-annexin V antibody (Abcam) was followed by goat anti-rabbit antibody–Texas Red X (Invitrogen) or, alternatively, by a rat anti-GP2 antibody (MBL) in combination with goat anti-rat antibody–Alexa Fluor 488 (Invitrogen). All incubations were for 1 h at room temperature (RT) in phosphate-buffered saline–0.1% bovine serum albumin (BSA). Sections were mounted in Vectashield containing 1 ng/ml 4′,6′-diamidino-2-phenylindole (DAPI). Images were acquired on an Axiovert 100 microscope (Zeiss).
Cryo-immunogold electron microscopy.
Peyer's patches were dissected from the intestine and perfuse-fixed in 2% paraformaldehyde plus 0.2% glutaraldehyde in 0.1 M PHEM buffer (60 mM PIPES, 25 mM HEPES, 2 mM MgCl2, 10 mM EGTA; pH 6.9) embedded in gelatin, cryoprotected in 2.3 M sucrose, and frozen in liquid nitrogen. Sample blocks were first trimmed using a diamond Cryotrim 90 knife (Diatome, Switzerland) at −100°C, and ultrathin sections of 70 nm were cut at −120°C with a Cryoimmuno knife (Diatome, Switzerland). Samples were cryosectioned with a Leica FCS cryoultratome and labeled with biotin-conjugated Ulex europaeus agglutinin 1 (UEA-1; Sigma). Bound lectin was detected with rabbit antibiotin (Rockland) followed by protein A conjugated to 15-nm gold particles (UMC, Utrecht, The Netherlands).
Scanning electron microscopy.
The pieces of small intestine containing Peyer's patches were fixed in 4% glutaraldehyde in 0.2 M PIPES (pH 7.2) for 1 h and dehydrated in ethanol and hexamethyldisilazane (Fluka, Switzerland) at RT. The dissected Peyer's patches were then coated thinly with platinum using a JEOL (Tokyo, Japan) fine-coat JFC-11000 sputtering device and examined with a Zeiss 962 digital scanning microscope.
Bone marrow reconstitution.
Mouse wild-type donor bone marrow cells were isolated from femora and tibias. The cell concentration was adjusted to 4 × 107 cells/ml in medium containing 0.2% BSA and 10 U heparin. Recipient SpiB mutant animals were irradiated with 10 MeV photons (equivalent to 7.0 Gy γ irradiation) delivered by a linear accelerator (Elekta, Crawley, United Kingdom). After 6 h, 107 donor cells were intravenously injected into the lateral tail vein of recipient mice.
RankL stimulation.
Small intestinal organoids were derived from WT and SpiB mutant mice as described previously (22). Recombinant mouse RankL (BioLegend) was added to the organoid culture medium at concentrations of 50 to 200 ng/ml, and fresh medium was added at day 2 and day 5. At various time points, organoids were harvested, and RNA was isolated with an RNeasy minikit (Qiagen) and quantified by optical density. cDNA was synthesized from 1 μg RNA by reverse-transcription PCR (iScript; Bio-Rad). mRNA abundances were determined by real-time reverse transcription-PCR using validated primer pairs (Table 1) by the SYBR green method (Bio-Rad).
Table 1.
Primers
| Primer | Sequence |
|---|---|
| mGapdh-F | GCCTTCCGTGTTCCTACCC |
| mGapdh-R | TGCCTGCTTCACCACCTTC |
| mSpiB-F | GGGGGCCTTGACTCTA |
| mSpiB-R | CTCTGGGGGGTACACC |
| mGP2-F | CCTGCGTTCTGACACTG |
| mGP2-R | GCCGTGCAGGTTATCA |
| mRank-F | ATGCGAACCAGGAAAGT |
| mRank-R | TGCCTGCATCACAGACT |
| mAnnexinV-F | AGGGCTGATGCAGAAGT |
| mAnnexinV-R | TCCCTGCCAAACACAG |
Glyceraldehyde-3-phosphate dehydrogenase (GADPH) mRNA abundance was used to normalize the data.
Bead uptake assay.
Polystyrene nanoparticles 500 nm in diameter were microinjected into the lumens of mouse small intestinal wild-type and SpiB mutant organoids, cultured in the presence or absence of RankL (100 ng/ml). After a 2-h culture period, the portion of nanoparticles endocytosed from the organoid lumen into the cytoplasm of organoid cells was quantified. For each condition, three histological sections were analyzed. Each section contained on average 300 beads (±10). Organoids were processed for cryo-immunogold EM as described above. Polyclonal annexin V antibody (Abcam) labeling was detected with 15-nm gold particles conjugated to protein A.
Microarray analysis.
Agilent microarrays (44K whole-mouse-genome dual-color arrays; G4122F) were used. Two replicates were performed in dye swaps to compensate for dye bias. Array data were normalized using Feature Extraction (v.9.5.3; Agilent), and data analysis was performed using Excel (Microsoft). Features were flagged and not further analyzed if signal intensities for both the Cy3 and Cy5 channels did not pass the Feature Extraction filter “Significant and Positive” or “Well above Background.”
RESULTS
M cells derive from Lgr5 stem cells.
