Abstract
Plants are constantly exposed to microbes, for this reason they have evolved sophisticated strategies to perceive and identify biotic interactions. Thus, plants have large collections of so-called resistance (R) proteins that recognize specific microbe factors as signals of invasion. One of these proteins is codified by the Arabidopsis thaliana HR4 gene in the Col-0 ecotype that is homologous to RPW8 genes present in the Ms-0 ecotype. In this study, we investigated the expression patterns of the HR4 gene in Arabidopsis seedlings interacting with the beneficial fungus Trichoderma atroviride. We observed the induction of the HR4 gene mainly at 96 hpi when the fungus interaction was established. Furthermore, we found that the HR4 gene was differentially regulated in interactions with the beneficial bacterium Pseudomonas fluorescens and the pathogenic bacterium P. syringae. When hormone treatments were applied to A. thaliana (Col-0), each hormone treatment induced changes in HR4 gene expression. On the other hand, the expression of the RPW8.1 and RPW8.2 genes of Arabidopsis ecotype Ms-0 in interaction with T. atroviride was assessed. Interestingly, these genes are interaction-responsive; in particular, the RPW8.1 gene shows a very high level of expression in the later stages of interaction. These results indicate that HR4 and RPW8 genes could play a role in the establishment of Arabidopsis interactions with beneficial microbes.
Keywords: Arabidopsis thaliana, Col-0, Ms-0, HR4, R genes, RPW8, Trichoderma atroviride
1. Introduction
Interactions between plants and microbes result in pathogenic or in beneficial associations. It has been revealed that in both interactions a common panel of signaling pathways might participate in the establishment of the equilibrium between plant and microbes or their break-up. Plants appear to detect both pathogenic and symbiotic microbes by a similar set of genes [1]. Beneficial microbes are initially recognized as strange organisms activating the plant’s immune system, which culminates in the establishment of intimate mutualistic relationships. Two strategies have been described to detect microbes, the first involves pattern recognition receptors (PRRs) and the second one disease resistance (R) proteins. Immune signaling in plants is initiated upon perception of non-self molecules or elicitors that are often conserved among different classes of microbes, called pathogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns (MAMPs) [2]. Plants also respond to endogenous molecules released by microbe invasion, such as cell wall or cuticular fragments called danger-associated molecular patterns (DAMPs) [3–5]. PAMPs, MAMPs or DAMPs are recognized by PRRs. The PRRs are divided into two classes: transmembrane receptor kinases (RKs) and transmembrane receptor-like proteins (RLPs). The stimulation of PRRs leads to MAMP-triggered immunity (MTI) [6,7]. The second strategy of perception involves recognition by plant intracellular receptors (R proteins) of the microbe’s effectors; this recognition induces effector-triggered immunity (ETI), and has led to co-evolutionary dynamics between the plant and the microbe. The effectors are characteristically variable and dispensable in contrast to PAMPs and MAMPs [6,8,9].
The R proteins detect the microbe’s effectors and activate strong defenses. Recently, plant R genes have been classified into eight groups based on their amino acid motif organization and their membrane spanning domains. In particular, the Arabidopsis RPW8 proteins are an example of the sixth class of R genes, which contains a coiled coil domain (CC), and an N-terminal transmembranal domain (TrD) helix that is assumed to localize RPW8 proteins to the endomembrane [8,10]. The two paralogous Arabidopsis Ms-0 R genes, RPW8.1 and RPW8.2, confer broad spectrum resistance to multiple powdery mildew isolates that belong to distinct Erysiphe spp., and cause diseases in numerous plant species [11]. In contrast, most characterized R genes confer resistance to one or a few isolates of a particular pathogen. All tested Arabidopsis accessions contain three homologs of RPW8, (HR1, HR2 and HR3) that are closely linked to the RPW8 locus [11,12]. Two basic Arabidopsis haplotypes have been identified at the RPW8 locus based on the presence or absence of RPW8.1 and RPW8.2 genes: One comprises both RPW8.1 and RPW8.2 genes and the other is found in susceptible ecotypes that contain the HR4 gene instead of the RPW8.1 and RPW8.2 genes [13,14].
Trichoderma species are soil-borne fungi that present a high activity of interaction with soil pathogens and plant roots. Trichoderma spp. can reduce the severity of plant diseases by inhibiting plant pathogens in the soil through their highly potent antagonistic activity [15,16]. Moreover, some Trichoderma strains can interact directly with roots; as a result, they increase plant growth, resistance to disease and tolerance to abiotic stresses. Like other beneficial microbes, Trichoderma elicits Induced Systemic Resistance (ISR) by jasmonic acid/ethylene (JA/ET) dependent pathways and triggers priming responses in the plant. However, the Trichoderma-plant cross-talk is dynamic and the expression of defense-related genes of the JA/ET and/or salicylic acid (SA) pathways may overlap, depending on several conditions [17,18]. Trichoderma also produces the phytohormones ET and indole-3-acetic acid (IAA), which play roles in interconnecting plant development and defense response [19,20]. Considering the large number of metabolites secreted and the intimate contact with the root epidermis by Trichoderma, as well as the expanding list of diverse elicitors produced by fungi, Trichoderma’s elicitors may be part of a signaling cascade resulting in greater colonization or plant resistance induction [21].
For a better understanding of plant-microbe interactions, it is essential to study the molecular recognition events. In this sense, we selected the HR4 gene for this study. Here, we analyzed the gene expression patterns of the Arabidopsis thaliana HR4 gene under different conditions of interaction with the beneficial fungus Trichoderma atroviride. Furthermore, RPW8.1 and RPW8.2 genes (from Ms-0 ecotype) paralogs of the HR4 gene (Col-0 ecotype) were examined during the interaction with T. atroviride. Moreover, the analysis of HR4 gene was extended to bacterial interactions, using beneficial Pseudomonas fluorescens and the plant-pathogenic Pseudomonas syringae pv. tomato DC3000 (Pst DC3000). Finally, HR4 gene transcriptional profiling in response to hormones such as ET, SA and methyl jasmonate (MeJA) was carried out in order to determine the response to hormones. Despite the fact that the ecotype Col-0 of A. thaliana is one of the more studied, the HR4 gene has not been previously characterized. Thus the data obtained on the HR4 gene provides valuable information regarding this type of R proteins in Col-0 background.
2. Results
2.1. The HR4 Gene (Ecotype Col-0) Expression in Interaction with Trichoderma
The HR4 gene was selected from a cDNA microarray experiment obtained after 48 h of interaction of 25-day-old A. thaliana roots with T. atroviride. From the results of microarray bioinformatics analysis, we focused on R genes that showed a significant induction during this interaction. From ten induced R genes, the HR4 gene showed the highest Z-score (5.1) on the microarray experiment.
qRT-PCR analyses were performed to corroborate the microarray data on HR4 expression after 48 h of interaction, using 25-day-old plants grown on MS (Figure 1B). An induction of 1.4 times of the HR4 gene was observed in A. thaliana roots after 48 h post-inoculation (hpi) with Trichoderma spores, in comparison to control plants without fungus.
