Abstract
Over 400 Krüppel-associated box zinc finger proteins (KRAB-ZFPs) are encoded in mammalian genomes. While KRAB-ZFPs strongly repress transcription in vitro, little is known about their biological function or gene targets in vivo. Regulator of sex limitation 1 (Rsl1), one of the first KRAB-Zfp genes assigned a physiological role, accentuates sex-biased liver gene expression, most dramatically for mouse sex-limited protein (Slp), which provides an in vivo reporter of KRAB-ZFP function. Slp is induced in males in the liver and kidney by growth hormone (GH) and androgen, respectively. In the liver but not kidney, the Rsl1 genotype correlates with methylation of a CpG dinucleotide in the Slp promoter that is demethylated at puberty. RSL1 binds 2 kb upstream of the Slp promoter, both in vitro and in vivo, within an enhancer containing response elements for STAT5b. Chromatin immunoprecipitation (ChIP) assays demonstrate that RSL1 recruits KAP1/TRIM28, the corepressor for KRAB action in vitro, to this enhancer. Slp induction requires rapid cycling of STAT5b in chromatin. Remarkably, RSL1 simultaneously binds adjacent to STAT5b with a reciprocal binding pattern that limits hormonal response. These experiments demonstrate a surprisingly dynamic interplay between a hormonal activator, STAT5b, and a KRAB-ZFP repressor and provide unique insights into KRAB-ZFP epigenetic mechanisms.
INTRODUCTION
Nearly half of all transcription factors encoded in the human genome are C2H2 zinc finger (ZNF) proteins (ZFPs), and more than 400 have an N-terminal Krűppel-associated box (KRAB) domain that acts to repress gene expression (12, 29). KRAB-Zfp genes arose in tetrapods and have amplified dramatically in mammals (6). In genetic terms, KRAB-Zfp genes are modifier loci, recognized by their effects on other genes. They are also excellent disease gene candidates, with substantial individual variation, shared molecular mechanisms, and broad expression (29). Their sheer number and rapid evolution argue that KRAB-ZFPs are critical architects of highly conserved as well as species-specific traits (12, 29).
Insights into KRAB-ZFP repression have been deduced largely from in vitro studies. ZNFs bind DNA with high specificity and affinity, and the ∼75-amino-acid KRAB domain interacts with KRAB-associated protein 1 (KAP1/TIF1β/TRIM28) (7) to recruit a complex of chromatin-modifying enzymes that are associated with transcriptional inhibition, including histone methyltransferases (e.g., SETDB1), histone deacetylases (HDACs), and DNA methyltransferases (DNMTs) (1, 34, 37, 49). KRAB-ZFPs are thought to silence gene expression by recruiting complexes that catalyze heterochromatin formation at specific sites in the genome. However, little is known about how KRAB-ZFPs select genomic targets for repression or how this state is reversed upon gene activation. Moreover, the role of KAP1 is incompletely understood because colocalization of KAP1 and KRAB-ZFP binding has been clearly demonstrated only in cell lines, with chimeric genes, or to the 3′ ends of KRAB-Zfp genes themselves, suggesting that they cross-regulate (14, 27, 30). In addition, KAP1 has functions that may be independent of KRAB-ZFPs (13).
Despite the huge size of the KRAB-Zfp family and a detailed view of in vitro activity, few biological roles of individual genes have been identified and even fewer cellular target genes are known. Genes with identified functions include Zfp568 (chato), which regulates extraembryonic tissue morphogenesis (9); Zfp809, which restricts retroviral transposition in embryonic stem cells (45); and Zfp57, which confers some parental genomic imprints (23) and causes transient neonatal diabetes in humans upon missense mutation (25). Our studies of the paralogous KRAB-Zfp gene regulator of sex limitation 1 (Rsl1) and Rsl2 have revealed roles in sexually dimorphic liver gene expression (19, 41). Furthermore, rsl mice, carrying homozygous null mutations, display subtle reproductive and metabolic phenotypes (17, 20), suggesting that KRAB-ZFPs may be important contributors to complex traits.
Rsl-null mice (rsl) were first noted by their increased expression in females of otherwise male-biased liver genes, including those for the hallmark sex-limited protein (Slp) and members of the major urinary protein (MUP) and cytochrome P450 (Cyp) families (41). Sexually dimorphic liver gene expression, well studied in rodents, initiates at puberty (42). In males, testosterone acts on the pituitary to direct growth hormone (GH) secretory oscillations over 3- to 4-h periods, leading to peaks of high serum GH followed by undetectable GH troughs. In females, GH secretion is more continuous, resulting in relatively low constant levels. In liver, the transcription factor STAT5b transduces the GH pulse to the nucleus and in association with hepatocyte nuclear factors (HNFs), peroxisome proliferation activators (PPARs), and GA-binding proteins (GABPs) induces male-biased genes and represses female-biased genes (4, 22, 44, 46). Dimorphism exists in the human liver as well. Of sex-biased human liver genes, half of the murine orthologs are sex biased and GH regulated, suggesting that this control pathway is evolutionarily conserved (48). Rsl accentuates sex-biased gene expression independently of either steroid or GH control (41).
The capacity of KRAB-ZFP repressors to modulate gene expression is clearly evident for Rsl genetic variants where Slp (gene symbol, C4a) serves as a sensitive in vivo reporter. In wild-type (WT) mice, Slp expression is male biased in the liver and kidney by two distinct tissue-specific mechanisms, unlike the tandemly duplicated paralog complement component 4B (C4b) (28). Natural mutations in both Rsl1 and Rsl2 in congenic rsl mice lead to high female expression of Slp in the liver after puberty (19). Bacterial artificial chromosome (BAC) transgenes that express Rsl1 at physiological levels suppress Slp in rsl females, restoring male-specific expression. In contrast to BAC transgenes, high Rsl1 expression from a liver-directed, overexpressed cDNA transgene extinguishes Slp in males as well as females (17). Physiological analysis reveals differences in dietary stress response and pubertal timing in Rsl-variant mice, despite their grossly normal appearance (17, 20). Rsl thus provides access to a broad spectrum of KRAB-ZFP roles.
Here we dissect the mechanism of RSL1 in vivo as a model for the biological action of other KRAB-ZFPs. We first correlate the known promoter CpG methylation of Slp in the liver with the presence of Rsl1, suggesting that RSL1 plays an active role in establishing and/or maintaining this epigenetic mark. Instead of binding at this CpG, RSL1 binds ∼2 kb upstream of the Slp transcriptional start site, near the hormone-dependent enhancer. Chromatin immunoprecipitation (ChIP) experiments demonstrate concordant binding of RSL1 and KAP1, the putative KRAB-ZFP corepressor. Remarkably, in the adult male liver, a dynamic interplay of STAT5b and RSL1 in chromatin modulates Slp expression. Together, these results provide compelling in vivo evidence for the KRAB-ZFP repression mechanism postulated from in vitro studies, correlate KRAB-ZFP binding with epigenetic regulation of a bona fide target gene in liver and reveal unexpected rapid oscillation in KRAB-ZFP binding in opposition to hormonal activation.
MATERIALS AND METHODS
Mice.
B10.D2 mice (WT) were purchased from Jackson Laboratory (stock number 000463). Congenic B10.D2.PL-rsl (rsl) and Rsl1-transgenic (Rsl1tg; Rsl−/−) mice were described previously (the gene used to create Rsl1tg mice was carried on the rsl background) (17). For GH treatment, recombinant human GH (Nutropin AQ; Genentech) was diluted to 200 μg/ml in Dulbecco modified Eagle medium (DMEM; Invitrogen), and 1.5 μg/g body weight was injected intraperitoneally. Control mice received DMEM alone. Livers were harvested 30 min after injection.
RNA analysis.
Total RNA was isolated by the guanidinium isothiocyanate method (5) or with RNeasy minikits (Qiagen) according to the manufacturer's instructions. For real-time quantitative reverse transcription-PCR (qRT-PCR), cDNA was synthesized using 2.5 μg total RNA with a high-capacity cDNA archive kit (Applied Biosystems, Warrington, United Kingdom). cDNA was amplified by PCR in an Applied Biosystems 7500 thermocycler with Power Sybr green master mix (Applied Biosystems) using the manufacturer's protocol. Primers were as follows: for Slp, forward primer 5′-GCAAACTGTCCCCTGAGACC-3′ and reverse primer 5′-ACTGCTGGATCCGAACGTG-3′; for C4, forward primer 5′-GCAAACTGTCCCCTGAAACA-3′ and reverse primer 5′-GAAACTGCTGGATCCTCATGTA-3′; and for 18S rRNA, forward primer 5′-CGCCGCTAGAGGTGAAATTC-3′ and reverse primer 5′-CCAGTCGGCATCGTTTATCG-3′. Reactions were performed in duplicate, and cycle threshold (CT) values were converted to relative expression levels using the 2−ΔΔCT method (32) with 18S rRNA as the normalization standard and an arbitrarily chosen WT male as the calibrator.
Antibodies.
Rabbit polyclonal anti-RSL1 antibody was synthesized at Rockland Immunochemicals (Gilbertsville, PA) with the peptide DMKRQGTAPVYPGI, which maps to the C-terminal end of the KRAB-B domain. The peptide was used for affinity purification before the antibody was used in Western blot and ChIP assays. Anti-KAP1 antibody (ab10484) was purchased from Abcam (Cambridge, MA). Anti-STAT5b antibodies used in Western blotting (71-2500) and ChIP (sc-836X) were from Invitrogen (Camarillo, CA) and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. Normal rabbit IgG (sc-2027) for ChIP was from Santa Cruz Biotechnology.
