Abstract
Functional redundancy is a pivotal mechanism that supports the robustness of biological systems at a molecular, cellular, and organismal level. The extensive prevalence of redundancy in molecular networks has been highlighted by recent systems biology studies; however, a detailed mechanistic understanding of redundant functions in specific signaling modules is often missing. We used affinity purification of protein complexes coupled to tandem mass spectrometry to generate a high-resolution protein interaction map of the three homologous p38 mitogen-activated protein kinases (MAPKs) in Drosophila and assessed the utility of such a map in defining the extent of common and unique functions. We found a correlation between the depth of integration of individual p38 kinases into the protein interaction network and their functional significance in cultured cells and in vivo. Based on these data, we propose a central role of p38b in the Drosophila p38 signaling module, with p38a and p38c playing more peripheral, auxiliary roles. We also present the first in vivo evidence demonstrating that an evolutionarily conserved complex of p38b with glycogen synthase links stress sensing to metabolic adaptation.
INTRODUCTION
The p38 mitogen-activated protein kinase (MAPK) pathway is a key evolutionarily conserved mediator of an organism's response to stressful environmental stimuli (reviewed in references 13 and 31). Similar to other members of the canonical MAPK family, p38 kinases are activated as part of a conserved three-tier kinase cascade by dual phosphorylation of the threonine and tyrosine residues in the activation loop. Active p38 kinases phosphorylate a wide spectrum of both nuclear and cytoplasmic substrates, including a limited number of subordinate kinases (e.g., MAPK-activated protein kinase 2 [MK2], p38-regulated/activated protein kinase [PRAK], and mitogen and stress-activated kinase 1 [MSK1] and MSK2), thus extending the cascade past the MAPK level (reviewed in reference 34). Through its substrates, p38 regulates a range of cellular processes, such as transcription (Mef2A [10], Mef2C, and ATF1, -2, and -6), cell cycle (cyclin D1 and cyclin-dependent kinase [CDK] inhibitors), proliferation (epidermal growth factor receptor [EGFR] and fibroblast growth factor receptor 1 [FGFR1]), protein turnover (Siah2), and apoptosis (p53 and Bcl-2) (reviewed in references 12 and 40). Given the extent and complexity of the resulting regulatory network, it is not surprising that the effects of p38 signaling vary drastically, depending on cell type, cellular microenvironment, and the nature and duration of stressful stimuli. Heterogeneity of the p38 signaling network becomes even more elaborate as most cells express more than one member of the p38 family, suggesting the possibility of both redundant and unique upstream regulatory mechanisms and downstream substrate pools.
In mammals, the p38 family consists of four genes, coding for p38α, -β, -γ, and -δ. p38α mRNA can undergo alternative splicing that produces additional isoforms of the kinase, including Mxi2 (5), Exip (49), and CSBP1 (26). Gene-targeting studies in mice are typically interpreted to suggest some degree of functional compensation between the four p38 kinases. Indeed, mice carrying null alleles of p38β (1), -γ (36), and -δ (36, 44) lack easily discernible phenotypes, suggesting that most, if not all, critical functions of these kinases can be carried out by ubiquitously expressed p38α. Nonetheless, evidence for the unique activities of various p38 isoforms is also beginning to accumulate. For instance, p38γ appears to antagonize differentiation of satellite cells during adult skeletal muscle regeneration (20). Conversely, p38α is a well-established activator of the myogenic differentiation program in a variety of cellular contexts, including normal development (23) and differentiation of rhabdomyosarcoma cells (33).
Clearly, a systems biology approach would provide a more complete view of redundant and unique functions of p38 family kinases within a given cellular context. We sought to examine whether protein-protein interaction (PPI) mapping using affinity purification coupled to mass spectrometry (AP-MS) could yield initial data of sufficient resolution to define the extent of the regulatory network(s) in this family of closely related kinases. We chose to test this approach using Drosophila melanogaster as a well-defined model system. AP-MS has been successfully used to build proteome-scale interaction maps in yeast (Saccharomyces cerevisiae) (18, 24), and Drosophila (22). However, large-scale data sets often lack the resolution necessary to firmly establish interaction preferences within families of closely related proteins.
The fly p38 module is much simpler compared to its mammalian counterpart. The family consists of three highly homologous proteins, p38a, -b, and -c, and only the first two members are believed to be bona fide p38 kinases. Coding sequences of all three kinases are contained within a single exon, eliminating the possibility of the added complexity of alternatively spliced variants. Previous genetic analyses firmly established the roles of p38a and p38b in stress response and antimicrobial immunity (6, 11). More recent studies began to reveal the roles of p38 signaling in other aspects of Drosophila development, including cell size regulation (14), epigenetic inheritance (38), and life span (47). As both p38a and p38b mutants are viable under normal conditions, whereas double mutants die at the larval and pupal stages, it has been suggested that the two kinases exhibit functional compensation, although an alternative possibility (i.e., synthetic lethality) has not been ruled out. The AP-MS interaction map presented in this study reveals that the three p38 proteins share a limited number of common interactors, arguing against extensive redundancy and suggesting nonoverlapping function. Moreover, the PPI map demonstrates that only p38b forms a high-affinity complex with evolutionarily conserved binding partners, such as MK2 and glycogen synthase, suggesting the central role of this kinase in the Drosophila p38 signaling module.
MATERIALS AND METHODS
Plasmid construction.
A plasmid for constitutive expression in S2 cells was generated by inserting a PCR-amplified 2.5-kb Ac5C promoter into the SacI and NotI sites of the pNTAP-A vector (Stratagene), followed by the removal of the tandem affinity purification (TAP) tag and the insertion of the 3×FLAG tag or hemagglutinin (HA) tag between NotI and BamHI sites. Total mRNA was extracted from S2 cells using TRIzol reagent (Invitrogen) and used for cDNA generation using the SuperScript III kit (Invitrogen). The following primers were used to PCR amplify open reading frames with S2 cDNA as the template (restriction enzyme sites are underlined): for p38a, CTAGGATCCATGTCAGTGTCCATTACAAA and CTAGGATCCTCACTTTACATCCTTTAGAAC; for p38b, CTAGGATCCATGTCGCGCAAAATGGCCAA and CTAGGATCCTTACTGCTCTTTGGGCAGGAG; for p38c, CTAGGATCCATGCCGGAGTTCGTGAGAG and CTAGGATCCTTAAAAATGCAAATCAAGTTGG; for myocyte-specific enhancer factor 2 (Mef2), TAGAATTCATGGGCCGCAAAAAAATTCA and ACAAGCTTCTATGTGCCCCATCCGCC; for MAPK-activated protein kinase 2 (MK2), ATAGCGGCCGCAATGCTTTCTCTGCAGAATC and AGAAAGCTTCTAGTTGCGCGTCGCATTG; for licorne (lic), ATAGCGGCCGCAATGTCCAAACGCCACCGC and AGAAAGCTTCTACTGCGCCGGCTGCGC; for glycogen synthase (GS), ATAGCGGCCGCAATGAATCGTCGCTTTTCGAG and AGAAAGCTTCTACTTAATCCCCAATTCC; for glycogenin (GYG), ATAGCGGCCGCAATGAGCAAATTCGCTTGGG and ACAGAATTCCTATGAGCTACCAGTTTCTTC; and for Caliban (Clbn), ATAGCGGCCGCAATGAAGACACGTTTCAATACC and ACAGAATTCTTATTTGTGATACTTCTGAAG. Digested PCR products were inserted into the respective sites of the Ac5C expression vector, and all constructs were sequenced.
Cell culture and transfections.
S2 cells were maintained as semiadherent cultures at 22°C in HyClone SFX serum-free insect medium (Thermo Scientific) supplemented with 5% heat-inactivated fetal bovine serum (FBS) (Sigma), 100 U/ml penicillin, and 100 μg/ml streptomycin. Plasmids were prepared using QIAfilter Mega kit (Qiagen) and transfected into cells at 50 μg per 15-cm plate using a standard calcium phosphate protocol.
Western blotting, subcellular fractionation, and co-IP.
Standard procedures were followed for SDS-PAGE and Western blotting. The following antibodies were used: phospho-p38 MAPK (Thr180 Tyr182) (Cell Signaling), Drosophila p38 dN-20 (Santa Cruz Biotechnology), FLAG M2 (Sigma), HA-tagged rabbit monoclonal antibody (Cell Signaling), actin I-19-R (Santa Cruz Biotechnology), and histone H3 (Cell Signaling). Subcellular fractionation of S2 cells was performed using the NE-PER nuclear and cytoplasmic extraction kit (Thermo Scientific). Immediately following the extraction, fractions were boiled in the presence of SDS-PAGE sample buffer. Coimmunoprecipitations (co-IPs) were done using one 15-cm plate of transfected cells per assay. Cells were lysed in the following buffer: 50 mM HEPES [pH 8.0], 100 mM KCl, 2 mM EDTA, 10% glycerol, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.1% NP-40, and protease inhibitor cocktail (Sigma) used at a 1:500 dilution. Lysates were incubated with anti-FLAG (M2) affinity beads (Sigma) at 4°C for 2 h, washed with lysis buffer, and boiled in the SDS-PAGE buffer to elute bound proteins.
