Abstract
The Cbl family proteins function as both E3 ubiquitin ligases and adaptor proteins to regulate various cellular signaling events, including the insulin/insulin-like growth factor 1 (IGF1) and epidermal growth factor (EGF) pathways. These pathways play essential roles in growth, development, metabolism, and survival. Here we show that in Drosophila melanogaster, Drosophila Cbl (dCbl) regulates longevity and carbohydrate metabolism through downregulating the production of Drosophila insulin-like peptides (dILPs) in the brain. We found that dCbl was highly expressed in the brain and knockdown of the expression of dCbl specifically in neurons by RNA interference increased sensitivity to oxidative stress or starvation, decreased carbohydrate levels, and shortened life span. Insulin-producing neuron-specific knockdown of dCbl resulted in similar phenotypes. dCbl deficiency in either the brain or insulin-producing cells upregulated the expression of dilp genes, resulting in elevated activation of the dILP pathway, including phosphorylation of Drosophila Akt and Drosophila extracellular signal-regulated kinase (dERK). Genetic interaction analyses revealed that blocking Drosophila epidermal growth factor receptor (dEGFR)-dERK signaling in pan-neurons or insulin-producing cells by overexpressing a dominant-negative form of dEGFR abolished the effect of dCbl deficiency on the upregulation of dilp genes. Furthermore, knockdown of c-Cbl in INS-1 cells, a rat β-cell line, also increased insulin biosynthesis and glucose-stimulated secretion in an ERK-dependent manner. Collectively, these results suggest that neuronal dCbl regulates life span, stress responses, and metabolism by suppressing dILP production and the EGFR-ERK pathway mediates the dCbl action. Cbl suppression of insulin biosynthesis is evolutionarily conserved, raising the possibility that Cbl may similarly exert its physiological actions through regulating insulin production in β cells.
INTRODUCTION
Upon ligand stimulation, activation of receptor tyrosine kinases (RTKs) initiates downstream signaling responses to control many physiological processes (50). Evolutionarily conserved from invertebrates to mammals, insulin/insulin-like growth factor 1 (IGF1) and epidermal growth factor (EGF) act through RTK-mediated signaling cascades, which play central roles in the regulation of growth, development, metabolism, and survival (3, 15, 29, 36, 56, 69). Sophisticated regulatory mechanisms are at work to regulate the duration and intensity of RTK signaling. The Cbl (Casitas B-lineage lymphoma) proteins, a family of E3 ubiquitin ligases and adaptor proteins (60), are key regulators of RTK signaling, and this is best exemplified by the negative control of the EGF pathway through Cbl-mediated ubiquitylation and endocytic destruction of the EGF receptor (EGFR) (11, 26, 27, 51, 55, 67). However, the functional evolution of Cbl's regulatory action with respect to the physiological interconnection and cooperation of multiple RTK pathways remains poorly understood.
Cbl proteins are known to regulate a diverse range of cellular events through promoting ubiquitylation-directed degradation of target proteins or acting as adaptors within the signaling complexes (51). A growing body of evidence has established that Cbl-dependent downregulation of the EGFR pathway is evolutionarily conserved from Caenorhabditis elegans to vertebrates (14, 17, 27, 64). In mammals, there are three Cbl homologues, c-Cbl, Cbl-b, and Cbl-3, which possess highly conserved TKB (tyrosine-kinase-binding) and RING finger domains in their N-terminal regions, allowing them to function as E3 ubiquitin ligases. c-Cbl and Cbl-b are ubiquitously expressed, and both contain proline-rich domains in their extended C-terminal portions that can mediate interactions with a plethora of proteins (51, 57). Interestingly, the Cbl orthologue in the fruit fly, Drosophila melanogaster (dCbl), exists as the long and short isoforms as a result of alternative splicing (47). The long form of dCbl has a domain structure identical to that of mammalian c-Cbl and Cbl-b, whereas the short version contains solely the TKB and RING finger domains. Both isoforms have been shown to downregulate EGFR signaling (32, 41), and recent studies have documented that the long isoform of dCbl regulates the EGFR pathway, while the short one preferentially controls notch signaling (62).
The evolutionarily conserved insulin/IGF1 signaling through their RTKs regulates multiple physiological processes, including metabolic homeostasis, stress resistance, and longevity (15, 56). The insulin signaling pathway is also subject to both positive and negative regulation (9, 53, 54, 58). Emerging evidence suggests an unanticipated complexity with respect to the functional effects of mammalian Cbl proteins upon insulin actions. In 3T3-L1 adipocytes, Cbl was demonstrated to act as an adaptor molecule and play a positive regulatory part in insulin-controlled glucose transport (30, 31, 49). On the other hand, it was reported that c-Cbl could promote the ubiquitylation of both insulin and IGF1 receptors (1, 52). Moreover, whole-body ablation of c-Cbl in mice led to reduced adiposity, presumably through increased energy expenditure, thus improving peripheral insulin sensitivity (37). These observations indicate that c-Cbl negatively regulates insulin signaling by promoting insulin receptor ubiquitylation and degradation (38). c-Cbl and Cbl-b also serve as key modulators of immune responses (10, 19, 44), and genetic deletion of Cbl-b was shown to enhance infiltration and activation of adipose tissue macrophages, resulting in peripheral insulin resistance in mice (18). Given the different roles of insulin signaling in diverse tissues or cell types, it has yet to be elucidated how Cbl exerts its cell type- or tissue-specific functions in the control of insulin action in systemic metabolism.
The Drosophila genome contains seven insulin-like peptide (dILP) genes (dilp1 to dilp7) and one single insulin receptor (dInR) gene (4). dILP2, dILP3, and dILP5, three primary forms of dILPs, are expressed and secreted by the median neurosecretory insulin-producing cells (IPCs) in the brain (5, 20). IPCs are thought to be functionally analogous to mammalian pancreatic islet β cells (48). The expression of dilp genes has been shown to be regulated independently by nutritional signals and developmental stages (5, 20); however, the underlying mechanisms remain largely elusive.
We investigated the physiological functions of dCbl using genetic models of Drosophila. Here we report that neuronal dCbl participates in the biosynthetic regulation of dILPs through downregulation of the EGFR signaling. Our findings indicate that Cbl-mediated modulation of insulin production may represent an evolutionarily conserved, cell type-specific mode of Cbl actions in the coordinate control of growth, life span, and metabolism.
MATERIALS AND METHODS
Fly strains.
The P{SUPor-P}dCblEY11427 line (designated dCblEY/+ for the heterozygote) and the P{EPgy2}dCblKG03080 line (designated dCblKG/+ for the heterozygote), each with a P-element inserted in the 5′ untranslated region (UTR) for disruption of the dCbl gene, were obtained from the Bloomington Drosophila Stock Center and backcrossed into a w1118 background for six generations. The green fluorescent protein (GFP)-expressing line P{GAL4-Kr.C}DC2,P{UAS-GFP.S65T}DC (Bloomington Drosophila Stock Center) was used to cross with the P{SUPor-P}dCblEY11427 or P{EPgy2}dCblKG03080 line for generating GFP-labeled heterozygous dCblEY/+ and dCblKG/+ lines, respectively. These two heterozygous lines were subsequently crossed to produce the identifiable homozygous dCblEY/KG larvae for further studies.
The UAS-dCbl-RNAi line (where UAS is upstream activating sequence and RNAi is RNA interference) (transformant accession number 22335) was obtained from the Vienna Drosophila RNAi Center. For all life span experiments, UAS-dCbl-RNAi flies were used after backcrossing into the w1118 background for six generations. The elav-Gal4 and dominant-negative (DN) UAS-EGFR-DN lines were from the Bloomington Drosophila Stock Center. UAS-EGFR-DN was backcrossed into the w1118 background for five generations before crossing with the UAS-dCbl-RNAi line. The dilp2-Gal4 line was a generous gift from Eric Rulifson (University of California at San Francisco [UCSF]). Heterozygous UAS-dCbl-RNAi animals were crossed with elav-Gal4 or dilp2-Gal4 to generate the strains with neuronal or IPC-specific dCbl knockdown by making use of the cyo balancer. For IPC imaging, the UAS-srcGFP line (Bloomington Drosophila Stock Center) was crossed with UAS-dCbl-RNAi or UAS-EGFR-DN flies before further crossing with the dilp2-Gal4 drivers to generate dilp2G4>GFP, dCbl-RNAi or dilp2G4>GFP, EGFR-DN flies. IPC cells were visualized by GFP fluorescence with confocal microscopy. Flies were maintained at 25°C with 12-h dark/12-h light cycles on standard food medium (20 g inactivated yeast powder, 60 g corn flour, 10 g agar, 100 g sucrose, and 15 ml 10% Tego in 75% ethanol per liter).
Starvation and paraquat sensitivity assays.
Flies at 3 days of age (10 flies/vial) were maintained on starvation medium containing 1% agar or on starvation medium supplemented with 5% sucrose and 30 mM paraquat. Flies were transferred daily to fresh medium, with the number of dead flies recorded every 12 h.
Body weight and life span.
The body weight of individual 3-day-old flies (90 flies/batch) was measured using a precision balance. For life span determination, newly eclosed flies were placed in vials (10 flies/vial). Flies were transferred to fresh food every other day, and the number of dead flies was monitored.
Glycogen and trehalose measurements.
Flies (8 to 10/group) were anesthetized using CO2 and homogenized in 250 μl of 0.25 M Na2CO3 buffer. After incubation at 95°C for 2 h, 150 μl of 1 M acetic acid and 600 μl of 0.25 M sodium acetate (pH 5.2) were added. The mixture was centrifuged for 20 min at 12,000 rpm at room temperature. To convert trehalose and glycogen into glucose, aliquots (50 μl) of the supernatant were incubated with 0.05 ml porcine trehalase (T8778; Sigma) at 37°C overnight or 5 units/ml amyloglucosidase (A7420; Sigma) at 55°C for 4 h. Total glucose was measured by glucose assay reagent (G3293; Sigma). For normalization, protein content in the supernatant was determined using a Bio-Rad protein assay reagent.
