Abstract
The health of the offspring depends on the genetic constitution of the parental germ cells. The paternal genome appears to be important; e.g., de novo mutations in some genes seem to arise mostly from the father, whereas epigenetic modifications of DNA and histones are frequent in the paternal gonads. Environmental contaminants which may affect the integrity of the germ cells comprise the polycyclic aromatic hydrocarbon, benzo[a]pyrene (B[a]P). B[a]P has received much attention due to its ubiquitous distribution, its carcinogenic and mutagenic potential, and also effects on reproduction. We conducted an in vitro fertilization (IVF) experiment using sperm cells from B[a]P-exposed male mice to study effects of paternal B[a]P exposure on early gene expression in the developing mouse embryo. Male mice were exposed to a single acute dose of B[a]P (150mg/kg, ip) 4 days prior to isolation of cauda sperm, followed by IVF of oocytes from unexposed superovulated mice. Gene expression in fertilized zygotes/embryos was determined using reverse transcription-qPCR at the 1-, 2-, 4-, 8-, and blastocyst cell stages of embryo development. We found that paternal B[a]P exposure altered the expression of numerous genes in the developing embryo especially at the blastocyst stage. Some genes were also affected at earlier developmental stages. Embryonic gene expression studies seem useful to identify perturbations of signaling pathways resulting from exposure to contaminants, and can be used to address mechanisms of paternal effects on embryo development.
Key Words: gene expression, benzo[a]pyrene, mouse embryo.
Reduced male fertility and sperm quality are reported for many developed economies, suggesting that male reproductive health is challenged by environmental factors associated with a modern lifestyle (Auger et al., 1995; Carlsen et al., 1992). There are indications that the genetic constitution of the paternal genome may be challenged by environmental agents (Olsen et al., 2010), potentially affecting the offspring (Marchetti and Wyrobek, 2005). Elevated exposure to such agents including polycyclic aromatic hydrocarbons (PAHs) has been reported in animal studies to affect reproduction and also the development of the fetus (Edwards et al., 2006; Somers et al., 2002). It is believed that transmittable effects may arise from accumulated DNA damage in mouse germ cells, which—beyond a critical stage of spermatogenesis—have a limited capacity for DNA repair (Marchetti and Wyrobek, 2008; Olsen et al., 2005, 2010). There are concerns that undesired environmental exposure may lead to transgenerational effects in humans. Transmittable genetic damage in humans has been studied in populations exposed to radiation (Dubrova et al., 1996), and there are reports indicating that children of smoking fathers have a higher risk of contracting childhood cancer (Ji et al., 1997).
Benzo[a]pyrene (B[a]P) is a PAH, which has received much attention due to its carcinogenic and reprotoxic potential and its ubiquitous distribution. B[a]P was determined in 1933 as a component of coal tar and is recognized as the first compound being responsible for occupation-associated cancers, in the form of the sooty warts (cancers of the scrotum) suffered by chimney sweeps in 18th century England (Phillips, 1983). B[a]P is found in coal tar, automobile exhaust fumes (especially from diesel engines), and all smoke resulting from the combustion of organic material (including cigarette smoke). The dominant route of exposure in humans is through ingestion of contaminated food such as charbroiled meat (Hattemer-Frey and Travis, 1991). Cooked meat products have been shown to contain up to 4ng/g of B[a]P (Kazerouni et al., 2001), fried chicken contains up to 5.5ng/g (Lee and Shim, 2007), and up to 62.6ng/g have been measured in overcooked charcoal barbecued beef (Aygün and Kabadayi, 2005).
Phase I CYP450 enzymes convert B[a]P into reactive metabolites (Pelkonen and Nebert, 1982), e.g., benzo[a]- pyrene-7,8-diol-9,10-epoxide (BPDE), which bind covalently to DNA (Jeffrey et al., 1977). B[a]P can also be metabolized via the aldo-keto reductase pathway, leading to increased levels of oxidative stress and formation of catechol DNA adducts (Penning, 2004). The metabolites react with a broad range of cellular molecules, and it follows that exposure to B[a]P may affect a number of cellular processes including gene transcription.
Our previous findings indicate that, although the sperm chromatin is highly compacted, B[a]P-derived DNA lesions may form in sperm after in vivo exposure of mice (Olsen et al., 2010) and in vitro exposure of human sperm (Sipinen et al., 2010). B[a]P exposure leads to the formation of DNA adducts in testis and sperm of exposed male mice (Verhofstad et al., 2010) and humans (Zenzes et al., 1999a). In a study of surplus human embryos from in vitro fertilization (IVF) treatment, the amount of B[a]P-derived DNA adducts in the embryo seemed to be more dependent on paternal than maternal smoking (Zenzes et al., 1999b). This study also suggested that paternally transmitted BPDE-DNA adducts can persist past the first embryonic cleavage divisions. Furthermore, exposure of male mice to high doses of B[a]P in the week before fertilization has been shown to induce dominant lethal mutations (Generoso et al., 1982; Shukla and Taneja, 2001).
Based on the ability of B[a]P to form DNA and protein adducts, and the reported multigenerational impact of chronic paternal B[a]P exposure (Mohamed et al., 2010) on sperm production and sperm parameters, we hypothesized that a single acute paternal preconceptional exposure to B[a]P could affect early embryonic transcription and the activation of the embryonic genome. It is important if such exposure can affect the fetus even when administered to the epididymal (cauda) sperm. Following paternal acute exposure of male mice to B[a]P 4 days prior to fertilization, we studied the effect of B[a]P-exposed sperm cells on the expression of key DNA damage response genes and genes important to embryo development, in early embryonic stages. We demonstrate that early embryonic transcription of multiple genes is affected by paternal preconceptional exposure to B[a]P.
MATERIALS AND METHODS
Exposure of male mice. Male mice (B6D2F1 from Charles River Laboratories, 8–12 weeks of age) received one ip injection of B[a]P (150mg/kg body weight) dissolved in corn oil, or corn oil only for controls, 4 days prior to isolation of their cauda sperm used for IVF treatment. Timing of the exposure to B[a]P was based on pilot studies and knowledge about the most susceptible stage of spermatogenesis with respect to dominant lethal mutations (Generoso, 1986). At the day of the IVF experiment males were killed by cervical dislocation. Cauda were surgically removed and transferred to an Eppendorf tube containing M2 medium (500 µl, Sigma). Using microscissors, a few incisions were made in the cauda and the sperm was allowed to disperse for 10min into ~250 µl HTF medium (EmbryoMax, Millipore) under liquid paraffin (MediCult), before transfer of sperm to the IVF dishes. Experiments comprise sperm from 14 males (7 exposed and 7 controls) and oocytes from 84 females.