We previously generated mice carrying an Lgr5 knock-in allele expressing an enhanced green fluorescent protein (EGFP)-ires-creERT2 cassette (3). In these mice, EGFP and creERT2 are specifically expressed in Lgr5 stem cells. These mice were crossed with Rosa26-LacZ Cre reporter mice (3). In the adult offspring, tamoxifen injection induces excision of a roadblock sequence at the Rosa26-LacZ locus uniquely in Lgr5-expressing cells. This leads to an irreversible activation of LacZ expression in the pertinent Lgr5 stem cell and all its subsequent Lgr5-negative descendants. Three days after a tamoxifen pulse, mice were sacrificed and the intestines fixed. Regions of the small intestine containing PPs were isolated, and paraffin sections were prepared. Microscopic analysis showed ribbons of LacZ+ cells in the epithelium emanating from crypts adjacent to the PP and extending into the FAE of the dome (Fig. 1). A similar analysis, performed approximately 6 months after Cre induction, showed that the FAE retained LacZ+ cells. To detect M cells within these ribbons of blue cells, sections were stained with the lectin Ulex europaeus agglutinin 1 (UEA-1), a specific M cell marker (9), (5). Although the lectin also detects glycoproteins on Paneth and goblet cell membranes, these cell types are never seen in the FAE when sections are stained for lysozyme or periodic acid Schiff's, respectively. Multiple M cells were observed within the blue LacZ+ FAE ribbons, implying that M cells derive from crypt-based Lgr5 stem cells. Of note, no LacZ expression was seen in uninduced mice of the same genotype.
Fig 1.
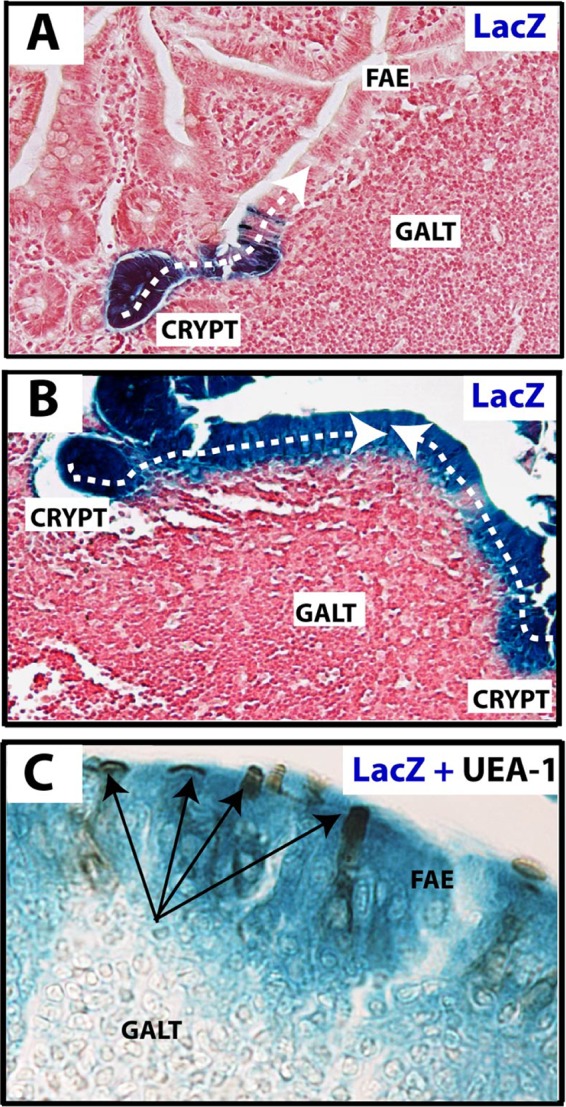
M cells are LGR-5 stem cell derived. (A) Lineage tracing of LGR-5–EGFP–IRES–creERT2 knock-in mice crossed with Rosa26 LacZ reporter mice 3 days after tamoxifen injection. The section shows that the LGR-5+ stem cell-derived LacZ genetic mark is inherited by all epithelial cells in the induced crypt. The immediate neighboring follicle-associated epithelial (FAE) cells of a Peyer's patch also stain blue (dashed line) and are therefore descendants of the same induced clone. (B) Analysis of a similar lineage-tracing experiment 6 months postinduction reveals continuous renewal of all FAE cells by crypt-resident LGR-5+ stem cells (dashed line). (C) Costaining for activity of LacZ (blue) and the presence of substrate for the lectin UEA-1 (dark brown [arrows]) within the FAE confirms the presence of M cells within the progeny of LGR-5+ crypt stem cells.
SpiB is expressed in M cells of adult wild-type animals and in prospective Peyer's patches during embryonic development.
To determine if any known transcriptional regulator is involved in the M cell phenotype, we evaluated reported microarray data derived from small intestinal M cells that had undergone fluorescence-activated cell-sorting (FACS) (27). We observed significant expression of SpiB, an Ets transcription factor closely related to PU.1. SpiB is expressed in lymphoid and myeloid cell types but is not known to be expressed outside the immune system (6). Of note, while the hematopoietic system is of mesodermal origin, the intestinal epithelium is of endodermal origin. Constitutive knockout of SpiB causes subtle defects in B cell receptor signaling but no major developmental abnormalities (25). By in situ hybridization (ISH) and immunohistochemistry (IHC), we compared SpiB expression (Fig. 2) to that of PU.1 (data not shown). The latter is a well-established marker for lymphocytes (Fig. 2A). This experiment consistently detected SpiB and PU.1 expression in the GALT. Moreover, it revealed expression in a subset of cells within the FAE, most likely M cells. In contrast, PU.1 yielded a signal exclusively in the immune cells of the follicles.
Fig 2.