DNA methylation analysis.
Liver and kidney DNAs were isolated by proteinase K digestion, followed by phenol-chloroform extraction and ethanol precipitation. One microgram DNA per sample was treated with sodium bisulfite and isolated with an EpiTect kit (Qiagen) according to the manufacturer's instructions. For methylation-specific qPCR, bisulfite-converted DNA was amplified in two independent reactions with the common forward primer (5′-GAGATTTTGGTGTTTGGGTTGTTTTT-3′) and a primer distinguishing the methylated from the unmethylated form [5′-CTCCCTTTAACCCCATAAACCC(G/A)-3′] at the position −66 bp from the Slp CpG transcription start site (−66 Slp CpG). For the −65 C4 CpG, the same forward primer was paired with reverse primers positioned similarly to those for Slp. Quantitative PCR used three mice per sex and genotype with the Power Sybr green master mix (Applied Biosystems) and an ABI 7500 real-time PCR system. To compare relative methylation with gene expression, results were plotted as the difference between nonmethylated and methylated CT values (i.e., methylation ratio). Mice with greater methylation levels had lower ratios.
For bisulfite sequencing, 1 μg DNA, pooled from three mice per sex and genotype, was bisulfite converted and isolated as described above. Primers 5′-GAGATTTTGGTGTTTGGGTTGTTTTT-3′ and 5′-GGGACTAGTCCCTCCCTTTAACCCCATAA-3′ amplify both Slp and C4 promoters. PCR products were digested with SpeI and cloned into the SpeI and EcoRV sites of pCR2.1Topo. Figure 2B shows bases that distinguish Slp from C4 clones. At least 20 clones per DNA pool were sequenced to assess the methylation status of −66/−65 and −75 CpGs simultaneously.
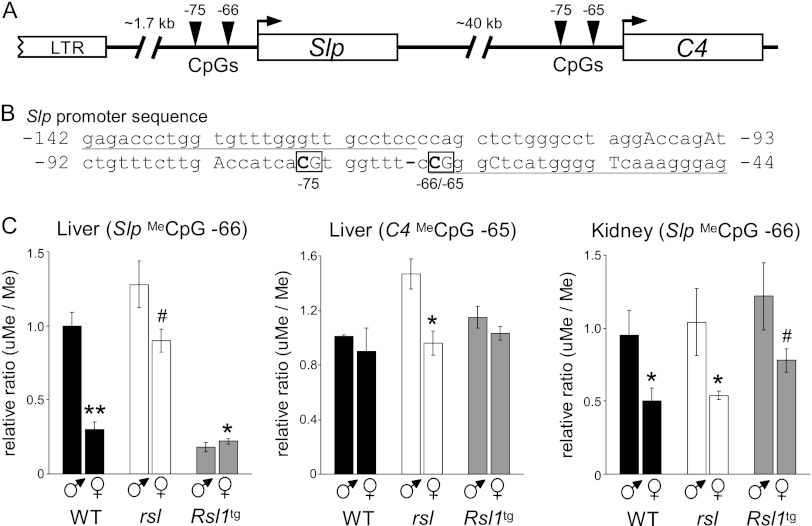
Fig 2.
Analysis of Slp and C4 promoters by quantitative methylation-specific PCR. (A) Slp/C4 locus. Triangles indicate CpG dinucleotides near the transcription start of each gene (bp −75 and −66/65). The ancient retrovirus upstream of Slp (LTR) is not present upstream of C4. (B) Slp promoter sequence amplified by methylation-specific primers. The −66 and −75 CpGs are boxed; bases in capital letters differ between Slp and C4; hyphen, a 1-bp deletion in C4 relative to the Slp sequence. (C) Relative ratio of unmethylated (uMe) to methylated (Me) PCR product for Slp and C4 in WT, rsl, and Rsl1tg mouse liver and kidney. Data were normalized to one WT male in each assay. Bars indicate the mean ± SEM; n = 3 mice per group. Sex differences within genotypes are indicated: **, P = 0.002; #, P < 0.1; *, P < 0.05.
In vitro DNA binding. (i) GST fusion protein isolation.
The Rsl1 cDNA was cloned by RT-PCR (forward primer, 5′-GGTCTGTACTCGTGCGTCTTT-3′; reverse primer, 5′-ACAACATCCATTTGCCTGGTA-3′) from WT liver RNA into the pGEM-T Easy vector (Promega). The resulting plasmid was the PCR template to create full-length (FL) glutathione S-transferase (GST)-Rsl1 fusion plasmid pGEXRsl1 and a 5′ and 3′ deletion series of the ZNFs. PCR products were cloned into the BamHI and SmaI sites of pGEX-2TK (GE Healthcare Life Sciences). Primers for Rsl1 FL (see Fig. 4A) were forward primer 5′-AAAAGGATCCATGCGGCTATCGATTCC-3′ and reverse primer 5′-TTATCACATGTGTGTGGATTT-3′. Plasmids were used to transform Escherichia coli BL21 (GE Healthcare Life Sciences), and protein expression was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). SDS-PAGE analysis of bacterial lysates revealed that ZNFs 2 to 4 inhibited Rsl1FL synthesis in E. coli (see Fig. 4B). To remove inhibition, plasmid pGEXRsl1dZF-1, which terminates 109 bp into the ZNF domain, was linearized with EcoRI (New England BioLabs) and filled in with the Klenow fragment (Roche). The 10 C-terminal ZNFs were obtained from pGEXRsl1 FL as an 884-bp EcoRI fragment that was filled in with the Klenow fragment and ligated into the modified pGEXRsl1dZF-1 to create pGEXRsl1 KBZF10, which was sequenced across cloning junctions for verification. GST-Rsl1 KBZF10 protein (see Fig. 4A) was isolated from lysates on glutathione-Sepharose (GE Healthcare Life Sciences). The plasmid for STAT5b synthesis (pcDNA3V5mSTAT5b) was created by cloning an RT-PCR product from WT liver into the XbaI/SacII sites of pcDNA3 modified with a V5 epitope (from Jorge Iniguez, University of Michigan) using the primers 5′-AAATCTAGAGCTATGTGGATACAGGCTCA-3′ and 5′-AAACCGCGGAGGTGGTGAGGTCTGGTCAT-3′.
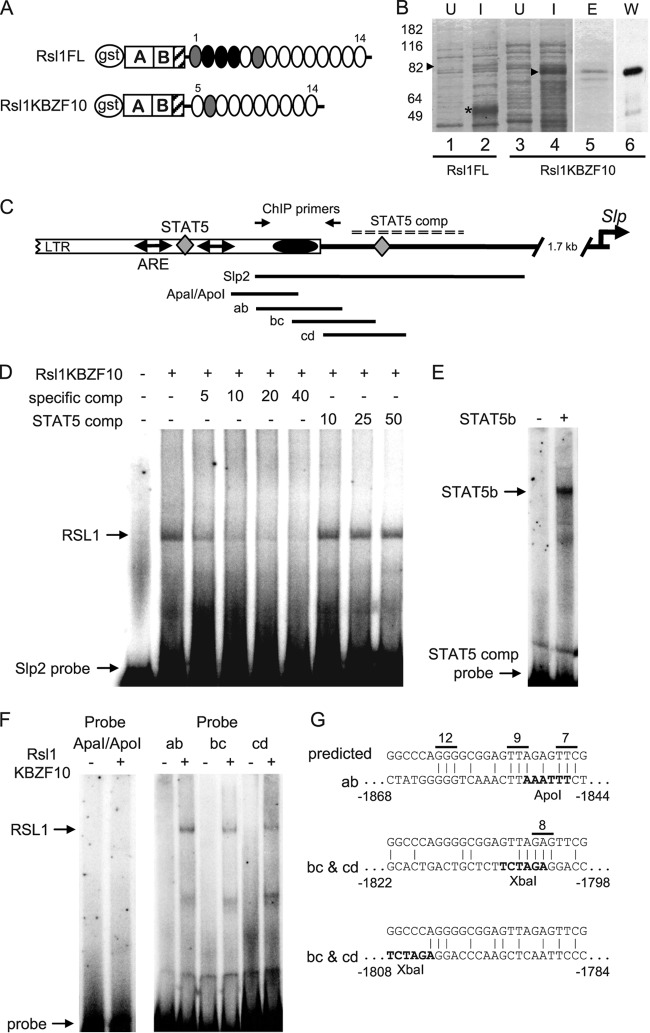
Fig 4.