RNAi in S2 cells and MTT assay.
For RNA interference (RNAi), cDNA from S2 cells (see “Plasmid construction”) was used to amplify double-stranded RNA (dsRNA) templates using the following primer pairs (with the T7 promoter sequence shown in lowercase): taatacgactcactatagggaGGCCGAACTAATTACCAGAC and taatacgactcactatagggaTCACTTTACATCCTTTAGAAC for p38a, taatacgactcactatagggaAAGCTTGCCAGGCCCTTCC and taatacgactcactatagggaCCGCCGTCTGGTTGTAGTG for p38b, taatacgactcactatagggaATGCCGGAGTTCGTGAGAG and taatacgactcactatagggaTTAAAAATGCAAATCAAGTTGG for p38c, and taatacgactcactatagggaATGGTGAGCAAGGGCGAGG and taatacgactcactatagggaGAACATGTCGAGCAGGTACG for green fluorescent protein (GFP). The resulting PCR products were used to synthesize dsRNAs using the T7 MEGAscript kit (Ambion). dsRNAs were treated with DNase, precipitated with LiCl, and quantified according to the manufacturer's protocol. S2 cells were seeded in 96-well plates in standard medium and allowed to attach for 3 h. Medium was then replaced with HyClone SFX (Thermo) without serum or antibiotics, and 2 μg of dsRNA were added per well. After a 3-h incubation, the serum concentration was restored, and cells were incubated for 72 h. Cells were then exposed to 37°C heat shock or 200 μM NaAsO2 and subsequently incubated with 10 μl of the MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide) reagent (ATCC) for 3 h. Medium was removed, and the dye was extracted from cells with 100 μl dimethyl sulfoxide (DMSO) by repeated pipetting and quantified by measuring the optical density at 570 nm (OD570).
Affinity purification and LC-MS/MS.
The AP-MS protocol was carried out as previously described (21), with the following modifications. Four 15-cm plates with S2 cells transfected with the bait-containing or empty vector were used per purification. Cells were harvested 72 to 96 h after transfection by centrifugation at 1,800 × g, and cell pellets were weighed. Cells were lysed in lysis buffer at a ratio of 1:5 (g cell pellet per ml buffer) by one freeze-thaw cycle on dry ice. Centrifugation-clarified lysates were incubated with the FLAG M2 beads at 4°C for 2 h. Beads were washed as described previously (21), and isolated complexes were digested using trypsin singles (Sigma) directly on beads for 4 h at 37°C. Proteins released by this initial digestion were separated from the beads and subjected to an additional overnight digestion at 37°C. Peptides were acidified with formic acid and loaded onto 75-μm-inside-diameter columns (Polymicro Technologies) manually packed with Zorbax 300SB C18 matrix (Agilent). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was carried out on a ThermoFinnigan LTQ mass spectrometer.
MS data analysis.
Acquired RAW files were converted to the mgf format and searched using Mascot software (Matrix Sciences) with the following settings: Drosophila RefSeq database (release 44), precursor ion mass tolerance of 3.0 Da, fragment ion mass tolerance of 0.6 Da, methionine oxidation and asparagine deamidation permitted as variable modifications, and a single missed trypsin cleavage site per peptide allowed. All MS data were deposited in the Tranche repository. Mascot-identified proteins were analyzed using the Analyst module of ProHits (28) to match the lists of proteins present in the p38 bait samples with those from the control sample set. The resulting list of matched protein identifications (IDs) was exported from ProHits as an Excel file (see Data set S1 in the supplemental material). All proteins present in both in the p38 bait samples and control samples were discarded, regardless of the Mascot score. The remaining proteins that were uniquely present in the p38 bait samples were considered specific interactor candidates. Only the candidates found in more than one replicate, from either the same or different baits, were analyzed further. Finally, we removed the candidates with Mascot scores below 50 and those identified by a single short (<10-amino-acid-long) peptide.
Fly stocks and crosses.
Fly stocks were maintained at 25°C on semidefined medium prepared according to the Bloomington Drosophila Stock Center (BDSC) recipe. All fly stocks for the RNAi experiments were obtained from BDSC, unless otherwise indicated. The gut-specific Gal4 driver line had the following genotype: P{w+mC = UAS-Dcr-2.D }, w1118; P{w+mW.hs = GawB}c601c601. The following RNAi responder lines were used: for p38a, y1 v1; P{y+t7.7 v+t1.8 = TRiP.JF02625}attP2, y1 sc* v1; P{y+t7.7 v+t1.8 = TRiP.HMS01224}attP2, and y1 sc* v1; P{y+t7.7 v+t1.8 = TRiP.GL00131}attP2 lines; for p38b, y1 v1; P{y+t7.7 v+t1.8 = TRiP.JF03341}attP2 and y1 sc* v1; P{y+t7.7 v+t1.8 = TRiP.GL00140}attP2 lines and VDRC line 108099; for p38c, VDRC line 105173; for GFP a y1 v1; P{y+t7.7 v+t1.8 = UAS-GFP.VALIUM10}attP2 line. Driver-carrying females were crossed with responder males, and the third-instar larvae were collected for stress survival tests. Canton-S, w1118, and Mpk1 flies (11) were obtained from BDSC; p38bΔ25, p38bΔ45, and the precise excision control line are described in reference 47.
Glycogen quantification assay.
Five larvae for each genotype-treatment condition combination were homogenized in 200 μl phosphate-buffered saline (PBS), heated at 85°C for 5 min to inactivate endogenous enzymes, and cleared by centrifugation at 14,000 × g for 5 min. Glycogen quantification was performed as described in reference 32. Briefly, glucose levels in larval extracts were measured using the GO glucose assay kit (Sigma) in the presence or absence of 1 U amyloglucosidase (Sigma) per 50 μl extract. Glycogen levels were determined by subtracting the glucose measurements in samples untreated with amyloglucosidase from those treated with the enzyme.
Statistical analysis.
All in vivo and cell culture experiments were performed in triplicate, and 200 to 300 larvae per test were scored in survival assays. One-way analysis of variance (ANOVA) and Dunnett’s test were used to compare values among various genotype-treatment combinations. A P value of <0.05 was considered to represent a statistically significant difference.
RESULTS
Distinct activation patterns of the Drosophila p38 MAPKs.
p38 MAPKs are known to be activated by a wide variety of stressful stimuli, and dual phosphorylation of the TGY tripeptide motif is believed to be essential for maximal activity. We tested six physiologically relevant, yet mechanistically distinct, stresses to determine activation profiles of the p38a and p38b MAPKs. As the two proteins comigrate on SDS-PAGE gels and the residues surrounding the TGY site are highly conserved, the phosphorylation analysis of endogenous kinases was not feasible. To circumvent this limitation, we generated plasmid constructs containing a constitutive actin 5C promoter driving the expression of the N-terminally 3×FLAG-tagged p38a, -b, and -c. We reasoned that the addition of a short 3×FLAG sequence would be unlikely to perturb the molecular function of p38 and would be sufficient to produce species of slower electrophoretic mobility. Because in p38c the canonical dual-phosphorylation TGY motif is replaced by the TDH sequence that is not recognized by the available phospho-p38 antibody, this kinase was excluded from stress-induced activation experiments.
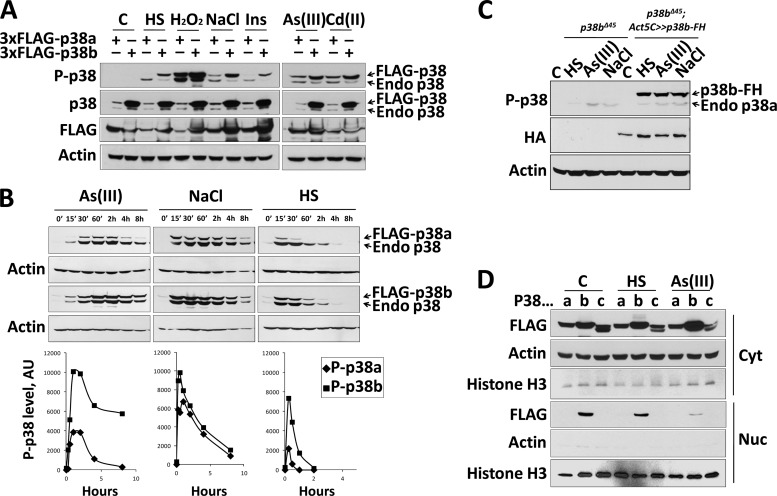
Schneider S2 cells transfected with the FLAG-p38 constructs were subjected to various stressful stimuli, and the activation state of individual kinases was measured using an antibody against a doubly phosphorylated TGY peptide (Fig. 1A). All six tested stimuli robustly activated FLAG-p38b. In contrast, FLAG-p38a was not activated by the 37°C heat shock or insulin treatment, and its activation by osmotic stress was noticeably weaker than that of p38b. Interestingly, the three stimuli that strongly activated p38a [i.e., hydrogen peroxide and metal ions As(III) and Cd(II)] are known to cause oxidative stress via the induction of intracellular reactive oxygen species (35).