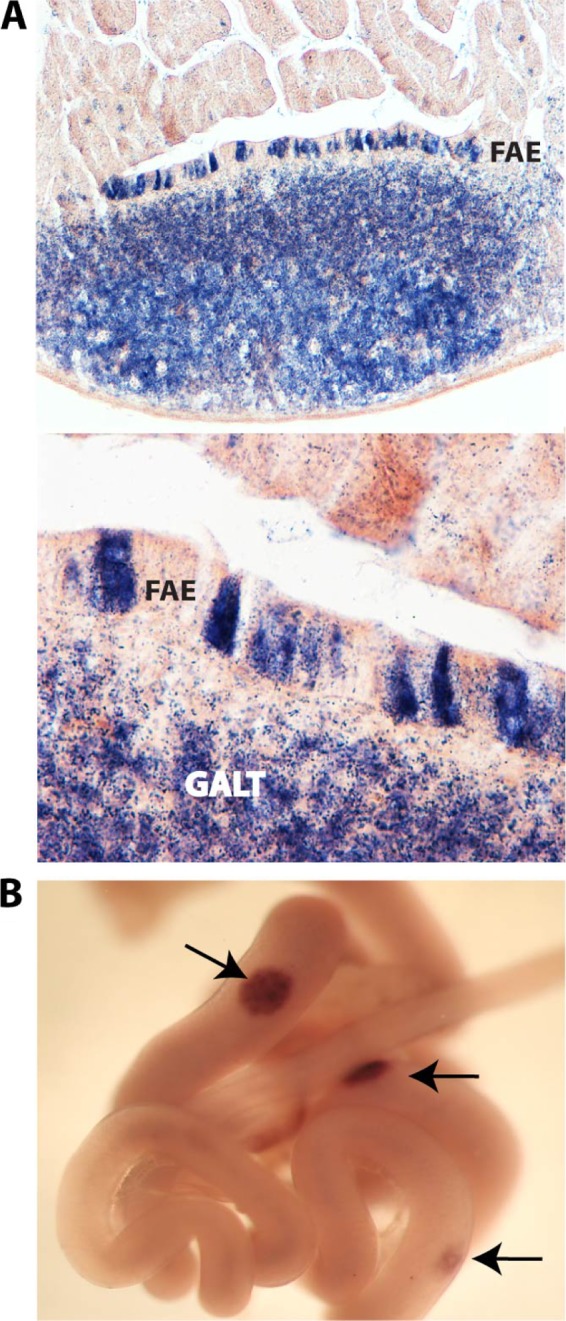
SpiB is expressed in M cells of adult wild-type animals and in Peyer's patches during normal embryonic development. (A) In situ hybridization for SpiB mRNA demonstrates expression in part of the follicle-associated epithelial cells (FAE) (arrow) in addition to the predicted presence in lymphoid cells (GALT). (B) Whole-mount in situ hybridization on E15.5 mouse small intestines for expression of SpiB suggests a role for this transcription factor during early Peyer's patch organogenesis.
We then documented the onset of SpiB expression in the intestine during embryonic development. Current models for the ontogeny of PPs, based mainly on knockout experiments, involve a lymphoid initiator cell (LTin), inducer cells (LTi), and organizer (LTo) cells (21). LTin cells secrete lymphotoxin (LT), whereas LTo cells respond to LT by upregulating VCAM expression. This adhesion molecule is the earliest detectable marker for PPs (1). We tested SpiB expression in the embryonic intestine at embryonic day 14.5 (E14.5), E15.5, and E16.5 by whole mount in situ hybridization (Fig. 2), and compared it to VCAM (data not shown). Clear patches of SpiB+ cells were first detected at E15.5. These patches are strikingly similar in appearance and number to VCAM+ patches, also detectable at this stage, which represent the earliest signs of PP development.
SpiB mutant animals lack M cells.
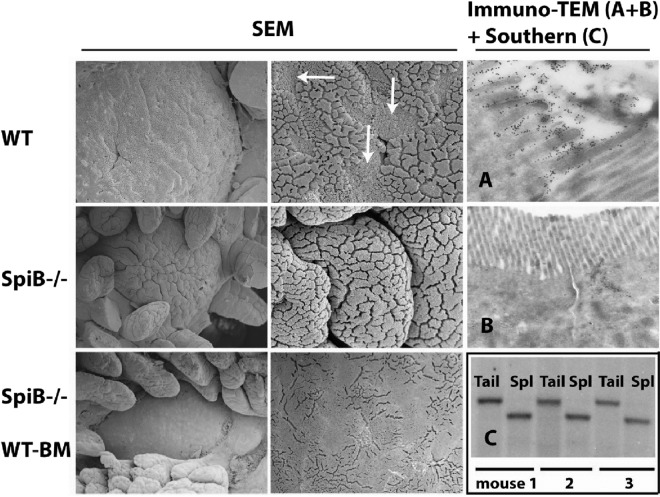
The observed epithelial expression of SpiB in the FAE cells of adult animals suggested a role in M cell fate determination. To investigate this, we studied PPs and their M cells in intestinal samples from SpiB−/− animals (25). Whereas in wild-type (WT) littermates, 5 to 10 PPs were found, 0 to 3 PPs were macroscopically detectable in mutant animals. Moreover, the few PPs in mutant animals were much smaller than those in WT mice. To test the putative absence of M cells in SpiB mutant animals, we performed scanning electron microscopy (SEM) and transmission electron microscopy (TEM) in WT and SpiB mutant Peyer's patches. Individual M cells could be easily identified in WT mice as “holes” in the regular brush border lawn, with fewer, short, and somewhat irregular microvilli (Fig. 3). Of note, FAE enterocytes are characterized by a very well-developed brush border. No M cells could be detected in the remaining small PPs of SpiB mutant mice by SEM or TEM (Fig. 3). The absence of M cells, based on these morphological criteria, was confirmed by incubation of EM sections with gold-labeled UEA-1 lectin (Fig. 3).
Fig 3.