In vitro identification of an RSL1 binding site upstream of Slp. (A) Gst-Rsl1 fusion proteins expressed in E. coli. FL, full-length protein. ZNFs 1 to 4 are deleted in Rsl1KBZF10. GST and KRAB A and B domains are indicated; striped box, peptide antigen for RSL1 antisera; ovals, ZNFs; gray ovals, degenerate ZNFs lacking C or H residue for zinc coordination (18); black ovals, ZNFs inhibiting RSL1 expression in E. coli. (B) SDS-PAGE analysis of lysates from bacteria transformed with Gst-Rsl1 FL (lanes 1 and 2) or Gst-Rsl1KBZF10 (lanes 3 to 6). U and I, uninduced and induced with IPTG, respectively; E, eluted protein; W, Western blot of eluted Rsl1KBZF10 probed with RSL1 antibody; arrowheads, expected protein positions; asterisk, truncated or degraded fusion protein upon induction of Gst-Rsl1FL. (C) Sequence elements 1.7 to 2.0 kb 5′ to Slp. Open rectangle, proviral 5′ LTR; double-headed arrows and diamonds, AREs and STAT5 sites, respectively; black oval at the LTR end, the RSL1 binding site; arrows, primers used for ChIP; dashed line, STAT5 competitor. The locations of other EMSA probes are shown below the diagram. (D) EMSA with Rsl1KBZF10 and Slp2 probes. A molar excess of nonradioactive competitors (comp) is indicated. The specific competitor was Slp2 (bp −1696 to −1874 from Slp position bp +1) or an 80-bp sequence from bp −1728 to −1807 that contains the proximal STAT5 consensus (dashed line in panel C). (E) To verify STAT5b binding, labeled 80-bp competitor was incubated with in vitro-transcribed/translated STAT5b for EMSA. (F) To delimit RSL1 binding, EMSA was performed with the ApaI/ApoI probe and overlapping 75-bp probes ab, bc, and cd. A shift occurs with each 75-bp probe but is most evident with probe ab. (G) The predicted RSL1 binding site based on the ZNF code (16) (top sequence) is aligned to regions with the best identity within Slp2 (bottom sequence). Lines above the predicted sequence indicate putative contacts for ZNFs 7, 8, 9, and 12. The 75-bp EMSA probe that contains the site is indicated. The location of each site relative to Slp position bp +1 is indicated.
(ii) Electrophoretic mobility shift assay (EMSA).
Radioactive probes were end labeled with T4 DNA kinase (New England BioLabs) and [γ-32P]ATP (MP Biomedicals). For each reaction, DNA probes (30,000 to 50,000 cpm) were incubated with 50 ng Gst-Rsl1 KBZF10 in binding buffer with poly(dI-dC) (12.5 ng/μl) at 30°C for 30 min, followed by electrophoresis at 4°C on 5% nondenaturing polyacrylamide gels. Similar reactions were performed with STAT5b synthesized with plasmid pcDNA3V5mSTAT5b in a rabbit reticulocyte in vitro transcription/translation kit (Promega). For competition, competitor DNA was added at the beginning of the binding reaction. Gel images were obtained by PhosphorImager (Molecular Dynamics).
ChIP.
ChIP reactions were performed as described by Boyd and Farnham (2) with the following modifications. Mouse liver was dissected, and 500 to 600 mg was rinsed in ∼3 ml ice-cold phosphate-buffered saline (PBS) and immediately minced on ice. Minced liver was cross-linked by shaking at room temperature for 15 min in 1.5% formaldehyde in PBS and stopped by adding 0.125 M glycine in PBS. To remove formaldehyde, liver was washed 5 times with a total of 50 ml ice-cold PBS. The cell pellet was disrupted in a Dounce homogenizer in 2.0 ml hypotonic buffer (10 mM Tris [pH 8.0], 10 mM NaCl, 3 mM MgCl2, 30 mM sucrose) with protease inhibitors and lysed to completion in 6.0 ml hypotonic buffer on ice for 10 min. Chromatin was pelleted (1,000 × g) and resuspended in 2.0 ml radioimmunoprecipitation assay (RIPA) buffer for sonication (10 pulses [1 pulse was 20 s sonication plus 30 s cooling]) with the power setting at 4 (Branson Sonifier 185 cell disrupter with a 2.5-mm tip). Sonicated chromatin was aliquoted (∼180 μl), diluted up to 800 μl with RIPA buffer, and stored at −80°C until immunoprecipitation (IP). One aliquot of sonicated lysate per IP was precleared with 60 μl protein A-agarose (Roche) and incubated at 4°C with rotation overnight with 1 to 10 μg antibody, according to the supplier. For RSL1, 5 to 10 μl antigen-purified antibody was used per IP. Two micrograms IgG (sc-2027; Santa Cruz Biotechnology, Santa Cruz, CA) was used as a negative control. Antibody-bound chromatin was isolated on 30 μl blocked protein A-agarose (Roche) and eluted in ChIP elution buffer (50 mM Tris [pH 8.0], 10 mM EDTA, 200 mM NaCl, 1% SDS, 2 mM dithiothreitol, 1.0 μg/ml proteinase K). Proteins were digested at 42°C for 1 h, and cross-linking was reversed at 65°C for 4 h. DNA was isolated by phenol-chloroform extraction and ethanol precipitation and was resuspended in 50 μl H2O. One to 5 μl DNA was analyzed in triplicate by real-time PCR as described above. ChIP primers for RSL1 and STAT5b binding to Slp were Slp2-F (5′-CTTGGTCTATGGGGGTCAAA-3′) and Slp17-R (5′-GGGTCCTCTAGAAGAGCAGTCA-3′), for binding to SOCS2 they were mSOCS2ChIP2-F (5′-GGCCTAAAGGTTCCCTCCTA-3′) and mSOCS2ChIP2-R (5′-AGCCAATGCCTATTAAGCCA-3′), and for binding to the negative-control intronic sequence they were Slp7intqPCR-F (5′-ACCCCCACCTCTGTGCTCCC-3′) and Slp7intqPCR-R)5′-AGAAATGCCCGGTGGCGTGG-3′).
For sequential ChIP (re-ChIP), 100 μl cross-linked liver lysate, as described above, was assayed with a ReChIP-IT kit from Active Motif. Alternatively, 350 μl lysate was immunoprecipitated as described above, except that proteinase K was omitted from the first elution buffer. Eluted chromatin was diluted 66 times in RIPA buffer and reprecipitated with the second antibody and processed for analysis as described above.
RESULTS
Rsl1 accentuates sex-specific expression of Slp in a tissue-specific manner.
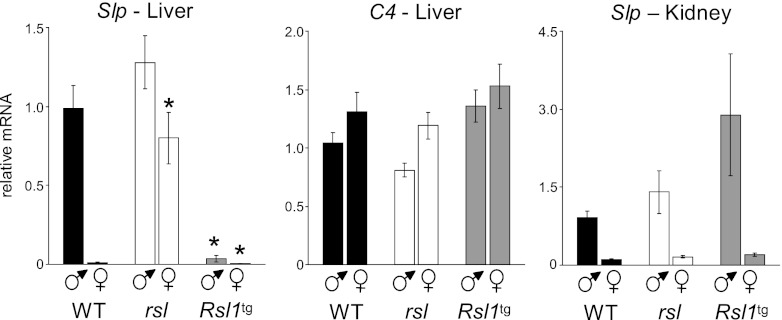
Mutations in Rsl1 lead to reduced sex bias in the expression of many mouse liver genes (19). In particular, Slp, the hallmark of Rsl regulation, provides a sensitive endogenous reporter of Rsl1 activity in the liver. Slp is also expressed in the kidney, as is Rsl1 (see Fig. S1 in the supplemental material), but the hormonal response is under direct androgen regulation rather than GH control (28). To discern an Rsl1 effect in kidney, we compared Slp expression in WT, rsl (Rsl1−/−), and transgenic mice with liver-restricted Rsl1 cDNA overexpressed on the rsl background (Rsl1tg). Liver Slp expression occurred in WT males and rsl mice of both sexes, as expected, and was fully suppressed by excess Rsl1 in Rsl1tg mice (Fig. 1, left). In contrast to the strong effect of Rsl1 on Slp, there was no significant effect on the tandemly duplicated homolog C4 (Fig. 1, center), indicating target gene selectivity rather than the existence of a large repressive domain.
Fig 1.
Rsl1 affects sex- and tissue-specific expression of Slp. Slp and C4 RNAs in liver and kidney of WT, rsl, and Rsl1tg mice were measured by qRT-PCR. To compare relative expression across sex and genotype, data were normalized to those for WT males. Bars indicate means ± SEMs; n ≥ 3 mice per group. *, significant differences (P < 0.001) from WT mice of the same sex. ♂, male; ♀, female.
In the kidney, the presence or absence of Rsl1 did not affect Slp expression as it did in the liver. Males expressed Slp in kidney at least 8-fold more than females, regardless of Rsl genotype, with consistently low expression in females (Fig. 1, right). These results may indicate differences in the chromatin environment around Slp in the liver versus the kidney that are established at puberty by STAT5b and the androgen receptor (AR), respectively (11, 28, 41).
Rsl1 affects CpG methylation of the Slp promoter.
To link Rsl1 to its molecular actions, we first examined differential Slp methylation, as a close association between KRAB-ZFP function and methylation of CpGs in DNA and histones has been shown in vitro (1, 23, 49). Previously, a CpG dinucleotide located at bp −66 from the Slp transcription start site (Fig. 2) was shown to be unmethylated in WT liver to a greater extent in males than females in accord with pubertal induction (47). Whether this sexually dimorphic mark was Rsl1 dependent was tested using methylation-specific quantitative PCR of DNA from WT, rsl, and Rsl1tg adults. Liver and kidney DNAs were treated with sodium bisulfite, and methylated and unmethylated DNAs were separately quantified by real-time PCR. The ratio of unmethylated to methylated DNA was plotted (Fig. 2C) to allow a direct comparison to Slp expression in Fig. 1. In liver, the relative level of methylation at the −66 CpG correlated with repression of Slp. This was most evident in WT females (i.e., ∼3 times more methylation in females than males) and in Rsl1tg mice, where there was substantially less unmethylated DNA (∼20% of WT male levels), in accord with less Slp expression (∼2% of WT male levels) (Fig. 2C; cf. Fig. 1). A similar site in the homologous C4 promoter (−65 CpG) is largely unmethylated in all mice, indicating that RSL1 does not influence methylation of Slp's adjacent paralog. In contrast, in the kidney, where Slp expression is not influenced by Rsl1 (Fig. 1) (41), methylation at the −66 CpG did not vary significantly with genotype (Fig. 2C). However, there was more unmethylated DNA in males, which likely reflects demethylation at puberty coincident with AR induction of Slp in kidney (28).