Fig 1.
Drosophila p38 kinases respond differently to stressful stimuli. (A) S2 cells transiently transfected with 3×FLAG-tagged p38a and p38b were stimulated with 37°C heat shock, 10 mM H2O2, 200 mM NaCl, 10 μg/ml insulin, 1 mM Cd(CH3COO)2, and 200 μM NaAsO2. The duration of all treatments was 45 min. Note that the total p38 antibody displays significantly higher affinity for p38b than p38a. FLAG antibody was used to confirm comparable expression levels of tagged kinases upon different treatments. (B) S2 cells transfected with 3×FLAG-tagged p38a and p38b were treated with 37°C heat shock, 200 mM NaCl, and NaAsO2 and harvested at the indicated time points. Western blots were probed by phospho-p38 antibody, and bands corresponding to dual-phosphorylation-tagged p38 were quantified and plotted below. The lower bands reflecting the phosphorylation status of endogenous p38a and p38b were used as an internal control to ensure consistency of treatments between transfected sets. AU, arbitrary units. (C) The activation profile of p38a is unaffected in the presence of overexpressed p38b. Homozygous p38b mutant third-instar larvae (p38bΔ45-null allele) and the counterparts ubiquitously expressing p38b-FLAG-HA were subjected to the following conditions: C, control normal food; HS, 37°C heat shock; As(III), food supplemented with 200 μM NaAsO2; NaCl, food supplemented with 400 mM NaCl. Activation of the endogenous p38a and overexpressed p38b-FH was examined by Western blotting using the phospho-specific p38 antibody. (D) Drosophila p38 isoforms display distinct patterns of nucleocytolasmic distribution. S2 cells transiently transfected with 3×FLAG-tagged p38a, p38b, and p38c were stimulated for 45 min with a 37°C heat shock or 200 μM NaAsO2 and subsequently fractionated into cytoplasmic (Cyto) and nuclear (Nuc) fractions along with untreated control (C) cells. The efficiency of fractionation of each sample was judged by assaying the levels of actin and histone H3, cytoplasmic and nuclear markers, respectively. Cross-contamination of the two fractions was determined to be less than 10%.
We next examined the dynamics of p38 activation in S2 cells in response to oxidative stress [As(III)], osmotic shock (NaCl), and heat treatment. As described above, cells were transfected with the FLAG-tagged p38 constructs and analyzed by Western blotting using the phospho-p38 antibody. Subsequent band quantification revealed pronounced differences in the levels and duration of stress-induced activation of the two kinases (Fig. 1B). p38b was more responsive to oxidative stress and heat shock, and its activation state appeared to persist for a longer period of time. For instance, As(III) stimulation only transiently activated p38a, and its phosphorylation level at 4 h was near the background level. In contrast, the same treatment potently activated p38b, with activity persisting past the 8-h time point. Interestingly, in the case of osmotic shock the activation level and the dynamics of the phosphorylated state were almost indistinguishable between the two kinases (Fig. 1B). Overall, these results demonstrate that p38a and p38b share some, but not all, modes of activation.
The observed differences in the activation profiles of the two p38 kinases may be due to competition for upstream activators. To test this possibility in vivo, we examined whether the activation pattern of p38a, a potentially weaker competitor, could be affected by the presence of a more robust p38b. p38b-null third-instar larvae were subjected to heat shock, oxidative stress, and osmotic shock, and the levels of phosphorylated p38a were measured by Western blotting. In a parallel set of experiments, p38b-null larvae ubiquitously expressing a tagged version of p38b were treated with the same stressful stimuli (Fig. 1C). The extent of p38a activation was the same regardless of the presence of p38b, arguing against direct competition between the two kinases.
Nucleocytoplasmic distribution of the Drosophila p38 kinases.
As mammalian p38 MAPKs are known to phosphorylate both cytoplasmic and nuclear targets, subcellular localization is recognized as an important aspect of p38 regulation and function. To compare the nucleocytoplasmic distributions of the fly p38 MAPKs, we expressed the FLAG-tagged isoforms in S2 cells, followed by stimulation with heat shock or oxidative stress and subcellular fractionation (Fig. 1D). We found that only p38b is present both in the cytoplasm and the nucleus in unstimulated cells and that p38b is exported out of the nucleus upon heat shock and more profoundly under oxidative stress. Conversely, p38a and p38c are present exclusively in the cytoplasm of unstimulated cells, and no redistribution is apparent upon induction with either stimulus.
Differential requirements of p38 kinases for stress resistance in cultured cells and Drosophila larvae.
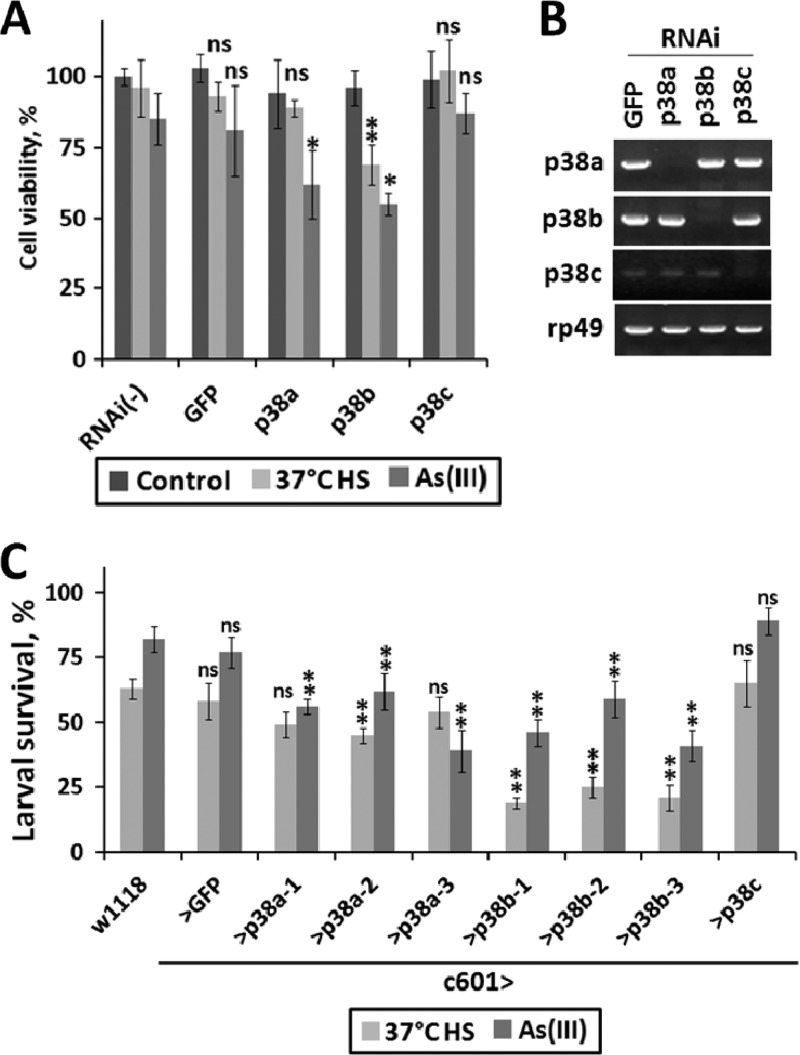
We next examined whether the differences in the activation of p38 isoforms upon exposure to various stressors are related to cellular stress resistance in culture and in vivo. First, we knocked down the expression of individual isoforms in S2 cells by RNAi and exposed cells to heat shock and oxidative stress. The resistance of cells to these treatments was measured using the MTT cell viability assay (Fig. 2A). After an 8-h heat shock treatment, cell viability was found to be almost the same in cells pretreated with dsRNAs against p38a and p38c compared to that in control cells treated with the GFP dsRNA. In contrast, cells with p38b knocked down displayed a moderate but statistically significant reduction in viability. In the case of oxidative stress induced by As(III), the viabilities of both p38a and p38b RNAi-treated cells were reduced but to a comparable extent. The viability of the p38c RNAi-treated cells was the same as that for the GFP control.
Fig 2.
Roles of p38 kinases in stress resistance. (A) S2 cells preincubated in the presence of dsRNAs targeting GFP or p38a, -b, and -c were exposed to a 37°C heat shock or 200 μM NaAsO2 for 12 h. Cell viability posttreatment was measured using the MTT assay as described in Materials and Methods. Triplicate samples were examined for each condition. All values were normalized to unstimulated S2 cells in the absence of RNAi treatment. Statistical significance was determined using ANOVA and Dunnett's test with a 95% confidence interval (*). ns, not significant. (B) The efficiency and specificity of RNAi-mediated knockdown of the p38 isoforms were examined by semiquantitative reverse transcription (RT)-PCR. (C) Early third-instar larvae of the indicated genotypes were placed in individual vials at 100 larvae per vial and either fed normal food or exposed to heat shock or food containing 200 μM NaAsO2. Three transgenic lines carrying constructs targeting different regions of p38a and p38b were used. At the end of treatment, larvae were washed out of the vials with 30% glycerol and the surviving animals were counted under the microscope. Statistical significance was determined using ANOVA and Dunnett's test with a 99% confidence interval (**). ns, not significant. A significant difference was observed in the survival rates of p38a versus p38b knockdown animals exposed to heat shock but not oxidative stress.