Superovulation and IVF. Female mice (B6D2F1 from Charles River Laboratories, 4–6 weeks of age) were injected ip with pregnant mare serum hormone gonadotropin (Folligon; Intervet) (5 IU) 3 days prior to the IVF procedure, to induce superovulation. Two days later (i.e., the day before IVF), animals received an additional ip injection of human chorionic gonadotropin (Ovitrelle; Serono) (5 IU). Mice were killed by cervical dislocation and oviducts were collected in M2 medium (Sigma). Egg clutches (10–20 oocytes) embedded in cumulus cells were extracted from each oviduct. Oocytes were transferred to IVF dishes (35-mm petri dish, Corning) and incubated in a droplet of HTF medium containing sperm under liquid paraffin for 4.5h (37°C). Oocytes from one side of the animal were combined with sperm from B[a]P-exposed animals, and oocytes from the other side where combined with sperm from control animals. Hence, oocytes from all animals were present in both the control group and the exposed group. After 4.5h the fertilized oocytes (zygotes) were washed 5 times in potassium simplex optimization medium (KSOM) (EmbryoMax, Millipore) before they were transferred to a drop of KSOM (200 µl) under liquid paraffin, in a petri dish (35mm). Samples from the 1-cell stage were collected immediately after fertilization. Isolation of successful fertilizations was based on the appearance of polar bodies on the oocyte cell surface of healthy looking oocytes, when these were visible, and based on experience from previous experiments in which the majority of the oocytes were shown to be fertilized. The remaining zygotes were allowed to grow to be harvested at the 2-, 4-, 8-, and blastocyst cell stages. Upon harvest, zygotes/embryos were collected separately in micro tubes filled with 5 µl lysis medium (CelluLyser, TATAA Biocenter) and then frozen at −70°C.
Gene expression analysis—reverse transcription-qPCR. The reverse transcription (RT)-qPCR analysis of cell lysates was performed as previously described (Bergkvist et al., 2010). In brief, cDNA synthesis was performed using a transcriptor first strand cDNA synthesis kit according to the manufacturer’s recommendation (Roche, Cat. no. 04896866001). Hence, no RNA extraction was conducted. Samples were incubated in a thermal cycler unit (Eppendorf Mastercycler) according to the following protocol: 10min at 25°C, 30min at 50°C, 5min at 85°C, and then held at 4°C. After RT reaction, samples were stored at −70°C until use. A preamplification step of the cDNA was included: preamplification PCR was run in 50 μl reactions containing 5 μl of cDNA, 5 μl of a mixture of all forward and reverse primers (250nM each), 25 μl of iQ Supermix (Bio-Rad), and water. PCR primers were designed using the online Universal Probe Library System from Roche, and all primers showed PCR efficiency higher than 90% (Supplementary table 1). A CFX 96 cycler (Bio-Rad) was used for the preamplification with the cycling conditions: polymerase activation at 95°C for 3min, followed by 18 cycles (each of 95°C 15 s, 57°C 4min, and 72°C 20 s). The product of the preamplification reaction was diluted 10 times and stored at −20°C. Then, a high throughput qPCR was performed as previously described in detail (Brevik et al., 2011) using the BioMark qPCR platform (Fluidigm). The robustness of the preamplification was validated by comparing qPCR expression levels of Chek1 (highly expressed) and Mlh3 (typically low expression) with and without preamplification. The relative expression of the two genes was similar when analyzed with and without preamplification (Brevik et al., 2011).
Data preprocessing. Prior to normalization, the raw data (Cq values) generated from qPCR experiments were preprocessed to ensure that measurements at low levels were well within the linear area of detection; all Cq values above 26 were coded as missing values. In addition, embryos with gene expression pattern radically different from the overall group mean were classified as outliers. The excluded outliers were considered to represent deteriorating embryos that were about to die. Filtering criteria for missing values was set to 70%, which is the minimum percentage of existing values, and all the patterns with less than 70% existing values were removed. All estimates are mean values based on 5–33 single zygotes/embryos. Missing values were replaced by using the information contained in the biological replicates when available. Because the highest measured Cq of a truly positive sample can be assumed to reflect the limit of detection (LOD) for that particular gene, assigning Cq(LOD) + 1 to the off-scale samples corresponds to a concentration that is half of the LOD. Following the preprocessing, we ended up with 51 genes out of 78 genes and 184 samples (embryos) passing the quality assurance criteria described above.
Identification of stably expressed normalizing genes. Following the raw data preprocessing steps, we chose an optimal normalization strategy. The gene expression stability of the 51 genes that passed our filtering criteria was analyzed using the geNorm algorithm (Vandesompele et al., 2002), in order to identify the most stably expressed and putative normalizer genes. Raw Cq values were transformed to linear scale (relative quantities [RQs]), as shown in the equation below, before analysis.