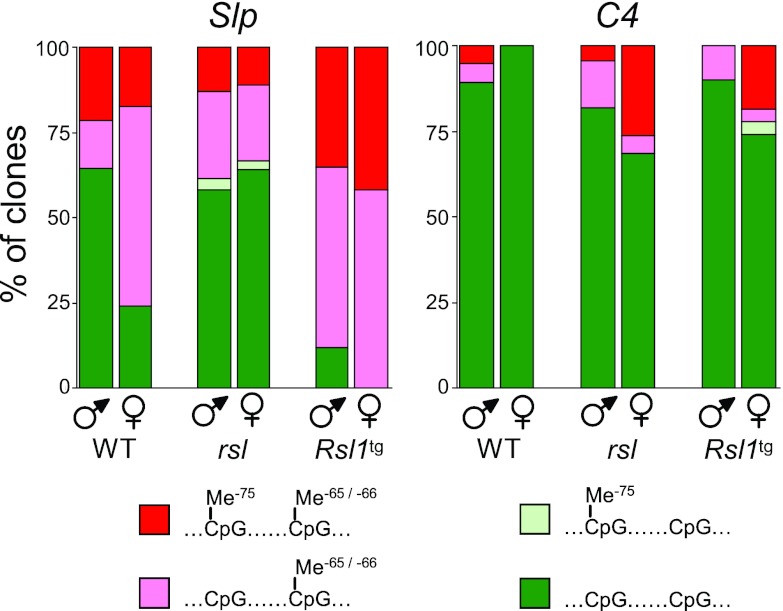
To test whether RSL1-associated methylation of the −66 CpG Slp site was stringently site specific or might spread to neighboring CpGs, bisulfite sequencing was used to assess simultaneously the −66 CpG and nearest neighbor CpG at bp −75. Consistent with previous findings, Slp's −66 CpG was specifically methylated and was methylated to a higher degree in WT female than male liver (75% versus 33%) (Fig. 3, pink plus red bars). This site was only about 37% methylated in male and female rsl mice but highly methylated (85% to 100%) in both sexes of Rsl1tg mice, corroborating that methylation was at least in part Rsl1 dependent. Methylation at the −75 CpG only weakly correlated with the presence or level of Rsl1, ranging from 15% in rsl mice to 40% in Rsl1tg mice. Methylation at the −75 CpG was not sexually dimorphic and largely present only when the −66 CpG was methylated. As expected, the C4 promoter was largely unmethylated regardless of sex or genotype (Fig. 3, right). Together with the methylation-specific PCR data (Fig. 2), there is a striking correlation between methylation at Slp's −66 CpG and the relative level of Rsl1 in liver, suggesting that DNA methylation is a component of the RSL1 repression mechanism.
Fig 3.
Analysis of Slp and C4 promoter methylation by bisulfite sequencing. Liver DNA from each sex of WT, rsl, and Rsl1tg mice was treated with sodium bisulfite. A 100-bp region encompassing CpG −65/−66 and CpG −75 was PCR amplified and cloned into pCR2.1Topo. At least 20 clones per sex and genotype were sequenced, and the percentage of clones methylated or not at one or both CpGs was charted.
RSL1 binds in vitro within proviral sequences upstream of Slp.
Surprisingly few KRAB-ZFP DNA binding sites have been identified in cellular genes, even for those repressors for which biological roles are known, in part due to a lack of strong candidate target genes. Moreover, binding site prediction is ambiguous because KRAB-ZFPs may have more than 30 ZNFs and their differential usage may lead to recognition of diverse sites (29). Hence, binding specificities may be highly variable. Evidence supporting this notion comes from a recent genome-wide study of ZNF263 that indicates that the current ZNF recognition code derived from in vitro assays is not sufficient by itself to predict in vivo binding (8). To biochemically identify an RSL1 binding site in its genetic target, Slp, we purified a GST-RSL1 fusion protein from bacteria for protein-DNA interaction studies. Fingers 1 to 4 (Fig. 4A) were deleted to obtain optimal expression (Rsl1KBZF10; Fig. 4B, lanes 3 to 5). The integrity of this fusion is demonstrated by Western blotting (Fig. 4B, lane 6) with an RSL1 antibody raised against the C-terminal region of the KRAB-B domain that recognizes RSL1 but not the paralog, RSL2 (see Fig. S2 in the supplemental material).
Three regions in Slp's 5′-flanking sequence were candidates for RSL1 binding. First was the −66 CpG site itself, in which methylation seems to be Rsl1 dependent (Fig. 2 and 3). Second was the putative tRNAPro primer binding site of a provirus ∼2 kb upstream of the transcription start site of Slp (38). Retroviral silencing has been suggested to be the primordial role of KRAB-ZFPs (40). This ancient provirus conferred hormonal regulation on Slp upon insertion and contains a tRNAPro primer binding site similar to that of murine leukemia viruses (MLVs) that are bound by ZFP809 in embryonic stem cells (45). The third candidate region is the enhancer within the proviral long terminal repeat (LTR) (Fig. 4C) that is associated with Slp hormonal control (24, 38). Multiple transcription factor binding sites have been identified here, including androgen response elements (AREs) and STAT5 binding sites, both of which can confer male-biased expression.
Gel shift (EMSA) probes approximately 200 bp in length were synthesized to sequences from the 5′ flank of Slp and incubated with Rsl1KBZF10. No RSL1 binding was observed with probes overlapping the −66 CpG or the viral primer binding site (data not shown). However, RSL1 binding was evident with a probe, Slp2, which overlaps the hormone-responsive enhancer (Fig. 4C and D; see Fig. S3 in the supplemental material). A canonical STAT5 binding site (5′-TTCNNNGAA-3′) also is contained within Slp2 (Fig. 4E; see Fig. S3 in the supplemental material). To test the possibility that RSL1 and STAT5b recognize the same site in Slp2 and alternate to confer repression or induction, the STAT5 binding sequence was used as a competitor. RSL1 binding was undiminished by the STAT5 competitor (Fig. 4D, right lanes), suggesting that RSL1 bound to the 5′ portion of Slp2 rather than the STAT5 site (Fig. 4C). To narrow the site of RSL1 binding, Slp2 was divided into three restriction fragments (ApaI/ApoI, ApoI/XbaI, Xba/3′ end) that were used individually as EMSA probes. None were shifted by RSL1 (Fig. 4F; see Fig. S3 in the supplemental material), suggesting that cleavage at one or more of these sites disrupts binding. To ensure the integrity of a complete binding site, three overlapping 75-bp probes (probes ab, bc, and cd) were synthesized (Fig. 4C; see the sequences in Fig. S3 in the supplemental material). All three bound RSL1, with probe ab having the highest apparent affinity (Fig. 4F).
To complement the gel shift data, a zinc finger code was applied to the eight C-terminal fingers of RSL1 found in Rsl1KBZF10 (16). Amino acids at positions 6, 3, 2, and −1 within each ZNF contact three consecutive bases, dependent on the amino acid side chains. This code predicted a 25-bp idealized binding site for RSL1 using each base specified at the highest probability (Fig. 4G). A sliding-window approach to localize maximum identity revealed a sequence in probe ab with 12 bases of identity, including three putative contacts for ZNFs 7, 9, and 12 (Fig. 4G). Two matches of 10 bases, one with a predicted ZNF8 contact, may explain the weaker RSL1-DNA complexes seen with probes bc and cd. Together, the preferred binding sequence determination and EMSA data suggest that RSL1 binds within the proviral LTR to exert effects on the Slp gene 2 kb downstream. This is the first identification of a KRAB-ZFP binding site in a genetically defined target gene. The location of this site within a proviral LTR hints at how KRAB-ZFP actions may have evolved from retroviral silencing to regulating endogenous genes.
RSL1 binds within the hormonal enhancer of Slp in the liver.
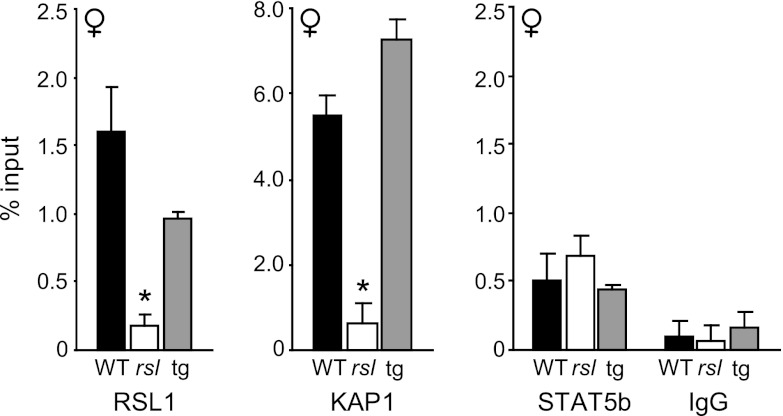
To test whether RSL1 binding occurs at the LTR in vivo, we used ChIP assays of liver chromatin with primers flanking the binding site identified in our in vitro assays (Fig. 4C). Females were analyzed first because they allow a clear distinction for RSL1-mediated repression due to the lack of hormonal induction that occurs in males. RSL1 binding was observed in WT and Rsl1tg females but not in rsl females (Fig. 5, left). This result validates the in vitro binding of RSL1 to the Slp upstream region and further confirms the specificity of the antibody since no signal was detected in Rsl−/− mice. Binding in WT and Rsl1tg females was approximately equivalent by ChIP, despite differing in Rsl1 mRNA levels by 50-fold, presumably because the number of their RSL1 binding sites is limiting. KAP1 was detected at the RSL1 binding site as well, but only when RSL1 was also present (Fig. 5, center), supporting its recruitment to chromatin by RSL1, as expected for the presumed universal corepressor of KRAB-ZFPs (33).
