Abstract
Histone methylation at specific lysine residues is a crucial regulatory process in transcriptional regulation. Using chromatin immunoprecipitation with microarray technology (ChIP-chip) analysis, we found that the H3K9-me2 target gene JAK2 was an important factor during differentiation of the HL-60 promyelocytic leukemia cell line by all-trans-retinoic acid (ATRA) treatment. Here, we report that the H3K9 methyltransferase G9a negatively regulated JAK2 transcription in histone methyltransferase activity and in a YY1-dependent manner during ATRA-mediated leukemia cell differentiation. We found that G9a knockdown repressed ATRA-mediated HL-60 cell differentiation. We demonstrated that G9a interacts with YY1 and is recruited to the JAK2 promoter along with corepressors, including histone deacetylase, that induced H3K9-me2. Repression of JAK2 transcription by G9a decreased H3Y41 phosphorylation and promoted inhibition of the recently identified JAK2-H3Y41P-HP1α pathway-mediated leukemogenesis.
INTRODUCTION
Eukaryotic chromatin consists of DNA and highly conserved histone proteins, which form the nucleosomes. Posttranslational modifications of histone N-terminal tails, notably acetylation, phosphorylation, methylation, and ubiquitination, influence the folding and functional status of chromatin and eventually control gene expression (15, 18). These modifications are highly dynamic and are important to the transcriptional outcome of a particular gene. Furthermore, numerous results have demonstrated that histone modifications can cross talk with each other and act in a combined manner to activate or repress chromatin-mediated transcription (23).
Among the various modifications, lysine methylation regulates diverse substrates, including histones and nonhistone proteins, and correlates with distinct biological outcomes. G9a and GLP are homologous histone methyltransferases (HMTases) and mediate histones H3K9-me1 and H3K9-me2 and methylation at H3K27 (36). G9a or GLP knockout causes embryonic lethality in mice, indicating that these factors play a crucial role in early mammalian development (37, 38). G9a/GLP-dependent DNA methylation in G9a or GLP knockout mice has been reported, although catalytically inactive G9a partially restores the aberrant DNA methylation pattern in G9a−/− cells (7, 10, 35). G9a and GLP also methylate nonhistone proteins, including p53, CDYL1, and Reptin, and automethylate (17, 22, 29, 32). Recent reports have shown that G9a/GLP forms a complex with other HMTases, Suv39h1 and ESET, indicating that G9a/GLP may have functional significance with the same substrate specificity for multiple HMTases (12, 34).
A member of the family of nonreceptor tyrosine kinases, Janus kinase 2 (JAK2), is involved in cytokine growth factor signal transduction (42). Previous studies have revealed that the JAK2-STAT signaling pathway is critical in normal hematopoiesis and leukemogenesis (24). JAK2 associates with various cytokine and growth factor receptors and phosphorylates each upon association. The activated JAK2 phosphorylates STAT proteins, which translocate to the nucleus to upregulate STAT target genes (33). The oncogenic potential of JAK2 was demonstrated after identification of a constitutively active JAK2V617F mutant, which is responsible for most cases of polycythemia (2, 20, 25). A recent study reported that JAK2 can activate the lmo2 leukemogenic gene via phosphorylation of histone H3Y41 and exclusion of HP1α from chromatin (4).
Yin Yang 1 (YY1) is a multifunctional mediator of different signaling pathways regulating cell proliferation and differentiation (13). As a transcription factor, YY1 either activates or represses the expression of multiple genes, and this activity depends on its association with cofactors (21, 43). YY1 deficiency in mice results in peri-implantation lethality during embryonic development (8). YY1 interacts with a number of proteins regulating cell proliferation and apoptosis, such as p53, Mdm2, Ezh2, Rb, caspases, and histone deacetylases (HDACs) (6). Additionally, YY1 interacts with a number of protein modifiers, including histone methyltransferases (HMTases). Early studies demonstrated that YY1 associates with p300 and HDACs, regulating histone acetylation and deacetylation, respectively (21, 43). Other studies have shown that the recruitment of Ezh2 and PRMT1 by YY1 mediates histone methylation (3, 30).
We identified JAK2 repression in a chromatin immunoprecipitation with microarray technology (ChIP-chip) screen for H3K9-me2 target promoters during leukemia cell differentiation. Among different H3K9 HMTases, we found that G9a negatively regulates JAK2 transcription. Recruitment of G9a to the JAK2 promoter is facilitated by association with the YY1 transcription factor. We also showed that G9a knockdown directly increased H3Y41 phosphorylation via the induction of JAK2 expression, which further supports the hypothesis that G9a may play an upstream regulatory role in JAK2-H3Y41P-HP1α-mediated leukemogenesis.
MATERIALS AND METHODS
Plasmid constructs.
For the luciferase assay, genomic DNA was prepared, and the JAK2 promoter region (−1980 to −981) was inserted into the pGL4.12 [luc2CP] vector (Promega, Madison, WI). The JAK2 promoter sequence was amplified using specific primers. The following PCR primers were used: the EcoRV site-linked primer, 5′-GATATCGGGCCCCTCACAAAATATCACATT-3′, as a forward primer and the HindIII site-linked primer, 5′-AAGCTTCCGACACACTCCTTGCCCTACTGG-3′, as a reverse primer. For the expression of YY1, the full length of YY1 cDNA was purchased from KUGI. The partial regions (amino acids [aa], 1 through 160, 150 through 300, and 290 through 414) of the YY1 cDNA were inserted into the pGEX-4T1 bacterial expression vector (Amersham Biosciences, Uppsala, Sweden) to construct glutathione S-transferase (GST) fusion proteins. Full-length YY1 cDNA was inserted into the pGEX-4T1 bacterial expression vector and the pEGFP-C1 eukaryotic expression vector. pcDNA3-Flag-hG9a (1,001 aa), pEGFP-hG9a (1,210 aa), pEGFP-hG9a-ΔSET, pCMV-HA-Ezh2, pCMV-Suv39h1, pCMV-myc-Smyd2, pCMX-HDAC1, and pcDNA3.1-HDAC2 were used in each experiment. The short hairpin RNAs (shRNAs) against G9a (catalog no. RHS4533-NM-006709) were purchased from Open Biosystems (Lafayette, CO). The small interfering RNAs (siRNAs) against YY1 (catalog no. sc-36863 and sc-44330) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Antibodies.
Antibodies against G9a (catalog no. 07-551; Millipore, Bedford, MA), JAK2 (sc-278; Santa Cruz Biotechnology), YY1 (sc-7341X; Santa Cruz Biotechnology), Suv39h1 (sc-025366; Santa Cruz Biotechnology), GFP (sc-9996; Santa Cruz Biotechnology), H3K9-me2 (07-441; Millipore), H3K4-me2 (07-030; Millipore), acetyl-H3 (06-599; Millipore), acetyl-H4 (06-598; Millipore), HDAC1 (05-614; Millipore), HDAC2 (ab12169; Abcam, Cambridge, MA), SMRT (sc-20778X; Santa Cruz Biotechnology), Ezh2 (3147S; Cell Signaling Technology, Danvers, MA), Smyd2 (sc-130879; Santa Cruz Biotechnology), histone H3 (sc-8654; Santa Cruz Biotechnology), H3Y41P (ab26310; Abcam), IgG (sc-2028; Santa Cruz Biotechnology), and β-actin (sc-47778; Santa Cruz Biotechnology) were employed for Western blot, immunoprecipitation, and ChIP analyses. Anti-human-CD14-APC (17-0149; eBioscience, San Diego, CA) and anti-CD11B-PE (FCMAB178P; Millipore) were employed for fluorescence-activated cell sorting (FACS) analysis.
Chemicals.
All-trans-retinoic acid (ATRA), dimethyl sulfoxide (DMSO), hemin, trichostatin A (TSA), and nicotinamide (NIA) were purchased from Sigma (St. Louis, MO). BIX 01294 was purchased from Cayman Chemical (Ann Arbor, MI).
Cell culture and transient transfection.
K562 and HL-60 cells were grown in RPMI 1640, and HEK293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal bovine serum and 0.05% penicillin-streptomycin at 37°C in a 5% CO2 atmosphere. K562 and HEK293T cells were transfected with each construct using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and polyethylenimine (Sigma), respectively.