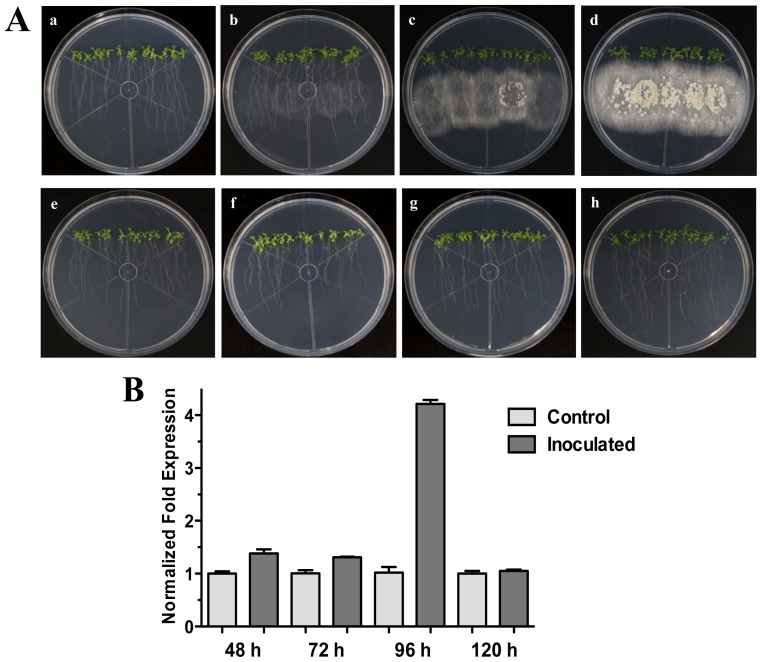
Figure 1.
Direct Interaction. Time course development of direct interaction between Arabidopsis thaliana plantlets and Trichoderma atroviride in MS plates. (A) Photographs were taken for 25-day-old Arabidopsis (Col-0) plants inoculated with T. atroviride at different time points of interaction: 48 h (a), 72 h (b), 96 h (c) and 120 h (d) post-inoculation. Photographs of 25-day-old Arabidopsis (Col-0) control non-inoculated plantlets at different times are indicated: 48 h (e), 72 h (f), 96 h (g) and 120 h (h); (B) Expression analysis of HR4 gene in direct interaction. Quantification of HR4 gene by qRT-PCR, expressed as relative mRNA level compared with control conditions, was calculated after normalization to the Arabidopsis actin 8 gene using the comparative threshold method. Analyses were performed by triplicate. Control conditions or non-inoculated plantlets (light grey bars) and tester conditions or inoculated plantlets (dark bars) at 48, 72, 96 and 120 hpi, respectively.
Our next step was to analyze additional times of interaction (72, 96 and 120 hpi) when the fungus is more grown and developed (Figure 1A). The results showed that HR4 highest induction was at 96 hpi, reaching a relative expression of 4.2-fold change (Figure 1B), when the fungus is already well established and beginning to sporulate (Figure 1A).
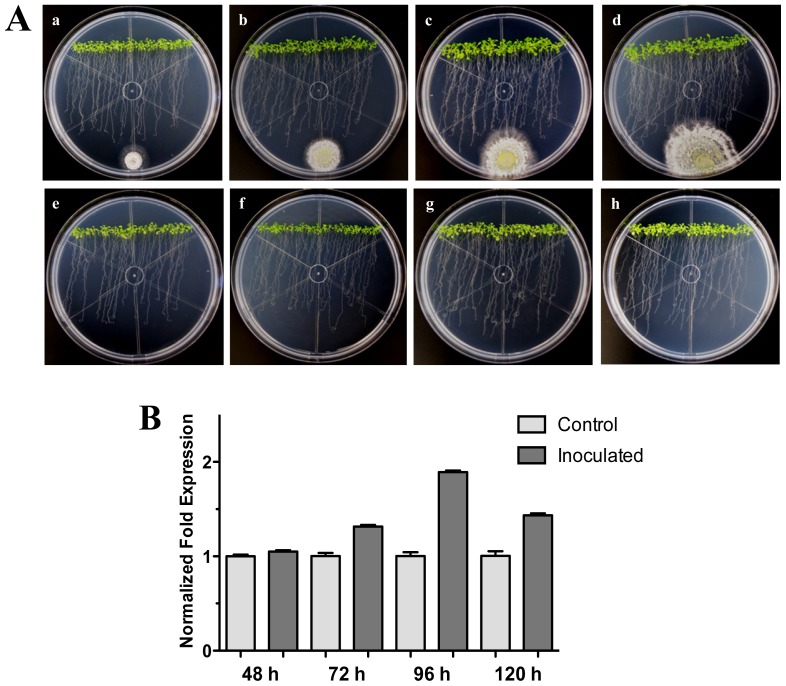
We further examined the effect of distance between the Arabidopsis-Trichoderma interaction. For this, an experiment where T. atroviride was inoculated at 3 cm distance from the root tips was carried out. As with the previous interactions, the expression levels for the HR4 gene were higher at the later stages (96 and 120 hpi) evaluated, when the fungus was more established, and contact was already evident in the Arabidopsis roots (Figure 2A). The maximum expression was at 96 hpi, reaching 1.9-fold in comparison to the control plants (Figure 2B). All these results suggest that the contact of the fungus with the plant roots could activate the recognition system and/or could promote a beneficial interaction establishment in the plant.
Figure 2.
Distant interaction. (A) Development of distant interaction of Arabidopsis thaliana plantlets with Trichoderma atroviride. (a), (b), (c) and (d) photographs of 17-day-old Arabidopsis (Col-0) plantlets inoculated with T. atroviride, the plantlets were photographed at 48 h (a), 72 h (b), 96 h (c) and 120 h (d) post-inoculation. (e), (f), (g) and (h) photographs of 17-day-old Arabidopsis (Col-0) control non-inoculated plantlets of 48 h (e), 72 h (f), 96 h (g) and 120 h (h); (B) Induction of HR4 gene by T. atroviride at a distance. Quantification of HR4 gene by qRT-PCR, expressed as relative mRNA level compared with control conditions, was calculated after normalization to the Arabidopsis actin 8 gene using the comparative threshold method. Analyses were performed by triplicate. Control conditions or non-inoculated plantlets (light grey bars) and tester conditions or inoculated plantlets (dark bars) at 48, 72, 96 and 120 hpi, respectively.
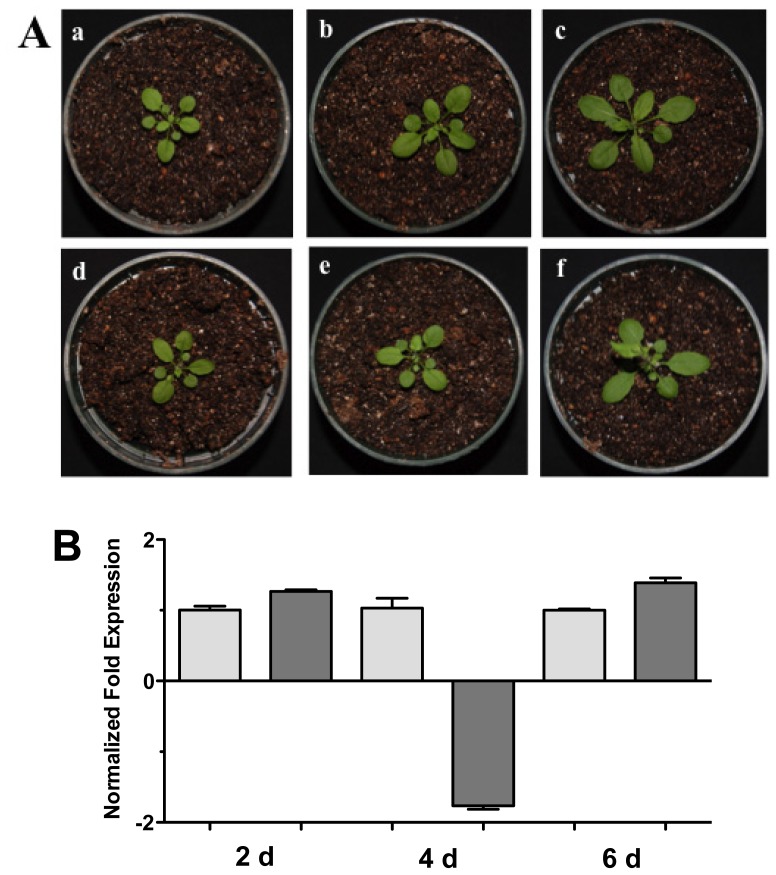
Complementary to this, we examined the HR4 expression of the Arabidopsis-Trichoderma interaction in soil. Arabidopsis plants in pots with soil were inoculated and the aerial part was used for expression studies at 2, 4, and 6 days post-inoculation (dpi) (Figure 3). Plants inoculated with T. atroviride increased aerial parts in comparison to the control plants (Figure 3A). The HR4 gene expression was measured at these points in time, observing a slight increase in gene transcription at 2 and 6 dpi (Figure 3B).