Next, we further investigated these effects in vivo. Although several studies previously examined the sensitivity of p38 mutant flies to various stresses (11, 14), we reasoned that using a tissue-specific knockdown of individual p38 isoforms in transgenic animals offers a more direct way to compare the effects these kinases exert on tissue viability. We chose the gut as a model tissue for these tests for the following reasons: (i) the gut is a simple epithelial tissue with direct exposure to chemical stressors in the food, (ii) p38 signaling has been shown to be critically involved in enterocyte survival under stress (37), and (iii) in larvae, the gut is the only tissue in which the three p38 isoforms are coexpressed at comparable levels, as judged by microarray analysis (7). We generated larvae with the gut-specific knockdown of the p38 kinases and control larvae expressing a GFP dsRNA construct. When these larvae were exposed to a 2-h 37°C heat shock, ∼48% (average of three different RNAi constructs) of animals with the p38a knockdown survived, compared to only ∼21% of p38b knockdown animals (Fig. 2C). These differences were clearly attributable to differential stress resistance as almost 100% of larvae of either genotype survived in the absence of heat shock. The survival rates of the p38c knockdown larvae were indistinguishable from those of control larvae. To examine the effects of p38 knockdown under oxidative stress, we exposed larvae to NaAsO2-supplemented food for a period of 24 h. The knockdown of both p38a and p38b significantly compromised the resistance of larvae to As(III). Survival rates based on an average of three RNAi constructs were ∼52% and 49% for p38a and p38b knockdown larvae, respectively, compared to an ∼80% survival rate observed in control larvae. Interestingly, As(III)-fed p38c knockdown larvae exhibited slightly elevated resistance (89%) compared to controls.
Proteomic analysis of the Drosophila p38 interactome.
For the AP-MS identification of p38-interacting proteins, we used the same 3×FLAG-tagged constructs used in the functional tests (Fig. 1). We chose a one-step affinity capture strategy (3, 16), as opposed to a more traditional tandem affinity purification (TAP), because multistep procedures in our experience (9), and according to several published studies (reviewed in reference 19), result in losses of low-affinity specific interacting proteins. Transient constitutive expression of the p38 baits in S2 cells was chosen over a stable inducible system as both antibiotic selection and typical expression inducers (e.g., heat shock or heavy metals) result in p38 activation and preclude examination of the unstimulated interactome (data not shown).
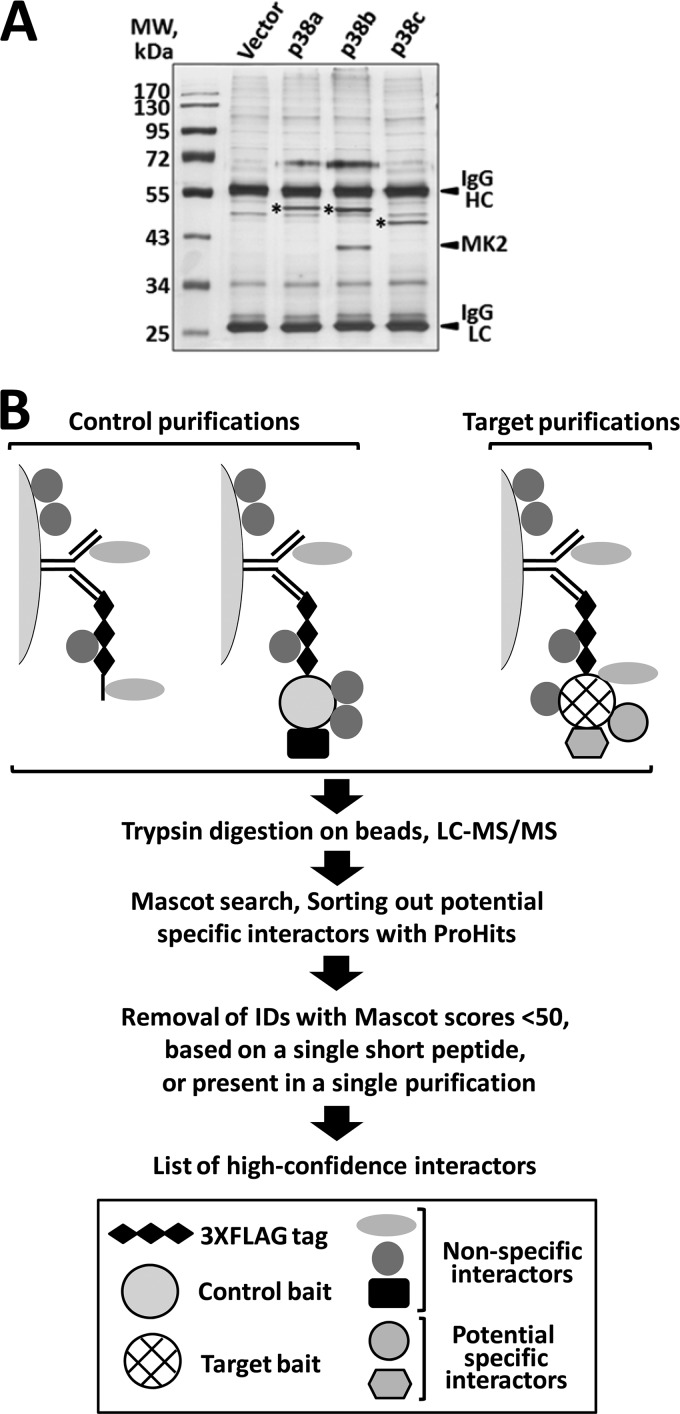
Protein complexes associated with the three p38 baits were purified from the lysates of transfected S2 cells using anti-FLAG affinity beads (Fig. 3A), and the isolated proteins were subjected to trypsin digestion directly on beads. Four independently generated transfected samples were used in AP-MS analyses of each bait. Recognizing that a one-step AP procedure is likely to produce a preparation contaminated with nonspecific binding proteins, we assembled an extensive set of control purifications that was later used to filter out these nonspecific contaminants. Our control set contained a total of 10 samples of two types: (i) S2 cells transfected with the empty expression vector that were either untreated or treated with heat shock or oxidative stress; and (ii) S2 cells transfected with an unrelated bait, Drosophila Mef2.
Fig 3.
One-step affinity capture and MS identification of the p38-interacting proteins. (A) The three 3×FLAG-tagged p38 baits and the interacting proteins isolated from transfected S2 cells using the AP-MS protocol described in Materials and Methods. The silver-stained gel of the material dissociated from the affinity matrix illustrates that the baits (marked with asterisks) are expressed as single bands of the expected molecular masses and at comparable levels. MK2, a high-affinity interactor of p38b, is easily detectable. HC, antibody heavy chain; LC, antibody light chain. (B) Schematic representation of the work flow in the AP-MS analysis of the p38 interactome. Anti-FLAG antibody affinity beads are depicted as semicircles.
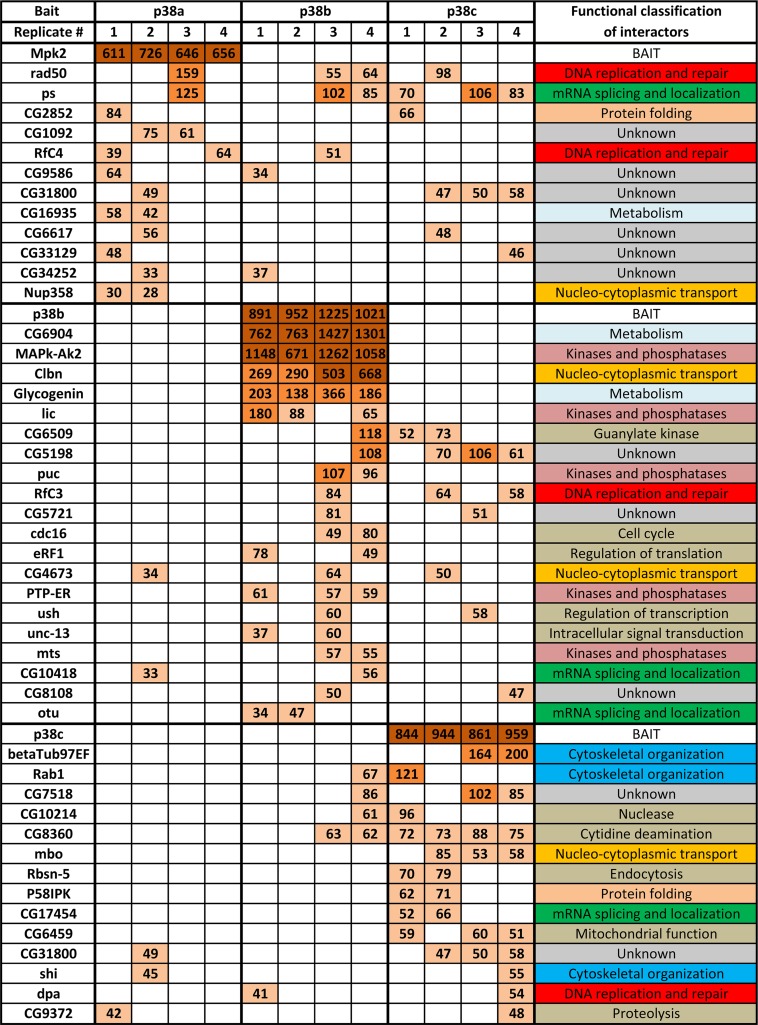
Peptides derived from the p38 complexes were analyzed by LC-MS/MS, followed by multistep bioinformatic data processing (diagrammed in Fig. 3B, and described in more detail in Materials and Methods). The resulting list of 46 high-confidence p38-interacting proteins and their functional classification are shown in Fig. 4A and in Fig. S2A in the supplemental material. Interestingly, the p38b PPI map is qualitatively distinct from the ones of p38a and -c, in that it contains a “core” of five strong interactors, MK2, CG6904, which represents Drosophila glycogen synthase (GS), Caliban (Clbn), glycogenin (GYG), and licorne (lic). Other interactors appear to be less abundant, which is suggestive of a possibility that these may be transiently binding kinase substrates. Only 3 interacting proteins are shared by all three p38 kinases—rad50, pasilla (ps), and CG4673 (see Fig. S2B)—supporting our hypothesis of limited functional overlap.