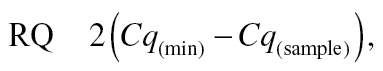 |
where Cq(min) is the sample with lowest Cq value. The geNorm calculates an expression stability M value (default limit, M < 1.5) and sample specific normalization factors (NFs). NFs were calculated as the geometric mean of the expression of the most stable normalizer. The selection of the optimal number of stably expressed genes was based on geNorm’s pairwise variation analyses between subsequent NFs using a default cutoff value of 0.15 for the inclusion of additional normalizers (Vandesompele et al., 2002). We also evaluated the suitability of the average gene expression value as a NF alongside of the 51 genes. For each individual sample, the average expression value was calculated based on the 51 genes that passed our filtering criteria. The three most stably expressed genes identified by geNorm were Carm1, Kat2a, and Lig3 (Supplementary fig. 1). The geometric mean of the Carm1, Kat2a, and Lig3 genes was used to calculate the NFs. The normalized relative expression quantity (NRQ) was generated by dividing the RQ of a sample by the sample specific NF, as follows:
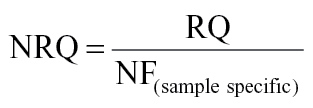 |
The fold difference (FD) between B[a]P-exposed and untreated control samples was then calculated by dividing the normalized RQ of B[a]P-exposed sample by the average of normalized RQ of cell stage–matched untreated control samples, as shown below:
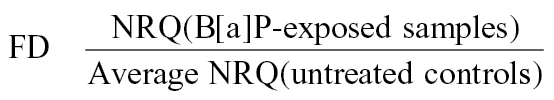 |
The FD was then log base 2 transformed; log2-transformed FDs are preferred for visualization. Using the approach outlined above, we compared gene expression in developing mouse embryos at the 1-, 2-, 4-, 8-, and blastocyst cell stages of embryo development, originating from fertilization with sperm from either B[a]P-exposed fathers or control fathers.
Statistical analysis. Gene expression data often deviate from normal distribution, but after log transformation they are normally distributed. Statistical significance between mean (log2-transformed) values was assayed by Student’s t-test. Values were considered to be significantly different when p < 0.05. Statistical tests were performed using Stata, version 9.
RESULTS
Zygotes and early embryos from five preimplantation stages (1-, 2-, 4-, 8-, and blastocyst cell stages), fertilized either with sperm from B[a]P-treated male or from control mice, were used to study the effect of paternal exposure to B[a]P on the expression of a panel of 78 mouse mRNAs. It is well known that embryonic cell cycle duration is subject to considerable variation, not only among different embryos but also among blastomers within the same embryo (Bowman and McLaren, 1970). In the present experiment, samples were selected exclusively from the pool of healthy looking embryos representative of that particular stage.
A total of 184 in vitro fertilized embryos (derived from 92 B[a]P exposed and 92 control sperm) at five different developmental stages were used for the final analysis. A set of gene expression data was obtained from each embryo. Sample extraction was timed according to 24h cycles, with the 1-, 2-, 4-, 8-cell, and blastocyst time points referring to 1, 24, 48, 72, and 120h, respectively. The stages can be readily identified up to the 8-cell stage.
Gene Expression Analysis
We used only those genes that passed the quality assurance criteria described in the Materials and Methods section. After data normalization and filtering, we ended up with a matrix of 51 genes × 184 samples (embryos), and this matrix was used for downstream analysis. The normalized data were used to calculate FD between B[a]P exposed and control samples. A complete list of genes included in the downstream analysis is shown in Supplementary table 1.
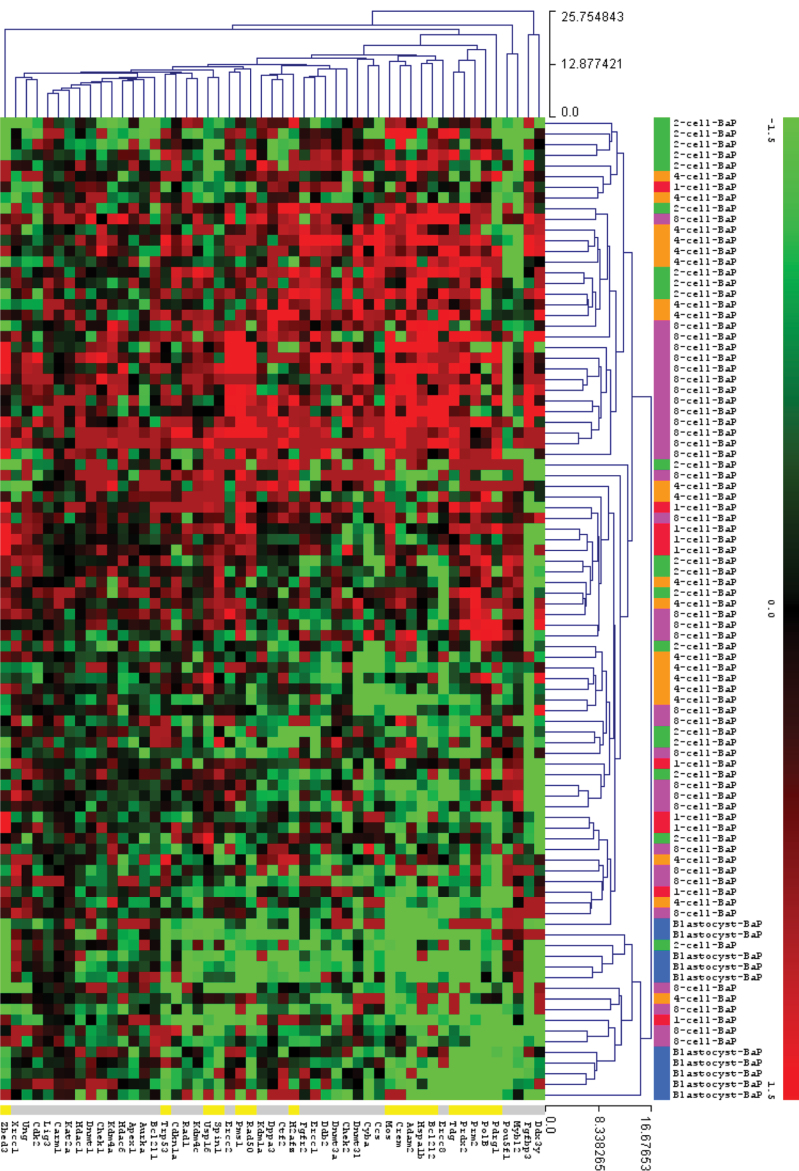
We then conducted hierarchical clustering analysis (average linkage and Euclidean distance similarity measurement) in order to group samples from different embryonic stages based on their transcript level similarity using the MeV software version 4.7 (Saeed et al., 2006). The hierarchical clustering analyses results were visualized in a dendrogram and are presented in Figure 1. By visual inspection of the clustering dendrogram, we observed that blastocyst stage samples clustered close to each other and most of the genes at this stage were downregulated following paternal B[a]P exposure; there were however a few samples from the earlier stages, which clustered together with the blastocysts (Fig. 1). The slight upregulation or lack of downregulation of three genes, Mybl2, Pou5f1, and Xrcc1, in five samples is responsible for the segregation of the blastocysts. Many of the 8-cell stage embryos (41%) cluster together in one group characterized by a marked upregulation of several genes; however the majority of the 8-stage embryos cluster together with samples from other developmental stages. In a subanalysis of the 13 samples that cluster together in the heatmap at the 8-cell stage (Fig. 1), the following genes were significantly upregulated more than twofold in embryos of B[a]P-exposed fathers: Adam2, Bcl2l2, Chek2, Crem, Ddb2, Ercc1, Ercc8, Fgfbp3, Hspa1b, Mos, Pms1, Pdrg1, Prm2, Rad50, Tdg, Ercc2, Zbed3 (data not shown).