RNA isolation and real-time PCR analysis.
Total RNA was isolated from transfected or 30 μM hemin-treated K562 cells and 1 μM ATRA- or DMSO-treated HL-60 cells using RNAiso Plus (TaKaRa Bio, Ohtsu, Japan). RNA (2.5 μg) was reverse transcribed with oligo(dT) primer (Fermentas, Burlington, ON, Canada) and M-MLV reverse transcriptase (Enzynomics, Daejeon, South Korea). The quantified cDNA was subjected to JAK2, G9a, Suv39h1, and YY1 mRNA expression analysis. The following PCR primers were used: 5′-TGCACTGGCAGCAACAGAGCC-3′ (forward) and 5′-TTCAAGCACGGCTGGAGGTGC-3′ (reverse) (JAK2), 5′-CCGGCGCAAGGCCAAGAAGA-3′ (forward) and 5′-CGGTGGGCCACACGGAAGTC-3′ (reverse) (G9a), 5′-ACGAGTGCAACTCCCGCTGC-3′ (forward) and 5′-GGCAGCCGCTCGTCAAGGTT-3′ (reverse) (Suv39h1), and 5′-GGATCCCTGGTCACCGTGGCGGCGGCCG-3′ (forward) and 5′-CTCGAGATGAGGGCAAGCTATTGTTCTT-3′ (reverse) (YY1). Disassociation curves were generated after each PCR run to ensure that a single product of appropriate length was amplified. The mean threshold cycle (CT) and standard error were calculated from individual CT values obtained from triplicates per stage. The normalized mean CT was estimated as the ΔCT by subtracting the mean CT of β-actin. The ΔΔCT value was calculated as the difference between the control ΔCT and values obtained for each sample. The n-fold change in gene expression relative to the untreated control was calculated as 2−ΔΔCT.
Western blot analysis.
Total proteins were prepared from lysed cells using radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate (SDC), 1% NP-40, 0.5× protease inhibitor cocktail, 1 mM EDTA), fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membranes. The membranes were probed overnight with the indicated antibodies at 4°C. The blots were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit, anti-mouse, or anti-goat antibodies (Santa Cruz Biotechnology) and detected using an enhanced chemiluminescence (ECL) system (Santa Cruz Biotechnology).
Histone extraction.
Transfected K562 cells were lysed in reticulocyte standard buffer (RSB) solution (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride [PMSF]) for 30 min at 4°C. Lysed cells were centrifuged, the supernatant was discarded, and the pellets were resuspended in RSB solution. The solution was then sonicated five times for 10 s each. Then, the solution was centrifuged, the supernatant was discarded, and the remaining pellet was washed with RSB solution six times for 10 min each at 4°C. Pellets were finally resuspended in high-speed buffer (20 mM HEPES, 0.65 M NaCl, 1 mM EDTA, 0.34 M sucrose, 1 mM β-mercaptoethanol, and 0.5 mM PMSF).
Luciferase assay.
The luciferase assay was conducted using the JAK2 promoter reporter. K562 cells were transfected with the pGL4.12-JAK2 promoter and the indicated DNA constructs using Lipofectamine 2000 (Invitrogen) or treated with 30 μM hemin for different times. After transfection, the cells were harvested and assayed for luciferase activity using a luciferase assay system (Promega). Each value was expressed as the mean of three replicates from a single assay, and the results were confirmed by performing the experiment at least three times.
Immunoprecipitation.
For the interaction assays, K562 and transfected HEK293T cells were lysed for 1 h in lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.5× protease inhibitor cocktail, and 1 mM PMSF) at 4°C. The lysates were immunoprecipitated with anti-G9a and anti-YY1 antibodies overnight at 4°C, and protein A/G agarose beads (GenDEPOT, Barker, TX) were added for 2 h with rotation at 4°C. The bound proteins were analyzed via Western blotting with anti-GFP, anti-YY1, and anti-G9a antibodies.
GST pulldown assay.
The GST-YY1 wild type (WT) (amino acids 1 through 414), GST-YY1 activation domain (AD) (amino acids 1 through 160), GST-YY1 repression domain (RD) (amino acids 150 through 300), GST-YY1 DNA binding domain (DBD) (amino acids 290 through 414) and GST-purified proteins were incubated overnight at 4°C with protein lysate from G9a-overexpressed HEK293T cells. The precipitated proteins were separated using 8% SDS-PAGE and then analyzed via Western blotting with anti-G9a antibody.
Chromatin immunoprecipitation assay.
HL-60 cells were treated with 1 μΜ ATRA or DMSO and harvested 72 h later. K562 cells were transfected with the indicated DNA constructs or 100 nM si-control (si-CTL) RNA and 100 nM si-YY1 RNAs. Cells were cross-linked with 1% formaldehyde, which was added to the medium for 10 min at room temperature, followed by the addition of 125 mM glycine for 5 min at room temperature and then lysis in SDS lysis buffer. The samples were sonicated and immunoprecipitated using the indicated antibodies. The immunoprecipitates were eluted and reverse cross-linked, after which the DNA fragments were purified using a PCR purification kit (Qiagen, Valencia, CA). A 200-bp fragment corresponding to nucleotides −1230 to −1031 of the JAK2 proximal promoter region and a 315-bp fragment corresponding to nucleotides −3946 to −3632 of the JAK2 distal promoter region were amplified by PCR. The following PCR primers were used: 5′-ACTGGTTCATTCTCATCCTTCAG-3′ (forward) and 5′-CGGGGGCTCGAAGCCCTGAA-3′ (reverse) (JAK2 proximal promoter region), 5′-GCCTCAGTGTGTCCCAATTT-3′ (forward) and 5′-TCTGTCCTGCTATCCCTGCT-3′ (reverse) (JAK2 distal promoter region), 5′-CCAGACAAACTCAAATAACGTACACA-3′ (forward) and 5′-AGTGGGTACCATTGTCCCTGTT-3′ (reverse) (LMO2 proximal promoter region), and 5′-CAGGCTTCTCCCGTGTAACTG-3′ (forward) and 5′-AGGACCTCACACGTTGAAGACA-3′ (reverse) (LMO2 distal promoter region). Disassociation curves were generated after each PCR run to ensure that a single product of the appropriate length was amplified. The mean threshold cycle (CT) and standard error were calculated from individual CT values obtained from duplicates per stage. The normalized mean CT was estimated as the ΔCT by subtracting the mean CT of the input.
FACS analysis.
HL-60 cells were transfected with G9a or G9a shRNA. After 24 h, transfected HL-60 cells were treated with 1 μM ATRA for 72 h. CD11B and CD14 expression was detected by FACS analysis, and Wright-Giemsa staining was performed to confirm cell morphology. The cells were stained with the CD14-APC (eBiosience) and CD11B-PE (Millipore) antibodies, which were measured using a FACS Aria I (BD Biosciences, San Diego, CA) at the National Instrumentation Center for Environmental Management. The results were analyzed using the FACScan Cell Quest software (BD Biosciences).
Statistical analysis.
Data are expressed as means ± standard deviations of results of three or more independent experiments. Statistical significance (P < 0.05) was evaluated using Microsoft Excel software (Redmond, WA). Differences between groups were evaluated with a one-way analysis of variance, followed by Student's t tests.
RESULTS
Identification of H3K9-me2 target gene JAK2 during differentiation of a leukemia cell line by ATRA.