Fig 5.
RSL1, KAP1, and STAT5b bind upstream of Slp in female liver. ChIP assays were performed for RSL1, KAP1, and STAT5b on liver from pairs of adult female WT, rsl, and Rsl1tg mice using primers flanking the RSL1 binding site, as shown in Fig. 4. IgG served as a negative control. Bars are the mean of the percent input ± SEM. RSL1, STAT5b, and IgG were plotted on the same scale to demonstrate low and similar binding of STAT5b in all genotypes. *, significant difference (P < 0.05) from WT.
Nuclear STAT5b is detectable in female liver at a low constant level insufficient to induce male-biased genes (21). ChIP assays revealed STAT5b binding to the Slp enhancer in females at similar levels regardless of genotype (Fig. 5). That STAT5b binding did not vary with the presence or absence of RSL1/KAP1 suggests that RSL1 and STAT5b colocalize upstream from Slp independently in females, likely due to distinct binding sites in the DNA, consistent with the EMSA results (Fig. 4). No STAT5b or RSL1 binding was detected with primers designed to a region ∼10 kb away within intron 7 (see Fig. S4 in the supplemental material). Together these results indicate that RSL1 binds upstream from Slp and recruits KAP1 to silence gene expression. Furthermore, this mechanism is sufficient to counter a low level of STAT5b present in female hepatocytes.
Dynamic interplay of inducer and repressor.
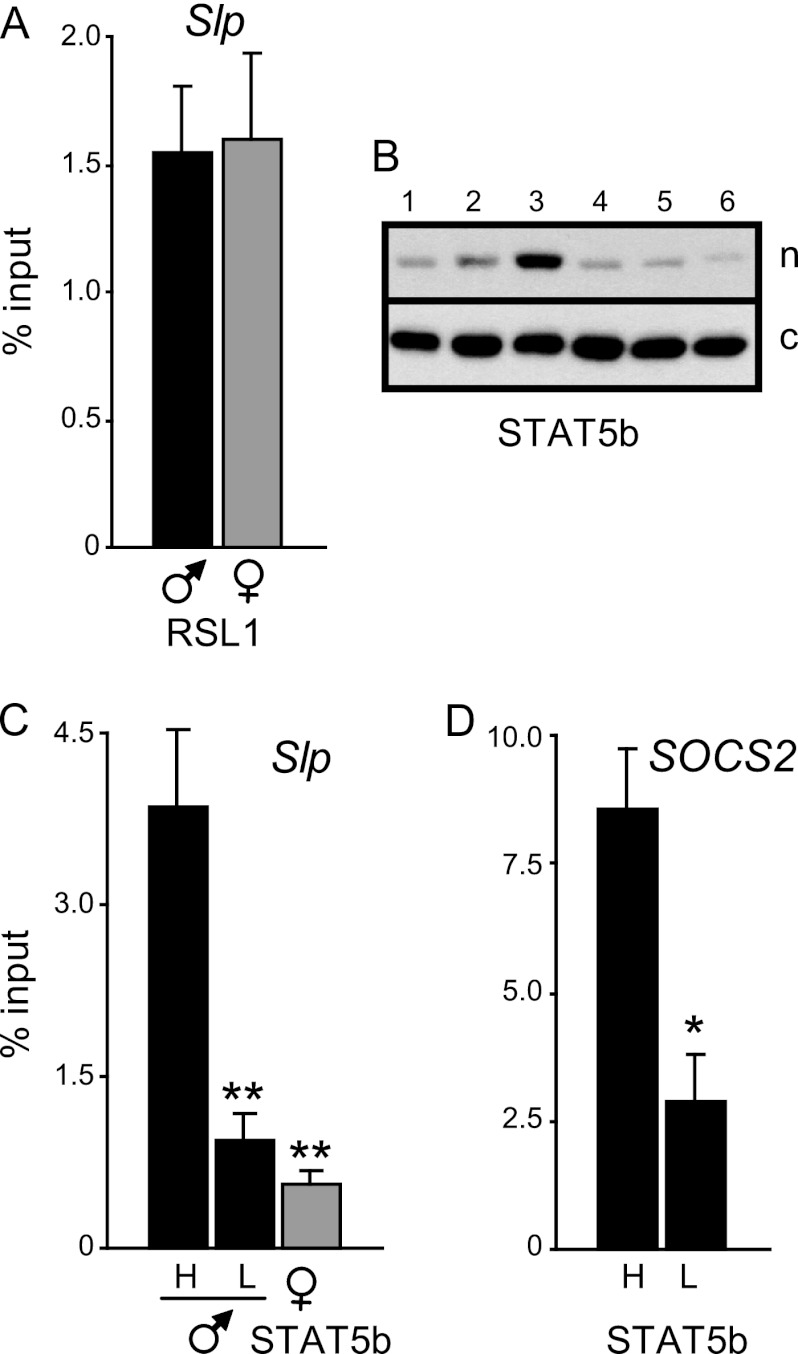
RSL1's effects are notable in female liver, but some repression also occurs in males, as evidenced by the higher level of Slp expression in rsl males than WT males (Fig. 1). How KRAB-Zfp repression limits hormonal activation may be uniquely examined in this physiological model. In WT mice, RSL1 binding by ChIP was the same, on average, in males (1.82% ± 0.35% input) and females (1.60% ± 0.33% input) (Fig. 6A), supporting the notion that repression does indeed occur in both sexes. However, exploring RSL1 interactions in males must take into account the nuclear cycling of STAT5b in response to GH pulses. Nuclear STAT5b levels were analyzed by Western blotting in males euthanized over a 3-h period (9 a.m. to noon). As expected, due to male GH pulsing, nuclear STAT5b levels varied severalfold (Fig. 6B). The extent of variation was the same regardless of genotype (data not shown), indicating that GH signaling to STAT5b is likely unaffected by Rsl directly. Males were designated “high” (e.g., Fig. 6B, lane 3) or “low” (e.g., Fig. 6B, lane 6) to assess binding in accord with their relative level of STAT5b. As expected, ChIP analysis correlated with a 4-fold differential in STAT5b binding dependent on nuclear levels (Fig. 6C). The SOCS2 promoter served as a positive control in ChIP to further verify in these mice that STAT5b binding correlated with high nuclear levels that are driven by the GH pulse (Fig. 6D), as shown previously in rats (3). Together, these data demonstrate the transient binding of STAT5b in chromatin in response to peaks of GH.
Fig 6.
Sex- and hormone-dependent binding of RSL1 and STAT5b to the Slp enhancer in WT mouse liver. (A) ChIP assays were performed for RSL1 on liver from WT males (n = 5; inputs ranged from 1.26% to 2.98%) and females (n = 3; inputs ranged from 1.01% to 2.14%). Bars are the mean of the percent input ± SEM. (B) Western blots of male liver nuclear (n) and cytoplasmic (c) extracts probed for STAT5b to assess the range in STAT5b levels due to nuclear cycling. (C) ChIP result for STAT5b binding to Slp. H, high nuclear STAT5b (e.g., lane 3 in panel B; n = 3); L, low nuclear STAT5b (e.g., lanes 5 and 6 in panel B; n = 5). Bars are the mean of the percent input ± SEM. ChIP results for STAT5b in WT female liver highlight sex-specific binding. (D) STAT5b ChIP for SOCS2 in WT males. H, GH-injected males (n = 4). Significant differences from WT males are indicated: *, P < 0.05; **, P < 0.005.