FIG. 1.
Unsupervised hierarchical clustering analysis of the relative expression of 51 genes after filtering and normalization of the data. The hierarchical clustering analysis is based on similarities in gene expression. Genes are coded according to their expression patterns (the expression scale spans from -6.0 to 4.5). Samples are horizontally labeled based on the developmental stage they belong to. Vertically, labeled genes indicate more than twofold significantly downregulated expression levels at the blastocyst stage. The covariance value was used as distance metric in this complete hierarchical linkage clustering analysis.
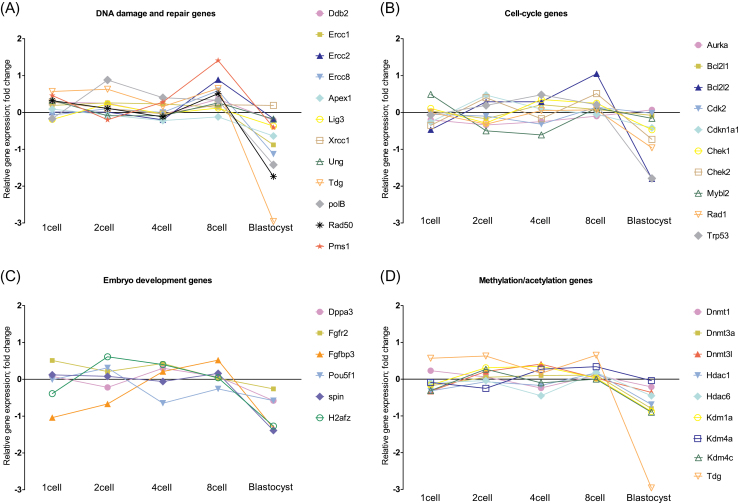
To investigate if the effects of paternal B[a]P exposure were strongly manifested in particular pathways, we looked at relative gene expression based on functional classification of the genes. The results are shown in Figure 2. There is a tendency towards an upregulation of many genes at the 8-cell stage. Still, we found no sign of statistically significant pathway-based enrichment in the effects of B[a]P exposure. The most obvious pattern that emerges is the downregulation of gene expression in embryos of exposed fathers at the blastocyst stage. Table 1 shows the 16 significantly differentially expressed genes (p < 0.05 and ± 2-fold cutoff value) between B[a]P-exposed and control samples in one or several embryonic development stages. With the exception of Pms1, all of these genes were downregulated more than 2 times at the blastocyst stage following paternal B[a]P exposure.
FIG. 2.
Relative gene expression at various developmental stages for (A) genes involved in DNA damage repair; (B) genes involved in cell-cycle regulation; (C) genes involved in embryo development; and (D) genes involved in methylation and acetylation pathways. Expression in embryos of exposed fathers is shown relative to expression in control embryos.
TABLE 1.
Differentially Expressed Genes (Based on Twofold Cutoff Value) Between Control Embryos and Embryos of Exposed Fathers at Various Stages of Embryo Development
| Genes | 1 cell | 2 cell | 4 cell | 8 cell | Blastocyst |
|---|---|---|---|---|---|
| Adam2 | −0.5 | 0.9 | 0.7 | 0.7 | −2.6 |
| Bcl2l2 | −0.5 | 0.3 | 0.3 | 1.0 | −1.8 |
| Crem | −0.9 | 0.6 | 0.1 | 1.0 | −1.4 |
| Cyba | −0.5 | 0.1 | −0.1 | 0.3 | −1.7 |
| H2afz | −0.4 | 0.6 | 0.4 | 0.0 | −1.3 |
| Hsp1b | −0.2 | 0.2 | 0.2 | 0.6 | −1.9 |
| Mos | 0.4 | 0.7 | 0.0 | 1.0 | −1.9 |
| Pms1 | 0.5 | −0.2 | 0.3 | 1.4 | −0.4 |
| PolB | −0.2 | 0.9 | 0.4 | 0.3 | −1.4 |
| Pdrg1 | −0.6 | −0.5 | 0.3 | 1.7 | −2.0 |
| Prdx2 | 0.2 | 0.4 | 0.0 | 0.2 | −3.4 |
| Prm2 | 0.5 | 0.5 | 0.9 | 0.7 | −4.6 |
| Rad50 | 0.3 | 0.1 | −0.1 | 0.5 | −1.7 |
| Spin | 0.1 | 0.1 | −0.1 | 0.2 | −1.4 |
| Tdg | 0.6 | 0.6 | 0.2 | 0.6 | −3.0 |
| Trp53 | −0.1 | 0.2 | 0.5 | 0.2 | −1.8 |
| Usp16 | 0.3 | 0.0 | 0.2 | 0.3 | −1.1 |
| Zbed3 | 1.7 | −0.6 | −0.1 | 0.8 | −1.9 |
Note. Bold values represent statistically significant up- and downregulated genes.