Figure 3.
Soil interaction. (A) Photographs of Arabidopsis plants inoculated with Trichoderma in pots. (a), (b) and (c), 1 month-old Arabidopsis plants in pots inoculated with Trichoderma atroviride, the plants were photographed at (a) 2 days, (b) 4 days, and (c) 6 days post-inoculation. (d), (e) and (f), 1 month-old Arabidopsis control non-inoculated plants of (d) 2 days, (e) 4 days, and (f) 6 days; (B) Expression analysis of HR4 gene from Arabidopsis plants grown in soil pots. Quantification of HR4 gene from aerial part of plants by qRT-PCR, expressed as relative mRNA level compared with control conditions, was calculated after normalization to the Arabidopsis actin 8 gene using the comparative threshold method. Analyses were performed by triplicate. Control conditions or non-inoculated plants (light grey bars) and tester conditions or inoculated plants (dark bars) at 2, 4, and 6 dpi, respectively.
2.2. The RPW8.1 and RPW8.2 Genes (Ecotype Ms-0) Expression in Interaction with Trichoderma atroviride
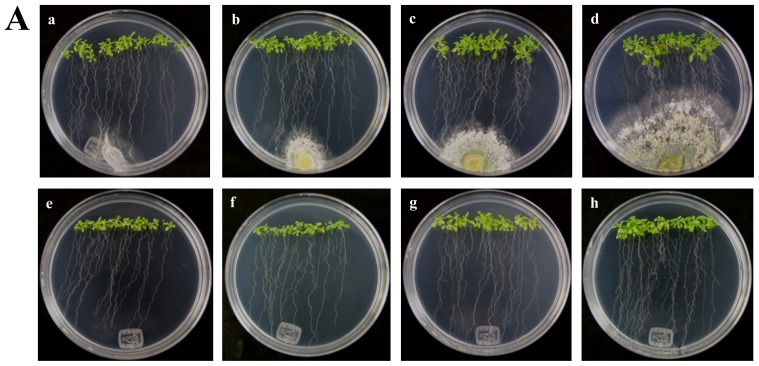
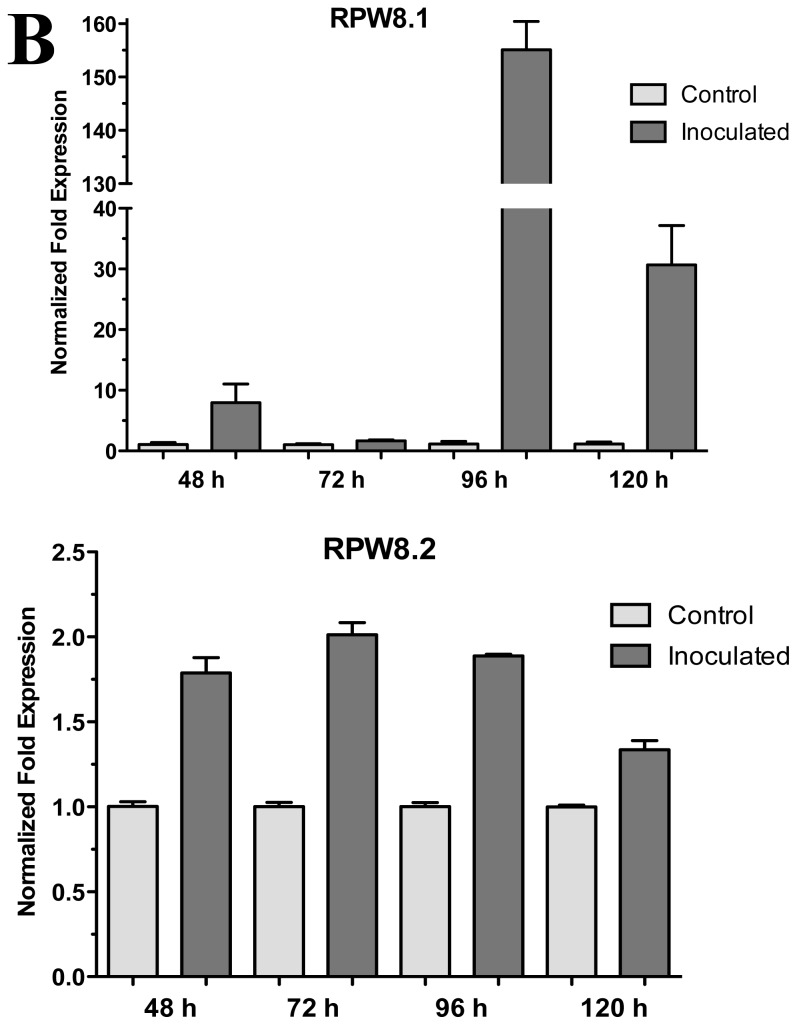
The A. thaliana RPW8 locus from ecotype Ms-0 contains two paralogous genes, RPW8.1 and RPW8.2, both of which confer resistance to powdery mildew fungi [14]. These genes have been extensively studied in interactions with pathogenic fungus (Powdery Mildew), but nothing is known about beneficial fungus interaction. According to the above, we used the A. thaliana ecotype Ms-0 in interaction with T. atroviride for the transcriptional characterization of the HR4 homologues genes, RPW8.1 and RPW8.2. The Trichoderma inoculation was made at the lower end of the Petri dish closed to the plantlets roots, and the gene expression was measure at 48, 72, 96 and 120 hpi (Figure 4A). Both, RPW8.1 and RPW8.2 genes were up-regulated, but the maximum expression was observed in the RPW8.1 gene reaching the highest expression level (155-fold) at 96 hpi, and the lowest expression level (1.67-fold) at 72 hpi. At 48 and 120 hpi the gene expression was 7.9 and 30.6-fold, respectively (Figure 4B RPW8.1). With respect to the RPW8.2 gene, it was up-regulated less than the RPW8.1, reaching the induction levels 1.7, 2.0, 1.8 and 1.3-fold at 48, 72, 96 and 120 hpi, respectively (Figure 4B RPW8.2). The inoculation with the beneficial fungus resulted in an increase in the lateral roots formation and growth of the aerial part in Arabidopsis seedlings ecotype Ms-0 (Figure 4A), this observation is consistent with that observed with Col-0 ecotype (Figure 2A).
Figure 4.
Expression of RPW8.1 and RPW8.2 genes from Arabidopsis thaliana ecotype Ms-0 in interaction with Trichoderma atroviride. (A) Development of interaction of Arabidopsis thaliana ecotype Ms-0 plantlets with Trichoderma atroviride. (a), (b), (c) and (d) photographs of 15-day-old Arabidopsis (Ms-0) plantlets inoculated with T. atroviride, the plantlets were photographed at (a) 48 h, (b) 72 h, (c) 96 h and (d) 120 h post-inoculation. (e), (f), (g) and (h), photographs of 15-day-old Arabidopsis (Ms-0) control non-inoculated plantlets of (e) 48 h, (f) 72 h, (g) 96 h and (h) 120 h; (B) Induction of RPW8.1 and RPW8.2 genes by T. atroviride interaction. Quantification of RPW8.1 and RPW8.2 genes by qRT-PCR, expressed as relative mRNA level compared with a control conditions, was calculated after normalization to the Arabidopsis actin 8 gene using the comparative threshold method. Analyses were performed by triplicate. Control conditions or non-inoculated plantlets (light grey bars) and tester conditions or inoculated plantlets (dark bars) at 48, 72, 96 and 120 hpi, respectively.