SpiB mutant animals and wild-type bone marrow chimeras derived from them lack M cells. M cells are morphologically characterized by short microvilli, an aspect easily detectable in scanning electron micrographs of the follicle-associated epithelium of normal Peyer's patches (arrows). Such cells were not detected in SpiB mutant animals. Transmission electron microscopy (TEM) combined with gold-labeled lectin (UEA-1) staining for α-l-fucose on wild-type and SpiB mutant PPs further validated the absence of M cells in the latter. Reconstitution of mutant animals (three examples shown) with wild-type bone marrow (BM chimeric) cells was monitored by Southern blot analysis detecting a 6.5-kb mutant allele in tail DNA and a 5.5-kb wild-type allele for SpiB in spleen (Spl) DNA. No recurrence of M cells was detected in these chimeras upon extensive analysis by SEM or TEM (data not shown).
The observed absence of M cells in SpiB mutant animals could represent a cell-autonomous effect of the mutation in the epithelial progenitor cell. Alternatively, it could be the indirect consequence of the absence of SpiB in the underlying lymphoid tissue. To address this issue, we generated bone marrow-chimeric mice. Twelve-week-old SpiB−/− animals were irradiated (7.0 Gy) and reconstituted with WT bone marrow. Twelve weeks after transplantation, a Southern blot analysis of the SpiB locus on genomic DNA derived from tail and spleen revealed a nearly complete reconstitution of the hematopoietic system of the SpiB−/− recipients (three animals) with WT donor immune cells (Fig. 3). Macroscopic analysis of the intestines revealed no change in number or size of the mutant PPs compared to animals without transplantation; i.e., no more than three small PPs were observed in each transplanted animal. In addition, neither SEM analysis (Fig. 3) nor TEM (data not shown) showed any recurrence of M cells.
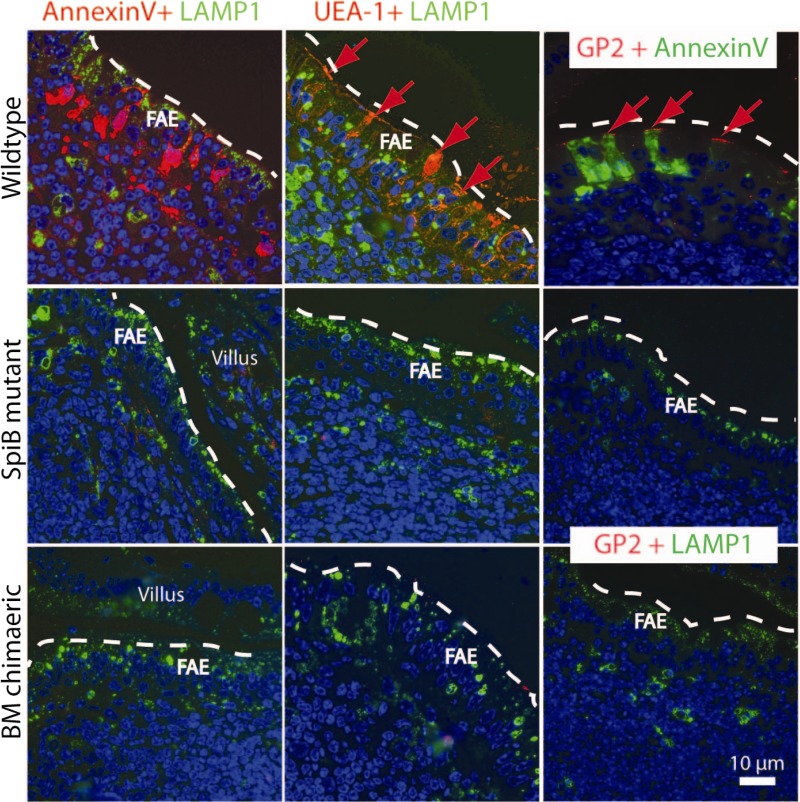
We then performed an immunofluorescence (IF) analysis on semithin cryosections (Fig. 4). We used three independent markers to distinguish M cells within the FAE: the classical membrane marker UEA-1 lectin (5, 9) and the more recently identified intracellular markers annexin V (28) and GP2 (11). Lamp1 staining (green), revealing a characteristic pattern of lysosomes in FAE, was used to visualize enterocytes on the domes (16). Lectin incubation resulted in a strong apical membrane signal of M cells within the FAE of WT PPs but not in the FAE of mutant animals or of mutant animals reconstituted with wild-type bone marrow. The total lack of M cells in these situations was further underscored by the simultaneous disappearance of the annexin V and GP2 markers. Together, these experiments indicated that introduction of WT SpiB+ immune cells did not restore M cell numbers in SpiB−/− epithelium.
Fig 4.
Immunofluorescent detection of two independent M cell-specific markers, UEA-1 and annexin V, establishes the absence of M cells in SpiB mutant animals and bone marrow chimeras. Incubation of 200- to 300-nm cryosections of Peyer's patches (PP) with the lectin UEA-1 (red) and the recently identified intracellular M cell-specific marker annexin V (red) shows that M cells are present in WT-derived PPs but absent in the follicle-associated epithelium (FAE) of mutant animals. Immunofluorescent staining for M cell markers in SpiB mutant animals, reconstituted with wild-type bone marrow (BM chimeric) cells (Fig. 3), shows that restoring SpiB expression in the follicle immune cells is insufficient to enable M cell differentiation in the FAE layer. Staining for LAMP-1 (late endosomal marker) was performed to determine the location of the FAE. FAE enterocytes have large Lamp1+ late endosomes that are absent in villus epithelial enterocytes.
RankL induces M cells in cultured small intestinal organoids.