Integrated Analysis of miRNA-mRNA Interaction
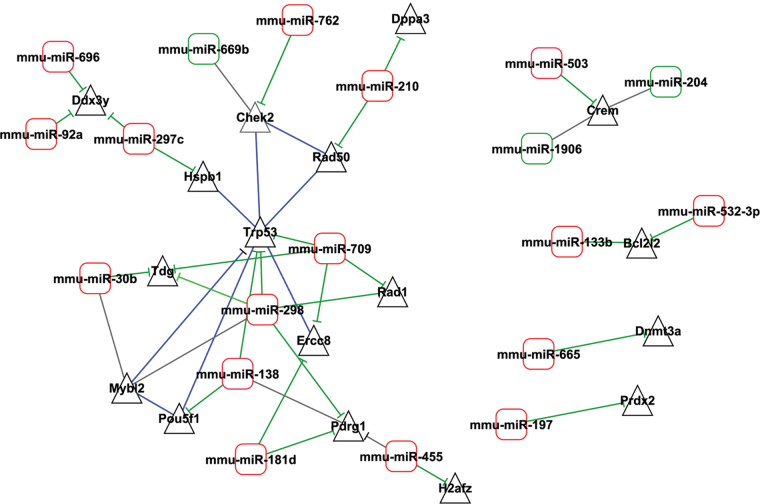
In a recent study, we investigated the effects of paternal exposure to B[a]P on global miRNA expression in the early developing mouse embryo and identified expression of 102 miRNAs (Brevik et al., 2012). In order to identify potential genes whose mRNA might be targeted by these 102 miRNAs, we conducted an inverse correlation analysis of the integrated expression profile data of the 102 miRNAs and the 51 gene list from this study. We first searched for predicted mRNA targets of the 102 miRNA list using the miRWalk database (Dweep et al., 2011). In order to determine how many of our B[a]P responsive genes could be considered predicted targets for B[a]P responsive miRNAs, we conducted similarity searches between the miRWalk database predicted target list, and our 51 membered gene list. We identified 37 genes from our list among the predicted gene target list targeted by 68 of the 102 B[a]P responsive miRNAs identified previously (Brevik et al., 2012). Most of the genes were targeted by more than one miRNA. The expression data for the 37 genes and the expression data for the 68 miRNAs were combined to identify inversely correlated miRNA-mRNA pairs. In particular, we were interested in those miRNA-mRNA pairs showing upregulated miRNA expression level in one or several embryonic development stages, and downregulated mRNA level in the corresponding embryonic stage following paternal B[a]P exposure. We found 33 inversely correlated miRNA-mRNA pairs (19 miRNAs and 17 mRNAs). The resulting network of miRNA-mRNA pairs, constructed using CytoScape, version 2.6 (Cline et al., 2007), is shown in Figure 3. A cluster consisting of miR-709, miR-455, miR-30b, miR-298 and miR-138, and Ercc8, Rad1, Tdg, Trp53, H2afz, Mybl2, Pdrg1, and Pou5f1 was evident from the network analysis.
FIG. 3.
miRNA-mRNA interaction network of 33 inversely correlated miRNA-mRNA pairs (19 miRNAs and 17 mRNAs). In this network, 16 out of 19 miRNAs (mmu-miR-133b, mmu-miR-138, mmu-miR-181d, mmu-miR-197, mmu-miR-210, mmu-miR-297c, mmu-miR-298, mmu-miR-30b, mmu-miR-455, mmu-miR-503, mmu-miR-532-3p, mmu-miR-665, mmu-miR-696, mmu-miR-709, mmu-miR-762 and mmu-miR-92a) were upregulated and the remaining three miRNAs (mmu-miR-1906, mmu-miR-204 and mmu-miR-669b) were downregulated, in previously determined of B[a]P dysregulated miRNAs (Brevik et al., 2012). These miRNAs are inversely correlated with the 17 mRNAs from this study. Connection lines represent miRNA-mRNA interaction; there are negative miRNA-mRNA pair correlation between the 16 upregulated miRNAs and downregulated mRNAs. There are also connection between genes, and inconsistently correlated miRNA-mRNA pairs.
To ascribe biological relevance to the selected 51 genes, we conducted a functional enrichment analysis using the online FunNet Transcriptional Networks Analysis tool, www.funnet.info (Prifti et al., 2008). The genes that passed our quality assurance criteria, and hence being the basis for Figure 1, were included in this enrichment analysis. The top seven biological processes represented by our gene list were DNA transcription, DNA damage response, cell cycle regulation, chromatin modification, oxidation-reduction processes, apoptosis, and embryo development. A complete table of 40 enriched GO biological processes is included as Supplementary table 2.
DISCUSSION
Changes in reproductive health has been reported for many Western countries and have been linked to exposure to environmental contaminants in association studies. Data supporting the importance of paternal exposure to testicular toxicants for fertility and health of the progeny have been reviewed by Hales and Robaire (2001). Recently, a study with mice documented paternally mediated transgenerational impact of B[a]P exposure on male mice fertility (Mohamed et al., 2010). Using 32P postlabeling, we recently showed very high levels of DNA adducts in human sperm exposed in vitro to the B[a]P metabolite BPDE (Sipinen et al., 2010). We also detected B[a]PDE-N(2)-dG adducts in mouse epididymal spermatozoa, i.e., in caput sperm 4 days after 3 × 50mg/kg ip B[a]P (Olsen et al., 2005, 2010). In the present report, we demonstrate that paternal B[a]P exposure of epididymal sperm significantly affects subsequent gene expression in the developing IVF mouse embryo. Although the mechanisms are far from being fully understood, this altered gene expression may be induced by the presence of DNA adducts in sperm DNA. Alternatively, epigenetic modifications may (also) be involved.