2.3. HR4 Gene Is Induced by Bacterial Interactions
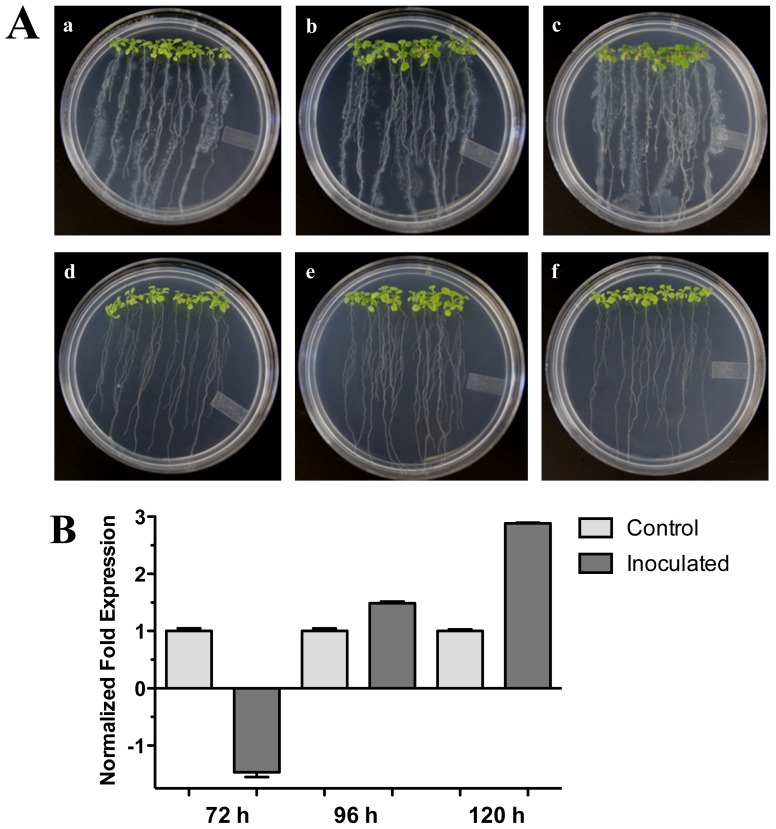
The effect of plant growth-promoting rhizobacterium (PGPR) Pseudomonas fluorescens on Arabidopsis (Col-0) 20-day-old seedlings was assessed (Figure 5A). We tested the expression of the Arabidopsis HR4 gene in interaction with P. fluorescens roots inoculated at 72, 96 and 120 hpi. As shown in Figure 5B, a gradual increase of the HR4 mRNA level with the progress of the interaction was observed, beginning with a repression at 72 h (−1.46), followed by inductions of 1.49 and 2.88-fold at the later stages of interaction.
Figure 5.
Pseudomonas fluorescens interaction. (A) Development of the interaction of Arabidopsis with P. fluorescens. (a), (b), and (c), photographs of 20-day-old Arabidopsis (Col-0) plantlets inoculated with P. fluorescens, the plantlets were photographed at (a) 72 h, (b) 96 h, and (c) 120 h post-inoculation. (d), (e) and (f), photographs of 20-day-old Arabidopsis (Col-0) control non-inoculated plantlets of (d) 72 h, (e) 96 h and (f) 120 h; (B) Expression analysis of the HR4 gene of Arabidopsis in interaction with beneficial bacterium P. fluorescens. Quantification of HR4 gene by qRT-PCR, expressed as relative mRNA level compared with a control condition, was calculated after normalization to the Arabidopsis actin 8. Control conditions or non-inoculated plantlets (light grey bars) and tester conditions or inoculated plantlets (dark bars) at 72, 96 and 120 hpi, respectively.
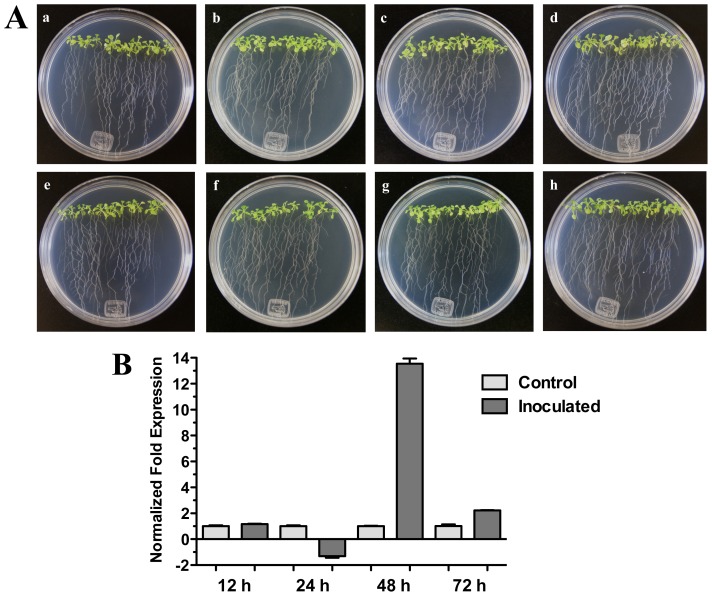
On the other hand, when Arabidopsis (Col-0) seedlings were inoculated with bacterial pathogenic Pseudomonas syringae pv. tomato DC3000 a significant increase of HR4 gene expression was observed at 48 h (13.54-fold) (Figure 6B). In this interaction, bacteria were inoculated in foliar area of 20-day-old seedlings, and initial characteristic symptoms of the P. syringae infection were observed at 72 h, with initial chlorosis and anthocyanin production in the center of the rosette seedlings (Figure 6A).
Figure 6.
Pseudomonas syringae interaction. (A) Development of the interaction of Arabidopsis with P. syringae pv. tomato DC3000. (a), (b), (c) and (d) photographs of 20-day-old Arabidopsis (Col-0) plantlets inoculated with P. syringae, the plantlets were photographed at 12 h (a), 24 h (b), 48 h (c) and 72 h (d) post-inoculation. (e), (f), (g), and (h), photographs of 20-day-old Arabidopsis (Col-0) control non-inoculated plantlets of 12 h (e), 24 h (f), 48 h (g) and 72 h (h); (B) Expression analysis of the HR4 gene of Arabidopsis in interaction with plant-pathogenic P. syringae. Quantification of the HR4 gene by qRT-PCR, expressed as relative mRNA level compared with a control condition, was calculated after normalization to the Arabidopsis actin 8. Control condition or non-inoculated plantlets (light grey bars) and tester conditions or inoculated plantlets (dark bars) at 12, 24, 48 and 72 hpi, respectively.
2.4. Effect of Phytohormones on the Expression of HR4 Gene
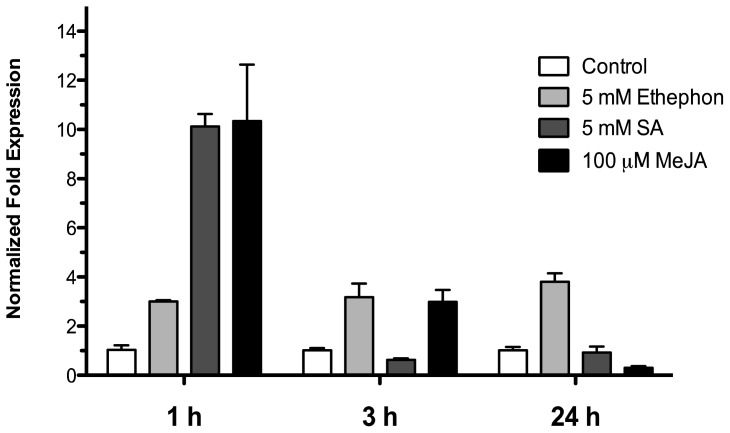
To examine the effect of phytohormones involved in biotic stress signaling on the HR4 gene expression, 15-day-old Arabidopsis (Col-0) seedlings were treated by spraying with 5 mM ethephon, 5 mM salicylic acid and 100 μM methyl jasmonate in a MS solution, control seedlings were treated with an equivalent amount of MS solution. The treated seedlings and controls were harvested at 1, 3, and 24 h after sprayed. As show in Figure 6, with ethephon (ET) treatment the HR4 gene was induced in early times at 1 and 3 h with 3.00 and 3.18-fold, respectively, and this induction was maintained at 24 h (3.79-fold). For SA and MeJA treatment, we observed a strong initial induction at 1 h of 10.1- and 10.3-fold, respectively. The next times, at 3 and 24 h, the level of expression was down-regulated for the two hormones (Figure 7).