Microscopic imaging analysis.
Fly brains were fixed in 4% paraformaldehyde (PFA) in sodium phosphate buffer (pH 7.4). After rinsing in sodium phosphate buffer, the brains were incubated in 0.01 M phosphate-buffered saline (PBS; pH 7.4). Specimens were analyzed using an Olympus FV1000 confocal microscope (Japan) with a ×60 water objective with a numerical aperture of 1.2. Confocal microscopic images were obtained at an optical section thickness of 0.31 to 0.37 μm and were processed using Olympus FV1000 software.
Reverse transcription-PCR (RT-PCR) analysis.
Total RNA was prepared from 3-day-old flies or INS-1 cells with TRIzol reagent (Invitrogen). First-strand cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase and random hexamer primers (Invitrogen). Regular PCR was performed using Taq kits (TaKaRa), and quantitative PCR (qPCR) was done with an ABI 7500 fast real-time PCR system using SYBR green PCR master mix (ABI). Ribosomal protein L32 (RPL32), α-tubulin 84B (α-Tub84B), or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an internal control for normalization. The oligonucleotide primers for the target genes analyzed are as follows: for dCbl, sense primer 5′-TCAAGGGCACCGAACAAAT-3′ and antisense primer 5′-TGCTCCGTCGTCCCAAAC-3′; for dCbl long form, sense primer 5′-ATGAGGACATCGTTGAGGTGG-3′ and antisense primer 5′-CAGAGTTGCGTGTAATGGGAG-3′; for dCbl short form, sense primer 5′-AACTAACATCCACAGACCAAC-3′ and antisense primer 5′-GAAGAACGAACAACTAAACGA-3′; for dilp2, sense primer 5′-ATCTGGACGCCCTCAATCC-3′ and antisense primer 5′-CCAAGATAGCTCCCAGGAAAGA-3′; for dilp3, sense primer 5′-AGAGAACTTTGGACCCCGTGAA-3′ and antisense primer 5′-TGAACCGAACTATCACTCAACAGTCT-3′; for dilp5, sense primer 5′-CGCTCCGTGATCCCAGTT-3′ and antisense primer 5′-GCTATCCAAATCCGCCAAG-3′; for CG2162, sense primer 5′-TGCCCAAAATGGACTATACGG-3′ and antisense primer 5′-GCTTCAACTTGACAAACGGAT-3′; for rat Ins-1, sense primer 5′-ACACCCAAGTCCCGTCGTGAAGTGG-3′ and antisense primer 5′-GGCGGGGAGTGGTGGACTCAGT-3′; and for rat Ins-2, sense primer 5′-TCATCCTCTGGGAGCCCCGC-3′ and antisense primer 5′-GGTCTGAAGGTCACCGGCCC-3′.
Antibodies and chemicals.
Polyclonal antibodies against p-Akt (synthetic phosphopeptide corresponding to residues surrounding Ser473 of mouse Akt), total Akt (a synthetic peptide corresponding to the carboxy-terminal sequence of mouse Akt), p-ERK (a synthetic phosphopeptide corresponding to residues surrounding Thr202/Tyr204 of human p44/42 mitogen-activated protein [MAP] kinase), and total ERK (a synthetic peptide corresponding to a sequence in the C terminus of rat p44/42 MAP kinase) were all purchased from Cell Signaling, and the α-tubulin antibody was purchased from Biomeda. ERK-specific kinase MEK inhibitor PD98059 (P215) and phosphoinositide 3-kinase (PI3K)-specific kinase inhibitor LY294002 (L9908) were from Sigma, and EGF was from R&D. PDX-1 antibody was from Upstate (07-696; Millipore).
Western immunoblotting.
Protein extracts were prepared from flies by homogenization in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl) using a Polytron homogenizer. Lysates were then sonicated for 30 s and centrifuged for 20 min at 13,000 × g to remove the debris. INS-1 cells were also lysed in RIPA buffer. Proteins (40 μg) were separated by SDS-PAGE and transferred to a polyvinylidene difluoride filter membrane (Amersham Biosciences). After incubation with the desired antibodies, the blots were developed with an Amersham Biosciences ECL Plus detection system.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were conducted with the PDX-1 antibody following the manufacturer's instructions (26156; Thermo). Quantitative PCR was done using an ABI 7500 Fast real-time PCR system. The primers used for amplification of the Ins-1 promoter sequence were 5′-CTGGGAAATGAGGTGGAAAA-3′ and 5′-AGGAGGGGTAGGTAGGCAGA-3′.
Cell culture, transfection, and luciferase reporter assay.
Rat insulinoma INS-1 cells were cultured in RPMI 1640 medium containing 2 mM l-glutamine supplemented with 10% fetal bovine serum, 16.7 mM glucose, 10 mM HEPES, 1 mM sodium pyruvate, 50 μM 2-mercaptoethanol, 100 IU ml/liter penicillin, and 100 μM streptomycin in 5% CO2 at 37°C. INS-1 cells were transfected with the rat insulin 1 promoter (RIP)-luciferase (Luc) reporter construct (kindly provided by M. German, UCSF) using Lipofectamine 2000 reagents (Invitrogen). Luciferase activities were analyzed using a dual-luciferase assay kit (Promega), according to the manufacturer's instructions.
c-Cbl knockdown by transient transfection.
Duplex small interfering RNA (siRNA) oligonucleotides used for silencing the expression of c-Cbl were purchased from Genepharma Ltd. (Shanghai, China) and had the following sequences: si-c-Cbl_#1, 5′-GCUUCCAAGGCUUCUUCUAdTdT-3′, directed against the coding region from nucleotide (nt) 1552 downstream of the start codon of rat c-Cbl; si-c-Cbl_#2, 5′-CCUUGGAAUGGGAGAGAAUdTdT-3′, directed against the coding region from nt 1819 downstream of the start codon of rat c-Cbl (GenBank accession number XM_576396); and si-CON, 5′-GCAGUAAGCGAUACGCAAAdTdT-3′, a nonspecific scrambled control. INS-1 cells were transfected with siRNA duplexes at 50 nM using Lipofectamine RNAiMAX reagent (13778; Invitrogen), and cells were cultured for 48 h before further analysis.
Plasmid and adenoviral c-Cbl overexpression.
The pKF plasmid for human c-Cbl expression was a generous gift from Jian Zhang (University of Chicago). The cDNA fragment for c-Cbl was subcloned into pcDNA6.0 (Invitrogen). The c-Cbl-C381A substitution mutant was generated using a Muta-direct site-directed mutagenesis kit (SBS, Shanghai, China). Recombinant adenoviruses for the overexpression of c-Cbl proteins were produced with an AdEasy adenoviral vector system (Stratagene, La Jolla, CA), following the manufacturer's instructions. Briefly, for control virus, the coding sequence of EGFP (pEGFP-N1; Clontech, Mountain View, CA) was subcloned into the pShuttle-cytomegalovirus (CMV) vector digested with SalI and NotI, and the cDNA fragments encoding c-Cbl proteins were likewise subcloned into pShuttle-CMV. The shuttle plasmids and pAdEasy-1 were subsequently cotransformed into BJ5183 bacteria to yield recombinant adenoviral DNA, which was then used for transfection of HEK293A cells. Recombinant viral particles were obtained from the cell lysates. INS-1 cells at 60% confluence were infected at a multiplicity of infection (MOI) of 20.
Measurement of insulin secretion and content.
Insulin secretion from INS-1 cells was determined as described previously (43, 66). Briefly, confluent cells in 24-well plates were washed with HEPES-balanced salt solution (HBSS), followed by incubation in the same buffer for 2 h. Insulin secreted from the medium was then measured after static incubation for 2 h in HBSS containing 2.8 mM or 16.7 mM glucose. To determine the intracellular insulin content, cells were lysed by incubation overnight at 4°C in 1 ml of a mixture of ethanol-water-concentrated HCl (750:235:15). After centrifugation for 5 min at 12,000 × g at 4°C, insulin was measured using an enzyme-linked immunosorbent assay (ELISA) kit (EZRMI-13K; Millipore).
Statistical analysis.
All data are presented as the mean ± standard error of the mean (SEM). Statistical analysis was performed using an unpaired two-tailed t test and one-way or two-way analysis of variance (ANOVA), followed by Bonferroni's posttest, with GraphPad Prism software (version 5.0). Log-rank tests were used for analysis of life span and stress survival curves. P values of <0.05 were considered statistically significant.
RESULTS
Characterization of dCbl expression and its importance in development and survival.
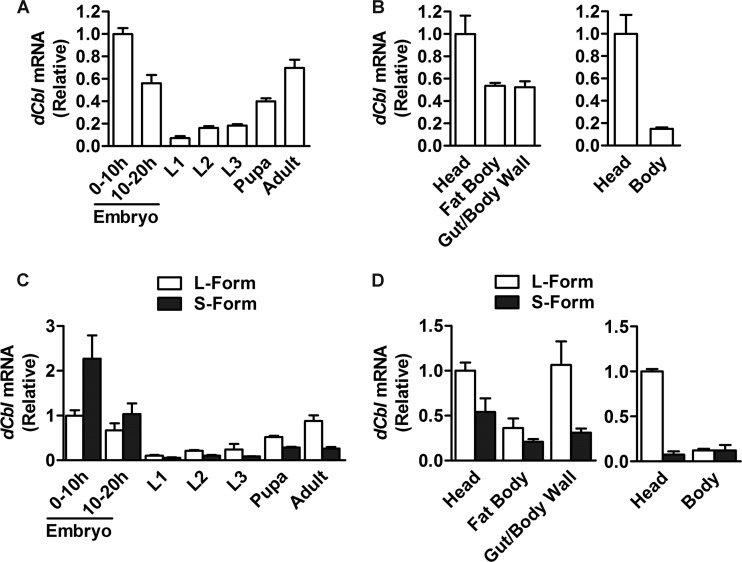
To gain insight into the functions of dCbl, we examined the mRNA expression patterns of dCbl in w1118 flies by quantitative RT-PCR (qRT-PCR). When assessed for transcripts encoding both the long and short forms, dCbl was expressed at high levels in embryos and adults but at low levels at the larval stages (Fig. 1A). Moreover, in both larvae and adult flies, the expression of dCbl was more abundant in the head than in the body (Fig. 1B). When detected for specific transcripts encoding each dCbl isoform, the short form was more efficiently expressed than the long form in embryos, whereas higher expression was observed for the long form in pupae and adults (Fig. 1C). This implies distinct functional roles for the two dCbl isoforms during development. Interestingly, the long form was expressed at a much higher level in the head of adult flies (Fig. 1D), suggesting a predominant role for this isoform in the brain.