Our main objective in this study has been to explore early embryonic transcriptional effects induced by paternal exposure to environmental chemicals. In a previous investigation (Brevik et al., 2011), we reported upregulated gene expression at the 8-cell stage of embryonic development after paternal exposure to glycidamide (GA), the active metabolite of acrylamide. We also noted an initial downregulation at the 1- and 2-cell stage of embryo development following paternal GA exposure. In this study, we further analyzed gene expression patterns in samples from the blastocyst stage. The effects of paternal exposure to B[a]P did not give rise to an initial downregulation at the 1- and 2-cell stage, as opposed to in the GA study. However, a common effect of both these environmental chemicals was to upregulate gene expression in developing embryos at the 8-cell stage. Most of the studied genes were similarly upregulated after both GA and B[a]P exposure (~88% of the genes were upregulated in embryos derived from B[a]P-exposed fathers compared with controls; for GA, the comparable gene ratio was 87%). Together these observations indicate that the 8-cell stage may represent a sensitive stage of mouse embryo development for chemically induced changes in gene expression. A striking observation in this experiment is the largely uniform downregulation of genes at the blastocyst stage (Figs. 1 and 2). Although no visible differences between the groups were evident upon careful morphological assessment in the microscope, the widespread downregulation of mRNAs in embryos of exposed fathers at the blastocyst stage could indicate that the number of cells was slightly different between the groups, or that the ratio between trophoblasts and embryoblasts was altered. For all practical purposes, counting cells in the microscope is virtually impossible at the blastocyst stage of embryo development without the use of nuclear staining, which was not employed in the present experiment. Previously it has been reported that paternal cyclophosphamide exposure causes decreased cell proliferation in developing embryos (Austin et al., 1994). Furthermore, Pou5f1 (Oct4) is required to develop a true inner cell mass at the blastocyst stage (Nichols et al., 1998); the relatively low expression of Pou5f1, which we observed in embryos of exposed fathers at the 4-, 8-, and blastocyst cell stages (Fig. 2), could be consistent with a compromised development of the inner cell mass in embryos of exposed fathers. Further studies are needed to address whether important phenotypical changes are manifest due to transcriptional dysregulation. Nevertheless, we believe that our rigorous data normalization procedures should be sufficiently stringent to adjust for the effect of potential differences in cell numbers between the groups. The clustering of 41% of the embryos exhibiting significantly upregulated gene expression at the 8-cell stage represents another interesting observation (Fig. 1). It is uncertain why this characteristic pattern of broad upregulation is manifested only in some embryos, whereas others express a more balanced pattern. One explanation could be that some of the exposed embryos develop their full transcriptional competency at slightly different times show a developmental delay. The B[a]P exposed sperm is assumed to inflict variable levels of damage on the cellular machinery in the developing embryos, thus the clustered samples could reflect embryos with more severe lesions than their IVF siblings.
During the first cell divisions after fertilization, extensive genome-wide demethylation of 5-methylcytosine of both the paternal and the maternal genome occurs, and interference with this process may interfere with embryo development or offspring phenotype. In this study, we included a set of genes related to DNA methylation; Dnmt1, Dnmt3a, Dnmt3l, and Tdg and indirectly Polβ. Tdg and Polβ showed significantly altered gene expression levels in early embryos of the B[a]P-exposed fathers. Tdg is believed to participate in the active demethylation of the paternal genome (reviewed by Nabel and Kohli [2011]). The altered expression of Tdg and Polβ during the first cell divisions after fertilization with B[a]P sperm may interfere with the epigenetic status in the early embryo; this warrants further studies.
Bcl2l2, Trp53, Pdrg1, and Pms1 were among the genes that were significantly upregulated at the 8-cell stage (Table 1). Bcl2l2 has antiapoptotic properties and belongs to a family of prosurvival molecules. It is targeted by two miRNAs, miR-133b and miR-582-3p, and the expression of Bcl2l2 was inversely correlated with miR-133b at the blastocyst stage (Fig. 3). Recently, a similar inverse correlation was reported in cancer cells, where overexpression of miR-133b was shown to induce apoptosis through downregulation of Bcl2l2 (Crawford et al., 2009). Another interesting response at the 8-cell stage was the upregulation of Pdrg1. Pdrg1 emerges as a novel cancer biomarker because the expression of this gene has been found to be upregulated in multiple malignancies including cancers of the colon, rectum, ovary, lung, stomach, breast, and uterus (Jiang et al., 2011). Jiang and coworkers also reported selective regulation of Pdrg1 by agents inducing genotoxic stress. Four different B[a]P dysregulated miRNAs were found to target Pdrg1 in the current experiment (Fig. 3). More research is however needed to fully characterize Pdrg1 modulation by its multiple B[a]P responsive miRNA associations. The upregulation of Pms1 at the 8-cell stage could indicate increased DNA mismatch repair activity. This protein belongs to the mutL/hexB family, and although its DNA repair function has not been proven, it is assumed to have a role in mismatch repair because it forms heterodimers with Mlh1, a commonly recognized mismatch repair protein. We recently observed a nonsignificant upregulation of Pms1 at the 8-cell stage in embryos of GA exposed fathers (Brevik et al., 2011).
From the matching of miRNAs with their target mRNAs, it is clear that several B[a]P responsive genes, e.g., Trp53, Pdrg1, Ddx3y, Crem, Check2, and Bcl2l2, are targeted by several B[a]P responsive miRNAs (Fig. 3). Based on the present data, we are unable to thoroughly assess the strength of each miRNAs’ contribution to the overall regulation of each gene. Rather than activation of a specific molecular mechanism, the matched miRNA and mRNA analyses suggest that the preconceptional paternal B[a]P exposure elicits broad genotoxic stress responses in the developing in vitro mouse embryo, with p53 protein as a central player. This study highlights some anticorrelated miRNA-mRNA pairs in the developing mouse embryo resulting from paternal exposure to B[a]P, but more studies are needed to thoroughly understand how key miRNA-mRNA regulatory modules in the developing embryo are affected by environmental contaminants. Considering the relatively narrow time window between exposure and sperm collection, the effects demonstrated in this experiment are mediated by sperm residing in the epididymis at the time of B[a]P treatment. The spermatogenesis is generally susceptible to xenobiotic exposure, but the most sensitive stages vary between different chemicals demonstrating different modes of action. We speculate that, in an evolutionary perspective, pathway-spanning effects that force the embryo to enter a recovery mode, is initiated by the B[a]P-exposed sperm; embryos of exposed fathers may require more time to cope with critical transitions. It is worth noting that the reported gene expression downregulation in embryos of exposed fathers is relative to controls. Hence, the observed downregulation may be interpreted as a delay or attenuation of a natural upregulation of several genes in control embryos.
Based on the present data, one cannot decide whether the B[a]P-induced effects are due to adducts in sperm DNA or may result from other sperm modifications. Our recent report showing BPDE adducts in highly compacted sperm DNA (Olsen et al., 2005, 2010), and also high levels of BPDE adducts in human sperm exposed to BPDE in vitro (Sipinen et al., 2010) suggest strongly that direct DNA adducts might be relevant.