Fig 4.
Drosophila p38 interactome. Shown is a list of high-confidence interactors of the three p38 kinases. Results of quadruplicate AP-MS analyses are shown for each bait. Mascot scores corresponding to each protein identification are color coded as follows: dark brown, >500; light brown, 100 to 500; beige, <100. The last column lists the functional classifications of identified binding partners derived from GO annotations using FlyBase and literature searches.
Having generated the PPI map of p38 kinases in unstimulated cells, we sought to gain initial insights into the dynamics of the network upon stressful stimulation. S2 cells transfected with the p38b bait were subjected to heat shock and oxidative stress (H2O2) and processed for the AP-MS analysis as described above. As total spectral counts (TSCs) have been shown to linearly correlate with protein abundance (29), we used this metric to estimate relative amounts of p38b-interacting proteins in complexes upon stimulation. Of particular interest to us were strong interactors MK2, GS, Clbn, and GYG (see Fig. S2C, top, in the supplemental material) and associated kinases and phosphatases (see Fig. S2C, bottom). TSCs of these interactors were normalized using the ratios of the bait TSCs in each sample to the average bait TSC across all samples. Although the low abundance of some of the p38b-binding proteins makes our analysis semiquantitative, several important trends emerge. Based on average TSC values, oxidative stress, but not heat shock, causes an ∼30% reduction of the amount of MK2 in the complex, concomitant with an ∼25% increase in the amount of GS. Both stimuli cause GYG to dissociate from the complex, with oxidative stress having a stronger effect. The p38b-Clbn interaction is unaffected by either stress. MAPK/extracellular signal-regulated kinase (ERK) kinase kinase 1 (Mekk1), undetectable in the complex in the absence of stress, becomes a prominent interactor upon heat shock stimulation, but not oxidative stress. Both stresses recruit small amounts of S6KII to the complex. Among associated phosphatases, the abundance of puckered (puc) is significantly elevated in the complex under stress, whereas the levels of protein tyrosine phosphatase-ERK/Enhancer of Ras1 (PTP-ER) and microtubule star (mts) become undetectable.
An important question arising from our data is whether the identified p38 interactors are kinase substrates or functional modulators. To address this broad question for p38b-interacting proteins, we used two experimentally derived phosphoproteomic databases that catalogue phosphopeptides present in early w1118 Drosophila embryos (49a) and Kc cells (4a). The w1118 data set is particularly suitable for the identification of p38b substrates as p38b is active in early developing embryos, as judged by Western blotting (see Fig. S3 in the supplemental material) and the abundance of the doubly phosphorylated TGY p38b peptide in the data set.
Phosphosites observed in the p38b-interacting proteins identified in our study are shown in Table S2 in the supplemental material. We reasoned that the phosphosites observed in both data sets are particularly abundant. Next, we performed computational prediction of the p38 phosphorylation sites in the interactors using Scansite. Finally, we determined the overlap between the predicted p38 phosphosites and the ones observed experimentally in the Drosophila data sets. Among the 20 p38b-interacting proteins 7 are likely substrates of the kinase as the predicted phosphorylation sites match the observed phosphopeptides. Importantly, this analysis confidently identifies MK2 and GS as p38b substrates, consistent with previous results. On the other hand, lic and puc are not expected to be p38 substrates. Indeed, neither of these interactors emerges as a substrate in our analysis. We therefore suggest that the remaining 13 p38b interactors participate in a native kinase complex(es) and likely serve as modulators of the kinase function, such as its activity, localization, etc.
Verification of high-affinity interactions of p38b by co-IP and their evolutionary conservation.
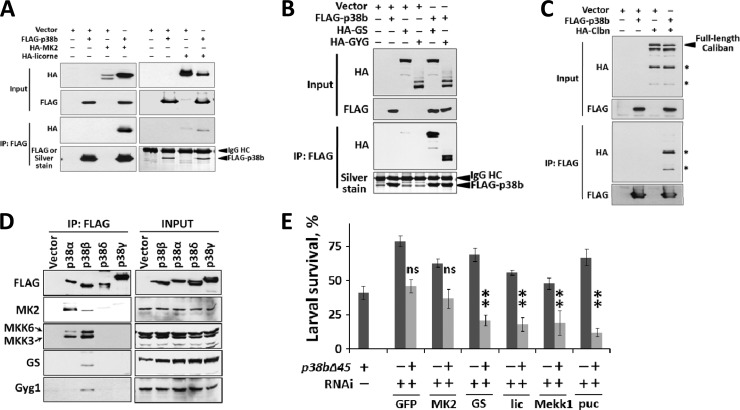
In unstimulated S2 cells, p38b strongly interacts with five proteins (Fig. 4). To test if these interactions can be detected by traditional co-IP, we generated HA-tagged versions of the interacting proteins and overexpressed them in the presence of FLAG-p38b in S2 cells. An evolutionarily conserved p38 binding partner, MK2, and lic specifically coprecipitate with p38b (Fig. 5A). We also observed that in the presence of overexpressed p38b, HA-MK2 is strongly stabilized, whereas lic levels become noticeably reduced. Furthermore, the levels of co-immunoprecipitated (co-IP) MK2 are much higher than those of lic, paralleling the MS data (Fig. 4; see Data set S1 in the supplemental material). Indeed, the p38b-MK2 interaction is so robust that MK2 is easily detectable on silver-stained gels of p38b immunoprecipitates (Fig. 3A).
Fig 5.
Evolutionarily conserved protein interactions within the p38b “core” complex confirmed by coimmunoprecipitation. (A, B, and C) S2 cells were transiently transfected with the constructs indicated on top of the blots. Co-IPs were performed using anti-FLAG affinity beads. Pulldown of FLAG-p38b was confirmed by either Western blotting or silver staining. The two proteolytic fragments of Caliban (approximately 60 and 40 kDa) that interact with p38b are indicated with stars in panel B. (D) HEK293T cells were transfected with 3×FLAG-tagged human p38α, -β, -δ, and -γ. Total cell lysates (INPUT) and anti-FLAG immunoprecipitates were analyzed by Western blotting using the antibodies against human homologs of the strong p38b interactors. (E) An in vivo assay was used to test the genetic interactions between the p38b-null allele (p38bΔ45) and RNAi-mediated knockdown alleles of strong p38b interactors found in the AP-MS screen. Third-instar larvae of indicated genotypes were exposed to oxidative stress, and their survival rates were scored as described above. The knockdown of MK2 fails to additively enhance the p38b mutation as no statistically significant (ns) difference is observed between the respective survival rates. Statistical significance was determined using ANOVA and Dunnett's test with a 99% confidence interval (**).
A novel p38 interactor, Clbn, is processed in S2 cells to produce several cleavage products (Fig. 5C). Only the N-terminal fragments of ca. 60 and 40 kDa, but not a larger fragment or the full-length Clbn, bind to p38b (V. E. Belozerov, K. W. M. Siu, A. C. Gingras, and J. C. McDermott, unpublished data).
Finally, both GS and GYG also specifically coimmunoprecipitate with p38b, and their expression levels are not influenced by the overexpression of the kinase. Interestingly, GYG expressed in S2 cells invariably appears on Western blots of total lysate as two main bands corresponding to the expected GYG molecular mass (35 kDa) followed by a smear of slower-migrating bands (Fig. 5B, top). These higher-molecular-mass species do not appear on the blot of coimmunoprecipitated GYG (Fig. 5C, bottom), even at longer film exposure times (not shown), indicating that these GYG species do not bind to p38b.