Fig 1.
Developmental and tissue expression patterns of dCbl mRNA. (A) Expression features of dCbl mRNA at different developmental stages. dCbl mRNA abundance in wild-type w1118 flies (60 embryos, 20 to 30 larvae, 10 pupae, and 10 adult flies at 3 days of age; n > 3 independent experiments) was assessed by real-time qRT-PCR using RPL32 as an internal control. L1, L2, and L3 first-, second-, and third-instar larvae, respectively. (B) Tissue distribution patterns of dCbl mRNA expression in w1118 third-instar larvae or adult flies (40 larvae and 20 adults, n > 3 independent experiments). (C) The abundance of the mRNA transcripts encoding the long (L) or short (S) form of dCbl was determined by qRT-PCR for w1118 flies at different developmental stages (60 embryos, 20 to 30 larvae, 10 pupae, and 10 adult flies; n > 3 independent experiments). (D) Tissue expression patterns of the mRNA transcripts for the long (L) or short (S) forms of dCbl in w1118 third-instar larvae and adult flies (30 larvae and 30 adults, n > 3 independent experiments).
Next, we investigated the importance of dCbl in development and survival using two mutant lines of flies (designated dCblEY and dCblKG; Bloomington Drosophila Stock Center) in which the dCbl gene was disrupted by insertion of the P-element in a 5′ UTR exon (Fig. 2A). Due to the poor viability of homozygous dCblEY/EY or dCblKG/KG flies, we crossed these two mutant lines and obtained viable heteroallelic dCblEY/KG flies that had markedly decreased dCbl mRNA transcripts (Fig. 2B). Disruption of dCbl expression resulted in an ∼24-h delay in larval pupation (Fig. 2C) and an ∼63% reduction of adult survival rate in male dCblEY/KG flies (Fig. 2D). Notably, the whole-body glycogen and trehalose (an insect disaccharide that is the main form of circulatory carbohydrates) levels in male dCblEY/KG flies decreased by ∼35% and ∼44%, respectively, compared to those in the w1118 control line (Fig. 2E). Moreover, male dCblEY/KG flies showed increased sensitivity to starvation, exhibiting an ∼20% decrease in the median survival time upon food deprivation (Fig. 2F) (3.6 days for dCblEY/KG flies and 4.5 days for w1118 flies). dCbl deficiency also led to a dramatically shortened life span, with an ∼67% reduction in the median life span observed in male dCblEY/KG flies (Fig. 2G; 23 days for dCblEY/KG flies and 70 days for w1118 control). These data suggest that dCbl plays an essential role in Drosophila development and survival, in agreement with previous reports (41, 42, 63).
Fig 2.
dCbl is essential for the development and survival of flies. (A) Two heterozygous dCbl mutant lines with P-element insertion at the indicated dCbl loci were crossed to generate the dCblEY/KG line. Genomic organization of the dCbl gene containing the P-element is shown (black boxes, coding exons; open boxes, UTRs; triangles, P-elements). (B) dCbl mRNA expression levels were analyzed by qRT-PCR in the whole body of 3-day-old adult flies of the indicated genotype (30 flies/group, n > 3 independent experiments). RPL32 was used as an internal control. (C) Disruption of dCbl resulted in a developmental delay. The pupation rate of wild-type w1118 or dCblEY/KG larvae was determined by recording the number of pupae from the third-instar larval stage (>120 animals/group) every 24 h. (D) Disruption of dCbl reduced the survival rate. The percentage of w1118 or dCblEY/KG flies that survived from the larval stage (>120 animals/group). Results are shown as the mean ± SEM; **, P < 0.01 versus w1118 by Student's t test. (E) Disruption of dCbl led to decreased carbohydrate levels. Whole-body glycogen and trehalose contents were measured in 3-day-old male adult w1118 or dCblEY/KG flies. Data are presented as the mean ± SEM; *, P < 0.05 versus the w1118 control by one-way ANOVA. (F) Sensitivity to starvation. Male adult flies at 3 days of age were deprived of food, and survival rates were determined (126 w1118 flies and 122 dCblEY/KG flies). χ2 = 66.91; P < 0.001 by log-rank test. (G) Life span. Survival curves of male w1118 (n = 225) and dCblEY/KG (n = 184) flies. χ2 = 405.1; P < 0.0001 by log-rank test.
Neuronal dCbl regulates Drosophila life span and stress tolerance.
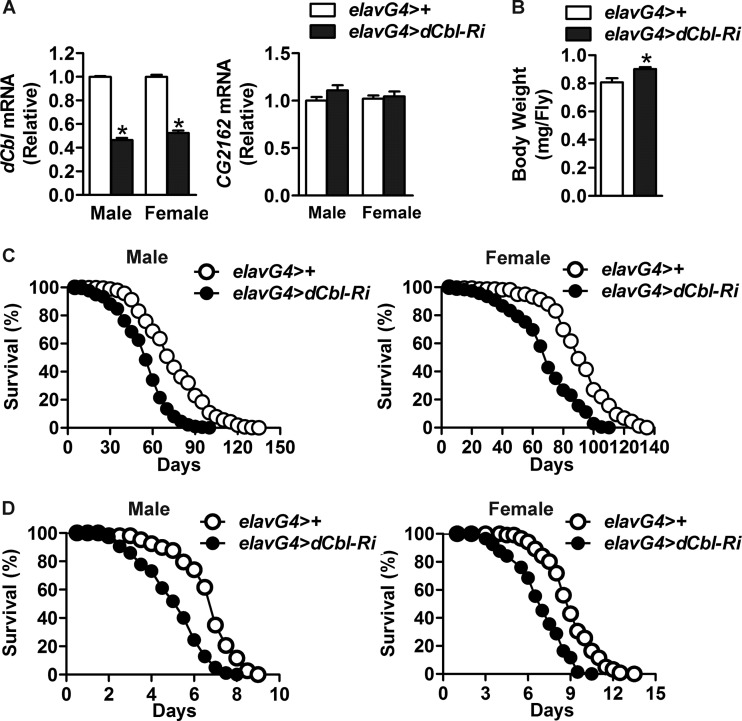
Given the high expression of dCbl in the heads of flies (Fig. 1B and D), we investigated the physiological functions of neuronal dCbl using the Gal4 and UAS-RNA interference (RNAi) system to knock down the expression of neuronal dCbl. We crossed UAS-dCbl-RNAi flies (Vienna Drosophila RNAi Center) with pan-neuron elav-GAL4 drivers to generate elav-GAL4/UAS-dCbl-RNAi (here denoted elavG4>dCbl-Ri) flies. Under fed or starved conditions, head dCbl mRNA abundance was decreased by ∼55% in male elavG4>dCbl-Ri flies and by ∼48% in female elavG4>dCbl-Ri flies compared with that in elavG4>+ control flies (Fig. 3A). In contrast, no changes in the mRNA levels of CG2162 (Fig. 3A), a potential off-target gene in UAS-dCbl-RNAi flies, were detected. Interestingly, knockdown of neuronal dCbl expression led to an ∼12% increase in the body weight of male adult flies (Fig. 3B). Moreover, neuronal dCbl suppression significantly reduced the life span of both male and female elavG4>dCbl-Ri flies (Fig. 3C), with ∼27% and ∼24% decreases, respectively, observed in their median life span (53 days for male elavG4>dCbl-Ri flies, 73 days for male elavG4>+ flies, 68 days for female elavG4>dCbl-Ri flies, and 90 days for female elavG4>+ flies). Consistent with their shorter life span, both male and female elavG4>dCbl-Ri flies also exhibited reduced tolerance to oxidative stress (Fig. 3D), displaying ∼25% and ∼26% decreases in their median survival time, respectively, upon treatment with paraquat (5.0 days for male elavG4>dCbl-Ri flies, 6.7 days for male elavG4>+ flies, 6.6 days for female elavG4>dCbl-Ri flies, and 8.9 days for female elavG4>+ flies). These data thus revealed a crucial role of neuronal dCbl in the regulation of longevity and stress tolerance of Drosophila.
Fig 3.
Knockdown of neuronal dCbl expression reduces oxidative stress resistance and life span. (A) Knockdown of neuronal dCbl expression was analyzed by qRT-PCR in the head of adult elavG4>dCbl-Ri flies relative to the control elavG4>+ flies in both male and female flies (90 flies/group, n > 3 independent experiments). The mRNA expression of CG2162, a potential off-target gene, was also assessed. (B) The body weight of adult flies of the indicated genotype at 3 days of age (90 flies/genotype). Error bars in panels A and B represent SEMs; *, P < 0.05 by Student's t test. (C) Life span. Survival curves of male and female elavG4>+ (300 male and 249 female) and elavG4>dCbl-Ri (252 male and 264 female) flies. χ2 = 220.2 for males and χ2 = 119.4 for females; P < 0.0001 by log-rank test. (D) Sensitivity to paraquat. Male and female adult flies at 3 days of age were treated with 30 mM paraquat. Survival rates were determined for elavG4>+ (165 male and 148 female) and elavG4>dCbl-Ri (200 male and 208 female) flies. χ2 = 113.8 for males and χ2 = 74.84 for females; P < 0.0001 by log-rank test.