Figure 7.
Phytohormonal treatment effects. Expression analysis of the HR4 gene from Arabidopsis by the addition of phytohormones, 5 mM ethephon, 5 mM SA, and 100 μM MeJA. Control conditions or without phytohormones (white bars) and tester conditions with 5 mM of ethephon (light grey bars), 5 mM SA (dark grey bars), and 100 μM MeJA (black bars) at 1, 3, and 24 h post sprayed, respectively.
3. Discussion
Plants have evolved in constant interaction with beneficial microorganisms, which influence plant growth and development and also plant health. These beneficial microorganisms are indispensable for a sustainable agriculture and environment. Although the plant-pathogen interaction is better understood, the plant-beneficial interaction is less known; hence, it is necessary to understand what is happening at the molecular level between plant and beneficial microorganisms [7]. It is a critical point for providing new strategies to improve plant productivity, while protecting the environment and biodiversity.
Some Trichoderma rhizosphere-competent strains have been shown to have direct effects on plants. These effects are: (i) the promotion of plant growth and development, and thus increased yields [22,23]; (ii) breaking seed dormancy and enhanced rates of seed germination [24–26]; (iii) improved tolerance to abiotic stresses during plant growth, in part due to improved root growth, and enhanced water-holding capacity of plants or enhanced nutrient uptake (phosphorus and several micronutrients) [26–29]; (iv) increased photosynthetic capacity [27]; and (v) systemic induction of plant defenses towards attack by pathogenic microorganisms [22,30]. Several global analyses have been published of the alteration of proteome [31–33] and transcriptome [30,34–36] of plants as a consequence of Trichoderma colonization.
Arabidopsis thaliana represents a functional system to study beneficial plant-microbe interactions. In particular, Contreras-Cornejo et al. in 2009 [19] reported the beneficial effects of Trichoderma species on plant growth and the development of A. thaliana. It has also been reported that the presence of Trichoderma primes the systemic resistance system, a complex signaling mechanism involving diverse hormones pathways. This complex signaling mechanism involving JA/ET-induced systemic resistance (ISR) and/or SA-dependent systemic acquired resistance (SAR) pathways [37]. The activation of ISR and/or SAR in plants is part of the response to microbes by the plants. In order to recognize microbes, plants have a large collection of R genes, which directly or indirectly recognize effectors from beneficial and pathogenic fungi or bacteria.
In a microarray of the interaction A. thaliana (Col-0)-T. atroviride [38] we analyzed the expression of R genes, where the HR4 gene was one of the most strongly induced. For this reason, in this study we focused on HR4 gene expression during the interaction with this beneficial fungus. At 48 hpi an induction of the HR4 gene was found; however, at 96 hpi the HR4 gene showed the highest peak of expression in comparison with the A. thaliana seedlings without fungus. The same behavior was observed when the fungus was inoculated at the end of the petri dishes (distance interaction), where the maximum peak of HR4 gene expression was also at 96 hpi. At this point in time, the fungus development was more evident and the interaction was established in both interactions. When the aerial part of 27-day-old Arabidopsis inoculated with Trichoderma in pots was analyzed transcriptionally, we observed a cyclic behavior of the HR4 gene with alternated transcript induction (2 and 6 dpi) and repression (4 dpi).
There are only a few reports about the HR4 gene. Xiao et al. [13] reported that the transcription of the HR4 gene from A. thaliana ecotype Col-0 was induced by powdery mildew and Peronospora parasitica. In another study, the HR4 gene induction from an interaction of A. thaliana (Col-0) with Botrytis cinerea was observed at 12 and 24 hpi [39]. In the present study, we report the first transcriptional regulation of the HR4 gene in interaction with a beneficial microbe.
The A. thaliana HR4 gene (ecotype Col-0) is homologue to RPW8 genes (ecotype Ms-0). The RPW8 locus from accession Ms-0 confers broad-spectrum resistance to powdery mildew. This locus contains two paralogous genes, RPW8.1 and RPW8.2, both of which contribute to resistance [12,13]. The origin of the RPW8 locus is relatively recent, probably after the separation of Arabidopsis from the Brassica lineages, and that RPW8.1 and RPW8.2 have evolved from an HR3-like predecessor gene by duplication and functional diversification. The HR4 gene is probably of most recent origin, which appears to have arisen from RPW8.1 [13,14].
Based on particular differences between Arabidopsis haplotypes (RPW8 locus and HR4), the expression of the RPW8.1 and RPW8.2 genes of Arabidopsis ecotype Ms-0 in interaction with T. atroviride was assessed. An important finding in the present study was the increased expression of RPW8.1 and RPW8.2 genes from Arabidopsis ecotype Ms-0 during the interaction with Trichoderma. The up-regulation of RPW8.2 gene was maintained in for the first three times with similar levels, which were similar to HR4 gene levels of ecotype Col-0. Furthermore, the expression profile of RPW8.1 was like that of HR4 gene, where at later stages there was an evident increase in mRNA level, especially at 96 hpi with a strong increase in both genes (Figures 1B and 3B). Xiao et al. [40] proposed that the increased transcription of RPW8s formed part of an amplification circuit that lead to the accumulation of SA. Therefore, SA formed during the expression of resistance to different pathogens also would induce the accumulation of transcripts of RPW8.1 and RPW8.2 via a feedback amplification circuit. Although there is much information about R genes in plant-pathogen interactions (e.g., RPW8 genes), only little is known about R genes in beneficial interactions.
In addition to fungal interactions, we analyzed the interaction between A. thaliana (Col-0) and bacteria. When the beneficial bacterium, the plant growth promoting rhizobacterium (PGPR) Pseudomonas fluorescens, was inoculated into the Arabidopsis roots, an induction of the HR4 gene specifically in the later stages (120 hpi) was noticed. This result is consistent with that reported by Wang et al. [41], who found using microarray analysis that the HR4 gene was up-regulated in the interaction A. thaliana-P. fluorescens (FPT9601-T5). On the other hand, in response to the infection by the pathogenic bacterium P. syringae pv tomato DC3000 on Arabidopsis seedling leaves (Col-0), the HR4 gene markedly increased the mRNA level at 48 hpi.
Plant hormones are essential for the regulation of plant defense, the importance of salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) as primary signals is well established [42–44]. The interaction of plants with diverse microbes results in changes in the level of these phytohormones, which are positive regulators of defense genes (PRs), transcription factors (WRKYs) and receptors (R genes). The R genes are the activators of immune response, and simultaneously the R gene promoters have cis-elements that increase the amplification by feedback positive regulation circuits stimulated by hormonal action [44]. Cross-talk among SA, JA and ET signaling pathways has emerged as an important regulatory mechanism of plant immunity [42,43]. There are different reports about what the hormonal activated pathway in the interaction Arabidopsis-Trichoderma [20] is. T. atroviride induces a delay and overlapping activation of the defense-related genes of the SA and JA/ET pathways against biotrophic and necrotrophic phytopathogens [19,45]. T. atroviride P1 and T. harzianum T22 are able to induce a long-lasting up-regulation of SA gene markers, after the infection with Botrytis cinerea the expression of defense genes increases induced through the JA pathway, culminating in ISR [17], and T. asperellum produces a clear ISR through SA signaling cascade [18]. In order to determinate the transcriptional response of the HR4 gene, expression was measured under hormonal treatment of Arabidopsis seedlings with the exogenous application of SA, MeJA and ethephon phytohormones, which have been implicated in the response to Trichoderma. The HR4 gene expression was up-regulated with SA and MeJA treatments observing an abrupt induction 1 h post-treatment, and a decrease in expression at later times. On the other hand, the ethephon treatment constantly maintained the up-regulated mRNA level three times. RPW8.1 and RPW8.2 genes involved a self-amplification mechanism via the SA-feedback circuit for activation of HR and resistance [40,46]. The response of the HR4 gene by SA reported in this study, and what was reported for RPW8.1 and RPW8.2 genes [40], could be due to W-box elements within their regulatory regions.