To test whether the components of the Drosophila p38b signaling complex are conserved in mammals, we expressed tagged versions of the four human p38 isoforms in HEK293T cells and examined immunoprecipitated kinase complexes by Western blotting for the presence of human homologs of the fly p38b complex (Fig. 5D). Interestingly, p38β is the only isoform that interacts with all four tested binding partners, MKK3 and -6, MK2, GS, and glycogenin 1, suggesting the possibility of functional similarities between this kinase and the Drosophila homolog.
Genetic interaction assay probes functional significance of physical interactions of p38b.
To test the involvement of biochemically confirmed p38b-interacting proteins in p38-mediated stress response, we used an in vivo genetic interaction assay. Briefly, we generated fly lines that express transgenic dsRNAs against strong p38b interactors specifically in larval gut (with the same Gal4 driver that was used in Fig. 2C). These transgenes were then introduced into the homozygous p38b-null background (p38bΔ45), or the isogenic wild-type background. Third-instar larvae bearing these allelic combinations were subjected to oxidative stress, and their survival was scored. We reasoned that among the RNAi knockdown alleles that compromise larval stress-resistance, two classes are likely to emerge: (i) the ones that attenuate resistance in an additive or synergistic fashion with the p38b mutation, indicative of the interactors that have p38b-independent roles in stress response; and (ii) the ones that fail to further sensitize p38b mutants (i.e., the interactors whose function is p38 dependent).
The results of these functional tests are shown in Fig. 5E. The only interactor that in knockdown larvae failed to display synthetic lethality with the p38b-null allele was MK2, suggesting that the function of this kinase is largely or entirely dependent on p38b signaling in the gut. Other interactors displayed additive phenotypes with the p38b mutation, consistent with their involvement in both p38b-dependent and parallel pathways.
p38b regulates glycogen metabolism through phosphorylation of GS.
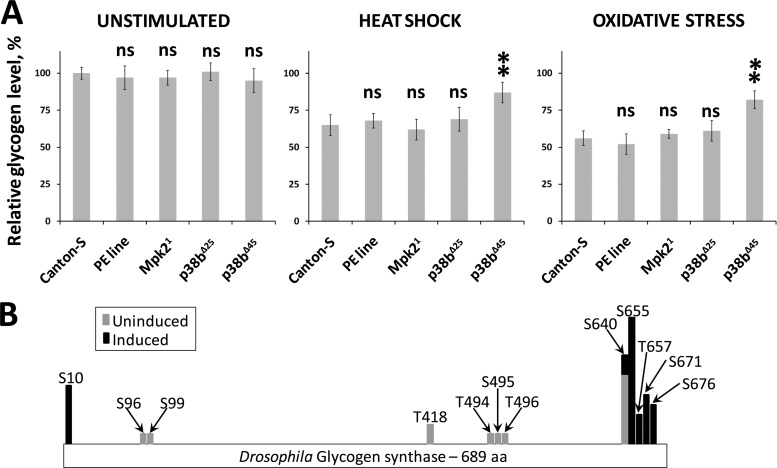
As GS forms a high-affinity complex with p38b that becomes more prominent under stress (see Fig. S2C in the supplemental material), we hypothesized that this interaction may serve as a direct link between stress signaling and metabolic adaptation. To test this idea in vivo, we examined the changes in glycogen levels induced by various stresses in p38 mutant larvae. When wild-type third-instar larvae were exposed to heat shock or oxidative stress, an ∼30% reduction in the level of stored glycogen was observed in surviving animals. Larvae homozygous for a p38a-null allele, Mpk21, exhibited a comparable rate of stress-induced glycogen loss. In the case of a hypomorphic p38b allele, p38bΔ25, larvae exposed to heat shock and As(III) lost an average of 19% and 24% glycogen, respectively, compared to untreated controls. Finally, p38b-null larvae (p38bΔ45) lost significantly less glycogen (6% and 14% of the levels in untreated controls) when subjected to the same stimuli (Fig. 6A).
Fig 6.
Stress-induced phosphorylation of GS by p38b correlates with metabolic adaptation in Drosophila larvae. (A) Glycogen metabolism is affected in p38b mutant larvae exposed to stress. Third-instar larvae of the indicated genotypes were exposed to a 1-h heat shock or fed As(III)-containing food for 8 h. Five surviving larvae in each group were homogenized and used in the glycogen assay as described in Materials and Methods. The results shown represent an average of three tests. The “PE line” refers to the precise excision of the transposon insertion used to generate p38bΔ25 (hypomorph) and p38bΔ45 (null) imprecise excision alleles (47). Glycogen levels in unstimulated larvae are presented relative to those in Canton-S larvae. Glycogen levels in stressed larvae are expressed relative to unstimulated levels in animals of the same genotype. Statistical significance was determined using ANOVA and Dunnett's test with a 99% confidence interval. ns, not significant. (B) Phosphoproteomic analysis of Drosophila GS. Phosphorylation sites in GS were compared between the samples derived from unstimulated or stress-stimulated S2 cells. A computationally predicted p38 phosphorylation site at S655 becomes abundant in samples from heat shock- and oxidative-stress-treated cells.
A possible mechanism underlying the above observation is an inhibitory phosphorylation of GS by p38b. The analysis of likely p38b substrates (see Table S2 in the supplemental material) pointed at S655 as the major p38b phosphorylation site within GS. If S655 is indeed specifically targeted by activated p38b under stress, the abundance of this phosphosite should increase significantly in p38b immunoprecipitates from stimulated samples. To test this possibility, we catalogued all phosphopeptides observed in our samples (4 unstimulated replicates and 5 replicates stimulated with heat shock and oxidative stress). The map of phosphosites demonstrating their dynamics under stress is shown in Fig. 6B. Indeed, S655 becomes the most prominent phosphorylation site in response to stress-induced p38b activation. These new findings provide a solid foundation for future investigation of p38-mediated GS phosphorylation in vivo.
DISCUSSION
It is universally recognized that in a cellular environment, protein function is crucially dependent on interactions with other biomolecules. PPIs underpin a highly interconnected and dynamic molecular network that governs every aspect of cellular activity, ranging from normal homeostasis to therapeutic treatment responses (43). Current models conceptualize scale-free PPI networks as consisting of nodes and hubs, depending on the number of functional connections of a given protein. The network robustness is primarily determined by redundancy, modularity, and feedback control (17, 42). In-depth understanding of these mechanisms is critically dependent on the availability of high-resolution PPI maps.
In this study, we used the Drosophila p38 MAPK family as a model to test whether a high-quality AP-MS-based PPI map of individual kinases can provide sufficient resolution to define the extent of their functional redundancy and reveal their unique functions.
Previous studies with Drosophila utilized null alleles of p38a and p38b and animal lethality as an assay readout to establish that both kinases mediate resistance to environmental stress and microbial infection in vivo. According to one study (11), p38a mutant flies are sensitive to dry starvation and oxidative stress, moderately sensitive to heat shock, and display no susceptibility to osmotic stress or bacterial infection. In contrast, p38b mutants exhibit pronounced sensitivity to all stresses tested (14). Some studies find enhanced sensitivity to osmotic stress and microbial infection in both p38a and p38b mutant animals (6, 37).
As systemic effects of p38 mutations are likely to mask the primary roles of these kinases at a cellular level, we attempted a more direct molecular and functional comparison in cultured S2 cells. Endogenous messages of all three p38 isoforms are expressed in this cell line, making it a particularly suitable system for comparative tests (7). Our experiments addressed three aspects of p38 activity. First, we examined activation profiles of p38a and p38b, as measured by dual phosphorylation at the TGY site, and found that p38b responds to a wide range of stresses, including heat shock, oxidative and metabolic stresses, and osmotic shock, whereas p38a response is limited to osmotic stress and partly to oxidative stress. A more detailed kinetic analysis of activation confirmed the observations presented above and revealed that apart from osmotic shock, other stimuli activated p38a in a less robust and more transient fashion than p38b.
Second, we examined the nucleocytoplasmic distribution of the p38 isoforms in unstimulated cells and upon activation. In mammals, nucleocytoplasmic shuttling of p38 is a crucial aspect of function (41). As none of the p38 kinases contains canonical nuclear localization signal (NLS) or nuclear export signal (NES) sequences, interaction partners, most prominently MK2 (2), serve as chaperones necessary for nucleocytoplasmic transport. We find that in S2 cells, p38b is the only isoform that is present in both cellular compartments, whereas p38a and p38c are exclusively cytoplasmic. Upon stimulation, p38b is exported from the nucleus, similar to mammalian p38α. The observed differences in subcellular distribution are consistent with our finding that MK2 interacts exclusively with p38b (see the PPI map and Fig. 3A). The common docking (CD) domain is well known to be a critical determinant of kinase interaction with cognate regulators and substrates (45). In p38 kinases, two residues, Asp316 and Asp319 (numbers correspond to Drosophila p38b [Dp38b]), form a negatively charged patch in the CD domain and are conserved from yeasts to humans. Both of these are mutated in p38a and p38c (see Fig. S1A in the supplemental material), likely explaining why these kinases fail to interact with MK2 and possibly other proteins. A recent study confirmed that the CD domain sequence in p38b is indeed necessary for the interaction with MK2 (37).