Neuronal dCbl deficiency is associated with upregulation of insulin-like peptides.
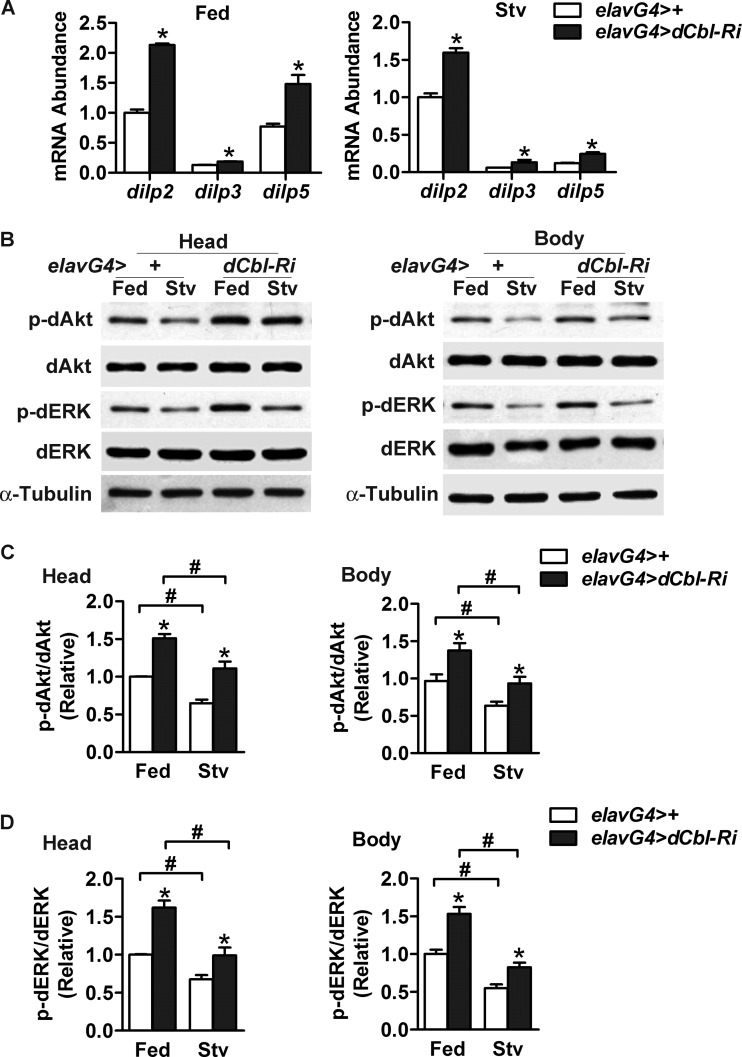
The insulin/IGF pathway is known to regulate stress resistance and longevity in flies (8, 59). We asked if neuronal dCbl regulates stress resistance and longevity by suppressing the production of dILPs in the brain. Indeed, in male elavG4>dCbl-Ri flies, knockdown of neuronal dCbl increased the mRNA expression levels of dilp2, dilp3, and dilp5 by ∼113%, ∼44%, and ∼92%, respectively, in the fed state and by ∼50%, ∼120%, and ∼100%, respectively, under the starved condition in comparison to those of elavG4>+ control flies (Fig. 4A). Female elavG4>dCbl-Ri flies also showed similarly upregulated expression of dilp2 (by ∼99%), dilp3 (by ∼102%), and dilp5 (by ∼76%) in the fed state (data not shown).
Fig 4.
Neuronal dCbl knockdown results in upregulated dilp gene expression in parallel with increased phosphorylation of dAkt and dERK. (A) RNA was extracted from the head of fed or starved (Stv; 24 h) male adult elavG4>+ or elavG4>dCbl-Ri flies (3 days of age, 90 flies/group). The abundance of dilp2, dilp3, or dilp5 mRNA was measured by qRT-PCR (n > 3 independent experiments). After normalization to the levels for RPL32, results are shown as means ± SEMs; *, P < 0.05 versus the value from control elavG4>+ flies by one-way ANOVA. (B) Western immunoblot analysis of dAkt and dERK phosphorylation using the indicated antibodies. Head or body protein extracts were analyzed for fed or starved (24 h) male adult flies (5 days of age, 90 flies/group). Representative results from three independent experiments are shown. Drosophila α-tubulin was used as an internal control. (C and D) Relative phosphorylation levels of dAkt (C) and dERK (D) were determined by densitometric quantification of the immunoblots, presented as the mean ± SEM (n = 3). *, P < 0.05 versus control elavG4>+ flies; #, P < 0.05 between fed and starved conditions by two-way ANOVA.
To determine the effects of neuronal dCbl knockdown on dILP signaling, we measured the phosphorylation levels of dAkt as well as the extracellular signal-related kinase dERK (Fig. 4B). In male elavG4>dCbl-Ri flies, phospho-dAkt levels in the head and body increased by ∼51% and ∼41%, respectively, in the fed state and by ∼50% and ∼42%, respectively, in the starved state compared with the level in elavG4>+ flies (Fig. 4C). Phospho-dERK levels in the head and body increased by ∼62% and ∼53%, respectively, under fed conditions and by ∼46% and ∼51%, respectively, under starved conditions (Fig. 4D). Therefore, knockdown of dCbl expression in neurons increased whole-body insulin/IGF signaling, most likely due to upregulation of dILP production.
dCbl in IPCs regulates dilp expression, life span, and stress resistance.
dILPs are expressed and secreted mainly through 7 pairs of neurons termed IPCs (4, 6). To further investigate whether dCbl regulates the production of dILPs in IPCs, we crossed UAS-dCbl-RNAi flies with dilp2-GAL4 drivers to generate dilp2-GAL4/UAS-dCbl-RNAi (here denoted dilp2G4>dCbl-Ri) flies in which dCbl expression was specifically knocked down in IPCs. In male dilp2G4>dCbl-Ri flies, the mRNA levels of dilp2, dilp3, and dilp5 increased by ∼71%, ∼43%, and ∼69%, respectively, in the fed state and by ∼74%, ∼66%, and ∼46%, respectively, under the starved condition compared with those in dilp2G4>+ control flies (Fig. 5A). Consistently, dilp2G4>dCbl-Ri flies also had an ∼9% increase in body weight (Fig. 5B) and exhibited a significant shortening of the life span (Fig. 5C), with an ∼14% decrease in the median life span observed (71 days for dilp2G4>dCbl-Ri flies and 83 days for dilp2G4>+ flies). Paraquat tolerance experiments showed an increased sensitivity to oxidative stress in male dilp2G4>dCbl-Ri flies (Fig. 5D), with an ∼42% reduction in median survival time observed (4.4 days for dilp2G4>dCbl-Ri flies and 7.6 days for dilp2G4>+ flies). These results suggest that dCbl may cell autonomously regulate dILP production in IPCs.
Fig 5.
IPC-specific dCbl knockdown leads to elevated dilp expression with reduced stress tolerance and longevity. (A) The mRNA abundance of dilp2, dilp3, or dilp5 was assessed by qRT-PCR in the heads of male adult dilp2G4>+ and dilp2G4>dCbl-Ri flies (3 days old, 90/group, n > 3 independent experiments) under the fed or starved state. Normalization was done using RPL32 as an internal control. Data are represented as the mean ± SEM; *, P < 0.05 versus dilp2G4>+ control flies by one-way ANOVA. (B) Photo of male adult dilp2G4>+ (left) and dilp2G4>dCbl-Ri (right) flies. The body weight was measured under fed states (3 days of age, 90 flies/group), shown as the mean ± SEM; *, P < 0.05 by Student's t test. (C) Life span. Survival curves of male dilp2G4>+ (n = 266) and dilp2G4>dCbl-Ri (n = 293) flies. χ2 = 68.10; P < 0.0001 by log-rank test. (D) Sensitivity to paraquat. Male adult flies at 3 days of age were treated with 30 mM paraquat, and survival rates were determined (264 dilp2G4>+ flies and 265 dilp2G4>dCbl-Ri flies). χ2 = 90.12; P < 0.0001 by log-rank test.
dCbl deficiency in neurons or IPCs affects carbohydrate metabolism and starvation resistance.
The insulin/IGF signaling pathway in Drosophila controls glucose homeostasis in response to nutritional states. To determine whether dCbl in neurons or IPCs regulates carbohydrate metabolism, we measured the total glycogen and trehalose levels in adult flies. Knockdown of neuronal expression of dCbl decreased both whole-body glycogen (by ∼28%) and trehalose (by ∼41%) in male elavG4>dCbl-Ri flies under fed conditions compared with those in elavG4>+ control flies (Fig. 6A). IPC-specific knockdown of dCbl expression similarly decreased glycogen (by ∼24%) and trehalose (by ∼27%) in male dilp2G4>dCbl-Ri flies compared with those in dilp2G4>+ control flies (Fig. 6B). Then, we examined the effect of dCbl deficiency upon the survival rates during starvation. Both neuronal and IPC-specific knockdown of dCbl expression significantly reduced the tolerance to food deprivation, leading to ∼15% (elavG4>dCbl-Ri) and ∼12% (dilp2G4>dCbl-Ri) decreases in their median survival times (Fig. 6C and D; 7.3 days for elavG4>dCbl-Ri flies and 8.6 days for elavG4>+ flies; 8.8 days for dilp2G4>dCbl-Ri flies and 10.0 days for dilp2G4>+ flies). These data indicate that dCbl in IPCs regulates carbohydrate metabolism and starvation resistance through upregulation of dILP production.
Fig 6.