In the interaction of T. harzianum T34 with Arabidopsis plants, a transcriptional regulation of SA-related genes, such as Enhanced Disease Susceptibility 1 (EDS1) and PR-1, was reported [35]. At an early stage (4 h), a strong down-regulation of EDS1 and PR-1 genes was detected, but after 48 h of Arabidopsis root colonization by T. harzianum an increase in the expression of these genes was reported [35]. Interestingly, RPW8-mediated resistance is associated with the expression of PR genes, and signaling in RPW8-mediated resistance occurs through a feedback amplification loop in the SA pathway, and requires signaling components such as EDS1, EDS5, PHYTOALEXIN DEFICIENT 4 (PAD4), and NONEXPRESSOR OF PATHOGENESIS-RELATED PROTEIN 1 (NPR1) [11,40,46,47].
The HR4 gene is classified as an early SA induced gene (early SAIG), as reported by Blanco et al. in 2009 [48], who analyzed the early genetic response to SA of Arabidopsis seedlings. Using microarray analysis, they identified 217 genes rapidly induced after 2.5 h SA treatment; 193 by a NPR1-dependent and 24 by a NPR1-independent pathway. The HR4 gene with a 3.0-fold-change ratio was included in the NPR1-dependent group. In another study by Galon, et al. in 2008 [49], they reported the elevate expression of 32 genes related to defense against pathogens in transcriptomic analysis using microarrays from A. thaliana and camta3 (insertion mutants in the Calmodulin binding transcription activators 3 gene) plants, amongst these genes there was the HR4 gene with a 6.7-fold change ratio, suggesting that CAMTA3 normally suppresses biotic defense responses. In accordance with these findings, our transcription data of HR4 and RPW8 genes support the idea that the SA pathway is a key player in the recognition, establishment and/or activation of plant systemic defense by the beneficial fungus T. atroviride.
4. Experimental Section
4.1. Plant Material and Growth Conditions
Arabidopsis thaliana (Col-0) seeds were surface-sterilized and sown on Petri dishes containing 0.5× Murashige and Skoog (MS) medium [1.4% (w/v) agar, 0.75% (w/v) sucrose] and placed at 4 °C for 2 days for vernalization, they then were placed in growth cabinets at 22 ± 1 °C for 7 days. Fifteen seedlings were transferred to Petri dishes containing 1× MS with 1.5% (w/v) sucrose, 1.4% (w/v) agar, the pH of the medium was adjusted to 7.0 and the seedlings were grown at 22 ± 1 °C for 16 days in a 16-h-light/8-h-dark cycle. After that period of time, Trichoderma atroviride was inoculated on the MS media as described below.
For Trichoderma distant interaction, pot interactions, ecotype Ms-0, and others microbe interactions, A. thaliana (Col-0 and Ms-0) seeds were surface-sterilized and sown on Petri dishes containing 0.2× MS medium [1% (w/v) agar, 0.75% (w/v) sucrose, the pH of the medium was adjusted to 7.0] and placed at 4 °C for 2 days for vernalization, they then were placed in growth cabinets at 22 ± 1 °C in a 16-h-light/8-h-dark cycle for 15–20 days. After that period of time, seedlings were inoculated with the respective culture of microbes on the MS media as described below.
4.2. Microorganism Growth Conditions and Seedling Inoculations
In a direct interaction experiment, T. atroviride (IMI206040) was grown on PDA plates for 8 days at 28 °C. Conidia were collected in distilled water and adjusted to a density of 1 × 103 spores per mL. Then 10 μL of the spore suspension were applied on roots of 25-day-old A. thaliana plantlets grown in Petri dishes with MS medium and cultured for different periods: 48, 72, 96 and 120 h post-inoculation (hpi), control non-inoculated plants were cultured at the same times points. On the other hand, in a distant interaction experiment, 10 μL of a suspension of 1 × 106 spores were applied at 3 cm of distance from the roots at the opposite ends of MS plates. Inoculated and non-inoculated plants were cultured for 48, 72, 96 and 120 h. After each treatment, plants were harvested and frozen in liquid nitrogen for RNA extraction. In soil experiments the Arabidopsis seedlings were sown in pots containing a soil mixture (one plant per pot). Each pot was maintained in a growth chamber (22 ± 1 °C) under a photoperiod of 16 h light/8 h dark until the plants were 1 month old. Then, three plants were inoculated with 1 mL of 1 × 106 spore suspension of T. atroviride and 1 mL of water was added to three uninoculated control plants. Aerial parts were harvested and frozen in liquid nitrogen at 2, 4 and 6 dpi.
For A. thaliana Ms-0 interaction with T. atroviride, the 15-day-old plantlets were inoculated with 10 μL of the spore suspension (1 × 106 spores) at the bottom of the plates close to the plantlets roots, inoculated, and control plantlets were harvested and frozen in liquid nitrogen at 48, 72, 96 and 120 h.
The plant pathogen Pseudomonas syringae pv. tomato DC3000 was cultivated on King’s B medium [50] with appropriate antibiotics at 28 °C, when bacterial culture reached mid to late log phase growth (OD600 = 0.6 to 1.0) the bacteria from liquid culture were harvested. Cells were washed once and resuspended in 10 mM MgCl2 solution, the OD600 was adjusted to 0.002 (1 × 106 cfu/mL) and 10 μL of bacterial suspensions were added to each of the 20-day-old seedlings on the foliar area. Then the plantlets were harvest and frozen at different times (12, 24, 48 and 72 hpi) of interaction for RNA extraction.
The plant growth-promoting rhizobacterium P. fluorescens was isolated in our research group from sugar cane rhizosphere, the bacterium was grown on LB medium at 28 °C until the early stationary phase. Just before inoculation, cells were washed once and resuspended in 10 mM MgCl2 solution; the OD600 was adjusted to 0.002 (1 × 106 cfu/mL), and 10 μL bacterial suspension were added to the roots of each of the 20-day-old plantlets. At different time points post-inoculation (72, 96 and 120 h) the plants were harvested and frozen; similarly, uninoculated control plants were harvest at the same times for RNA extraction.
4.3. Phytohormone Application
Phytohormones were applied as a spray to 15-day-old plantlets with 0.5× MS solution containing ethephon, SA and MeJA at the rate of 5 mM, 5 mM and 100 μM, respectively. The MS plates with treated seedlings were sealed and the plantlets were harvested at 1, 3 and 24 h after each treatment. Control plants were sprayed with 0.5× MS solution and sealed and harvest at the same time points as the treatment plants.