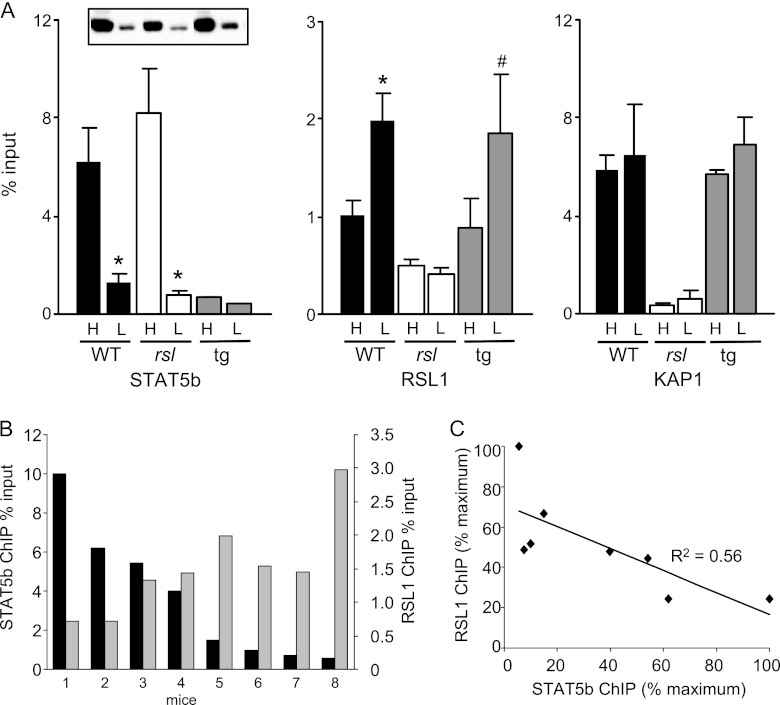
To determine whether RSL1 binding is affected by the presence or absence of STAT5b, chromatin was reproducibly obtained at peak STAT5b levels by injecting male mice with GH and harvesting livers after 30 min. For trough STAT5b levels, untreated mice were tested for low nuclear STAT5b by Western blotting. ChIP showed robust STAT5b binding to the Slp enhancer, in accord with nuclear levels in WT males (an ∼5-fold range from high to low) (Fig. 7A). A similar pattern was seen in rsl males. However, very little STAT5b binding was observed in Rsl1tg males, even at peak nuclear levels (Fig. 7A, inset). These data suggest that excess RSL1 in male Rsl1tg liver alters the chromatin structure to prevent STAT5b access, in accord with a complete loss of Slp expression. When RSL1 binding was assessed, no binding was detected in rsl males, as expected. KAP1 binding was also absent in rsl males, again highlighting the necessity of RSL1 to recruit KAP1 to chromatin. Intriguingly, in both WT and Rsl1tg males, RSL1 binding was evident and differed by about 2-fold in males with high versus low STAT5b levels (Fig. 7A, middle). The similarity of this pattern suggests that RSL1 binding in both genotypes is sensitive to GH, regardless of STAT5b binding. Furthermore, this GH sensitivity is opposite in direction from that of STAT5b. In contrast, KAP1 binding did not reflect GH levels in WT or Rsl1tg males. To further visualize the reciprocal nature of STAT5b and RSL1 binding, the WT male ChIP results were compared pairwise for these two factors in 8 males (Fig. 7B). ChIP data were normalized by converting percent input to percent maximum and plotted in descending order of STAT5b binding. The correlation coefficient of 0.56 indicates an inverse relationship between RSL1 and STAT5b binding (Fig. 7C), suggesting that antagonism between this repressor and activator may be due, at least in part, to reciprocal DNA binding. Together, these results suggest for the first time a dynamic oscillation between a KRAB-ZFP repressor and a hormonal activator in an adult tissue.
Fig 7.
Dynamic interaction of RSL1, KAP1, and STAT5b on the Slp enhancer in male liver. To assess factor binding relative to STAT5b levels, male mice were injected with GH 30 min prior to isolation of livers (peak) or not (trough); STAT5b levels were confirmed by Western blotting of nuclear lysates. (A) ChIP data graphed as mean of the percent input ± SEM (n = 2 to 4 mice per group). (Inset) Western blot showing relative levels of nuclear STAT5b in representative individuals in the same lane order as the ChIP data. H, high nuclear STAT5b; L, low nuclear STAT5b. Immunoprecipitation with RSL1 antibody in WT and Rsl1tg males was approximately 25% of that obtained with antibodies to STAT5b or KAP1 but was significantly above the IgG negative-control level (mean = 0.3 ± 0.04% input; P < 0.001). Significant differences between high and low nuclear STAT5b are indicated: *, P < 0.05; #, P = 0.11. (B) RSL1 and STAT5b bind reciprocally in WT males. STAT5b ChIP results from 8 WT males (dark bars) were arranged from highest to lowest and graphed next to the corresponding RSL1 ChIP data (light bars). (C) ChIP results were plotted as percent maximum, defined as the individual with the greatest percent input value. R2, correlation coefficient.
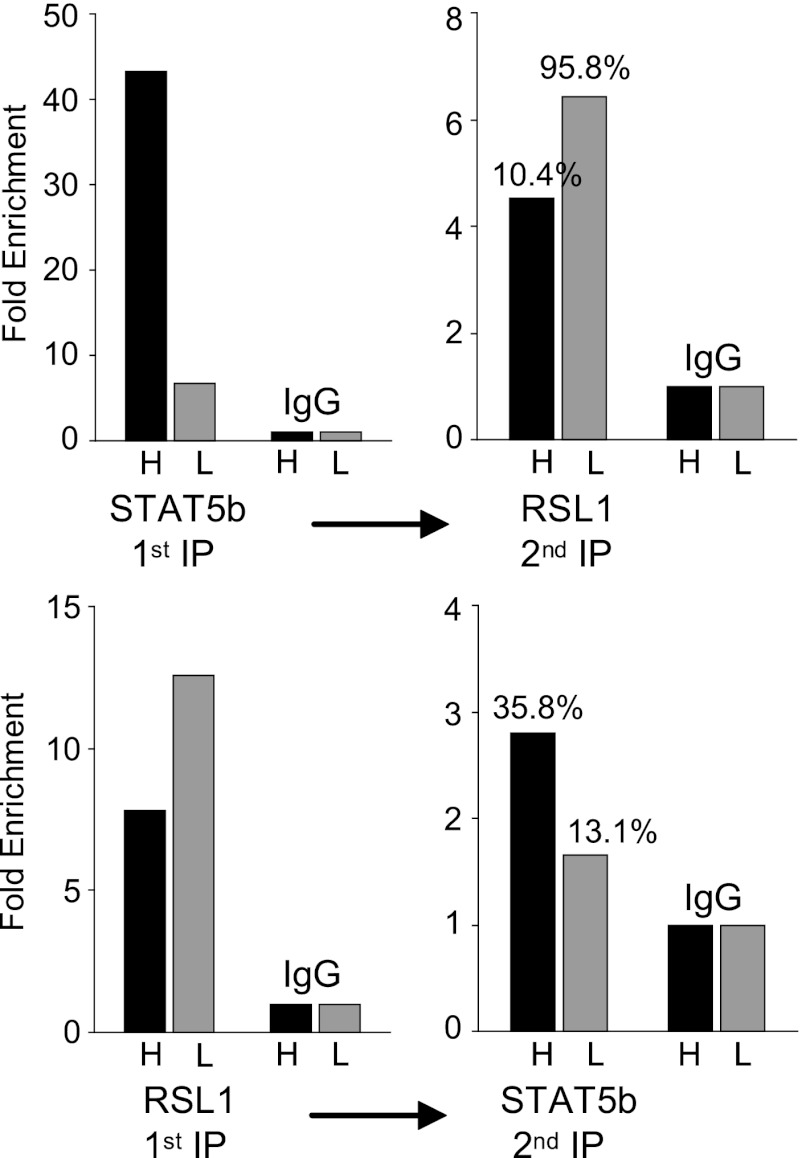
To verify the reciprocal binding of STAT5b and RSL1 and determine the proportion of complexes with one or both proteins, sequential ChIP (re-ChIP) was performed with chromatin from GH-treated or untreated WT males (Fig. 8; data are from a representative pair of mice). When the STAT5b IP was first, about 6 times more STAT5b bound to the Slp enhancer in response to GH, similar to the results obtained before (Fig. 7A). When these complexes were then immunoprecipitated for RSL1, the proportion containing both proteins differed depending on nuclear STAT5b levels: only 10% of complexes had both factors when STAT5b was high, whereas 95% had both factors when STAT5b was low. In a control experiment using the STAT5b antibody for both the first and second IPs, about 90% of complexes from the first IP were reprecipitable in the second IP (see Fig. S5 in the supplemental material). That precipitated complexes were efficiently reprecipitated suggests that the proportion of fragments bound only by the first antibody and therefore in the supernatant in the second reaction can be estimated by subtraction. Thus, during the STAT5b peak, most enhancers were bound by STAT5b and not RSL1. In contrast, although far fewer complexes were precipitated by anti-STAT5b when nuclear levels were low, almost all of these also contained RSL1. These results confirm that the inducer, STAT5b, and repressor, RSL1, do indeed cooccupy the Slp enhancer in WT male liver, but with a large differential that is dependent on the GH pulse.
Fig 8.
STAT5b and RSL1 cooccupancy at the Slp enhancer varies in accord with GH. Re-ChIP was performed on WT males that were treated with GH (high [H]) or not (low [L]), as described in the legend to Fig. 7. Results are fold enrichment relative to the IgG negative control. (Top) Re-ChIP results with the STAT5b antibody used first and the RSL1 antibody used second; (bottom) reciprocal experiment. Numbers denote the percentage of the first IP with cooccupied complexes.
To corroborate the results presented above, the re-ChIP experiment was performed in the inverse direction. When the RSL1 antibody was used first, RSL1 binding in the GH-treated male was about 50% of that in the untreated male, as seen in conventional ChIP (Fig. 8). The subsequent IP with the STAT5b antibody also revealed differential partitioning of complexes dependent on GH. In the GH-treated male, 36% of the precipitated Slp enhancers showed cooccupancy, while only 13% were cooccupied in the untreated male. Together, these data indicate a dramatic shift in binding at the Slp enhancer between STAT5b and RSL1 that occurs in WT male liver in accord with GH signaling (Fig. 9). Some of the reconfiguring was expected, given that the intracellular transition of STAT5b is well documented (4, 21). More remarkable was the subtle shift in RSL1 on and off the chromatin that must be occurring in response to GH to account for the different proportions of singly occupied or cooccupied enhancers.
Fig 9.