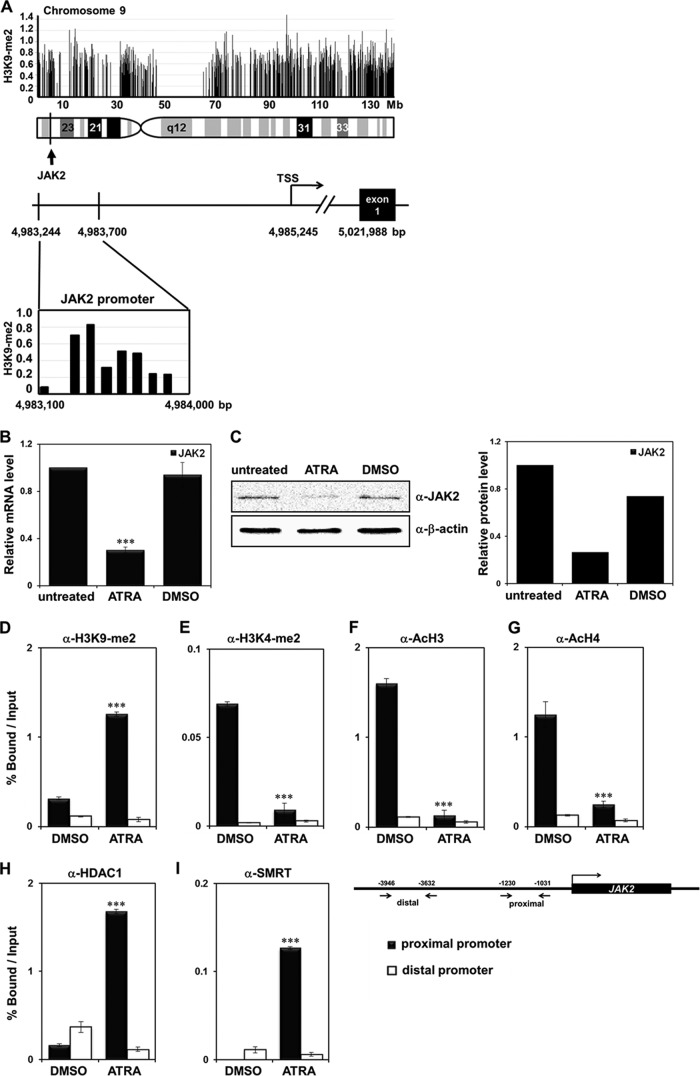
We previously performed ChIP-chip analysis on a human promyelocytic leukemia cell line, HL-60, after treatment with ATRA to identify H3K9-me2 targets during leukemia cell differentiation (18a). Among the profiled target transcriptional start site (TSS) regions, we focused on JAK2 as a potential target gene of H3K9-me2 modification during leukemia cell differentiation. JAK2 is located on human chromosome 9 (9p24), and the H3K9-me2 target site is located in the JAK2 promoter, indicating modification during differentiation (Fig. 1A).
Fig 1.
Identification of JAK2 as an H3K9-me2 target gene in ATRA-treated HL-60 cells. (A) An expanded view of the JAK2 locus from chromosome 9 is shown in the genuine 5′-3′ orientation. The locus of JAK2 TSS sequences used in the ChIP-chip analysis is shown with H3K9-me2 levels. (B) HL-60 cells were treated with 1 μM ATRA or DMSO and harvested 72 h later. JAK2 mRNA levels were measured using real-time PCR. The results are the average of three independent experiments, and error bars represent ± standard deviation (SD). ***, P < 0.001 compared with the value for the untreated control. (C) Western blot analysis of JAK2 expression levels in ATRA-treated HL-60 cells. JAK2 expression was quantified and normalized to the level of β-actin. (D to I) Recruitment levels of H3K9-me2, H3K4-me2, acetyl-H3, acetyl-H4, HDAC1, and SMRT in the JAK2 proximal and distal promoter regions during ATRA-mediated differentiation of HL-60 cells using the chromatin immunoprecipitation (ChIP) assay. HL-60 cells were treated with 1 μM ATRA or DMSO for 48 h. Cross-linked samples were immunoprecipitated with the indicated antibodies. The precipitated DNA fragments were subjected to real-time PCR. The results are averages of three independent experiments, and the error bars represent ± SD ***, P < 0.001 compared with the value for the DMSO control.
Verification of the H3K9-me2 target gene JAK2 promoter.
HL-60 cells were treated with ATRA, and epigenetic status and recruitment of chromatin remodelers to the JAK2 promoter were analyzed. First, we verified that JAK2 expression was downregulated during ATRA-mediated differentiation of the HL-60 cells (Fig. 1B and C). H3K9-me2 levels in the JAK2 promoter increased by ATRA treatment (Fig. 1D), whereas H3K4-me2 levels decreased during differentiation (Fig. 1E). As expected, H3 and H4 histone acetylation decreased significantly in the JAK2 promoter when cells were treated with ATRA (Fig. 1F and G). As histone modifications such as H3K9-me2 recruit various chromatin remodeling cofactors that alter target gene transcription, we examined the association of cofactors in the JAK2 promoter. H3K9-me2 is a known marker of repressed transcription, and enhanced association of the corepressors HDAC1 and SMRT on the JAK2 promoter was observed in the ChIP assay and real-time PCR after the ATRA treatment (Fig. 1H and I). Increased binding of this corepressor complex and repressive histone modification marks to the JAK2 promoter further confirmed that JAK2 was highly repressed during HL-60 cell differentiation by ATRA. No apparent changes in histone modifications or corepressor recruitment were observed in the JAK2 distal promoter (Fig. 1D through I). Based on these results, we concluded that the JAK2 promoter was H3K9 methylated and transcriptionally repressed by actively recruiting various corepressor complexes during ATRA-treated leukemia cell differentiation.
G9a regulates JAK2 expression.
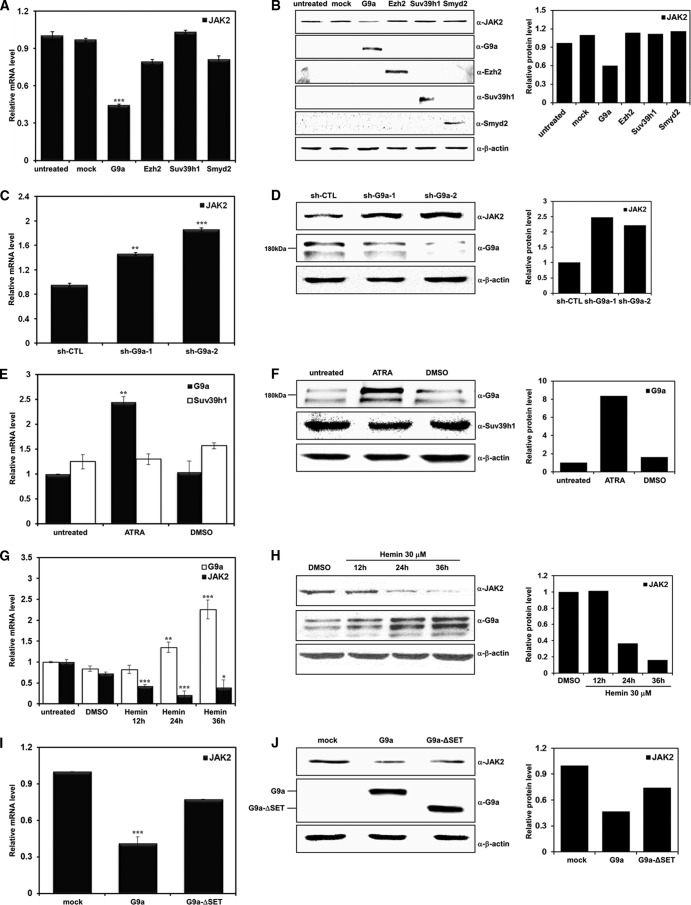
JAK2 downregulation during differentiation of the leukemia cell line by ATRA prompted us to hypothesize that epigenetic regulators are involved in this process. As JAK2 was screened as an H3K9-me2 target gene by ChIP-chip profiling, we concentrated on H3K9 HMTases among the histone modifiers and tested different H3K9 HMTases that are functionally related to transcriptional repression. Among the HMTases tested by real-time PCR, G9a significantly reduced JAK2 expression (Fig. 2A). In contrast, the other H3K9 HMTases tested failed to alter JAK2 expression. Downregulation of JAK2 by G9a was confirmed by Western blot analysis, indicating that G9a might be involved in the regulation of JAK2 transcription (Fig. 2B). Comparable expression levels of tested HMTases were verified by Western blotting. Negative regulation of JAK2 expression by G9a was further confirmed using two different G9a shRNAs (Fig. 2C and D). Different G9a isoforms, including splice variants, have been reported. Two G9a bands were detected with the antibodies that we used, and both bands were efficiently knocked down by different G9a shRNAs used in this study (Fig. 2D). When we tested G9a expression before and after ATRA treatment in HL-60 cells by real-time PCR and Western blot analysis, G9a expression was significantly upregulated after ATRA treatment (Fig. 2E and F). As mentioned above, ATRA treatment did not affect the protein expression level of the other H3K9 HMTase, Suv39h1. To investigate whether the increase in G9a expression during leukemia cell differentiation was cell-type specific, we differentiated another leukemia cell line, K562, which is an erythroleukemia cell type that can be differentiated with hemin, and monitored G9a expression patterns. As with HL-60 cells after ATRA treatment, G9a expression started to increase 24 h after hemin treatment and significantly increased in the 36 h after treatment (Fig. 2G and H). The JAK2 expression level showed the opposite pattern to that of G9a. As G9a expression increased, JAK2 expression decreased, indicating that JAK2 transcription might be regulated by G9a in the K562 cell line (Fig. 2G and H). The importance of HMTase activity in G9a-mediated JAK2 repression was confirmed when the SET domain-deleted G9a mutant G9a-ΔSET was evaluated, and no change in JAK2 expression was observed in real-time PCR and Western blot analysis (Fig. 2I and J). These results suggest that increased H3K9 HMTase G9a specifically repressed JAK2 expression in a SET domain-dependent manner during ATRA-mediated HL-60 differentiation.