4.4. RNA Extraction and Real-Time qRT-PCR
Total RNA was extracted from inoculated and non-inoculated plants using the Concert™ Plant RNA reagent (Invitrogen, Carlsbad, CA, USA). Real-time PCR was performed in 10 μL of reaction mixture made up of 5 μL of Power SYBR® Green RT-PCR Mix (2×), 200 nM of each oligonucleotide, 50 ng of RNA template and 0.08 μL of RT Enzyme Mix (125×) for one-step RT-PCR, using an StepOne Real-Time PCR Detection System and StepOne Software v2.1 (Applied Biosystems). The thermal cycling conditions consisted of 30 min at 48 °C (cDNA synthesis), 10 min at 95 °C (Activation of AmpliTaq Gold® DNA polymerase), followed by 40 cycles for PCR cycling of 15 s at 95 °C for denature and anneal/extend of 1 min at 60 °C. These PCR reactions were repeated by triplicate for each condition. Quantification of HR4, RPW8.1 and RPW8.2 gene expression was based on a cycle threshold value and normalized to the actin 8 (At1g49240) gene values. Absence of contaminant genomic DNA was confirmed by reactions in which no RT Enzyme Mix reverse transcriptase was added, and also primers for the actin 8 gene were designed between two exons. The primers used were: 5′-CATCTCGAGAGACGAGAGCTAA-3′ and 5′-CTGAAGCCGTCGTAAATGACTT-3′ for HR4 (At3g50480) transcript, 5′-GGACACTAAACTTGCTGAAGTTA-3′ and 5′-CAATAAT TATGGGGAATAAGAGAGA-3′ for RPW8.1 transcript, 5′-ACAAAATAATGCCTCAACCGAAG-3′ and 5′-TGAGTCGTTTGACACAATTGGG-3′ for RPW8.2 transcript and 5′-GCCAGTGGTCGT ACAACCG-3′ and 5′-TCATGAGGTAATCAGTAAGGTCAC-3′ for actin 8.
5. Conclusions
Whereas many R genes have been characterized in plant-pathogen interactions, such as RPW8 genes, little is known about R genes involved in mutualistic interactions. Our results indicate that the HR4 gene modulates their expression in interaction with the tested microbes. It was induced in particular in Trichoderma atroviride interaction, suggesting that it plays a role in the process of recognition and/or establishment of the interaction. When analyzing the Arabidopsis ecotype Ms-0, which has RPW8 genes instead of HR4, it was observed that these genes are also responsive to T. atroviride, suggesting that this type of R genes is regulated in beneficial interactions.
Acknowledgments
We are grateful to Jorge Ramírez-Salcedo, José L. Santillán-Torres, Simón Guzmán-León, and Lorena Chávez-González from the Unidad de Microarreglos de DNA-IFC-UNAM for Microarray hybridization assays. We would also like to acknowledge Shunyuan Xiao and Robert Berkey for providing the seeds of Arabidopsis thaliana ecotype Ms-0. This work was supported by grant from the Consejo Nacional de Ciencia y Tecnología (CONACYT-Fondos Sectoriales de Ciencia Básica 2008-1, México, grant no. 103106).
References
- 1.Zhao S., Qi X. Signaling in plant disease resistance and symbiosis. J. Integr. Plant Biol. 2008;7:799–807. doi: 10.1111/j.1744-7909.2008.00702.x. [DOI] [PubMed] [Google Scholar]
- 2.Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Lotze M.T., Zeh H.J., Rubartelli A., Sparvero L.J., Amoscato A.A., Washburn N.R., Devera M.E., Liang X., Tör M., Billiar T. The grateful dead: Damage-associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol. Rev. 2007;220:60–81. doi: 10.1111/j.1600-065X.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 4.Tör M., Lotze M.T., Holton N. Receptor-mediated signalling in plants: Molecular patterns and programmes. J. Exp. Bot. 2009;60:3645–3654. doi: 10.1093/jxb/erp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zipfel C., Robatzek S. Pathogen-associated molecular pattern-triggered immunity: Veni, Vidi…? Plant Physiol. 2010;154:551–554. doi: 10.1104/pp.110.161547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dodds P.N., Rathjen J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 7.Zamioudis C., Pieterse C.M.J. Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 2012;25:139–150. doi: 10.1094/MPMI-06-11-0179. [DOI] [PubMed] [Google Scholar]
- 8.Gururani M.A., Venkatesh J., Upadhyaya C.P., Nookaraju A., Pandey S.K., Park S.W. Plant disease resistance genes: Current status and future directions. Physiol. Mol. Plant Pathol. 2012;78:51–65. [Google Scholar]
- 9.Bent A.F., Mackey D. Elicitors, Effectors, and R Genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 2007;1:399–436. doi: 10.1146/annurev.phyto.45.062806.094427. [DOI] [PubMed] [Google Scholar]
- 10.Xiao S., Wang W., Yang X. Evolution of resistance genes. Nucleic Acids Mol. Biol. 2008;21:1–25. [Google Scholar]
- 11.Micali C., Göllner K., Humphry M., Consonni C., Panstruga R. The Arabidopsis Book. Vol. 6. American Society of Plant Biology; Rockville, MD, USA: 2008. The Powdery Mildew Disease of Arabidopsis: A Paradigm for the Interaction Between Plants and Biotrophic Fungi; pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao S., Ellwood S., Calis O., Patrick E., Li T.X., Coleman M., Turner J.G. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science. 2001;291:118–120. doi: 10.1126/science.291.5501.118. [DOI] [PubMed] [Google Scholar]
- 13.Xiao S., Emerson B., Ratanasut K., Patrick E., O’Neill C., Bancroft I., Turner J.G. Origin and maintenance of a broad-spectrum disease resistance locus in Arabidopsis. Mol. Biol. Evol. 2004;21:1661–1672. doi: 10.1093/molbev/msh165. [DOI] [PubMed] [Google Scholar]
- 14.Orgil U., Araki H., Tangchaiburana S., Berkey R., Xiao S. Intraespecific genetic variations, fitness cost and benefit of RPW8, a disease resistance locus in Arabidopsis thaliana. Genetics. 2007;176:2317–2333. doi: 10.1534/genetics.107.070565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoyos-Carvajal L., Ordua S., Bissett J. Growth stimulation in bean (Phaseolus vulgaris L.) by Trichoderma. Biol. Control. 2009;51:409–416. [Google Scholar]
- 16.Moran-Diez E., Hermosa R., Ambrosino P., Cardoza R.E., Gutiérrez S., Lorito M., Monte E. The ThPG1 endopolygalacturonase is required for the Trichoderma harzianum–plant beneficial interaction. Mol. Plant Microbe Interact. 2009;22:1021–1031. doi: 10.1094/MPMI-22-8-1021. [DOI] [PubMed] [Google Scholar]
- 17.Tucci M., Ruocco M., de Masi L., de Palma M., Lorito M. The beneficial effect of Trichoderma spp. on tomato is modulated by the plant genotype. Mol. Plant Pathol. 2011;12:341–354. doi: 10.1111/j.1364-3703.2010.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshioka Y., Ichikawa H., Naznin H.A., Kogure A., Hyakumachi M. Systemic resistance induced in Arabidopsis thaliana by Trichoderma asperellum SKT-1, a microbial pesticide of seedborne diseases of rice. Pest Manag. Sci. 2012;68:60–66. doi: 10.1002/ps.2220. [DOI] [PubMed] [Google Scholar]
- 19.Contreras-Cornejo H.A., Macías-Rodríguez L.I., Cortés-Penagos C., López-Bucio J. Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol. 2009;149:1579–1592. doi: 10.1104/pp.108.130369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermosa R., Viterbo A., Chet I., Monte E. Plant-beneficial effects of Trichoderma and of its genes. Microbiology. 2012;158:17–25. doi: 10.1099/mic.0.052274-0. [DOI] [PubMed] [Google Scholar]
- 21.Viterbo A., Wiest A., Brotman Y., Chet I., Kenerley C. The 18mer peptaibols from Trichoderma virens elicit plant defence responses. Mol. Plant Pathol. 2007;8:737–746. doi: 10.1111/j.1364-3703.2007.00430.x. [DOI] [PubMed] [Google Scholar]