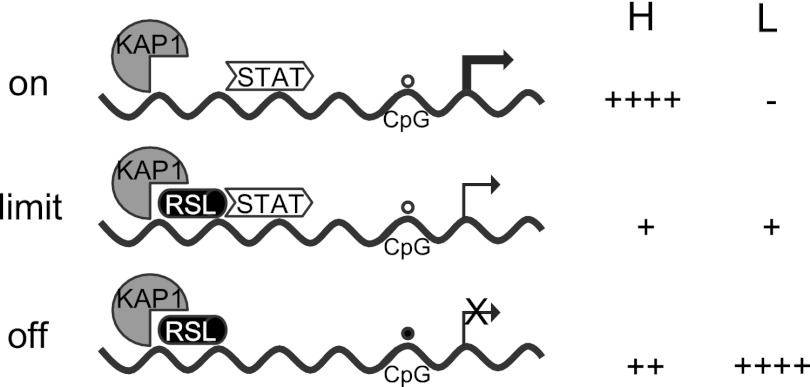
Interplay of opposing forces on the Slp enhancer. In males, STAT5b transitions from high to low chromatin binding at the peak (high [H]) and trough (low [L]) of the GH pulse. RSL1 binding is qualitatively reciprocal to that of STAT5b; KAP1 presence is more invariant. Although the STAT5b transition is complete, some RSL1 is always bound. Circles, methylation status (open, unmethylated; closed, methylated) of the −66 Slp CpG. The size of the arrow indicates transcription expected on the basis of the bound protein complex. +, relative proportion of the corresponding complexes on the basis of the re-ChIP data. Females bear similarity to males during the GH trough.
DISCUSSION
KRAB-Zfp genes have expanded and diverged in mammals under positive selection, implying the increasing influence of these transcriptional regulators during recent evolution (12, 29). A detailed KRAB-ZFP repression mechanism has been discerned from in vitro studies, but their biological function remains poorly characterized since in only a few cases have target genes or physiological roles been identified (e.g., see references 9, 19, and 36). Rsl1, originally identified as a modifier of the mouse Slp gene, provides a model of KRAB-ZFP action in a genetically rich system with experimental access in adult tissues. Here we show that Rsl1-dependent methylation of a dinucleotide in the Slp promoter directly links DNA modification to KRAB-ZFP repression in the liver. RSL1 binds 2 kb from the Slp promoter, both in vitro and in vivo, within a hormonally responsive enhancer, suggesting that KRAB-ZFPs may oppose activators to modulate hormonal control. KAP1 associates with this region of chromatin in RSL-expressing but not RSL-null mice, confirming in vivo that KRAB-ZFPs recruit KAP1 to mediate gene silencing. In males, RSL1 and STAT5b undergo reciprocal binding in accord with pulsatile GH stimulation, whereas KAP1, once recruited to chromatin by RSL1, shows little oscillation. Together, these findings offer in vivo proof of the KRAB-ZFP repression mechanism inferred from in vitro studies. Moreover, KRAB-ZFP repression not only can lead to formation of facultative heterochromatin but also can be dynamic and temper hormonal activation.
Methylation of CpG dinucleotides at cytosines is a hallmark of epigenetic regulation and heritably and reversibly integrates intrinsic and environmental cues, such as those from hormones and diet (15). The methylated −66 CpG in the Slp promoter in liver is demethylated in males at puberty, as is the −91 CpG in the Cyp2d9 promoter, providing a sexually dimorphic epigenetic mark of male-specific expression (46, 47). Methylation at this site prevents binding of the transcriptional activator GABP, in part underlying the lack of Slp expression in WT females. The presence of Rsl1 correlates strongly with methylation at the −66 CpG and coincident repression of Slp in liver: rsl mice have high female Slp expression, whereas RSL1 overexpression (Rsl1tg) leads to near complete methylation of the site and suppression of Slp in both sexes. This further evidences DNA methylation as a component of KRAB-ZFP action, as has been shown in cases of imprinting as well as repression (1, 49). RSL1 binds 2 kb upstream from the −66 CpG and recruits KAP1, with which DNA methyltransferases (DNMTs) can associate (49), likely facilitating the methylation that prevents GABP action. Alternatively, the methylation may be an effect rather than cause of transcriptional inactivity associated with RSL binding. Interestingly, Slp is also male biased in kidney, due to direct androgen rather than GH action (28), but is not impacted by Rsl1, despite higher expression levels than in liver (see Fig. S1 in the supplemental material). Regardless of Rsl genotype, methylation at the −66 CpG site is 2-fold greater in female than male kidney (Fig. 2D). This suggests that demethylation linked to hormonal activation in both tissues may differ in a manner dependent upon the inducer and regardless of the machinery that created the epigenetic mark. It is interesting to note that locus-specific DNA demethylation has recently been shown to be part of the response to transforming growth factor β signaling (39). Thus, in liver but not in kidney, Rsl1 is a component in a developmental switch whereby KRAB-ZFP-directed gene silencing, accomplished in part via promoter methylation, is overcome by hormone induction.
KRAB-ZFPs bind DNA via a C-terminal tandem array of C2H2 zinc fingers, each of which has potential to bind three consecutive bases (31). While engineering an array of fingers to target a specific sequence has become a powerful tool to manipulate DNA in situ, applying the same code to predict natural ZNF binding sites has been less successful. This is likely due to the difficulty in predicting which of many fingers may be used at any time, leading to potential variability and flexibility in natural binding sites. Since KRAB-ZFPs may contain dozens of ZNFs, binding sites may be quite large, making a consensus difficult to discern. In this study, RSL1 binds multiple sequences in vitro, but the one with the highest apparent affinity occurs within the proviral LTR upstream of Slp, consistent with a direct role in repression. Interestingly, weaker potential binding nearby is within the B2 repetitive element into which the provirus inserted (38). Since restricting viral activity may be the primordial role of KRAB-ZFPs (40), the effect on Slp expression may be secondary to the insertion. KRAB-ZFP silencing is effective over long distances (10), suggesting that other genes neighboring endogenous proviruses may be impacted by KRAB-ZFPs bound nearby. ChIP-sequencing with tools to distinguish individual KRAB-ZFPs to assess their genome-wide binding in a biological context will be informative in this regard.
The RSL1 binding site is centered between a pair of STAT5 consensus elements 129 bp apart in the Slp upstream region (Fig. 4). Clustering of STAT5 elements is common among sex-biased as well as unbiased STAT5-responsive genes. Comparisons of STAT5 binding sites of several target genes suggest that low-affinity sites are associated with male-specific genes, whereas high-affinity sites are more common in female-specific or non-sex-biased genes (21). The precise sequences that distinguish low- from high-affinity binding are unclear, but low-affinity sites may allow a more rapid response to changing STAT5b levels. Due to the proximity of sites in the Slp enhancer, conventional ChIP cannot distinguish binding of individual elements. However, twice as much STAT5b binding (as percent input) was detected in WT males with ChIP primers flanking the more distal rather than the more proximal site (data not shown). This intercalation of multiple RSL and STAT5 sites of different affinities may permit more sensitive transcriptional modulation and may enter into the cyclical repositioning of one or both factors in chromatin.
The Slp enhancer allows investigation of how transcriptional activation is limited by repression. RSL1 and STAT5b bind nonoverlapping sequences, permitting simultaneous occupancy of the Slp enhancer (Fig. 8 and 9). In females, STAT5b levels are low and RSL1 repression dominates. In males, STAT5b levels vary with pulsatile GH secretion and RSL1 binding declines as STAT5b binding increases. When STAT5b levels are low, RSL1 is present on the few enhancers bound by STAT5b and most enhancers are occupied only by RSL1 (Fig. 8 and 9). At peak STAT5b levels, some enhancers are cooccupied but the majority are bound by STAT5b alone, and interestingly, some are bound by RSL1 alone. RSL1 and STAT5b appear to bind independently since the GH-dependent 2-fold difference in RSL1 binding in WT males also occurs in Rsl1tg males in the absence of STAT5b binding (Fig. 7). While some relative binding differences may be a function of the antibodies, the ChIP and re-ChIP data are consistent with dynamic and reciprocal positioning of both RSL1 and STAT5b on the Slp enhancer (Fig. 9). Such cycling in KRAB-ZFP binding has not been previously noted and demonstrates how these factors may not just repress but may also modulate gene regulation.
Changes in RSL1 or STAT5b occupancy are likely affected by neighboring proteins and chromatin structure. WT males indicate that some actions of RSL1, mediated by recruitment of KAP1 and associated chromatin-modifying proteins, are reversible and largely overcome by the subsequent GH pulse and wave of STAT5b into the nucleus. That this does not occur in Rsl1tg males suggests that elevated RSL1 leads to a heterochromatin configuration that prevents binding and activation by STAT5b. Conversely, in rsl mice, the RSL1/KAP1 complex is not present and therefore does not limit STAT5b activation. Thus, Slp is derepressed in rsl males as well as females, leading to expression exceeding that in WT males (Fig. 1).
Despite the reciprocal interplay between RSL1 and STAT5b, a similar reorganization within chromatin was not detected for KAP1, partly due to a higher degree of variability in KAP1 ChIP data. KAP1 orchestrates numerous protein-protein interactions (13) that may affect its chromatin binding in a manner substantially different from that for either RSL1 or STAT5b, both of which are bound directly to DNA. The prolonged association of KAP1 with chromatin even in the absence of the recruiting KRAB-ZFP may be a component of epigenetic memory, a phenomenon observed in cell lines in which KRAB-ZFP-initiated heterochromatin persists through multiple cell divisions even after the KRAB-ZFP is no longer present (1).