Fig 2.
SET domain-dependent JAK2 downregulation by G9a. (A and B) K562 cells were transfected with pcDNA3 as a control and the indicated DNA constructs. Real-time PCR and Western blot analyses were performed using JAK2-specific primers and antibody. The expression of each construct is shown by Western blot analysis. (C and D) K562 cells were transfected with two different G9a shRNAs. JAK2 expression levels were confirmed by real-time PCR and Western blot analyses. (E and F) HL-60 cells were treated with 1 μM ATRA or DMSO and harvested 72 h later. G9a and Suv39h1 expression levels were confirmed in differentiated HL-60 cells via real-time PCR and Western blot analyses. (G and H) G9a and JAK2 expression patterns as a function of time after treatment with 30 μM hemin in K562 cells. (I and J) K562 cells were transfected with pEGFP-C1, pEGFP-G9a, and pEGFP-G9a-ΔSET. JAK2 expression levels were confirmed via real-time PCR and Western blot analyses. β-Actin was used as a loading control. All data are shown as means ± SD (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Results of the Western blot analysis were quantified and normalized to the level of β-actin.
G9a represses JAK2 transcription.
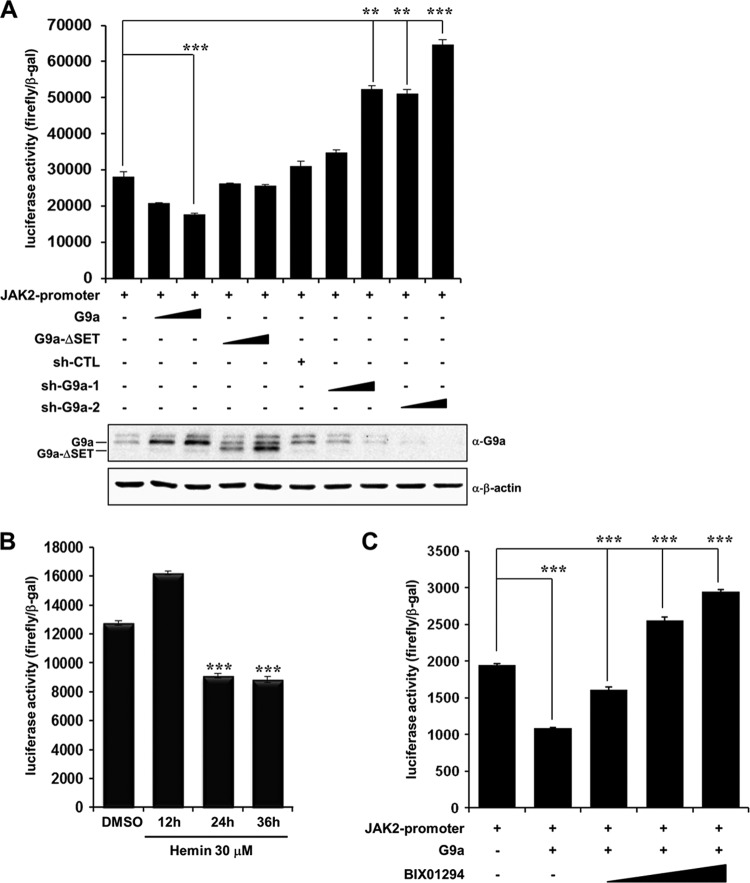
Because G9a repressed JAK2 expression during differentiation, we determined whether G9a directly inhibited JAK2 transcription by transient transfection of G9a in intact cells. We conducted a reporter assay using a JAK2-luc reporter system to examine G9a-mediated transcriptional regulation of JAK2. The JAK2 promoter region has various transcription factor binding sites, and we cloned the −1980 to −981 region into the pGL4.12 [luc2CP] vector to conduct the luciferase assay. Consistent with the real-time PCR and Western blot results, JAK2 transcription was repressed by G9a overexpression in K562 cells (Fig. 3A). Using the SET domain-deleted G9a mutant, we verified that transcriptional repression of JAK2 by G9a was dependent on HMTase activity. G9a knockdown by two independent G9a shRNAs eliminated transcriptional repression of JAK2 mediated by G9a (Fig. 3A). These findings indicate that G9a negatively regulated JAK2 transcription by directly acting on the JAK2 promoter. Differentiation of the K562 cell line by hemin treatment confirmed the downregulation of JAK2 transcription (Fig. 3B). To confirm the role of G9a for negative regulation of JAK2 transcription, we examined the effect of BIX 01294, a G9a-specific inhibitor. G9a-mediated JAK2 transcriptional repression was restored by increasing the concentration of BIX 01294 (Fig. 3C). These data further confirmed G9a-mediated transcriptional repression of JAK2. These results suggest that G9a transcriptionally repressed JAK2 during leukemia cell differentiation and that this repression was HMTase activity dependent.
Fig 3.
G9a represses JAK2 transcription. (A) K562 cells were cotransfected with the pGL4.12-JAK2 promoter (1 μg) and pcDNA3-Flag-G9a (0.5 and 1 μg), pEGFP-G9a-ΔSET (0.5 and 1 μg), pLKO.1 (1 μg) as sh-CTL, and G9a shRNAs (0.5 and 1 μg), along with the β-galactosidase expression plasmid. Cell extracts were assayed for luciferase activity. G9a overexpression or knockdown was confirmed by Western blot analysis. (B) K562 cells were transfected with the pGL4.12-JAK2 promoter and treated with 30 μM hemin at different times or with DMSO for 36 h as a control. The cell extracts were assayed for luciferase activity. (C) Restoration of G9a-mediated JAK2 transcriptional repression by BIX 01294. The pGL4.12-JAK2 promoter (1 μg) and pcDNA3-Flag-G9a (1 μg) were cotransfected into K562 cells. Twenty-four hours after transfection, BIX 01294 (1.5, 5, and 10 μM) was treated for 24 h, and the luciferase activity was measured. Luciferase activities were normalized to those of β-galactosidase. The pcDNA3 empty vector was used as a negative control and added to maintain equal amounts of total transfected DNA. All data are representative of at least three independent experiments and presented as means ± SD (n = 4). **, P < 0.01; ***, P < 0.001.
YY1 is involved in G9a-mediated JAK2 downregulation.