Knoop et al. recently reported that mice mutant for the cytokine RankL fail to develop M cells in their PPs (14). This defect could be restored by systemic administration of exogenous RankL (14). While intestinal epithelial cells express the Rank receptor irrespective of their position in the intestinal tract, expression of RankL is detected only in the reticular cells lining the outer rim of the PP dome. These reticular cells are in direct contact with the FAE (14). These observations support a model in which RankL, secreted from within the PP dome, instructs newly emerged epithelial cells that emanate from flanking crypts to adopt an M cell fate. To determine whether RankL signaling on its own is sufficient to induce the M cell fate in non-FAE intestinal epithelium, we utilized a recently developed in vitro culture system (22). In this culture system, crypts or single Lgr5 stem cells expand into organoids that recapitulate stem cell-driven crypt-villus self-renewal. Intestinal organoids can be expanded by weekly 1:5 passaging for at least a year. In these organoids, basic crypt-villus architecture and self-renewal kinetics are retained, while all cell types of the crypt-villus units are produced at approximately normal ratios. Importantly, intestinal organoids are grown under sterile conditions and contain only epithelial elements. This technology thus allows the study of intestinal epithelium in the absence of confounding effects deriving from the gut microbiome, the intestinal stroma, or the immune system.
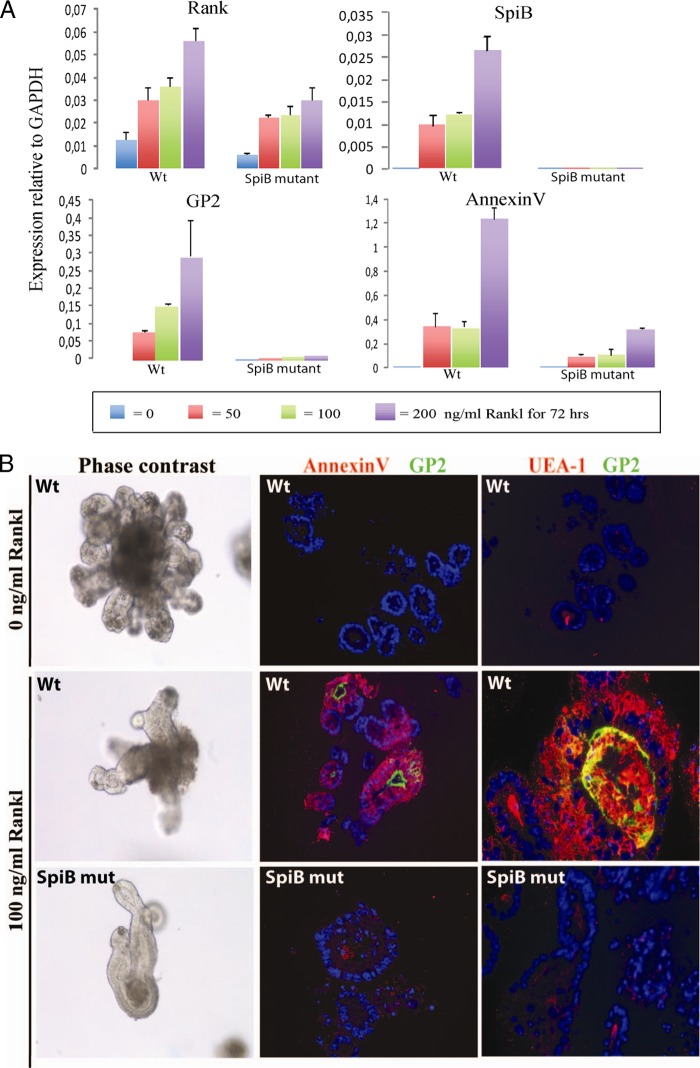
First, we confirmed Rank expression in the organoids. The epithelial cells showed a low level of expression, which was upregulated in response to RankL (Fig. 5A). We then incubated WT organoids with RankL and measured mRNA levels for SpiB and the newly described M cell markers annexin V and GP2 (11, 27). SpiB mRNA appeared within 24 h of stimulation, whereas GP2 mRNA was first seen after 3 days. Expression levels of SpiB, annexin V, and GP2 were maximal at 3 days of stimulation (Fig. 5A). When RankL was added to organoids derived from SpiB-null mice, annexin V induction was strongly reduced and GP2 induction completely absent (Fig. 5A), despite normal upregulation of Rank. Expression of the lectin UEA-1, annexin V, and the GP2 receptor was confirmed by immunofluorescent staining on organoids that were cultured for 7 days in medium containing 100 ng/ml of RankL (Fig. 5B). Proteins posttranslationally modified with UEA-1-reactive sugars were massively increased, aberrantly leading to secretion of UEA-1+ proteins also at the basal side. GP2 staining was confined to the apical membranes, in accordance with its transcytotic role of FimH+ bacteria (11). Annexin V localized to the cytoplasm, as was seen in M cells in vivo (Fig. 4). These data implicated SpiB as an essential component in the RankL-induced conversion of FAE enterocytes to M cells.
Fig 5.