- 22.Harman G.E., Howell C.R., Viterbo A., Chet I., Lorito M. Trichoderma species-opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2004;2:43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- 23.Lorito M., Woo S.L., Harman G.E., Monte E. Translational research on Trichoderma: From ‘omics to the field. Annu. Rev. Phytopathol. 2010;48:395–417. doi: 10.1146/annurev-phyto-073009-114314. [DOI] [PubMed] [Google Scholar]
- 24.Delgado-Sánchez P., Ortega-Amaro M.A., Rodríguez-Hernández A.A., Jiménez-Bremont J.F., Flores J. Further evidence from the effect of fungi on breaking Opuntia seed dormancy. Plant Signal. Behav. 2010;5:1229–1230. doi: 10.4161/psb.5.10.12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delgado-Sánchez P., Ortega-Amaro M.A., Jiménez-Bremont J.F., Flores J. Are fungi important for breaking seed dormancy in desert species? Experimental evidence in Opuntia streptacantha (Cactaceae) Plant Biol. 2011;13:154–159. doi: 10.1111/j.1438-8677.2010.00333.x. [DOI] [PubMed] [Google Scholar]
- 26.Mastouri F., Björkman T., Harman G.E. Seed treatment with Trichoderma harzianum alleviates biotic, abiotic, and physiological stresses in germinating seeds and seedlings. Phytopathology. 2010;100:1213–1221. doi: 10.1094/PHYTO-03-10-0091. [DOI] [PubMed] [Google Scholar]
- 27.Harman G.E. Myths and dogmas of biocontrol: Changes in perceptions derived from research on Trichoderma harzianum T-22. Plant Dis. 2000;84:377–393. doi: 10.1094/PDIS.2000.84.4.377. [DOI] [PubMed] [Google Scholar]
- 28.Bae H., Sicher R.C., Kim M.S., Kim S.H., Strem M.D., Melnick R.L., Bailey B.A. The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao. J. Exp. Bot. 2009;60:3279–3295. doi: 10.1093/jxb/erp165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yildirim E., Taylor A.G., Spittler T.D. Ameliorative effects of biological treatments on growth of squash plants under salt stress. Sci. Hortic. (Amst) 2006;111:1–6. [Google Scholar]
- 30.Alfano G., Ivey M.L.L., Cakir C., Bos J.I.B., Miller S.A., Madden L.V., Kamoun S., Hoitink H.A.J. Systemic modulation of gene expression in tomato by Trichoderma hamatum 382. Phytopathology. 2007;97:429–437. doi: 10.1094/PHYTO-97-4-0429. [DOI] [PubMed] [Google Scholar]
- 31.Marra R., Ambosino P., Carbone V., Vinale F., Woo S.L., Ruocco M., Ciliento R., Lanzuise S., Ferraioli S., Soriente I., et al. Study of the three-way interaction between Trichoderma atroviride, plant and fungal pathogens using a proteome approach. Curr. Genet. 2006;50:307–321. doi: 10.1007/s00294-006-0091-0. [DOI] [PubMed] [Google Scholar]
- 32.Segarra G., Casanova E., Bellido D., Odena M.A., Oliveira E., Trillas I. Proteome, salicylic acid, and jasmonic acid changes in cucumber plants inoculated with Trichoderma asperellum strain T34. Proteomics. 2007;7:3943–3952. doi: 10.1002/pmic.200700173. [DOI] [PubMed] [Google Scholar]
- 33.Shoresh M., Harman G.E. The molecular basis of shoot responses of maize seedlings to Trichoderma harzianum T22 inoculation of the root: A proteomic approach. Plant Physiol. 2008;147:2147–2163. doi: 10.1104/pp.108.123810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno C.A., Castillo F., González A., Bernal D., Jaimes Y., Chaparro M., González C., Rodriguez F., Restrepo S., Cotes A.M. Biological and molecular characterization of the response of tomato plants treated with Trichoderma koningiopsis. Physiol. Mol. Plant Pathol. 2009;74:111–120. [Google Scholar]
- 35.Morán-Diez E., Rubio B., Domínguez S., Hermosa R., Monte E., Nicolás C. Transcriptional response of Arabidopsis thaliana after 24 h incubation with the biocontrol fungus Trichoderma harzianum. Plant Physiol. 2012;169:614–620. doi: 10.1016/j.jplph.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 36.Brotman Y., Lisec J., Méret M., Chet I., Willmitzer L., Viterbo A. Transcript and metabolite analysis of the Trichoderma-induced systemic resistance response to Pseudomonas syringae in Arabidopsis thaliana. Microbiology. 2012;158:139–146. doi: 10.1099/mic.0.052621-0. [DOI] [PubMed] [Google Scholar]
- 37.Shoresh M., Harman G.E., Mastouri F. Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 2010;48:21–43. doi: 10.1146/annurev-phyto-073009-114450. [DOI] [PubMed] [Google Scholar]
- 38.Sáenz-Mata J., Jiménez-Bremont J.F. Unpublished work. Institute Potosino of Scientific and Technological Research; San Luis Potosí, SLP, México: 2009. [Google Scholar]
- 39.Mulema J.M.K., Denby K.J. Spatial and temporal transcriptomic analysis of the Arabidopsis thaliana-Botrytis cinerea interaction. Mol. Biol. Rep. 2012;39:4039–4049. doi: 10.1007/s11033-011-1185-4. [DOI] [PubMed] [Google Scholar]
- 40.Xiao S.Y., Brown S., Patrick E., Brealey C., Turner J.G. Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 occurs via a salicylic acid-dependent amplification circuit and is required for hypersensitive cell death. Plant Cell. 2003;15:33–45. doi: 10.1105/tpc.006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Ohara Y., Nakayashiki H., Tosa Y., Mayama S. Microarray analysis of the gene expression profile induced by the endophytic plant growth-promoting rhizobacteria, Pseudomonas fluorescens FPT9601-T5 in Arabidopsis. Mol. Plant Microbe Interact. 2005;18:385–396. doi: 10.1094/MPMI-18-0385. [DOI] [PubMed] [Google Scholar]
- 42.Verhage A., van Wees S.C.M., Pieterse C.M.J. Plant immunity: It’s the hormones talking, but what do they say? Plant Physiol. 2010;154:536–540. doi: 10.1104/pp.110.161570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pieterse C.M.J., Leon-Reyes A., van der Ent S., van Wees S.C.M. Networking by small-molecules hormones in plant immunity. Nat. Chem. Biol. 2009;5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- 44.Bari R., Jones J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 45.Salas-Marina M.A., Silva-Flores M.A., Uresti-Rivera E.E., Castro-Longoria E., Herrera-Estrella A., Casas-Flores S. Colonization of Arabidopsis roots by Trichoderma atroviride promotes growth and enhances systemic disease resistance through jasmonic acid/ethylene and salicylic acid pathways. Eur. J. Plant. Pathol. 2011;131:15–26. [Google Scholar]
- 46.Xiao S., Calis O., Patrick E., Zhang G., Charoenwattana P., Muskett P., Parker J.E., Turner J.G. The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 2005;42:95–110. doi: 10.1111/j.1365-313X.2005.02356.x. [DOI] [PubMed] [Google Scholar]
- 47.Feys B.J., Moisan L.J., Newman M.A., Parker J.E. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001;20:5400–5411. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanco F., Garreton V., Frey N., Dominguez C., Perez-Acle T., van der Straeten D., Jordana X., Holuigue L. Identification of NPR1-dependent and independent genes early induced by salicylic acid treatment in Arabidopsis. Plant Mol. Biol. 2005;59:927–944. doi: 10.1007/s11103-005-2227-x. [DOI] [PubMed] [Google Scholar]
- 49.Galon Y., Nave R., Boyce J.M., Nachmias D., Knight M.R., Fromm H. Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 2008;582:943–948. doi: 10.1016/j.febslet.2008.02.037. [DOI] [PubMed] [Google Scholar]
- 50.King E.O., Ward M.K., Raney D.E. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 1954;44:301–307. [PubMed] [Google Scholar]