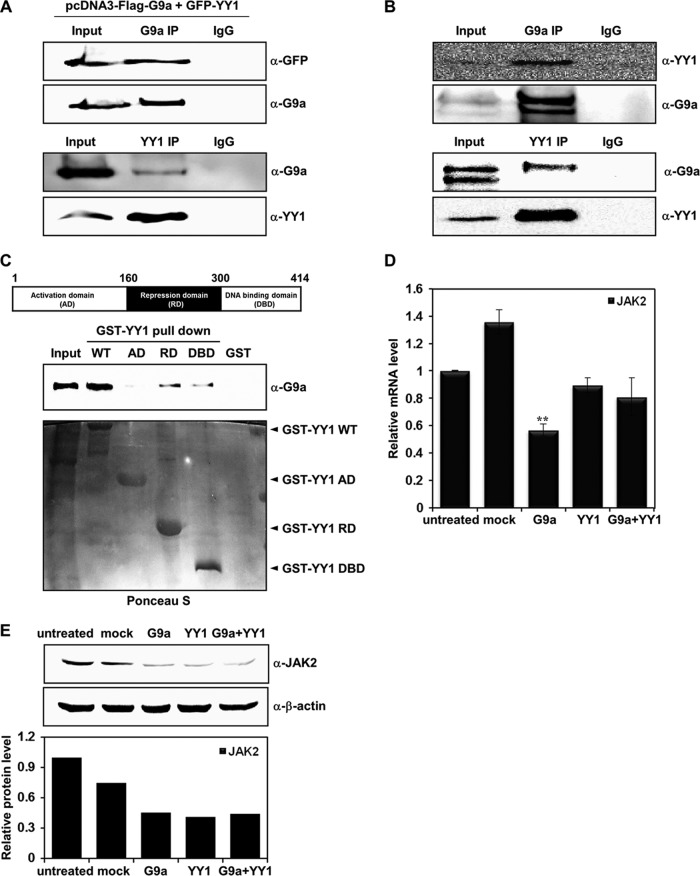
We further analyzed the JAK2 promoter sequence to search for possible transcription factor binding sites. Among the many binding sites for different transcription factors, we found that 6 YY1 binding sites were clustered between the −1230 to −1031 sequences of the JAK2 promoter region, indicating that YY1 may be involved in JAK2 transcription. YY1 plays an essential role in various biological processes such as embryogenesis, cell proliferation, differentiation, apoptosis, and tumorigenesis (13). As a ubiquitously expressed zinc finger transcription factor, YY1 has bifunctional activity in target gene transcription. YY1 regulates target gene expression by altering histone modification via recruitment of histone modifiers to the YY1 target gene promoter. Corepressors such as HDAC2 were recruited to the YY1 target promoter in a YY1-dependent manner (1). We attempted to determine the manner in which these proteins interact with one another to better understand the function of YY1 in G9a-mediated JAK2 transcriptional regulation. Interaction assays using coimmunoprecipitation were conducted to confirm complex formation between G9a and YY1. First, we cotransfected G9a and YY1, and immunoprecipitations were performed with both antibodies. The immunoprecipitation assays showed that YY1 interacted strongly with G9a (Fig. 4A). In addition, endogenous interaction assays between the two proteins clearly indicated that endogenous G9a and YY1 interacted with each other in vivo (Fig. 4B). These findings indicate that G9a regulated JAK2 transcription possibly through an interaction with YY1. YY1 is composed of an N-terminal activation domain (AD), a C-terminal DNA binding domain (DBD), and a repression domain (RD) in the middle. We constructed three YY1 deletion mutants containing each domain and performed a GST pulldown assay to identify which domain was responsible for this interaction. The pulldown assay revealed that the YY1 RD and DBD interacted with G9a, although the interactions were weak compared to that of the full-length YY1 (Fig. 4C).
Fig 4.
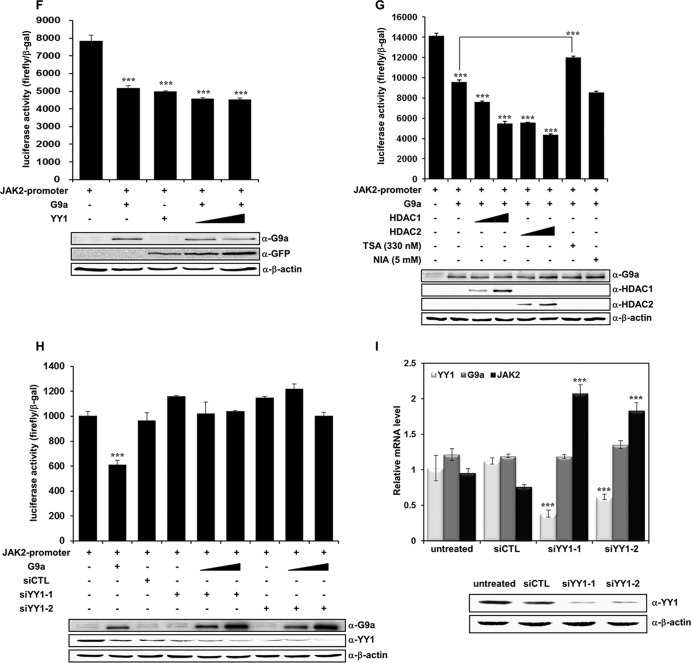
YY1 is involved in G9a-mediated JAK2 downregulation. (A) HEK293T cells were cotransfected with pcDNA3-Flag-G9a and pEGFP-YY1 constructs. Anti-G9a and anti-YY1 immunoprecipitates were analyzed by Western blot analysis with anti-GFP and anti-G9a antibodies. (B) G9a endogenously interacts with YY1 in K562 cells. Anti-G9a and anti-YY1 immunoprecipitates from K562 whole-cell extracts were analyzed by Western blot analysis with anti-YY1 and anti-G9a antibodies. (C) Cell lysates from G9a-overexpressed HEK293T cells were incubated with GST, the GST-YY1 wild type, or GST-YY1 deletion mutants and immunoblotted with anti-G9a antibody. The activation domain (AD), repression domain (RD), and DNA binding domain (DBD) of YY1 are shown. The levels of full-length YY1 and YY1 deletion mutants were determined by Ponceau S staining. (D) JAK2 mRNA levels were analyzed in transfected K562 cells by real-time PCR. K562 cells were transfected with pcDNA3, pcDNA3-Flag-G9a, and pEGFP-YY1 and cotransfected with pcDNA3-Flag-G9a and pEGFP-YY1. Results are shown as means ± SD (n = 3). **, P < 0.01 compared with the value for the untreated control. (E) K562 cells were transfected with the indicated DNA constructs, and JAK2 protein levels were confirmed via Western blot analysis. β-Actin was used as a loading control. Results of the Western blot were quantified and normalized to the level of β-actin. (F) K562 cells were transfected with the pGL4.12-JAK2 promoter (1 μg), pcDNA3-Flag-G9a (1 μg), and pEGFP-YY1 (1 and 1.5 μg). Following transfection, cells were grown for 48 h, and cell extracts were assayed for luciferase activity. Expression of the transfected constructs are shown in the Western blots. (G) pGL4.12-JAK2 promoter (1 μg), pcDNA3-Flag-G9a (0.5 μg), pCMX-HDAC1 (0.5 and 1 μg), and pcDNA3.1-HDAC2 (0.5 and 1 μg) were transfected into K562 cells. Twenty-four hours after transfection, 330 nM trichostatin A (TSA) or 5 mM nicotinamide (NIA) were added for 24 h, and luciferase activities were measured. G9a, HDAC1, and HDAC2 expressions were confirmed by Western blot analysis. (H) K562 cells were transfected with the pGL4.12-JAK2 promoter (1 μg), pcDNA3-Flag-G9a (0.5 μg, 1 μg), si-CTL RNA (100 nM), and si-YY1 RNAs (100 nM), along with the β-galactosidase expression plasmid. Luciferase activity was measured 48 h after the transfection. G9a overexpression and YY1 knockdown by two different si-YY1 RNAs are shown in the Western blots. (F, G, and H) Luciferase activities were normalized to that of β-galactosidase, and the results are presented as means ± SD (n = 4). ***, P < 0.001 compared with the value for the reporter plasmid alone. (I) K562 cells were transfected with si-CTL RNA (100 nM) or si-YY1 RNAs (100 nM). YY1, G9a, and JAK2 expression levels were confirmed by real time-PCR. YY1 knockdown by si-YY1 RNAs is shown in the Western blots.