Third, we used RNAi to demonstrate that the knockdown of individual p38 isoforms differentially sensitizes cells to mechanistically distinct stresses. Both p38a and p38b RNAi reduced cell resistance to oxidative stress induced by As(III), whereas only p38b knockdown sensitized cells to heat shock. Importantly, this functional difference was also observed in vivo, when the two kinases were knocked down in a tissue-specific manner by RNAi in the larval gut. The knockdown of p38c in either system did not result in decreased stress resistance, arguing against direct involvement of this kinase in the typical stress response. Consistent with this result, previous findings (6, 15) demonstrated that microbial resistance of p38c-null flies is unaffected. Although very likely a product of the p38a gene duplication, p38c shares only 50% protein sequence identity and 68% similarity with the parental gene. Moreover, several highly conserved motifs found in prototypic p38 kinases are mutated in p38c (see Fig. S1A in the supplemental material)—e.g., the dual-phosphorylation site in the activation loop (TGY to TDH), and the acidic patch in the CD domain (DPTD to EPHHHA)—all supporting the divergence of p38c from the canonical p38 signaling mechanisms.
Collectively, the above findings support two important conclusions: (i) Drosophila p38 isoforms differ considerably with respect to their involvement in cell survival under stress, and (ii) among the three family members, p38b plays a central role in stress signaling and shares key mechanistic features of p38 signaling in mammals. Evolutionary conservation of protein sequence is an unbiased measure of the functional significance of a protein for the survival of an organism. In this regard, it is noteworthy that at the level of protein sequence similarity, p38b diverged at an ∼2-fold-lower rate than p38a over the 40 million years of Drosophila evolution (see Fig. S1B in the supplemental material).
Assuming that functional significance of a protein is related to its integration into cellular PPI network, these conclusions are clearly reflected in the PPI map of the three p38 kinases. Only p38b forms a stable high-affinity complex or complexes with 5 other proteins, whereas p38a and p38c bind to a number of weaker-interacting partners. As reliable isolation of low-affinity interactions from background contaminants is a major challenge in interaction proteomics (19), we designed our study to include rigorous quality control. First, the AP-MS analyses of each bait were performed in quadruplicate using independently generated cell samples, and only the interactions present in more than one sample were considered. Second, an extensive 10-sample control data set was used to filter out nonspecific copurifying proteins. As a result, our p38 interaction map minimally overlaps with a recently published large-scale Drosophila protein interaction map (DPiM) PPI map (22) (see Table S2 in the supplemental material). Importantly, none of the high-affinity p38b-interacting proteins identified in our study, with the exception of MK2, was recovered by DPiM. In addition, we found that up to a third of the p38-interacting proteins in DPiM are present in our data set of nonspecific binding proteins, highlighting the limited resolution at some nodes in genome-wide PPI maps.
An important criterion for assessing the quality of a PPI map is the presence of known interacting proteins. In unstimulated cells, we recovered a strong evolutionarily conserved interaction of p38b with MK2 and a much weaker interaction with lic, a Drosophila MAP2K specific to the p38 signaling module. As these two proteins were expected to interact with p38, their consistent recovery offered an independent validation of our AP-MS procedure and the resulting PPI map. Furthermore, we observed the interactions of p38b with three phosphatases, puc, PTP-ER, and mts. We reasoned that stimulation-induced changes in the kinase-phosphatase network may reveal novel aspects of the mechanism of p38 regulation and used spectral counting to evaluate relative protein abundance in the complexes. Our analyses identified Mekk1 as a component of the complex in cells stimulated with heat shock. In contrast, no association with a canonical MAP3K was observed under oxidative stress (Fig. 4D). Several MAP3Ks have been implicated in the activation of the p38 cascade, depending on stress (50), including Mekk1, TAK1, ASK1, and Slipper; however the MAP3K transducing the oxidative stress signal is currently unknown. Both tested stimuli induced the association of small quantities of S6KII with the p38b complex, suggesting a potential point of molecular cross talk with the Akt pathway. Interestingly, p38 has been shown to be an upstream activator of S6K both in S2 cells (14) and in mouse osteoblasts (46), and our data provide the first indication that this epistatic relationship may be mediated by physical interaction.
Among the three phosphatases present in the unstimulated p38b interactome, only puc was found to associate more strongly with the complex upon stimulation, whereas PTP-ER and mts became undetectable. puc is a dual-specificity phosphatase previously thought to be specific to the JNK signaling pathway (30). However, the dynamics of p38b-puc interaction are consistent with the possibility that puc also serves to inactivate p38. Interestingly, p38b was shown to transcriptionally upregulate puc, thereby controlling JNK signaling (6). Therefore, if puc is indeed a p38b phosphatase, this transcriptional mechanism may contribute to negative feedback regulation.
The association of p38b with its two strongest binding partners, MK2 and GS, may also be affected by stressful stimuli. When cells are exposed to oxidative stress, the average TSC value of MK2 is reduced by ∼30% compared to that in unstimulated samples, concomitant with a similar increase in the average TSC of GS. These changes parallel the export of p38b into the cytoplasm that is most pronounced under oxidative stress (Fig. 1D). As GS is present exclusively in the cytoplasm of S2 cells (data not shown), this result may indicate that MK2 and GS form distinct complexes with p38b. Indeed, we biochemically confirmed that MK2 and GS bind to p38b in a mutually exclusive fashion (see Fig. S2 in the supplemental material).
As the Drosophila p38 signaling module is a genetically tractable model for the mammalian counterpart, an important question is which mammalian p38 isoform shares evolutionarily conserved functions with the fly p38b? Our data indicate that human p38β shares functionally important interaction partners with p38b, namely, GS, glycogenin, MK2, and MKK3/6. Interestingly, the other three mammalian p38 isoforms fail to form stable interactions with the glycogen synthesis enzymes, suggesting a possible unique role of p38β in metabolic adaptation in mammals.
The p38b-GS interaction is of particular interest as it appears conserved in metazoan evolution. This interaction received a high score in a yeast two-hybrid screen of the Caenorhabditis elegans proteome (27), and it was also identified in glutathione S-transferase (GST) pulldown experiments using mouse skeletal muscle lysate (25), as well as our co-IP experiments in HEK293T cells. It was suggested that by phosphorylating GS at several residues, most prominently Ser644, mammalian p38 primes GS for further phosphorylation by glycogen synthase kinase 3 (GSK3), thereby contributing to GS inactivation (25). This mechanism is likely conserved in Drosophila as our phosphoproteomic analyses identify an equivalent residue, Ser655, as the primary target of p38b in stress-stimulated cells. In a functional assay, we found that in wild-type or p38a mutant larvae, stress causes a decrease in the amount of stored glycogen, likely due to the energy mobilization response. Strikingly, in stress-exposed p38b mutant larvae, glycogen storage is affected to a lesser extent. As glycogen levels are maintained by balancing the rates of synthesis and degradation, and GS is the key enzyme determining the rate of glycogen synthesis, p38 shifts the balance toward free glucose by inhibiting GS in response to stress. Our data provide the first demonstration of the regulation of glycogen levels by p38 in vivo. Interestingly, GYG, a glycosyltransferase that nucleates the glycogen particle, also associates with p38b in unstimulated cells. Upon stimulation, GYG dissociates from the complex, although the functional significance of this dynamic interaction remains to be investigated.
In contrast to p38b, which binds to a variety of functionally important partners, the interactions formed by p38a and p38c tend to be weaker, and many of these interactors (50% in the case of p38a) are functionally uncharacterized. Only three proteins interact with all p38 isoforms: ps (39), an mRNA splicing regulator homologous to the mammalian NOVA proteins (4); rad50, a component of the MRN DNA repair complex (8); and CG4673. Although the functional significance of these interactions is currently unknown, it is conceivable that these proteins depend most on association with p38 and therefore contain the binding interfaces that are the least discriminating between the isoforms.
In broader terms, we suggest that the Drosophila p38 signaling module consists of a central “hub” kinase, p38b, and two auxiliary nodes, p38a, and p38c. The three kinases are coexpressed and share significant sequence homology, yet display minimal overlap in the PPI network and function. This type of module architecture is not surprising as it has been shown that functional degeneracy, rather than strict redundancy, strongly enhances evolvability and as such contributes to evolutionary fitness (48).
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Canadian Institutes of Health Research and Natural Sciences and Engineering Research Council of Canada grants to K.W.M.S. and J.C.M.
We thank Andre Bedard (McMaster University) for providing S2 cells and Alysia Vrailas-Mortimer and Subhabrata Sanyal for p38 mutant flies. We also thank Christina Pagiatakis for technical assistance and Srdjana Ratkovic, Helen McNeill, Arthur Hilliker, and members of the Gingras laboratory for helpful discussion.
V.E.B., J.C.M., and A.C.G. designed the experiments; V.E.B. and Z.Y.L. performed the MS analyses; V.E.B. performed all other experiments; V.E.B., J.C.M., and A.C.G. analyzed the results; and V.E.B., J.C.M., A.C.G., and K.W.M.S. wrote the manuscript.
We declare that they have no conflict of interest.