Metabolic effects of neuronal or IPC-specific suppression of dCbl expression. (A and B) Whole-body glycogen or trehalose levels were measured in male adult flies under fed conditions (3 days of age, 10 flies/group, n > 3 independent experiments). Values for elavG4>+ versus elavG4>dCbl-Ri flies (A) or dilp2G4>+ versus dilp2G4>dCbl-Ri flies (B) were normalized to total protein levels. Data are presented as the mean ± SEM; *, P < 0.05 versus the >+ control group by one-way ANOVA. (C and D) Sensitivity to starvation. Survival curves of starved male adult flies at 3 days of age were determined for elavG4>+ flies (n = 226) versus elavG4>dCbl-Ri flies (n = 205) (C) and dilp2G4>+ flies (n = 137) versus dilp2G4>dCbl-Ri flies (n = 133) (D). χ2 = 163.5 (C) and χ2 = 29.66 (D); P < 0.0001 versus >+ control group by log-rank test.
dCbl regulation of dilp expression is genetically coupled to the dEGFR signaling pathway.
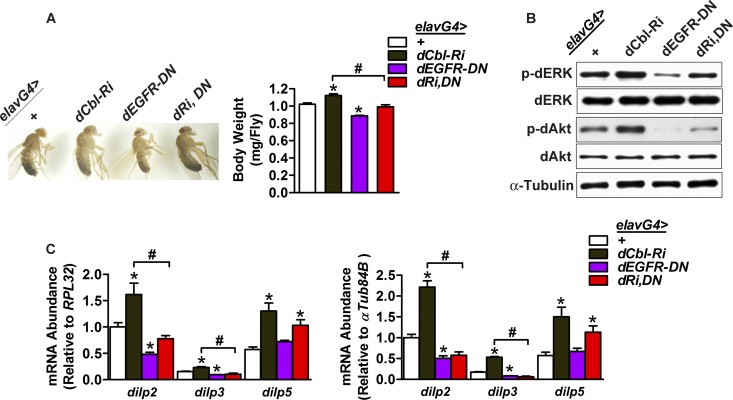
To investigate the mechanism by which dCbl suppresses dilp expression in IPCs, we examined the role of the dEGFR pathway by overexpressing a dominant-negative form of EGFR (dEGFR-DN) (12) in pan-neurons or IPCs. First, we crossed UAS-dEGFR-DN flies with UAS-dCbl-RNAi flies to generate UAS-dCbl-RNAi; dEGFR-DN flies. Subsequently, we crossed UAS-dCbl-RNAi; dEGFR-DN flies with elav-GAL4 drivers to produce elavG4>dCbl-RNAi; dEGFR-DN (here denoted elavG4>dRi, DN) flies in which dEGFR signaling was specifically inhibited in neurons through dEGFR-DN overexpression in the context of neuron-specific dCbl knockdown. In comparison with elavG4>+ controls, male elavG4>dEGFR-DN flies had an ∼13% body weight decrease, and neuronal dEGFR-DN overexpression in elavG4>dRi, DN flies blunted dCbl deficiency-dependent increases of body weight (Fig. 7A). In accordance with ERK phosphorylation activation as a key component of the EGFR signaling pathway (50), elavG4>dEGFR-DN flies exhibited marked reductions in the phosphorylation of dERK and dAkt in the head; and neuronal dEGFR-DN overexpression largely reversed dCbl deficiency-promoted increases of their phosphorylation levels (Fig. 7B). Importantly, male elavG4>dEGFR-DN flies showed significant reductions in the mRNA levels of dilp2 (by ∼52% and ∼49% when normalized to the level of RPL32 and α-Tub84B, respectively) and dilp3 (by ∼39% and ∼53% when normalized to the level of RPL32 and α-Tub84B, respectively) but not dilp5 (Fig. 7C). Furthermore, dCbl deficiency-dependent elevations in the expression of dilp2 and dilp3 but not dilp5 were significantly blocked by neuronal dEGFR-DN overexpression in male elavG4>dRi, DN flies (Fig. 7C).
Fig 7.
Blocking dEGFR signaling reverses the effects of neuronal dCbl deficiency. (A) Male adult elavG4>+, elavG4>dCbl-Ri, elavG4>dEGFR-DN, and elavG4>dCbl-Ri, EGFR-DN (dRi,DN) flies at 3 days of age. The body weight was measured under fed states (90 flies/group). (B) Western immunoblot analysis of the phosphorylation levels of dAkt or dERK for the head protein extracts from fed male adult flies of the indicated genotype (90 flies/group). Drosophila α-tubulin was used as a loading control. Representative results are shown from three independent experiments. (C) The mRNA abundance of dilp2, dilp3, or dilp5 in the head of male adult flies was determined by qRT-PCR (90 flies/group, n = 4 to 5 independent experiments). Normalization was done using RPL32 or α-Tub84B as the internal control. Data in panels A and C are shown as the mean ± SEM. *, P < 0.05 versus elavG4>+ control; #, P < 0.05 between elavG4>dCbl-Ri and elavG4>dRi, DN by one-way ANOVA.
To further confirm that dCbl regulation of dilp expression involves the dEGFR pathway specifically in IPCs, we crossed UAS-dCbl-RNAi; dEGFR-DN flies with dilp2-GAL4 drivers to generate dilp2G4>dCbl-Ri; dEGFR-DN (here denoted dilp2G4>dRi, DN) flies in which dEGFR-DN was overexpressed in the face of dCbl knockdown in IPCs. As dCbl and EGFR have been implicated in the regulation of cell survival and proliferation (28, 33, 63), we first utilized the UAS-srcGFP line to produce dilp2G4>GFP, dCbl-RNAi and dilp2G4>GFP, dEGFR-DN flies and tested whether dCbl suppression or blocking of dEGFR signaling affects the survival of IPCs. GFP-facilitated visualization showed no alterations in the morphology and number of IPCs in dilp2G4>GFP, dCbl-RNAi or dilp2G4>GFP, dEGFR-DN flies compared to dilp2G4>GFP controls (Fig. 8A). Consistently, IPC-specific overexpression of dEGFR-DN not only reduced the body weight (Fig. 8B) and the expression of dilp2 and dilp3 (Fig. 8C) in male dilp2G4>dEGFR-DN flies but also blunted dCbl deficiency-dependent increases of body weight (Fig. 8B) and upregulation of dilp2 and dilp3 (Fig. 8C) in male dilp2G4>dRi, DN flies.
Fig 8.
Inhibition of dEGFR signaling reverses the effects of IPC-specific dCbl knockdown without affecting IPCs. (A) The brain IPCs were visualized using GFP in male adult dilp2G4>GFP, dilp2G4>GFP, dCbl-RNAi, and dilp2G4>GFP, dEGFR-DN flies at 3 days of age. Shown are the IPC bodies localized medially in the pars intercerebralis. (B) The body weight of male adult flies was measured under fed states (90 flies/group). (C) The mRNA abundance of dilp2, dilp3, or dilp5 in the head of male adult flies (90 flies/group, n > 3 independent experiments) was determined by qRT-PCR using RPL32 or α-Tub84B as the internal control for normalization. Data in panels B and C are shown as the mean ± SEM; *, P < 0.05 versus dilp2G4>+ control; #, P < 0.05 between dilp2G4>dCbl-RNAi and dilp2G4>dRi, DN by one-way ANOVA.
Taken together, these genetic interaction analyses demonstrate that dCbl suppresses dilp2 and dilp3 expression by modulating EGFR signaling in IPCs without affecting the growth or survival of IPCs. In addition, the inability of dEGFR-DN to affect the expression of dilp5 suggests that additional pathways may be involved in dCbl-mediated suppression of dILP production.
Mammalian c-Cbl regulates insulin production and secretion in INS-1 β cells.
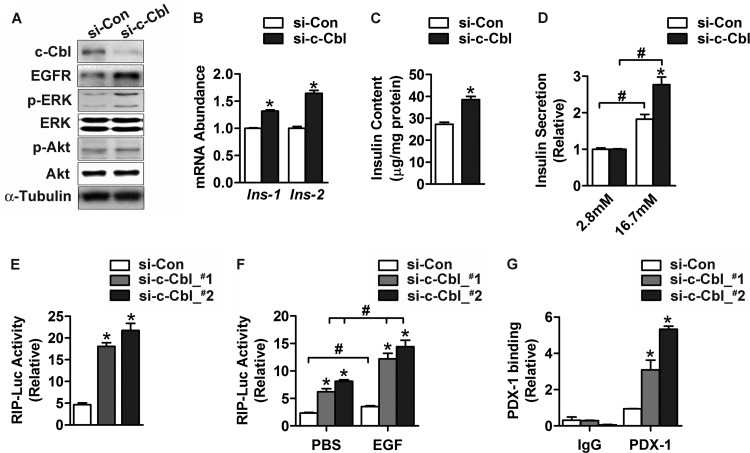
To determine whether dCbl regulation of dILP production is evolutionarily conserved, we examined the effect of suppression of c-Cbl expression on insulin production in rat insulinoma INS-1 cells. In accordance with the established role of c-Cbl in negatively regulating EGFR signaling, knockdown of c-Cbl by two siRNAs (si-c-Cbl) substantially increased the level of EGFR and the phosphorylation of ERK in INS-1 β cells (Fig. 9A). Importantly, knockdown of c-Cbl significantly increased the mRNA abundance of both Ins-1 and Ins-2 (Fig. 9B) as well as cellular insulin contents (by ∼40%) (Fig. 9C). Furthermore, knockdown of c-Cbl increased glucose-stimulated insulin secretion by ∼51% (from ∼1.83- to ∼2.77-fold) (Fig. 9D). To test whether c-Cbl inhibits insulin transcription, we cotransfected INS-1 cells with either of the two si-c-Cbl RNAs together with the plasmid expressing the luciferase reporter under the control of rat insulin 1 promoter (RIP). Suppression of c-Cbl expression resulted in 3- to 4-fold enhancement of the RIP transcriptional activity (Fig. 9E). While EGF indeed showed a significant stimulatory effect, c-Cbl knockdown substantially augmented the ability of EGF to increase the RIP transcriptional activity (Fig. 9F).