We next determined whether YY1 had a synergistic effect in the negative regulation of JAK2 expression by G9a. Interestingly, real-time PCR showed that YY1 did not produce additive effects on G9a-mediated JAK2 repression. YY1 alone or YY1 plus G9a expression did not result in a further decrease in JAK2 expression (Fig. 4D). Rather, JAK2 expression seemed to be unaffected by G9a and YY1 cotransfection compared to that of G9a alone in the Western blot analysis (Fig. 4E). We performed a JAK2-luc reporter assay with G9a and YY1 to further investigate whether the interaction between G9a and YY1 had any effect on G9a-mediated transcriptional repression of JAK2. Consistent with the other results, G9a or YY1 alone repressed JAK2 transcription. Again, G9a cotransfection and an increase in the amount of YY1 did not further repress JAK2 transcription, suggesting that YY1 functions as a mediator for G9a recruitment to the JAK2 promoter but may not function as a direct transcriptional repressor (Fig. 4F). To further characterize the manner in which G9a-mediated transcriptional repression of JAK2 could be associated with HDAC, we performed JAK2 reporter assays using HDACs and the HDAC inhibitor trichostatin A (TSA). Adding TSA significantly restored the JAK2 transcriptional repression by G9a, strongly suggesting that HDAC was involved in G9a-induced JAK2 transcriptional repression (Fig. 4G). Cotransfection of HDAC1 or HDAC2 with G9a further repressed G9a-mediated JAK2 repression (Fig. 4G). Adding nicotinamide (NIA) did not affect G9a-mediated JAK2 transcriptional repression, indicating that another class of histone deacetylases, sirtuins, are not involved in this process (Fig. 4G). The role of YY1 in G9a-mediated transcriptional repression of JAK2 was further investigated by performing the JAK2-luc reporter assay in the presence of si-YY1 RNA. Interestingly, YY1 knockdown by two independent si-YY1 RNAs completely abolished the JAK2 transcriptional repression induced by G9a (Fig. 4H). This strongly suggested that negative regulation of JAK2 transcription by G9a was dependent on the presence of YY1. We then tested whether the JAK2 expression was influenced by the absence or the presence of YY1. JAK2 downregulation was dependent on YY1, which was based on the increased JAK2 expression under two different YY1 knockdown conditions (Fig. 4I). Taken together, these results suggest that G9a interacts with YY1 in vivo and mediates transcriptional repression of JAK2 in a YY1-dependent manner.
G9a and YY1 are recruited to the JAK2 promoter and repress transcription.
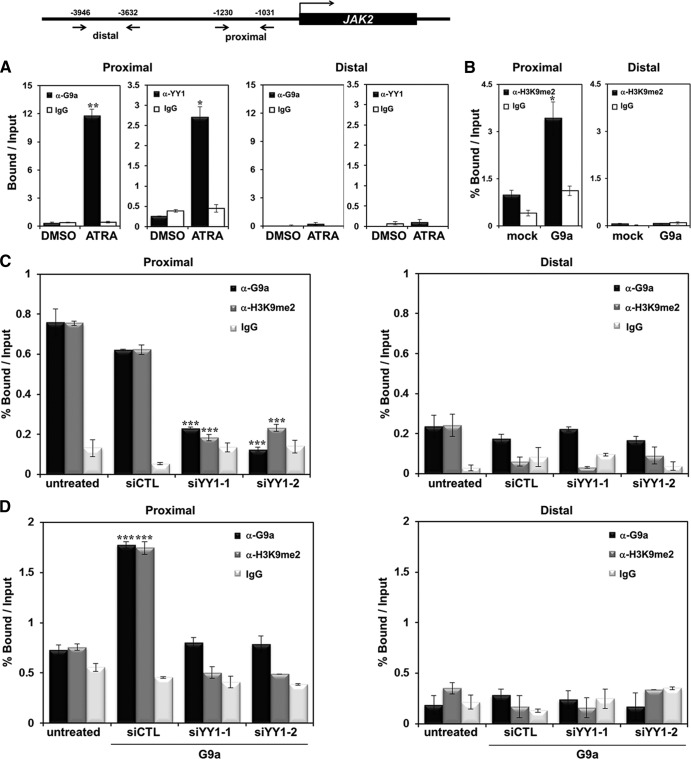
To further elucidate the mechanisms underlying JAK2 transcriptional regulation by G9a, we performed a ChIP analysis with real-time PCR. First, we observed significantly increased G9a recruitment to the JAK2 promoter in the presence of ATRA (∼12 fold), which was consistent with the initial results of the ChIP-chip analysis (Fig. 5A). Interestingly, the YY1 recruitment also increased (∼2.5 fold) when ATRA was added. No changes in G9a and YY1 levels were observed in the distal JAK2 promoter region. Then, we overexpressed G9a and observed that H3K9-me2 on the JAK2 promoter was increased significantly in the proximal promoter region but not in the distal region (Fig. 5B). Next, we evaluated whether YY1 played a role in G9a recruitment to the JAK2 promoter. Figure 5C and D show that G9a was highly recruited to the JAK2 promoter and that YY1 knockdown by two independent si-YY1 RNAs resulted in a significant reduction of G9a recruitment to the JAK2 promoter. These findings consistently demonstrate that G9a may repress JAK2 via binding of the YY1 transcription factor to the JAK2 promoter. The finding that the H3K9-me2 level on the JAK2 promoter was significantly downregulated in the absence of YY1 also confirmed that G9a-mediated JAK2 transcriptional repression is YY1 dependent (Fig. 5C and D). These results indicate that G9a repressed JAK2 expression via interacting with YY1 and recruitment to the promoter during HL-60 differentiation by ATRA.
Fig 5.
G9a and YY1 are recruited to the JAK2 promoter. Shown is a schematic diagram of primer pairs of the JAK2 proximal and distal promoter regions in the ChIP assay and real-time PCR analysis (top). (A) HL-60 cells were treated with 1 μM ATRA for 72 h. DMSO was used as a control. ChIP analysis was conducted using anti-G9a and anti-YY1 antibodies. Recruitment of G9a and YY1 to the JAK2 promoter region was normalized by input. Results are shown as means ± SD (n = 3). * and **, P < 0.05 and P < 0.01, respectively, compared with the value for the DMSO control. (B) K562 cells were transfected with pEGFP-C1 or pEGFP-G9a. Cross-linked samples were immunoprecipitated with anti-H3K9-me2 antibody, and the precipitated DNA fragments were subjected to real-time PCR in the JAK2 promoter regions. Results are representative of at least three independent experiments (± SD) *, P < 0.05. (C) K562 cells were transfected with 100 nM si-CTL RNA or 100 nM si-YY1 RNAs. ChIP analysis was performed using anti-G9a and anti-H3K9me2 antibodies, and results were confirmed by real-time PCR. Recruitment of G9a and H3K9me2 to the JAK2 promoter region was normalized by input. (D) si-CTL RNA (100 nM) or si-YY1 RNAs (100 nM) with pEGFP-G9a were cotransfected into K562 cells. Cross-linked samples were immunoprecipitated with anti-G9a and anti-H3K9-me2 antibodies, and the precipitated DNA fragments were subjected to real-time PCR in the JAK2 promoter regions. Results are shown as means ± SD (n = 3). ***, P < 0.001 compared with the value for the untreated control.
G9a decreases H3Y41 phosphorylation on the lmo2 promoter.