In conclusion, paternal exposure to B[a]P leads to altered gene expression in preimplantation mouse embryos. In particular, a significant downregulation of several genes at the blastocyst stage was observed. At the 8-cell stage a clear differential response to paternal B[a]P exposure was found, with nearly 41% of the embryos forming a distinct cluster with significantly upregulated expression of 13 genes. Analysis of the embryonic gene expression suggests a dysregulation of genes related to DNA repair, cell cycle, Trp53, and MAPK signaling. It is well known that post-translational modifications may affect the functionality of transcribed gene products. It is at his point, however, not clear to what extent dysregulated gene expression at these early stages of embryo development may result in phenotypic changes, possibly predisposing the individual to disease later in life. This is an intriguing question that clearly deserves further attention. This study represents a first exploration of paternally mediated transcriptomal changes in early embryo development, induced by a common environmental chemical.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org.
FUNDING
The Research Council of Norway (182048/V40); The Czech Republic Ministry of Education, Youth and Sports (AVOZ50520701).
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Jill Andersen for her help in handling experimental animals.
Authors’ Roles: A.B., B.L., G.B., N.D., and A.K.O. contributed to the study conception and design. B.L. and A.B. performed the animal experiments and sample extraction. V.R. performed the RT-qPCR. A.B. and N.D. carried out the data analysis and drafted the manuscript. All authors critically reviewed, contributed to and approved the final version of the manuscript.
REFERENCES
- Auger J.,, Kunstmann J. M.,, Czyglik F.,, Jouannet P. (1995).. Decline in semen quality among fertile men in Paris during the past 20 years. N. Engl. J. Med. 332, 281––285 [DOI] [PubMed] [Google Scholar]
- Austin S. M.,, Robaire B.,, Hales B. F.,, Kelly S. M. (1994).. Paternal cyclophosphamide exposure causes decreased cell proliferation in cleavage-stage embryos. Biol. Reprod. 50, 55––64 [DOI] [PubMed] [Google Scholar]
- Aygün S. F.,, Kabadayi F. (2005).. Determination of benzo[a]pyrene in charcoal grilled meat samples by HPLC with fluorescence detection. Int. J. Food Sci. Nutr. 56, 581––585 [DOI] [PubMed] [Google Scholar]
- Bergkvist A.,, Rusnakova V.,, Sindelka R.,, Garda J. M.,, Sjögreen B.,, Lindh D.,, Forootan A.,, Kubista M. (2010).. Gene expression profiling—Clusters of possibilities. Methods 50, 323––335 [DOI] [PubMed] [Google Scholar]
- Bowman P.,, McLaren A. (1970).. Cleavage rate of mouse embryos in vivo and in vitro. J. Embryol. Exp. Morphol. 24, 203––207 [PubMed] [Google Scholar]
- Brevik A., Lindeman B., Brunborg G., Duale N. (2012).. Paternal benzo[a]pyrene exposure modulates microRNA expression patterns in the developing mouse embryo. Intl. J. Cell Biol. 2012, 407 431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevik A.,, Rusnakova V.,, Duale N.,, Slagsvold H. H.,, Olsen A. K.,, Storeng R.,, Kubista M.,, Brunborg G.,, Lindeman B. (2011).. Preconceptional paternal glycidamide exposure affects embryonic gene expression: Single embryo gene expression study following in vitro fertilization. Reprod. Toxicol. 32, 463––471 [DOI] [PubMed] [Google Scholar]
- Carlsen E.,, Giwercman A.,, Keiding N.,, Skakkebaek N. E. (1992).. Evidence for decreasing quality of semen during past 50 years. BMJ 305, 609––613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M. S.,, Smoot M.,, Cerami E.,, Kuchinsky A.,, Landys N.,, Workman C.,, Christmas R.,, Avila-Campilo I.,, Creech M.,, Gross B.,, et al. (2007).. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366––2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford M.,, Batte K.,, Yu L.,, Wu X.,, Nuovo G. J.,, Marsh C. B.,, Otterson G. A.,, Nana-Sinkam S. P. (2009).. MicroRNA 133B targets pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem. Biophys. Res. Commun. 388, 483––489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrova Y. E.,, Nesterov V. N.,, Krouchinsky N. G.,, Ostapenko V. A.,, Neumann R.,, Neil D. L.,, Jeffreys A. J. (1996).. Human minisatellite mutation rate after the Chernobyl accident. Nature 380, 683––686 [DOI] [PubMed] [Google Scholar]
- Dweep H.,, Sticht C.,, Pandey P.,, Gretz N. (2011).. miRWalk–database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 44, 839––847 [DOI] [PubMed] [Google Scholar]
- Edwards T. M.,, Moore B. C.,, Guillette L. J., Jr. (2006).. Reproductive dysgenesis in wildlife: A comparative view. Int. J. Androl. 29, 109––121 [DOI] [PubMed] [Google Scholar]
- Generoso W. M. (1986).. Relationship between alkylation sites and induction of dominant lethals and heritable translocations in mice. Prog. Clin. Biol. Res. 209B, 493––500 [PubMed] [Google Scholar]
- Generoso W. M.,, Cain K. T.,, Hellwig C. S.,, Cacheiro N. L. (1982).. Lack of association between induction of dominant-lethal mutations and induction of heritable translocations with benzo[a]pyrene in postmeiotic germ cells of male mice. Mutat. Res. 94, 155––163 [DOI] [PubMed] [Google Scholar]
- Hales B. F.,, Robaire B. (2001).. Paternal exposure to drugs and environmental chemicals: Effects on progeny outcome. J. Androl. 22, 927––936 [DOI] [PubMed] [Google Scholar]
- Hattemer-Frey H. A.,, Travis C. C. (1991).. Benzo-a-pyrene: Environmental partitioning and human exposure. Toxicol. Ind. Health 7, 141––157 [DOI] [PubMed] [Google Scholar]