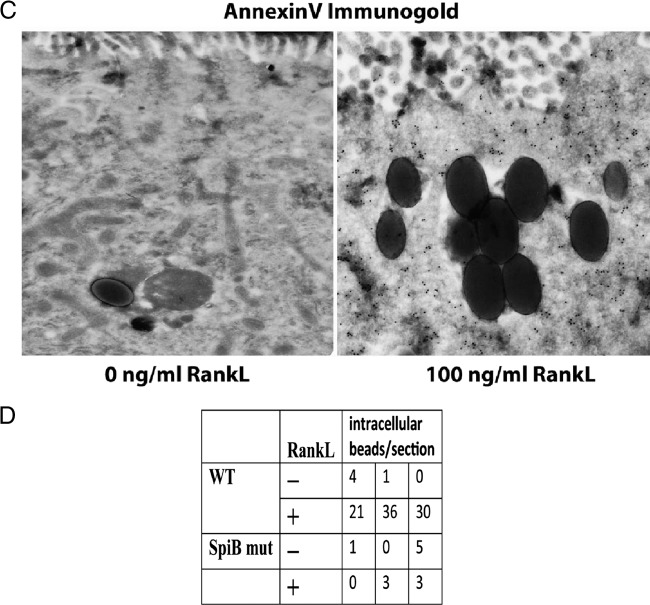
Incubation of WT organoids, but not SpiB mutant-derived organoids, with RankL induces functional M cells that express SpiB and the M cell-specific markers annexin V, GP2, and UEA-1. (A) Standard minigut cultures containing WT and SpiB mutant organoids were supplemented with 0, 50, 100, or 200 ng/ml of RankL for 72 h. Expression, as determined by qPCR, at 72 h is shown. (B) RankL-treated (7 days) and control organoids were stained for expression of the annexin V and GP2 receptors and for the lectin UEA-1 by immunofluorescence on semithin cryosections (right columns). Phase-contrast images of studied organoids are shown in the left column. (C) WT and SpiB mutant organoids were cultured in the presence and absence of 100 ng/ml RankL. Two hours after microinjection of 500 nm beads into the lumens of organoids, endocytosis was quantified by scoring the number of beads inside cells using EM. To visualize M cells, the sections were immunogold labeled for the marker annexin V. Typical examples of WT organoids are shown. (D) To quantify the endocytosis ability of in vitro-induced M cells, three EM sections containing approximately 300 beads (±10) were analyzed for each condition.
To functionally test the in vitro-generated M cells, we performed a bead uptake experiment. This assay reflects the typical transcytotic ability of M cells. Polystyrene beads were microinjected into the lumen of cultured organoids, and uptake of beads into annexin V+ cells was monitored using EM. Figure 5C shows that the amount of beads endocytosed by RankL-stimulated WT organoids is significantly increased compared to the amount endocytosed by unstimulated organoids. The SpiB mutant organoids proved completely insensitive to incubation with RankL.
The gene profile of intestinal organoid M cells resembles that of in vivo M cells.
The transcriptome of isolated PP M cells was recently described (27). To determine to what extent the RANKL-induced in vitro M cell phenotype is similar, we performed a microarray analysis of RankL-stimulated organoids. The results obtained were compared to microarray data obtained from PP-derived M cells (27). A total of 164 genes were found to be upregulated in RankL-activated organoids as well as in vivo M cells (data not shown). This array data set can be viewed at http://www.ncbi.nlm.nih.gov/geo under accession no. GSE38785. Strikingly, within this list, the SpiB gene is seen among the 25 genes showing the greatest change, together with the genes for the M cell markers annexin V and GP2 (Table 2).
Table 2.
| Gene name | Expression change (log2 ratio) |
Description | |
|---|---|---|---|
| Wt+RankL vs Wt | M cell vs intestinal epithelial cell | ||
| Ccl20 | 7.88 | 3.19 | Chemokine (C-C motif) ligand 20 (Ccl20), transcript variant 1, mRNA (NM_016960) |
| Ccl6 | 6.84 | 4.67 | Chemokine (C-C motif) ligand 6 (Ccl6), mRNA (NM_009139) |
| Prnp | 6.25 | 3.93 | Prion protein (Prnp), mRNA (NM_011170) |
| Gp2 | 6.09 | 6.09 | Glycoprotein 2 (zymogen granule membrane) (Gp2), mRNA (NM_025989) |
| Serpina1b | 6.07 | 8.18 | Serine (or cysteine) preptidase inhibitor, clade A, member 1b, mRNA (cDNA clone IMAGE:5125410) (BC037008) |
| Umod | 5.83 | 3.95 | Uromodulin (Umod), mRNA (NM_009470) |
| Ncf4 | 5.82 | 3.95 | Neutrophil cytosolic factor 4 (Ncf4), mRNA (NM_008677) |
| Siglec5 | 5.77 | 8.62 | Sialic acid binding Ig-like lectin 5 (Siglec5), mRNA (NM_145581) |
| Rac2 | 5.46 | 5.14 | RAS-related C3 botulinum substrate 2 (Rac2), mRNA (NM_009008) |
| Sh3kbp1 | 5.44 | 3.85 | SH3-domain kinase binding protein 1 (Sh3kbp1), transcript variant 2, mRNA (NM_021389) |
| Anxa5 | 5.32 | 5.88 | Annexin A5 (Anxa5), mRNA (NM_009673) |
| Ccl9 | 5.12 | 6.64 | Chemokine (C-C motif) ligand 9 (Ccl9), mRNA (NM_011338) |
| Scg5 | 5.00 | 3.23 | Secretogranin V (Scg5), mRNA (NM_009162) |
| Anxa10 | 4.98 | 3.19 | Annexin A10 (Anxa10), transcript variant 2, mRNA (NM_011922) |
| Eno3 | 4.72 | 1.38 | Enolase 3, beta muscle (Eno3), transcript variant 1, mRNA (NM_007933) |
| Gfi1b | 4.63 | 6.32 | Growth factor independent 1B (Gfi1b), transcript variant 1, mRNA (NM_008114) |
| Ppp1r14c | 4.62 | 3.33 | Protein phosphatase 1, regulatory (inhibitor) subunit 14c (Ppp1r14c), mRNA (NM_133485) |
| Dpysl3 | 4.53 | 2.74 | Dihydropyrimidinase-like 3 (Dpysl3), transcript variant 2, mRNA (NM_009468) |
| Itih5 | 4.33 | 5.52 | Inter-alpha (globulin) inhibitor H5 (Itih5), mRNA (NM_172471) |
| Spib | 4.24 | 6.14 | Spi-B transcription factor (Spi-1/PU.1 related) (Spib), mRNA (NM_019866) |
| Rab32 | 4.23 | 1.45 | RAB32, member RAS oncogene family (Rab32), mRNA (NM_026405) |
| Plk2 | 4.21 | 5.06 | Polo-like kinase 2 (Drosophila) (Plk2), mRNA (NM_152804) |
| Pfn2 | 4.10 | 3.41 | Profilin 2 (Pfn2), mRNA (NM_019410) |
| Kctd12 | 4.07 | 6.75 | Potassium channel tetramerization domain containing 12 (Kctd12), mRNA (NM_177715) |
| Gjb2 | 4.03 | 3.25 | Gap junction protein, beta 2 (Gjb2), mRNA (NM_008125) |
DISCUSSION
In this study, we document the origin of M cells, a cell type contained within the follicle-associated epithelium of Peyer's patches. The FAE is a continuation of the epithelium that forms the luminal lining of the small intestine and the colon. Outside the FAE, this lining consists of repeating crypt-villus units. At the bottoms of crypts, Lgr5+ stem cells are intermingled with Paneth cells. These cycling Lgr5 stem cells give rise to all epithelial cell types of the small intestine: enterocytes and much smaller numbers of accessory goblet, enteroendocrine, tuft, and Paneth cells (2). The quiescent +4 stem cell population, not studied here, is also capable of producing all cell types of the epithelium and appears to be directly connected to the Lgr5 stem cell population (26, 30).