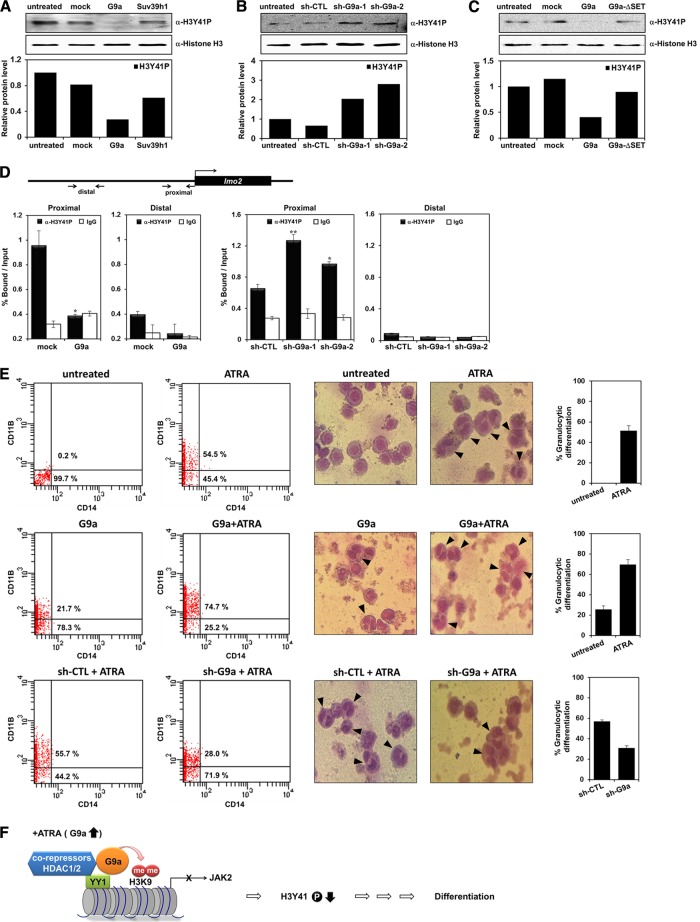
It was previously shown that JAK2 phosphorylates histone H3Y41 and excludes HP1α from the promoter of leukemogenic gene lmo2 (4). As we showed that G9a negatively regulates JAK2 transcription, we tested whether G9a directly influences the H3Y41 phosphorylation status. G9a overexpression decreased H3Y41 phosphorylation compared to that of Suv39h1 (Fig. 6A). Regulation of H3Y41 phosphorylation was further supported by two independent G9a shRNAs (Fig. 6B). Additionally, we confirmed that downregulation of H3Y41 phosphorylation was dependent on the HMTase activity of G9a (Fig. 6C). We next examined the changes in H3Y41 phosphorylation on the JAK2 target gene lmo2. When G9a was overexpressed, the H3Y41 phosphorylation level significantly decreased on the lmo2 promoter (Fig. 6D). G9a knockdown by two different G9a shRNAs showed elevated H3Y41 phosphorylation levels (Fig. 6D). We conclude that G9a repressed JAK2 transcription and decreased JAK2 expression, resulting in the downregulation of H3Y41 phosphorylation on the lmo2 promoter. Taken together, these results further support that JAK2 downregulation by G9a directly influenced H3Y41 phosphorylation status on the leukemogenic oncogene lmo2 promoter and indicate that G9a is involved in JAK2-H3Y41P-HP1α transcriptional signaling in leukemogenesis.
Fig 6.
G9a decreases H3Y41 phosphorylation. (A to C) Histone extracts were prepared from transfected K562 cells. The H3Y41 phosphorylation level was confirmed by Western blot analysis with anti-H3Y41P antibody. The histone H3 was used as a loading control. Western blot bands were quantified and normalized to the levels of histone H3. (D) Schematic diagram of primer pairs of lmo2 proximal and distal promoter regions in the ChIP assay and real-time PCR analysis (top). K562 cells were transfected with pEGFP-C1, pEGFP-G9a, pLKO.1, or two G9a shRNAs. ChIP analysis was performed using anti-H3Y41P antibody and confirmed by real-time PCR. H3Y41 phosphorylation levels at the lmo2 promoter regions were normalized by input. Results are shown as means ± SD (n = 3). * and **, P < 0.05 and P < 0.01 compared with the value for each control. (E) HL-60 cells were transfected with G9a or G9a shRNAs and treated with 1 μΜ ATRA for 72 h. The granulocytic differentiation of HL-60 cells was analyzed by FACS analysis using anti-CD11B and anti-CD14 antibodies. The percentage of CD11B-positive cells in the HL-60 cells is shown. Morphologies of transfected and ATRA-treated HL-60 cells were observed by Giemsa staining. The arrows represent differentiated HL-60 cells, and the graphs are presented to show the percentage of granulocytic differentiation. (F) Model of the mechanism of G9a-mediated repression of JAK2 transcription during leukemia cell differentiation.
G9a induces cell differentiation via negative regulation of JAK2 transcription.
To further investigate the role of G9a in ATRA-dependent differentiation of leukemia cells, we examined expression levels of the CD11b differentiation marker via FACS analysis. Interestingly, cells transfected with G9a alone showed increased CD11b expression (21.7%) compared to that in the untreated cells. Furthermore, G9a-transfected and ATRA-treated cells had significantly increased levels of the cell differentiation marker (74.7%) (Fig. 6E). Knockdown of endogenous G9a by shRNA slightly decreased ATRA-mediated cell differentiation. These results suggest that G9a negatively regulates JAK2 transcription and further induces ATRA-mediated granulocytic differentiation of HL-60 cells. The effect of G9a on HL-60 cell differentiation was further confirmed by morphological observation using Giemsa staining. The differentiated cells are marked with arrows in the ATRA-, sh-G9a- and ATRA-, and G9a- and ATRA-treated conditions (Fig. 6E, right panels). Taken together, these results suggest that G9a plays an important role in HL-60 cell differentiation.
DISCUSSION
G9a and GLP are members of the Suv39h subgroup of the SET domain containing proteins with H3K9-me1 and H3K9-me2 methylating specificity. Although roles of G9a in various biological processes and human diseases have been proposed, G9a function in hematopoiesis or leukemogenesis has not yet been systematically evaluated. Previous studies have suggested that lymphocyte development remains mostly intact in lymphocyte-specific G9a knockout, although some immune responses are impaired in B cell-specific G9a knockout mice and have reduced gene assembly and use of Ig(λ) chains (39). In the present study, we examined novel H3K9-me2 target genes during ATRA-mediated differentiation in HL-60 cells using ChIP-chip analysis. Among the target genes, JAK2 was downregulated during leukemia cell line differentiation via H3K9-me2 modification at the promoter. We hypothesized that H3K9 HMTase G9a functions as a negative regulator of JAK2 expression via YY1 interaction and is recruited to the JAK2 promoter during leukemia cell differentiation.
Previous profiling studies found that ATRA treatment modulates differential expression of a variety of genes (27). Initially, we confirmed that G9a was upregulated by ATRA treatment in HL-60 cells. ATRA induces an increase in H3K9 methylation during differentiation (11). In fact, the induction of G9a expression following ATRA treatment has been reported during germ cell differentiation (40). Although very little is known about G9a upregulation during ATRA treatment, we found that a highly conserved retinoic acid response element (RARE) was present in the G9a promoter (−1898 to −1893), suggesting that ligand-dependent retinoic acid receptor (RAR)-mediated G9a upregulation can occur during HL-60 differentiation. The detailed mechanism of ATRA-dependent RAR-mediated G9a transcriptional regulation remains to be investigated. We showed that H3K9 HMTase G9a negatively regulates transcription of the hematopoiesis- and leukemogenesis-related gene, JAK2, indicating that G9a is involved in ATRA-mediated leukemia cell line HL-60 differentiation. In a previous study, the JAK2-H3Y41P-HP1α signaling pathway in leukemogenesis was proposed and the role of JAK2-mediated epigenetic regulation of transcription was demonstrated (4). Furthermore, Griffiths et al. demonstrated JAK2-mediated regulation of H3Y41 phosphorylation on the Nanog promoter and suggested that JAK2 signaling is an important mediator of embryonic stem (ES) cell self-renewal (14). A recent study suggested that JAK2 gain-of-function mutations (JAK2-V617F and JAK2-K539L) phosphorylate PRMT5 and inhibit arginine methyltransferase activity and hence disrupt target gene expression (26). Amplification of chromosome-bound 9p24 induces cooperation between JAK2 and histone demethylase JMJD2C and increases H3Y41 phosphorylation and H3K9 methylation to target oncogenes in lymphomas (31). These observations, in combination with our current finding identifying G9a as a potentially important transcriptional regulator of JAK2 after ATRA treatment, suggest a significant role of JAK2 epigenetic regulation during leukemogenesis.