Fig 9.
Knockdown of c-Cbl expression enhances insulin production in INS-1 β cells. (A to D) INS-1 cells were maintained at 16.7 mM glucose and transfected for 48 h with an oligonucleotide control of random sequence (small interfering control [si-Con]) or two oligonucleotides (si-c-Cbl_#1 and si-c-Cbl_#2) that were directed against c-Cbl. (A) Protein abundance of c-Cbl and EGFR and phosphorylation levels of Akt or ERK were analyzed by Western immunoblotting using the indicated antibodies. α-Tubulin was used as a loading control. (B) The abundance of Ins-1 and Ins-2 mRNA was determined by qRT-PCR. GAPDH was used as an internal control for normalization. (C) Intracellular insulin content was measured by ELISA. For panels B and C, data are shown as the mean ± SEM (n = 3 independent experiments); *, P < 0.05 versus small interfering control by Student's t test. (D) Secreted insulin was measured by ELISA. Transfected cells were precultured at 2.8 mM glucose for 2 h and then subjected to stimulation by 16.7 mM glucose. Data represent the mean ± SEM (n = 3 independent experiments); *, P < 0.05 versus small interfering control; #, P < 0.05 versus values at 2.8 mM glucose by two-way ANOVA. (E and F) INS-1 cells cultured at 16.7 mM glucose were cotransfected for 48 h with small interfering control, si-c-Cbl_#1, or si-c-Cbl_#2, together with the plasmid expressing the RIP-Luc reporter. (E) Luciferase activities were directly measured from cell extracts. (F) Luciferase activities were determined after cells were preincubated in medium containing 2.8 mM glucose and 1% fetal bovine serum and then cultured with PBS or 200 ng/ml EGF for 6 h. Results are shown as the mean ± SEM (n = 3 independent experiments); *, P < 0.05 versus small interfering control; #, P < 0.05 versus PBS control by one-way ANOVA. (G) ChIP assays. INS-1 cells cultured at 16.7 mM glucose were transfected for 48 h with small interfering control, si-c-Cbl_#1, or si-c-Cbl_#2. Chromatin extracts were immunoprecipitated with anti-PDX-1 antibody or IgG. The precipitated genomic DNA was subjected to qPCR analysis in triplicate using the primers for the Ins-1 promoter. Cellular chromatin extract was used as the input control for normalization. Data are shown as the mean ± SEM (n = 3 independent experiments); *, P < 0.05 versus small interfering control by one-way ANOVA.
Insulin transcription is known to be upregulated by many transcription factors that are subject to modulation by ERK signaling (23), including PDX-1. We performed ChIP experiments to examine the effect of c-Cbl suppression on the binding of PDX-1 to the insulin promoter. Quantitative PCR assessment revealed significant increases of PDX-1 abundance bound to the Ins-1 promoter region as a result of c-Cbl knockdown in INS-1 cells that were transfected with si-c-Cbl RNAs (Fig. 9G).
In contrast to the observations from c-Cbl-knockdown cells, transient overexpression of the wild-type (WT) c-Cbl but not an E3 ligase-defective mutant (C381A) significantly reduced the transcriptional activity of RIP in INS-1 cells (Fig. 10A). Additionally, adenovirus-mediated overexpression of c-Cbl-WT but not c-Cbl-C381A decreased the insulin contents in INS-1 cells (Fig. 10B). Thus, these results suggest that c-Cbl also negatively regulates insulin biosynthesis and secretion through downregulating EGFR signaling in mammalian β cells, revealing a regulatory action of Cbl that is evolutionarily conserved.
Fig 10.
Effects of c-Cbl overexpression on insulin production in INS-1 β cells. (A) INS-1 cells maintained at 16.7 mM glucose were cotransfected for 48 h with the empty vector (control [Con]) or plasmids encoding the WT or an E3 ligase-deficient mutant (C381A) c-Cbl protein along with the RIP-Luc reporter. Cell lysates were analyzed by Western immunoblotting with the indicated antibodies. Luciferase activities were measured, and results are shown as the mean ± SEM (n = 3 independent experiments); *, P < 0.05 versus the control by one-way ANOVA. (B) INS-1 cells maintained at 16.7 mM glucose were infected for 48 h with control adenovirus-GFP (Ad-GFP; at an MOI of 20) or with adenoviruses expressing the indicated forms of c-Cbl protein (at an MOI of 20). Cell lysates were analyzed by immunoblotting, and intracellular insulin content was measured by ELISA. Values are shown as the mean ± SEM (n = 3 independent experiments); *, P < 0.05 compared to the adenovirus-GFP control by one-way ANOVA.
Activation of ERK mediates the effect of c-Cbl on insulin production in INS-1 cells.
Activation of ERKs constitutes a key signal transduction step in EGFR actions (65). To determine whether ERK mediates the c-Cbl action, we examined the effect of PD98059, an inhibitor of ERK-specific kinase MEK, in INS-1 cells. PD98059 blocked ERK phosphorylation, as expected (Fig. 11A); importantly, PD98059 also abolished c-Cbl knockdown-induced increases in insulin contents (Fig. 11B) and glucose-stimulated insulin secretion (Fig. 11C). Furthermore, inhibition of ERK (Fig. 11D) abrogated the ability of c-Cbl deficiency to enhance the RIP transcriptional activity (Fig. 11E). In contrast, blocking Akt phosphorylation by the PI3K inhibitor LY294002 (Fig. 11F) did not influence the effect of c-Cbl knockdown on the RIP activity (Fig. 11G). As ERKs have been reported to regulate the transcription of insulin genes (23, 68), these data indicate that c-Cbl controls insulin production through downregulation of ERK-mediated signaling in pancreatic β cells.
Fig 11.
c-Cbl affects insulin production in an ERK-dependent fashion in β cells. (A to C) INS-1 cells maintained at 16.7 mM glucose were transfected for 48 h with small interfering control or si-c-Cbl_#1 and si-c-Cbl_#2. (A and B) Transfected cells were treated for 2 h with dimethyl sulfoxide (DMSO) or 20 μM ERK-specific kinase MEK inhibitor PD98059. (A) Protein abundance of c-Cbl and EGFR as well as phosphorylation levels of Akt or ERK were analyzed by Western immunoblotting with the indicated antibodies. α-Tubulin was used as a loading control. (B) Intracellular insulin content was determined by ELISA. Data are shown as the mean ± SEM (n = 3 independent experiments); *, P < 0.05 versus small interfering control by two-way ANOVA. (C) Secreted insulin was measured by ELISA. Transfected cells were precultured at 2.8 mM glucose for 2 h and then subjected to stimulation by 16.7 mM glucose in the presence of dimethyl sulfoxide or 20 μM PD98059. Fold stimulation values are shown as the mean ± SEM (n = 3 independent experiments); *, P < 0.05 versus small interfering control by two-way ANOVA. (D to G) INS-1 cells cultured at 16.7 mM glucose were cotransfected for 48 h with the small interfering control, si-c-Cbl_#1, or si-c-Cbl_#2, along with the RIP-Luc reporter plasmid. Cells were then treated with dimethyl sulfoxide, 20 μM PD98059 (D and E), or 60 μM PI3K inhibitor LY294002 (F and G) for 16 h. Western immunoblot analysis for protein abundance of c-Cbl and phosphorylation levels of ERK (D) or Akt (F). Luciferase activities were measured from cell extracts (E and G). Results are shown as the mean ± SEM (n = 3 independent experiments); *, P < 0.05 versus small interfering control by one-way ANOVA.
DISCUSSION
In Drosophila, a single dCbl gene encodes two forms of dCbl proteins, which evolved into three homologues, c-Cbl, Cbl-b, and Cbl-c, in mammals (60). As multidomain adaptor proteins with intrinsic E3 ubiquitin ligase activities, c-Cbl and Cbl-b have been implicated in diverse physiological processes, displaying characteristics of functional redundancy under certain circumstances. However, our understanding of the detailed mechanisms that underlie the physiological actions of dCbl has been limited, largely due to the myriad of signaling pathways that are subject to the positive or negative influences of Cbl proteins. In the present study, utilization of the genetic models of Drosophila enabled us to examine the molecular evolution of Cbl actions. Our findings revealed the functional importance of dCbl in the IPCs in the brains of flies. dCbl cell autonomously inhibits the production of dILPs in IPCs and comprises an additional layer of coordination in insulin/IGF regulation of growth, carbohydrate metabolism, stress resistance, and longevity.
We found that dCbl in the IPCs of Drosophila controls the biosynthesis of dILPs. dILPs are the ligands of the insulin/IGF-like signaling pathway that regulates growth, development, metabolism, and life span (4, 5). We demonstrated that disruption of dCbl in neurons or IPCs resulted in upregulation of dILPs, increased body growth, higher sensitivity to oxidative stress or starvation, decreased levels of trehalose, and shortened life span. The longevity and metabolic phenotypes can be explained by increased dILP production and signaling. We also observed that c-Cbl, a mammalian homologue, similarly inhibited insulin production and secretion in rat pancreatic β cells. However, whole-body deletion of c-Cbl does not affect the blood insulin levels in mice (60), and abrogation of Cbl-b leads to an increase in insulin levels presumably due to insulin resistance resulting from adipose inflammation (18). Thus, both c-Cbl and Cbl-b are likely to suppress insulin expression and secretion, and deficiency of one form may be functionally compensated for by the other form in mice. Conditional gene-targeting studies in mouse models are needed to clarify this issue.