The FAE clearly differs in composition from the epithelial lining that is organized into crypts and villi. Differentiation in the FAE is limited to two morphologically and functionally distinguishable cell types: FAE-specific enterocytes and so-called M cells. The M cells specialize in monitoring the content of the intestinal lumen for immunogenic substances and deliver antigens at their basal side to the immune cells of the follicles. We asked whether the crypt-based Lgr5 stem cells are able to generate the typical FAE cell types. Lineage-tracing experiments (3), based on Lgr5-driven activation of the Rosa26-LacZ locus, demonstrate that Lgr5 stem cells residing in crypts adjacent to PP generate the FAE cells. A subset of the epithelial cells within the LacZ+ ribbons overlying PPs, moreover, are decorated with the M cell-specific substrate for the lectin UAE-1. This clearly shows that M cells constitute the sixth cell type derived from Lgr5 stem cells.
From our studies on SpiB null mice, it appears that this Ets family transcription factor is essential for development of M cells in a cell-autonomous fashion. SpiB, like the closely related PU.1 gene product, was known to induce effector functions in mesodermally derived lymphoid and myeloid cells (7, 10). In situ hybridizations on small intestinal sections showed expression of SpiB in M cells. The assumed role of SpiB in generating these immune-regulating endodermal cells is consistent with microarray data from FACS-sorted M cells (27). A complete absence of M cells, based on extensive EM analysis and staining for the lectin UEA-1, annexin V, and GP2, in SpiB mutant mice demonstrated the absolute requirement of this transcription factor in M cell development in adult animals. Wild-type bone marrow transplantations into SpiB-null mice did not rescue the M cell defect, showing that it is specifically due to the lack of SpiB expression in the epithelial layer.
The notion that M cells arise from Lgr5 stem cells and that the transcriptional regulator SpiB turns on M cell-specific genes could be confirmed in our recently developed 3-dimensional intestinal organoid culture system. We exploited the previously reported dependence of M cell development on the cytokine RankL (14). First, we found that the mere addition of this cytokine to the standard three-growth-factor (Rspondin1, epithelial growth factor, and Noggin) cocktail resulted in organoids in which the majority of the differentiated cells expressed M cell markers. Of note, M cells never appear in standard minigut cultures. Increase in the levels of mRNA for SpiB and Rank occurred within 24 h under these conditions. A concomitant increase of mRNA for the M cell marker annexin V was detected. GP2 messengers were first detected at day 3. When SpiB mutant organoids were exposed to RankL, M cell marker expression did not occur. The in vitro-induced M cells appeared to be functionally competent, as measured in a bead uptake experiment. In the absence of SpiB, no RankL-induced bead uptake was detected. Microarray analysis of RankL-activated organoids convincingly confirmed the upregulation of SpiB and the known M cell markers GP2 and annexin V. Moreover, it generated a list of 161 other genes whose expression was previously reported to be specifically enhanced in PP-derived M cells (27). Thus, SpiB is necessary for M cell differentiation. Unlike RankL, it appears not to be sufficient for M cell differentiation, as forced retroviral expression of SpiB in organoids did not induce GP2 expression (data not shown).
Taken together, these observations sketch the following scenario. Lgr5 stem cells at crypt bottoms generate naïve progenitors. When leaving the crypts, these progenitors adopt one of six differentiated cell fates. Only when progenitors move into FAE rather than a villus flank will these cells sense RankL secreted by underlying reticular cells. Exposure to this cytokine suffices for these cells to adopt the M cell fate. An early downstream regulator in this process is the SpiB transcription factor. If its expression cannot be induced, no M cells are formed. We predict that RankL will induce, parallel to SpiB, one or more regulator genes.
M cells have been difficult to study, based on their exceedingly low numbers within the intestinal epithelium and their presence in architecturally complex structures, i.e., the PPs. In this study, we describe a simple method to produce large numbers of M cells in vitro and in isolation of all nonepithelial elements of PPs. This technology will allow detailed functional studies on the mechanism of antigen recognition, processing, and transport by M cells.
ACKNOWLEDGMENTS
We acknowledge Jos Onderwater (electron microscopy, Leiden University Medical Center) for technical assistance with the scanning electron microscopy and Nico Ong (Netherlands Cancer Institute) for technical assistance with the images.