Studies using G9a-deficient ES cells identified MAGE-A genes as direct target genes through transcriptional repression by H3K9 HMTase activity (37). G9a interacts with transcription factor CDP/cut and is recruited to the human p21 promoter, where it represses transcription (28). When the JAK2 promoter sequences were screened, a number of YY1 transcription factor binding sites were identified. Indeed, G9a interacted with YY1 and was recruited to the JAK2 promoter, where it negatively regulated JAK2 transcription via H3K9 methylating activity. Interestingly, G9a-mediated transcriptional repression of JAK2 was dependent on the presence of YY1, suggesting that YY1 functions as a mediator between G9a and JAK2. YY1 associates with numerous histone-modifying proteins, including SUV39H2, EED/EZH2, ATF2, DNMT3, p300, SRC, and PRMT1 (6, 19, 44). Studies have suggested that YY1 recruits HDACs and mediates transcriptional repression of target genes (6, 13). G9a also associates with a corepressor complex including HDAC1/2, recruits to the target gene promoter, and represses transcription (9, 16). We showed that HDAC1 was recruited to the JAK2 promoter after ATRA treatment and further verified that both HDAC1/2 synergistically repressed JAK2 transcription along with G9a. These associations strongly suggest the possibility of a G9a and YY1 interaction. Identifying key repressor molecules that are responsible for G9a-mediated transcriptional repression of JAK2 is important for a better understanding of complicated epigenetic regulation during leukemogenesis. Although the roles of G9a in development have been reported, the link between G9a and carcinogenesis remains poorly understood. A recent report suggested a potential role for G9a in human cancers via methylation of the nonhistone protein p53 (17). Our study further expanded the potential roles of G9a during carcinogenesis by showing G9a upregulation of during ATRA-mediated leukemia cell differentiation and negative regulation of JAK2 transcription by G9a recruitment to the JAK2 promoter along with YY1 and corepressors (Fig. 6F). During leukemia cell differentiation by ATRA, G9a was recruited to the JAK2 promoter along with YY1 and the corepressor complex including HDAC, mediated H3K9-me2, and repressed transcription. A cell differentiation assay using FACS analysis further supported our conclusion that G9a plays an important role in ATRA-mediated cell differentiation. Cellular differentiation of HL-60 cells was inhibited in G9a knockdown cells, suggesting that G9a may modulate the differentiation of the HL-60 leukemic cell line by regulating JAK2 expression. Remarkably, G9a knockdown significantly increased H3Y41 phosphorylation on the leukemic oncogene lmo2 promoter by derepressing JAK2 transcription.
In summary, we show that HMTase G9a negatively regulates JAK2 transcription and H3Y41 phosphorylation on the lmo2 promoter. The data presented in this study provide new insights into G9a function in the regulation of hematopoiesis and leukemogenesis. Further elucidation of the molecular mechanisms of JAK2 regulation by other histone-modifying activities, including demethylases, will remain for future investigations.
ACKNOWLEDGMENTS
We thank Sung Hee Baek of Seoul National University for the pcDNA3-Flag-G9a and pCMV-HA-Ezh2 clones; Thomas Jenuwein of the Research Institute of Molecular Pathology (IMP) for pCMV-Suv39h1; Mark A. Brown of the University of Texas at Austin for pCMV-myc-Smyd2; and Martin J. Walsh of the Mount Sinai School of Medicine for pEGFP-hG9a and pEGFP-hG9a (ΔSET) clones.
This study was supported by the National R&D Program for Cancer Control, Ministry of Health and Welfare (1020150), and the Environmental Health Center for Childhood Leukemia and Cancer, Ministry of the Environment, Republic of Korea.
Footnotes
Published ahead of print 16 July 2012
H.-J.S. and J.-Y.K. contributed equally to this article.
REFERENCES
- 1. Aoyama T, et al. 2010. Histone modifiers, YY1 and p300, regulate the expression of cartilage-specific gene, chondromodulin-I, in mesenchymal stem cells. J. Biol. Chem. 285: 29842–29850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baxter EJ, et al. 2005. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365: 1054–1061 [DOI] [PubMed] [Google Scholar]
- 3. Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. 2004. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 18: 2627–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dawson MA, et al. 2009. JAK2 phosphorylates histone H3Y41 and excludes HP1α from chromatin. Nature 461: 819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6. Deng Z, Cao P, Wan M, Sui G. 2010. Yin Yang 1: a multifaceted protein beyond a transcription factor. Transcription 1: 81–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dong KB, et al. 2008. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 27: 2691–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Donohoe ME, et al. 1999. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 19: 7237–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. 2005. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell. Biol. 25: 10338–10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Epsztejn-Litman S, et al. 2008. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat. Struct. Mol. Biol. 15: 1176–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feldman N, et al. 2006. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 8: 188–194 [DOI] [PubMed] [Google Scholar]
- 12. Fritsch L, et al. 2010. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol. Cell 37: 46–56 [DOI] [PubMed] [Google Scholar]
- 13. Gordon S, Akopyan G, Garban H, Bonavida B. 2006. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25: 1125–1142 [DOI] [PubMed] [Google Scholar]
- 14. Griffiths DS, et al. 2011. LIF-independent JAK signalling to chromatin in embryonic stem cells uncovered from an adult stem cell disease. Nat. Cell Biol. 13: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grunstein M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389: 349–352 [DOI] [PubMed] [Google Scholar]
- 16. Gyory I, Wu J, Fejer G, Seto E, Wright KL. 2004. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 5: 299–308 [DOI] [PubMed] [Google Scholar]
- 17. Huang J, et al. 2010. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J. Biol. Chem. 285: 9636–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- 18a. Kim JY, et al. 2012. KDM3B is the H3K9 demethylase involved in transcriptional activation of lmo2 in leukemia. Mol. Cell. Biol. 32: 2917–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ko CY, Hsu HC, Shen MR, Chang WC, Wang JM. 2008. Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex. J. Biol. Chem. 283: 30919–30932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kralovics R, et al. 2005. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 352: 1779–1790 [DOI] [PubMed] [Google Scholar]
- 21. Lee JS, et al. 1995. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 9: 1188–1198 [DOI] [PubMed] [Google Scholar]
- 22. Lee JS, et al. 2010. Negative regulation of hypoxic responses via induced Reptin methylation. Mol. Cell 39: 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee JS, Smith E, Shilatifard A. 2010. The language of histone crosstalk. Cell 142: 682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levine RL, Pardanani A, Tefferi A, Gilliland DG. 2007. Role of JAK2 in the pathogenesis and therapy of myeloproliferative disorders. Nat. Rev. Cancer 7: 673–683 [DOI] [PubMed] [Google Scholar]
- 25. Levine RL, et al. 2005. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 7: 387–397 [DOI] [PubMed] [Google Scholar]
- 26. Liu F, et al. 2011. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell 19: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu TX, et al. 2000. Gene expression networks underlying retinoic acid-induced differentiation of acute promyelocytic leukemia cells. Blood 96: 1496–1504 [PubMed] [Google Scholar]
- 28. Nishio H, Walsh MJ. 2004. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc. Natl. Acad. Sci. U. S. A. 101: 11257–11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rathert P, et al. 2008. Protein lysine methyltransferase G9a acts on non-histone targets. Nat. Chem. Biol. 4: 344–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rezai-Zadeh N, et al. 2003. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 17: 1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rui L, et al. 2010. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell 18: 590–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sampath SC, et al. 2007. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol. Cell 27: 596–608 [DOI] [PubMed] [Google Scholar]
- 33. Schwaller J, et al. 2000. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol. Cell 6: 693–704 [DOI] [PubMed] [Google Scholar]
- 34. Shinkai Y, Tachibana M. 2011. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 25: 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. 2008. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 27: 2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. 2001. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276: 25309–25317 [DOI] [PubMed] [Google Scholar]
- 37. Tachibana M, et al. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16: 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tachibana M, et al. 2005. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3–K9. Genes Dev. 19: 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thomas LR, et al. 2008. Functional analysis of histone methyltransferase g9a in B and T lymphocytes. J. Immunol. 181: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Volle DH, et al. 2009. The orphan nuclear receptor small heterodimer partner mediates male infertility induced by diethylstilbestrol in mice. J. Clin. Invest. 119: 3752–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reference deleted.
- 42. Ward AC, Touw I, Yoshimura A. 2000. The Jak-Stat pathway in normal and perturbed hematopoiesis. Blood 95: 19–29 [PubMed] [Google Scholar]
- 43. Yang WM, Inouye C, Zeng Y, Bearss D, Seto E. 1996. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. U. S. A. 93: 12845–12850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou Q, Gedrich RW, Engel DA. 1995. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J. Virol. 69: 4323–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]