It has been well established that Cbl downregulates the EGFR signaling pathway via its E3 ubiquitin ligase activity in both insects and mammals (51, 55). EGFR signaling has been documented to play a crucial role in postnatal growth (34) as well as the expansion of pancreatic β-cell mass in response to feeding of a high-fat diet or during pregnancy (16). The Cbl family members likely negatively regulate insulin biosynthesis and secretion in both insects and mammals by downregulating the EGFR/ERK pathway via their E3 ubiquitin ligase activity. Cbl may also regulate the growth and proliferation of β cells by downregulating other RTK pathways, such as the platelet-derived growth factor (PDGF) receptor signaling pathway (7). PDGF signaling has been reported to be influenced by c-Cbl (35). Additional mechanisms may also contribute to the observed phenotypes of flies with dCbl deficiency in neurons or IPCs as well as the enhancement of insulin secretion in INS-1 cells.
The molecular mechanisms by which dilp genes are transcriptionally controlled are poorly understood in Drosophila. Whereas neuronal or IPC-specific dCbl suppression affected the expression of all three dilp genes examined, EGFR signaling was genetically coupled to the regulation of dilp2 and dilp3 but not dilp5. This further supports the notion that multiple signaling mechanisms may be involved in mediating dCbl regulation of dILP production. On an interesting note, distinct patterns of dilp gene expression have also been reported in a recent study that implicated dERK signaling in mediating SNF (short neuropeptide F) regulation of dilp expression within IPCs of Drosophila (25). In mammalian β cells, a number of transcription factors, such as PDX-1, MafA, and NFAT, act to regulate insulin expression in response to ERK activation (21, 24). While we observed a c-Cbl deficiency-elicited augmentation of PDX-1 binding to the insulin promoter in INS-1 cells, it remains unclear whether other factors are also implicated. In addition, it has yet to be defined whether transcription factors in the dEGFR pathway, e.g., PntP1 (13, 70), or a PDX-1 analogue can act to mediate dCbl suppression of dILP production in Drosophila. In this context, the molecular evolution of the regulatory factors involved in the transcriptional control of insulin/IGF1 represents an intriguing question that warrants more detailed investigations.
Recent studies suggest a common ancestral origin for both mammalian hypothalamic neurosecretory cells and the IPCs of flies (61). Global c-Cbl-knockout mice (37) and heterozygous mice expressing a C379A knock-in mutation within the RING finger domain of c-Cbl (38) all had higher energy expenditure and improved peripheral insulin sensitivity. Given the central importance of the hypothalamic circuitries in the control of whole-body energy balance (22), it is tempting to speculate that c-Cbl or Cbl-b may exert metabolic effects through modulating the hypothalamic neuroendocrine networks. Besides EGFR, neuronal Cbl may also target other signaling molecules, such as Src homology 2B (SH2B), that may interact with Cbl. We and others have previously shown that dSH2B, an adaptor protein that positively regulates insulin/IGF1 and leptin signaling pathways (40, 45, 46), plays crucial roles in metabolism, stress resistance, and longevity in flies (54). SH2B1, an SH2B family member, is required for the maintenance of normal body weight and glucose homeostasis in both humans and mice (2, 39). It is yet to be deciphered whether dCbl exerts its actions upon metabolism, stress tolerance, and life span through interacting with dSH2B to regulate the insulin/IGF1 pathway.
In summary, we show that knockdown of dCbl in neurons and IPCs results in increased dILP production and/or signaling activation, leading to reduced life span and stress tolerance. dCbl suppresses dILP production, at least in part, by downregulating the EGFR/ERK pathway through its E3 ligase activity. Moreover, Cbl suppression of insulin biosynthesis is evolutionarily conserved in mammalian β cells, suggesting that the Cbl pathway is critical for insulin production and secretion across animal species.
ACKNOWLEDGMENTS
We thank the Bloomington Drosophila Stock Center and Vienna Drosophila RNAi Center for providing the fly stocks and information and Eric Rulifson and M. German (UCSF) and Jian Zhang (University of Chicago) for the reagents. We also thank Jiansheng Kang and Wenxia Li (Institute for Nutritional Sciences) and Wei Song (Harvard Medical School) for technical assistance.
This work was supported by grants from the Ministry of Science and Technology (973 Program grants 2012CB524900 and 2011CB910900), the National Natural Science Foundation (grants 30970584, 81021002, 30988002, and 30830033), the Chinese Academy of Sciences (The Knowledge Innovation Programs, grants KSCX2-EW-R-09 and KSCX2-EW-Q-1-09, and the CAS/SAFEA International Partnership Program), and the Science and Technology Commission of the Shanghai Municipality (grant 10XD1406400) to Y.L. and W.L.
Y.Y., W.L., and Y.L. conceived and designed the experiments. Y.Y., Y.S., S.H., and C.Y. performed the experiments. Y.Y., Y.S., S.H., L.R., W.L., and Y.L. analyzed the data. Y.Y., L.R., W.L., and Y.L. wrote the paper.
Footnotes
Published ahead of print 9 July 2012
REFERENCES
- 1. Ahmed Z, Smith BJ, Pillay TS. 2000. The APS adapter protein couples the insulin receptor to the phosphorylation of c-Cbl and facilitates ligand-stimulated ubiquitination of the insulin receptor. FEBS Lett. 475: 31–34 [DOI] [PubMed] [Google Scholar]
- 2. Bachmann-Gagescu R, et al. 2010. Recurrent 200-kb deletions of 16p11.2 that include the SH2B1 gene are associated with developmental delay and obesity. Genet. Med. 12: 641–647 [DOI] [PubMed] [Google Scholar]
- 3. Barbieri M, Bonafe M, Franceschi C, Paolisso G. 2003. Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am. J. Physiol. Endocrinol. Metab. 285: E1064–E1071 [DOI] [PubMed] [Google Scholar]
- 4. Brogiolo W, et al. 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11: 213–221 [DOI] [PubMed] [Google Scholar]
- 5. Broughton SJ, et al. 2005. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. U. S. A. 102: 3105–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cao C, Brown MR. 2001. Localization of an insulin-like peptide in brains of two flies. Cell Tissue Res. 304: 317–321 [DOI] [PubMed] [Google Scholar]
- 7. Chen H, et al. 2011. PDGF signalling controls age-dependent proliferation in pancreatic [bgr]-cells. Nature 478: 349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng CL, Gao TQ, Wang Z, Li DD. 2005. Role of insulin/insulin-like growth factor 1 signaling pathway in longevity. World J. Gastroenterol. 11: 1891–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi Y, et al. 2002. PTEN, but not SHIP and SHIP2, suppresses the PI3K/Akt pathway and induces growth inhibition and apoptosis of myeloma cells. Oncogene 21: 5289–5300 [DOI] [PubMed] [Google Scholar]
- 10. Duan L, Reddi AL, Ghosh A, Dimri M, Band H. 2004. The Cbl family and other ubiquitin ligases: destructive forces in control of antigen receptor signaling. Immunity 21: 7–17 [DOI] [PubMed] [Google Scholar]
- 11. Ettenberg SA, et al. 1999. cbl-b inhibits EGF-receptor-induced apoptosis by enhancing ubiquitination and degradation of activated receptors. Mol. Cell Biol. Res. Commun. 2: 111–118 [DOI] [PubMed] [Google Scholar]
- 12. Freeman M. 1996. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87: 651–660 [DOI] [PubMed] [Google Scholar]
- 13. Gabay L, et al. 1996. EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development 122: 3355–3362 [DOI] [PubMed] [Google Scholar]
- 14. Galisteo ML, Dikic I, Batzer AG, Langdon WY, Schlessinger J. 1995. Tyrosine phosphorylation of the c-cbl proto-oncogene protein product and association with epidermal growth factor (EGF) receptor upon EGF stimulation. J. Biol. Chem. 270: 20242–20245 [DOI] [PubMed] [Google Scholar]
- 15. Giannakou ME, Partridge L. 2007. Role of insulin-like signalling in Drosophila lifespan. Trends Biochem. Sci. 32: 180–188 [DOI] [PubMed] [Google Scholar]
- 16. Hakonen E, et al. 2011. Epidermal growth factor (EGF)-receptor signalling is needed for murine beta cell mass expansion in response to high-fat diet and pregnancy but not after pancreatic duct ligation. Diabetologia 54: 1735–1743 [DOI] [PubMed] [Google Scholar]
- 17. Hime GR, Dhungat MP, Ng A, Bowtell DD. 1997. D-Cbl, the Drosophila homologue of the c-Cbl proto-oncogene, interacts with the Drosophila EGF receptor in vivo, despite lacking C-terminal adaptor binding sites. Oncogene 14: 2709–2719 [DOI] [PubMed] [Google Scholar]
- 18. Hirasaka K, et al. 2007. Deficiency of Cbl-b gene enhances infiltration and activation of macrophages in adipose tissue and causes peripheral insulin resistance in mice. Diabetes 56: 2511–2522 [DOI] [PubMed] [Google Scholar]
- 19. Huang F, Gu H. 2008. Negative regulation of lymphocyte development and function by the Cbl family of proteins. Immunol. Rev. 224: 229–238 [DOI] [PubMed] [Google Scholar]
- 20. Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. 2002. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 12: 1293–1300 [DOI] [PubMed] [Google Scholar]
- 21. Khoo S, et al. 2003. Regulation of insulin gene transcription by ERK1 and ERK2 in pancreatic beta cells. J. Biol. Chem. 278: 32969–32977 [DOI] [PubMed] [Google Scholar]
- 22. Lam TK. 2010. Neuronal regulation of homeostasis by nutrient sensing. Nat. Med. 16: 392–395 [DOI] [PubMed] [Google Scholar]