This work was supported by Koninklijke Nederlandse Akademie van Wetenschappen and Koningin Wilhelmina Fonds.
We have no conflicts of interest to report.
Footnotes
Published ahead of print 9 July 2012
REFERENCES
- 1. Adachi S, Yoshida H, Kataoka H, Nishikawa S. 1997. Three distinctive steps in Peyer's patch formation of murine embryo. Int. Immunol. 9: 507–514 [DOI] [PubMed] [Google Scholar]
- 2. Barker N, van de Wetering M, Clevers H. 2008. The intestinal stem cell. Genes Dev. 22: 1856–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barker N, et al. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 [DOI] [PubMed] [Google Scholar]
- 4. Bjerknes M, Cheng H. 2006. Intestinal epithelial stem cells and progenitors. Methods Enzymol. 419: 337–383 [DOI] [PubMed] [Google Scholar]
- 5. Clark MA, Jepson MA, Hirst BH. 1995. Lectin binding defines and differentiates M-cells in mouse small intestine and caecum. Histochem. Cell Biol. 104: 161–168 [DOI] [PubMed] [Google Scholar]
- 6. Dahl R, Ramirez-Bergeron DL, Rao S, Simon MC. 2002. Spi-B can functionally replace PU.1 in myeloid but not lymphoid development. EMBO J. 21: 2220–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Friedman AD. 2007. Transcriptional control of granulocyte and monocyte development. Oncogene 26: 6816–6828 [DOI] [PubMed] [Google Scholar]
- 8. Gerbe F, Brulin B, Makrini L, Legraverend C, Jay P, 2009. DCAMKL-1 expression identifies Tuft cells rather than stem cells in the adult mouse intestinal epithelium. Gastroenterology 137: 2179–2180, 2180–2181 [DOI] [PubMed] [Google Scholar]
- 9. Giannasca PJ, Giannasca KT, Falk P, Gordon JI, Neutra MR. 1994. Regional differences in glycoconjugates of intestinal M cells in mice: potential targets for mucosal vaccines. Am. J. Physiol. 267: G1108–1121 [DOI] [PubMed] [Google Scholar]
- 10. Goebl MK. 1990. The PU. 1 transcription factor is the product of the putative oncogene Spi-1. Cell 61: 1165–1166 [DOI] [PubMed] [Google Scholar]
- 11. Hase K, et al. 2009. Uptake through glycoprotein 2 of FimH(+) bacteria by M cells initiates mucosal immune response. Nature 462: 226–230 [DOI] [PubMed] [Google Scholar]
- 12. Hooper LV. 2009. Do symbiotic bacteria subvert host immunity? Nat. Rev. Microbiol. 7: 367–374 [DOI] [PubMed] [Google Scholar]
- 13. Kikuchi T, Mori S, Nishikawa SI, Yokota Y. 2000. Detection of mRNAs in Peyer's patches of the developing mouse embryo. J. Immunol. Methods 240: 15–22 [DOI] [PubMed] [Google Scholar]
- 14. Knoop KA, et al. 2009. RANKL is necessary and sufficient to initiate development of antigen-sampling M cells in the intestinal epithelium. J. Immunol. 183: 5738–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kraehenbuhl JP, Neutra MR. 2000. Epithelial M cells: differentiation and function. Annu. Rev. Cell Dev. Biol. 16: 301–332 [DOI] [PubMed] [Google Scholar]
- 16. Kujala P, et al. 2011. Prion uptake in the gut: identification of the first uptake and replication sites. PLoS Pathog. 7: e1002449 doi:10.1371/journal.ppat.1002449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muncan V, et al. 2006. Rapid loss of intestinal crypts upon conditional deletion of the Wnt/Tcf-4 target gene c-Myc. Mol. Cell. Biol. 26: 8418–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Niedergang F, Kraehenbuhl JP. 2000. Much ado about M cells. Trends Cell Biol. 10: 137–141 [DOI] [PubMed] [Google Scholar]
- 19. Ohno H, Hase K. 2010. Glycoprotein 2 (GP2): grabbing the FimH bacteria into M cells for mucosal immunity. Gut Microbes 1: 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peters PJ, Bos E, Griekspoor A. 2006. Cryo-immunogold electron microscopy. Curr. Protoc. Cell Biol. 32: 4.7.1–4.7.19. [DOI] [PubMed] [Google Scholar]
- 21. Randall TD, Carragher DM, Rangel-Moreno J. 2008. Development of secondary lymphoid organs. Annu. Rev. Immunol. 26: 627–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sato T, et al. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265 [DOI] [PubMed] [Google Scholar]
- 23. Srinivas S, et al. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stelzner M, et al. 2012. A nomenclature for intestinal in vitro cultures. Am. J. Physiol. Gastrointest. Liver Physiol. 302: G1359–G1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Su GH, et al. 1997. Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J. 16: 7118–7129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takeda N, et al. 2011. Interconversion between intestinal stem cell populations in distinct niches. Science 334: 1420–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Terahara K, et al. 2008. Comprehensive gene expression profiling of Peyer's patch M cells, villous M-like cells, and intestinal epithelial cells. J. Immunol. 180: 7840–7846 [DOI] [PubMed] [Google Scholar]
- 28. Verbrugghe P, et al. 2006. Murine M cells express annexin V specifically. J. Pathol. 209: 240–249 [DOI] [PubMed] [Google Scholar]
- 29. Wong VW, et al. 2012. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat. Cell Biol. 14: 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yan KS, et al. 2012. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. U. S. A. 109: 466–471 [DOI] [PMC free article] [PubMed] [Google Scholar]