Footnotes
Published ahead of print 16 July 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Beardmore VA, et al. 2005. Generation and characterization of p38beta (MAPK11) gene-targeted mice. Mol. Cell. Biol. 25: 10454–10464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ben-Levy R, Hooper S, Wilson R, Paterson HF, Marshall CJ. 1998. Nuclear export of the stress-activated protein kinase p38 mediated by its substrate MAPKAP kinase-2. Curr. Biol. 8: 1049–1057 [DOI] [PubMed] [Google Scholar]
- 3. Breitkreutz A, et al. 2010. A global protein kinase and phosphatase interaction network in yeast. Science 328: 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brooks AN, et al. 2011. Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res. 21: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a. Brunner E, et al. 2007. A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 25: 576–583 [DOI] [PubMed] [Google Scholar]
- 5. Casar B, et al. 2007. Mxi2 promotes stimulus-independent ERK nuclear translocation. EMBO J. 26: 635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen J, et al. 2010. Participation of the p38 pathway in Drosophila host defense against pathogenic bacteria and fungi. Proc. Natl. Acad. Sci. U. S. A. 107: 20774–20779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chintapalli VR, Wang J, Dow JA. 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720 [DOI] [PubMed] [Google Scholar]
- 8. Ciapponi L, et al. 2004. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr. Biol. 14: 1360–1366 [DOI] [PubMed] [Google Scholar]
- 9. Cox DM, Du M, Guo X, Siu KW, McDermott JC. 2002. Tandem affinity purification of protein complexes from mammalian cells. Biotechniques 33: 267–268, 270 [DOI] [PubMed] [Google Scholar]
- 10. Cox DM, et al. 2003. Phosphorylation motifs regulating the stability and function of myocyte enhancer factor 2A. J. Biol. Chem. 278: 15297–15303 [DOI] [PubMed] [Google Scholar]
- 11. Craig CR, Fink JL, Yagi Y, Ip YT, Cagan RL. 2004. A Drosophila p38 orthologue is required for environmental stress responses. EMBO Rep. 5: 1058–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cuadrado A, Nebreda AR. 2010. Mechanisms and functions of p38 MAPK signalling. Biochem. J. 429: 403–417 [DOI] [PubMed] [Google Scholar]
- 13. Cuenda A, Rousseau S. 2007. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta 1773: 1358–1375 [DOI] [PubMed] [Google Scholar]
- 14. Cully M, et al. 2010. A role for p38 stress-activated protein kinase in regulation of cell growth via TORC1. Mol. Cell. Biol. 30: 481–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davis MM, Primrose DA, Hodgetts RB. 2008. A member of the p38 mitogen-activated protein kinase family is responsible for transcriptional induction of Dopa decarboxylase in the epidermis of Drosophila melanogaster during the innate immune response. Mol. Cell. Biol. 28: 4883–4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dunham WH, et al. 2011. A cost-benefit analysis of multidimensional fractionation of affinity purification-mass spectrometry samples. Proteomics 11: 2603–2612 [DOI] [PubMed] [Google Scholar]
- 17. Friedman A, Perrimon N. 2007. Genetic screening for signal transduction in the era of network biology. Cell 128: 225–231 [DOI] [PubMed] [Google Scholar]
- 18. Gavin AC, et al. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- 19. Gavin AC, Maeda K, Kuhner S. 2011. Recent advances in charting protein-protein interaction: mass spectrometry-based approaches. Curr. Opin. Biotechnol. 22: 42–49 [DOI] [PubMed] [Google Scholar]
- 20. Gillespie MA, et al. 2009. p38-{gamma}-dependent gene silencing restricts entry into the myogenic differentiation program. J. Cell Biol. 187: 991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goudreault M, et al. 2009. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell. Proteomics 8: 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guruharsha KG, et al. 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keren A, Tamir Y, Bengal E. 2006. The p38 MAPK signaling pathway: a major regulator of skeletal muscle development. Mol. Cell. Endocrinol. 252: 224–230 [DOI] [PubMed] [Google Scholar]
- 24. Krogan NJ, et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- 25. Kuma Y, Campbell DG, Cuenda A. 2004. Identification of glycogen synthase as a new substrate for stress-activated protein kinase 2b/p38beta. Biochem. J. 379: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee JC, et al. 1994. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature 372: 739–746 [DOI] [PubMed] [Google Scholar]
- 27. Li S, et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu G, et al. 2010. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat. Biotechnol. 28: 1015–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu H, Sadygov RG, Yates JR., III 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76: 4193–4201 [DOI] [PubMed] [Google Scholar]
- 30. McEwen DG, Peifer M. 2005. Puckered, a Drosophila MAPK phosphatase, ensures cell viability by antagonizing JNK-induced apoptosis. Development 132: 3935–3946 [DOI] [PubMed] [Google Scholar]
- 31. Ono K, Han J. 2000. The p38 signal transduction pathway: activation and function. Cell. Signal. 12: 1–13 [DOI] [PubMed] [Google Scholar]
- 32. Palanker L, Tennessen JM, Lam G, Thummel CS. 2009. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab. 9: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Puri PL, et al. 2000. Induction of terminal differentiation by constitutive activation of p38 MAP kinase in human rhabdomyosarcoma cells. Genes Dev. 14: 574–584 [PMC free article] [PubMed] [Google Scholar]
- 34. Roux PP, Blenis J. 2004. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 68: 320–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ryabinina OP, Subbian E, Iordanov MS. 2006. D-MEKK1, the Drosophila orthologue of mammalian MEKK4/MTK1, and Hemipterous/D-MKK7 mediate the activation of D-JNK by cadmium and arsenite in Schneider cells. BMC Cell Biol. 7: 7 doi:10.1186/1471-2121-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sabio G, et al. 2005. p38gamma regulates the localisation of SAP97 in the cytoskeleton by modulating its interaction with GKAP. EMBO J. 24: 1134–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seisenbacher G, Hafen E, Stocker H. 2011. MK2-dependent p38b signalling protects Drosophila hindgut enterocytes against JNK-induced apoptosis under chronic stress. PLoS Genet. 7: e1002168 doi:10.1371/journal.pgen.1002168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seong KH, Li D, Shimizu H, Nakamura R, Ishii S. 2011. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell 145:1049–1061 [DOI] [PubMed] [Google Scholar]
- 39. Seshaiah P, Miller B, Myat MM, Andrew DJ. 2001. pasilla, the Drosophila homologue of the human Nova-1 and Nova-2 proteins, is required for normal secretion in the salivary gland. Dev. Biol. 239: 309–322 [DOI] [PubMed] [Google Scholar]
- 40. Shi Y, Gaestel M. 2002. In the cellular garden of forking paths: how p38 MAPKs signal for downstream assistance. Biol. Chem. 383: 1519–1536 [DOI] [PubMed] [Google Scholar]
- 41. Shiryaev A, Moens U. 2010. Mitogen-activated protein kinase p38 and MK2, MK3 and MK5: menage a trois or menage a quatre? Cell. Signal. 22: 1185–1192 [DOI] [PubMed] [Google Scholar]
- 42. Stelling J, Sauer U, Szallasi Z, Doyle FJ, III, Doyle J. 2004. Robustness of cellular functions. Cell 118: 675–685 [DOI] [PubMed] [Google Scholar]
- 43. Stelzl U, Wanker EE. 2006. The value of high quality protein-protein interaction networks for systems biology. Curr. Opin. Chem. Biol. 10: 551–558 [DOI] [PubMed] [Google Scholar]
- 44. Sumara G, et al. 2009. Regulation of PKD by the MAPK p38delta in insulin secretion and glucose homeostasis. Cell 136: 235–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tanoue T, Adachi M, Moriguchi T, Nishida E. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2: 110–116 [DOI] [PubMed] [Google Scholar]
- 46. Tokuda H, et al. 2003. p38 MAP kinase regulates BMP-4-stimulated VEGF synthesis via p70 S6 kinase in osteoblasts. Am. J. Physiol. Endocrinol. Metab. 284: E1202–E1209 [DOI] [PubMed] [Google Scholar]
- 47. Vrailas-Mortimer A, et al. 2011. A muscle-specific p38 MAPK/Mef2/MnSOD pathway regulates stress, motor function, and life span in Drosophila. Dev. Cell 21: 783–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Whitacre J, Bender A. 2010. Degeneracy: a design principle for achieving robustness and evolvability. J. Theor. Biol. 263: 143–153 [DOI] [PubMed] [Google Scholar]
- 49. Yagasaki Y, Sudo T, Osada H. 2004. Exip, a splicing variant of p38alpha, participates in interleukin-1 receptor proximal complex and downregulates NF-kappaB pathway. FEBS Lett. 575: 136–140 [DOI] [PubMed] [Google Scholar]
- 49a. Zhai B, et al. 2008. Phosphoproteome analysis of Drosophila melanogaster embryos. J Proteome Res. 7: 1675–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhuang ZH, Zhou Y, Yu MC, Silverman N, Ge BX. 2006. Regulation of Drosophila p38 activation by specific MAP2 kinase and MAP3 kinase in response to different stimuli. Cell. Signal. 18: 441–448 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.