KRAB-Zfp genes are thought to have arisen to repress retroviral expression but may have been retained in part for their ability to regulate host genes (40). The breadth and diversity of their biological roles are suggested by the evolutionary expansion and divergence of this huge gene family. The ability of KRAB-ZFPs to fine-tune as well as suppress regulatory networks demonstrated here hints that they may broadly contribute to quantitative variation in gene expression at both species and individual levels. Evidence for this is found in the phenotypic variability shown by mice with mutations in KRAB-Zfp genes, such as chato and Zfp57 (9, 23). Similarly, hypomorphic mutations in KAP1/TRIM28 increase phenotypic noise for many traits, and KAP1 knockout leads to phenotypic and epigenetic variability during embryogenesis (26, 35, 43). The profound developmental defects in chato and Zfp57 mutants contributed to their identification but complicate molecular analysis. For Rsl, phenotypes in adult liver permit ready experimental access to the fluid chromatin landscape in vivo. This allowed us to demonstrate the novel dynamic role of a KRAB-ZFP in modifying transcription in a physiological context. RSL may be the first of many KRAB-ZFPs that will prove to serve as modifier loci for a broad spectrum of phenotypes ultimately impacting differential susceptibility to complex diseases.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grant DK053998 (to D.M.R.) and a pilot and feasibility grant from the Michigan Diabetes Research and Training Center (NIH 5P60-DK020572) (to C.J.K.). Core support was from the MDRTC (5P60-DK020572) and the Cancer Center Support Grant (5P30-CA46592).
We thank Doug Engel, Sundeep Kalantry, John Moran, and Andy Lieberman for comments on the manuscript, Lei Yin for advice on ChIP assays, and Chris LaPensee for advice on GH treatment. Meredith Sorenson, Ania Owczarczyk, Megan Lung, and Alicia Philippou provided excellent technical assistance.
We have no conflicting interests to disclose.
Footnotes
Published ahead of print 16 July 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Ayyanathan K, et al. 2003. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 17: 1855–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boyd KE, Farnham PJ. 1999. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol. Cell. Biol. 19: 8393–8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chia DJ, Rotwein P. 2010. Defining the epigenetic actions of growth hormone: acute chromatin changes accompany GH-activated gene transcription. Mol. Endocrinol. 24: 2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choi HK, Waxman DJ. 2000. Pulsatility of growth hormone (GH) signalling in liver cells: role of the JAK-STAT5b pathway in GH action. Growth Horm. IGF Res. 10(Suppl B): S1–S8 [DOI] [PubMed] [Google Scholar]
- 5. Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159 [DOI] [PubMed] [Google Scholar]
- 6. Emerson RO, Thomas JH. 2009. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 5: e1000325 doi: 10.1371/journal.pgen.1000325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Friedman JR, et al. 1996. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 10: 2067–2078 [DOI] [PubMed] [Google Scholar]
- 8. Frietze S, Lan X, Jin VX, Farnham PJ. 2010. Genomic targets of the KRAB and SCAN domain-containing zinc finger protein 263. J. Biol. Chem. 285: 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garcia-Garcia MJ, Shibata M, Anderson KV. 2008. Chato, a KRAB zinc-finger protein, regulates convergent extension in the mouse embryo. Development 135: 3053–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Groner AC, et al. 2010. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 6: e1000869 doi: 10.1371/journal.pgen.1000869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hemenway C, Robins DM. 1987. DNaseI-hypersensitive sites associated with expression and hormonal regulation of mouse C4 and Slp genes. Proc. Natl. Acad. Sci. U. S. A. 84: 4816–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huntley S, et al. 2006. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 16: 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iyengar S, Farnham PJ. 2011. KAP1 protein: an enigmatic master regulator of the genome. J. Biol. Chem. 286: 26267–26276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iyengar S, Ivanov AV, Jin VX, Rauscher FJ III, Farnham PJ. 2011. Functional analysis of KAP1 genomic recruitment. Mol. Cell. Biol. 31: 1833–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jaenisch R, Bird A. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33 (Suppl): 245–254 [DOI] [PubMed] [Google Scholar]
- 16. Kaplan T, Friedman N, Margalit H. 2005. Ab initio prediction of transcription factor targets using structural knowledge. PLoS Comput. Biol. 1: e1 doi: 10.1371/journal.pcbi.0010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krebs CJ, Khan S, MacDonald JW, Sorenson M, Robins DM. 2009. Regulator of sex-limitation KRAB zinc finger proteins modulate sex-dependent and -independent liver metabolism. Physiol. Genomics 38: 16–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krebs CJ, Larkins LK, Khan SM, Robins DM. 2005. Expansion and diversification of KRAB zinc-finger genes within a cluster including regulator of sex-limitation 1 and 2. Genomics 85: 752–761 [DOI] [PubMed] [Google Scholar]
- 19. Krebs CJ, et al. 2003. Regulator of sex-limitation (Rsl) encodes a pair of KRAB zinc-finger genes that control sexually dimorphic liver gene expression. Genes Dev. 17: 2664–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krebs CJ, Robins DM. 2010. A pair of mouse KRAB zinc finger proteins modulates multiple indicators of female reproduction. Biol. Reprod. 82: 662–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laz EV, Sugathan A, Waxman DJ. 2009. Dynamic in vivo binding of STAT5 to growth hormone-regulated genes in intact rat liver. Sex-specific binding at low- but not high-affinity STAT5 sites. Mol. Endocrinol. 23: 1242–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leuenberger N, Pradervand S, Wahli W. 2009. Sumoylated PPARalpha mediates sex-specific gene repression and protects the liver from estrogen-induced toxicity in mice. J. Clin. Invest. 119: 3138–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li X, et al. 2008. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell 15: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loreni F, Stavenhagen J, Kalff M, Robins DM. 1988. A complex androgen-responsive enhancer resides 2 kilobases upstream of the mouse Slp gene. Mol. Cell. Biol. 8: 2350–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mackay DJ, et al. 2008. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat. Genet. 40: 949–951 [DOI] [PubMed] [Google Scholar]
- 26. Messerschmidt DM, et al. 2012. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335: 1499–1502 [DOI] [PubMed] [Google Scholar]
- 27. Meylan S, et al. 2011. A gene-rich, transcriptionally active environment and the pre-deposition of repressive marks are predictive of susceptibility to KRAB/KAP1-mediated silencing. BMC Genomics 12: 378 doi: 10.1186/1471-2164-12-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nelson SA, Robins DM. 1997. Two distinct mechanisms elicit androgen-dependent expression of the mouse sex-limited protein gene. Mol. Endocrinol. 11: 460–469 [DOI] [PubMed] [Google Scholar]
- 29. Nowick K, et al. 2011. Gain, loss and divergence in primate zinc-finger genes: a rich resource for evolution of gene regulatory differences between species. PLoS One 6: e21553 doi: 10.1371/journal.pone.0021553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Geen H, et al. 2007. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 3: e89 doi: 10.1371/journal.pgen.0030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pavletich NP, Pabo CO. 1991. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science 252: 809–817 [DOI] [PubMed] [Google Scholar]
- 32. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45 doi: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ryan RF, et al. 1999. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 19: 4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schultz DC, et al. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shibata M, Blauvelt KE, Liem KF, Jr, Garcia-Garcia MJ. 2011. TRIM28 is required by the mouse KRAB domain protein ZFP568 to control convergent extension and morphogenesis of extra-embryonic tissues. Development 138: 5333–5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shin JH, et al. 2011. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell 144: 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sripathy SP, Stevens J, Schultz DC. 2006. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 26: 8623–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stavenhagen JB, Robins DM. 1988. An ancient provirus has imposed androgen regulation on the adjacent mouse sex-limited protein gene. Cell 55: 247–255 [DOI] [PubMed] [Google Scholar]
- 39. Thillainadesan G, et al. 2012. TGF-beta-dependent active demethylation and expression of the p15(ink4b) tumor suppressor are impaired by the ZNF217/CoREST complex. Mol. Cell 46: 636–649 [DOI] [PubMed] [Google Scholar]
- 40. Thomas JH, Schneider S. 2011. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 21: 1800–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tullis KM, Krebs CJ, Leung JY, Robins DM. 2003. The regulator of sex-limitation gene, rsl, enforces male-specific liver gene expression by negative regulation. Endocrinology 144: 1854–1860 [DOI] [PubMed] [Google Scholar]
- 42. Wauthier V, Sugathan A, Meyer RD, Dombkowski AA, Waxman DJ. 2010. Intrinsic sex differences in the early growth hormone responsiveness of sex-specific genes in mouse liver. Mol. Endocrinol. 24: 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Whitelaw NC, et al. 2010. Reduced levels of two modifiers of epigenetic gene silencing, Dnmt3a and Trim28, cause increased phenotypic noise. Genome Biol. 11: R111 doi: 10.1186/gb-2010-11-11-r111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wiwi CA, Waxman DJ. 2005. Role of hepatocyte nuclear factors in transcriptional regulation of male-specific CYP2A2. J. Biol. Chem. 280: 3259–3268 [DOI] [PubMed] [Google Scholar]
- 45. Wolf D, Goff SP. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yokomori N, Kobayashi R, Moore R, Sueyoshi T, Negishi M. 1995. A DNA methylation site in the male-specific P450 (Cyp 2d-9) promoter and binding of the heteromeric transcription factor GABP. Mol. Cell. Biol. 15: 5355–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yokomori N, Moore R, Negishi M. 1995. Sexually dimorphic DNA demethylation in the promoter of the Slp (sex-limited protein) gene in mouse liver. Proc. Natl. Acad. Sci. U. S. A. 92: 1302–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y, et al. 2011. Transcriptional profiling of human liver identifies sex-biased genes associated with polygenic dyslipidemia and coronary artery disease. PLoS One 6: e23506 doi: 10.1371/journal.pone.0023506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zuo X, et al. 2012. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J. Biol. Chem. 287: 2107–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.