- 23. Lawrence M, Shao C, Duan L, McGlynn K, Cobb MH. 2008. The protein kinases ERK1/2 and their roles in pancreatic beta cells. Acta Physiol. 192: 11–17 [DOI] [PubMed] [Google Scholar]
- 24. Lawrence MC, McGlynn K, Park BH, Cobb MH. 2005. ERK1/2-dependent activation of transcription factors required for acute and chronic effects of glucose on the insulin gene promoter. J. Biol. Chem. 280: 26751–26759 [DOI] [PubMed] [Google Scholar]
- 25. Lee KS, et al. 2008. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat. Cell Biol. 10: 468–475 [DOI] [PubMed] [Google Scholar]
- 26. Levkowitz G, et al. 1999. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 4: 1029–1040 [DOI] [PubMed] [Google Scholar]
- 27. Levkowitz G, et al. 1998. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 12: 3663–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li L, et al. 2011. Cbl-regulated Akt and ERK signals are involved in beta-elemene-induced cell apoptosis in lung cancer cells. Mol. Med. Report 4: 1243–1246 [DOI] [PubMed] [Google Scholar]
- 29. Liu G, Rogers J, Murphy CT, Rongo C. 2011. EGF signalling activates the ubiquitin proteasome system to modulate C. elegans lifespan. EMBO J. 30: 2990–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu J, DeYoung SM, Hwang JB, O'Leary EE, Saltiel AR. 2003. The roles of Cbl-b and c-Cbl in insulin-stimulated glucose transport. J. Biol. Chem. 278: 36754–36762 [DOI] [PubMed] [Google Scholar]
- 31. Liu J, Kimura A, Baumann CA, Saltiel AR. 2002. APS facilitates c-Cbl tyrosine phosphorylation and GLUT4 translocation in response to insulin in 3T3-L1 adipocytes. Mol. Cell. Biol. 22: 3599–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meisner H, et al. 1997. Interactions of Drosophila Cbl with epidermal growth factor receptors and role of Cbl in R7 photoreceptor cell development. Mol. Cell. Biol. 17: 2217–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miettinen P, Ormio P, Hakonen E, Banerjee M, Otonkoski T. 2008. EGF receptor in pancreatic beta-cell mass regulation. Biochem. Soc. Trans. 36: 280–285 [DOI] [PubMed] [Google Scholar]
- 34. Miettinen PJ, et al. 2006. Downregulation of EGF receptor signaling in pancreatic islets causes diabetes due to impaired postnatal beta-cell growth. Diabetes 55: 3299–3308 [DOI] [PubMed] [Google Scholar]
- 35. Miyake S, Mullane-Robinson KP, Lill NL, Douillard P, Band H. 1999. Cbl-mediated negative regulation of platelet-derived growth factor receptor-dependent cell proliferation. A critical role for Cbl tyrosine kinase-binding domain. J. Biol. Chem. 274: 16619–16628 [DOI] [PubMed] [Google Scholar]
- 36. Moghal N, Sternberg PW. 1999. Multiple positive and negative regulators of signaling by the EGF-receptor. Curr. Opin. Cell Biol. 11: 190–196 [DOI] [PubMed] [Google Scholar]
- 37. Molero JC, et al. 2004. c-Cbl-deficient mice have reduced adiposity, higher energy expenditure, and improved peripheral insulin action. J. Clin. Invest. 114: 1326–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Molero JC, et al. 2006. Genetic ablation of the c-Cbl ubiquitin ligase domain results in increased energy expenditure and improved insulin action. Diabetes 55: 3411–3417 [DOI] [PubMed] [Google Scholar]
- 39. Morris DL, Cho KW, Rui L. 2010. Critical role of the Src homology 2 (SH2) domain of neuronal SH2B1 in the regulation of body weight and glucose homeostasis in mice. Endocrinology 151: 3643–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morris DL, Cho KW, Zhou Y, Rui L. 2009. SH2B1 enhances insulin sensitivity by both stimulating the insulin receptor and inhibiting tyrosine dephosphorylation of insulin receptor substrate proteins. Diabetes 58: 2039–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pai LM, Barcelo G, Schupbach T. 2000. D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell 103: 51–61 [DOI] [PubMed] [Google Scholar]
- 42. Pai LM, et al. 2006. Differential effects of Cbl isoforms on Egfr signaling in Drosophila. Mech. Dev. 123: 450–462 [DOI] [PubMed] [Google Scholar]
- 43. Qiu Y, et al. 2010. A crucial role for RACK1 in the regulation of glucose-stimulated IRE1alpha activation in pancreatic beta cells. Sci. Signal. 3: ra7 doi:10.1126/scisignal.2000514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rao N, Dodge I, Band H. 2002. The Cbl family of ubiquitin ligases: critical negative regulators of tyrosine kinase signaling in the immune system. J. Leukoc. Biol. 71: 753–763 [PubMed] [Google Scholar]
- 45. Ren D, Li M, Duan C, Rui L. 2005. Identification of SH2-B as a key regulator of leptin sensitivity, energy balance, and body weight in mice. Cell Metab. 2: 95–104 [DOI] [PubMed] [Google Scholar]
- 46. Ren D, et al. 2007. Neuronal SH2B1 is essential for controlling energy and glucose homeostasis. J. Clin. Invest. 117: 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robertson H, Hime GR, Lada H, Bowtell DD. 2000. A Drosophila analogue of v-Cbl is a dominant-negative oncoprotein in vivo. Oncogene 19: 3299–3308 [DOI] [PubMed] [Google Scholar]
- 48. Rulifson EJ, Kim SK, Nusse R. 2002. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296: 1118–1120 [DOI] [PubMed] [Google Scholar]
- 49. Saltiel AR, Pessin JE. 2003. Insulin signaling in microdomains of the plasma membrane. Traffic 4: 711–716 [DOI] [PubMed] [Google Scholar]
- 50. Schlessinger J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103: 211–225 [DOI] [PubMed] [Google Scholar]
- 51. Schmidt MH, Dikic I. 2005. The Cbl interactome and its functions. Nat. Rev. Mol. Cell Biol. 6: 907–918 [DOI] [PubMed] [Google Scholar]
- 52. Sehat B, Andersson S, Girnita L, Larsson O. 2008. Identification of c-Cbl as a new ligase for insulin-like growth factor-I receptor with distinct roles from Mdm2 in receptor ubiquitination and endocytosis. Cancer Res. 68: 5669–5677 [DOI] [PubMed] [Google Scholar]
- 53. Slack C, et al. 2010. Regulation of lifespan, metabolism, and stress responses by the Drosophila SH2B protein, Lnk. PLoS Genet. 6: e1000881 doi:10.1371/journal.pgen.1000881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Song W, et al. 2010. SH2B regulation of growth, metabolism, and longevity in both insects and mammals. Cell Metab. 11: 427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Swaminathan G, Tsygankov AY. 2006. The Cbl family proteins: ring leaders in regulation of cell signaling. J. Cell. Physiol. 209: 21–43 [DOI] [PubMed] [Google Scholar]
- 56. Taguchi A, White MF. 2008. Insulin-like signaling, nutrient homeostasis, and life span. Annu. Rev. Physiol. 70: 191–212 [DOI] [PubMed] [Google Scholar]
- 57. Tanaka S, Neff L, Baron R, Levy JB. 1995. Tyrosine phosphorylation and translocation of the c-cbl protein after activation of tyrosine kinase signaling pathways. J. Biol. Chem. 270: 14347–14351 [DOI] [PubMed] [Google Scholar]
- 58. Taniguchi CM, Emanuelli B, Kahn CR. 2006. Critical nodes in signalling pathways: insights into insulin action. Nat. Rev. Mol. Cell Biol. 7: 85–96 [DOI] [PubMed] [Google Scholar]
- 59. Tatar M, Bartke A, Antebi A. 2003. The endocrine regulation of aging by insulin-like signals. Science 299: 1346–1351 [DOI] [PubMed] [Google Scholar]
- 60. Thien CBF, Langdon WY. 2001. Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2: 294–307 [DOI] [PubMed] [Google Scholar]
- 61. Wang S, Tulina N, Carlin DL, Rulifson EJ. 2007. The origin of islet-like cells in Drosophila identifies parallels to the vertebrate endocrine axis. Proc. Natl. Acad. Sci. U. S. A. 104: 19873–19878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang Y, Chen Z, Bergmann A. 2010. Regulation of EGFR and Notch signaling by distinct isoforms of D-cbl during Drosophila development. Dev. Biol. 342: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang Y, et al. 2008. Drosophila cbl is essential for control of cell death and cell differentiation during eye development. PLoS One 3: e1447 doi:10.1371/journal.pone.0001447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Waterman H, Levkowitz G, Alroy I, Yarden Y. 1999. The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J. Biol. Chem. 274: 22151–22154 [DOI] [PubMed] [Google Scholar]
- 65. Wetzker R, Bohmer FD. 2003. Transactivation joins multiple tracks to the ERK/MAPK cascade. Nat. Rev. Mol. Cell Biol. 4: 651–657 [DOI] [PubMed] [Google Scholar]
- 66. Yang L, et al. 2010. Deficiency in RNA editing enzyme ADAR2 impairs regulated exocytosis. FASEB J. 24: 3720–3732 [DOI] [PubMed] [Google Scholar]
- 67. Yoon CH, Lee J, Jongeward GD, Sternberg PW. 1995. Similarity of sli-1, a regulator of vulval development in C. elegans, to the mammalian proto-oncogene c-cbl. Science 269: 1102–1105 [DOI] [PubMed] [Google Scholar]
- 68. Zhang SS, et al. 2009. Coordinated regulation by Shp2 tyrosine phosphatase of signaling events controlling insulin biosynthesis in pancreatic beta-cells. Proc. Natl. Acad. Sci. U. S. A. 106: 7531–7536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang W, Thompson BJ, Hietakangas V, Cohen SM. 2011. MAPK/ERK signaling regulates insulin sensitivity to control glucose metabolism in Drosophila. PLoS Genet. 7: e1002429 doi:10.1371/journal.pgen.1002429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhu S, Barshow S, Wildonger J, Jan LY, Jan YN. 2011. Ets transcription factor Pointed promotes the generation of intermediate neural progenitors in Drosophila larval brains. Proc. Natl. Acad. Sci. U. S. A. 108: 20615–20620 [DOI] [PMC free article] [PubMed] [Google Scholar]