- Jeffrey A. M.,, Weinstein I. B.,, Jennette K. W.,, Grzeskowiak K.,, Nakanishi K.,, Harvey R. G.,, Autrup H.,, Harris C. (1977).. Structures of benzo(a)pyrene-nucleic acid adducts formed in human and bovine bronchial explants. Nature 269, 348––350 [DOI] [PubMed] [Google Scholar]
- Ji B. T.,, Shu X. O.,, Linet M. S.,, Zheng W.,, Wacholder S.,, Gao Y. T.,, Ying D. M.,, Jin F. (1997).. Paternal cigarette smoking and the risk of childhood cancer among offspring of nonsmoking mothers. J. Natl. Cancer Inst. 89, 238––244 [DOI] [PubMed] [Google Scholar]
- Jiang L.,, Luo X.,, Shi J.,, Sun H.,, Sun Q.,, Sheikh M. S.,, Huang Y. (2011).. PDRG1, a novel tumor marker for multiple malignancies that is selectively regulated by genotoxic stress. Cancer Biol. Ther. 11, 567––573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazerouni N.,, Sinha R.,, Hsu C. H.,, Greenberg A.,, Rothman N. (2001).. Analysis of 200 food items for benzo[a]pyrene and estimation of its intake in an epidemiologic study. Food Chem. Toxicol. 39, 423––436 [DOI] [PubMed] [Google Scholar]
- Lee B. M.,, Shim G. A. (2007).. Dietary exposure estimation of benzo[a]pyrene and cancer risk assessment. J. Toxicol. Environ. Health Part A 70, 1391––1394 [DOI] [PubMed] [Google Scholar]
- Marchetti F.,, Wyrobek A. J. (2005).. Mechanisms and consequences of paternally-transmitted chromosomal abnormalities. Birth Defects Res. C Embryo Today 75, 112––129 [DOI] [PubMed] [Google Scholar]
- Marchetti F.,, Wyrobek A. J. (2008).. DNA repair decline during mouse spermiogenesis results in the accumulation of heritable DNA damage. DNA Repair (Amst.) 7, 572––581 [DOI] [PubMed] [Google Scholar]
- Mohamed E.,, Song W. H.,, Oh S. A.,, Park Y. J.,, You Y. A.,, Lee S.,, Choi J. Y.,, Kim Y. J.,, Jo I.,, Pang M. G. (2010).. The transgenerational impact of benzo(a)pyrene on murine male fertility. Hum. Reprod. 25, 2427––2433 [DOI] [PubMed] [Google Scholar]
- Nabel C. S.,, Kohli R. M. (2011).. Molecular biology. Demystifying DNA demethylation. Science 333, 1229––1230 [DOI] [PubMed] [Google Scholar]
- Nichols J.,, Zevnik B.,, Anastassiadis K.,, Niwa H.,, Klewe-Nebenius D.,, Chambers I.,, Schöler H.,, Smith A. (1998).. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379––391 [DOI] [PubMed] [Google Scholar]
- Olsen A. K.,, Andreassen A.,, Singh R.,, Wiger R.,, Duale N.,, Farmer P. B.,, Brunborg G. (2010).. Environmental exposure of the mouse germ line: DNA adducts in spermatozoa and formation of de novo mutations during spermatogenesis. PLoS One 5, e11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A. K.,, Lindeman B.,, Wiger R.,, Duale N.,, Brunborg G. (2005).. How do male germ cells handle DNA damage? Toxicol. Appl. Pharmacol. 207, 521––531 [DOI] [PubMed] [Google Scholar]
- Pelkonen O.,, Nebert D. W. (1982).. Metabolism of polycyclic aromatic hydrocarbons: Etiologic role in carcinogenesis. Pharmacol. Rev. 34, 189––222 [PubMed] [Google Scholar]
- Penning T. M. (2004).. Aldo-keto reductases and formation of polycyclic aromatic hydrocarbon o-quinones. Meth. Enzymol. 378, 31––67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D. H. (1983).. Fifty years of benzo(a)pyrene. Nature 303, 468––472 [DOI] [PubMed] [Google Scholar]
- Prifti E.,, Zucker J. D.,, Clement K.,, Henegar C. (2008).. FunNet: An integrative tool for exploring transcriptional interactions. Bioinformatics 24, 2636––2638 [DOI] [PubMed] [Google Scholar]
- Saeed A. I.,, Bhagabati N. K.,, Braisted J. C.,, Liang W.,, Sharov V.,, Howe E. A.,, Li J.,, Thiagarajan M.,, White J. A.,, Quackenbush J. (2006).. TM4 microarray software suite. Meth. Enzymol. 411, 134––193 [DOI] [PubMed] [Google Scholar]
- Shukla Y.,, Taneja P. (2001).. Antimutagenic effect of black tea extract using ‘rodent dominant lethal mutation assay’. Toxicology 168, 269––274 [DOI] [PubMed] [Google Scholar]
- Sipinen V.,, Laubenthal J.,, Baumgartner A.,, Cemeli E.,, Linschooten J. O.,, Godschalk R. W.,, Van Schooten F. J.,, Anderson D.,, Brunborg G. (2010).. In vitro evaluation of baseline and induced DNA damage in human sperm exposed to benzo[a]pyrene or its metabolite benzo[a]pyrene-7,8-diol-9,10-epoxide, using the comet assay. Mutagenesis 25, 417––425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers C. M.,, Yauk C. L.,, White P. A.,, Parfett C. L.,, Quinn J. S. (2002).. Air pollution induces heritable DNA mutations. Proc. Natl. Acad. Sci. U.S.A. 99, 15904––15907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J.,, De Preter K.,, Pattyn F.,, Poppe B.,, Van Roy N.,, De Paepe A.,, Speleman F. (2002).. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhofstad N.,, van Oostrom C. T.,, van Benthem J.,, van Schooten F. J.,, van Steeg H.,, Godschalk R. W. (2010).. DNA adduct kinetics in reproductive tissues of DNA repair proficient and deficient male mice after oral exposure to benzo(a)pyrene. Environ. Mol. Mutagen. 51, 123––129 [DOI] [PubMed] [Google Scholar]
- Zenzes M. T.,, Bielecki R.,, Reed T. E. (1999a). Detection of benzo(a)pyrene diol epoxide-DNA adducts in sperm of men exposed to cigarette smoke. Fertil. Steril. 72, 330––335 [DOI] [PubMed] [Google Scholar]
- Zenzes M. T.,, Puy L. A.,, Bielecki R.,, Reed T. E. (1999b). Detection of benzo[a]pyrene diol epoxide-DNA adducts in embryos from smoking couples: Evidence for transmission by spermatozoa. Mol. Hum. Reprod. 5, 125––131 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.