Abstract
The Mga regulator of Streptococcus pyogenes directly activates the transcription of a core regulon that encodes virulence factors such as M protein (emm), C5a peptidase (scpA), and streptococcal inhibitor of complement (sic) by directly binding to a 45-bp binding site as determined by an electrophoretic mobility shift assay (EMSA) and DNase I protection. However, by comparing the nucleotide sequences of all established Mga binding sites, we found that they exhibit only 13.4% identity with no discernible symmetry. To determine the core nucleotides involved in functional Mga-DNA interactions, the M1T1 Pemm1 binding site was altered and screened for nucleotides important for DNA binding in vitro and for transcriptional activation using a plasmid-based luciferase reporter in vivo. Following this analysis, 34 nucleotides within the Pemm1 binding site that had an effect on Mga binding, Mga-dependent transcriptional activation, or both were identified. Of these critical nucleotides, guanines and cytosines within the major groove were disproportionately identified clustered at the 5′ and 3′ ends of the binding site and with runs of nonessential adenines between the critical nucleotides. On the basis of these results, a Pemm1 minimal binding site of 35 bp bound Mga at a level comparable to the level of binding of the larger 45-bp site. Comparison of Pemm with directed mutagenesis performed in the M1T1 Mga-regulated PscpA and Psic promoters, as well as methylation interference analysis of PscpA, establish that Mga binds to DNA in a promoter-specific manner.
INTRODUCTION
Regulation of gene expression in response to changing stimuli allow bacteria to rapidly adapt to their constantly changing environment. Control of transcription is often mediated by direct interactions between target gene promoters and specialized DNA-binding proteins that either enhance (activate) or inhibit (repress) RNA polymerase-mediated initiation (10). Transcription factors possess DNA-binding domains that allow them to recognize and specifically bind to a conserved DNA sequence (binding site) within their target promoters. A conserved family of DNA binding motifs found within many prokaryotic transcription factors, as well as in eukaryotic cells, is the helix-turn-helix (HTH) domain (4). The third helix in the HTH fold is often called the “recognition” helix because it forms the principal DNA-protein interface by inserting into the major groove of the DNA to interact with specific nucleotides; however, DNA contacts may vary across the fold (4). HTH domains can be quite diverse in structure, with the winged HTH (wHTH) possessing an additional C-terminal β-strand hairpin (4). In order to differentially regulate gene expression, DNA-binding proteins must be able to discriminate specific sequences. These sequences often contain a dyad symmetry reflecting that dimers and other multimers of the DNA-binding protein interact with the DNA (21).
Streptococcus pyogenes (the group A Streptococcus [GAS]) is a Gram-positive obligate human pathogen that is the causative agent of both benign diseases (e.g., pharyngitis and impetigo) and life-threatening infections (e.g., necrotizing fasciitis). GAS infects hundreds of millions of people each year, resulting in over 500,000 deaths and contributing to a significant burden on the health of the human population in the entire world (5). Importantly, GAS is able to adapt to diverse niches within the human body in order to obtain nutrients, adhere to tissues, evade the immune system, and replicate. It has become evident that host adaptation by GAS occurs through broad changes in its transcriptome in response to the diverse in vivo environments encountered during infection (20, 25). Not surprisingly, the GAS genome encodes a wide array of known and putative global transcriptional regulators, including 13 two-component systems (TCS) and several “stand-alone” regulators (e.g., RofA-like proteins [RALPs], Rgg, and Mga) that control virulence gene expression in response to external stimuli (13).
Mga, the multiple gene activator of GAS, regulates expression of approximately 10% of the genome (22). The core regulon is composed of a small number of key virulence factors that Mga activates through binding to their promoter DNA, including genes encoding M protein (emm), M-like proteins (arp and mrp), C5a peptidase (scpA), and the streptococcal inhibitor of complement (sic) (22). The secondary Mga regulon represents genes that have low levels of activation or repression and include operons involved in carbohydrate metabolism and other metabolic processes (22). Whether Mga also directly interacts with the promoters of these operons is currently unknown, although it is hypothesized to act indirectly through its influence on other regulatory networks (e.g., CcpA, RALPs, and Rgg). Mga, which is ubiquitous in GAS, can be found in all genomes as one of two divergent alleles (mga-1 and mga-2) associated with serum opacity factor (SOF)-positive and -negative strains, respectively (8a).
Mga is a 62-kDa protein with two distinct domains that show homology to phosphotransferase system (PTS) regulatory domain (PRD) activators of alternative PTS sugar operons, such as the Geobacillus stearothermophilus MtlR and Bacillus subtilis LicR (8a). At the N terminus are domains involved in DNA binding, including a conserved Mga domain (CMD), a classical helix-turn-helix domain (helix-turn-helix 3 [HTH-3]), and a winged helix-turn-helix domain (wHTH-4) (17, 27). The wHTH-4 is required for DNA binding to all Mga-regulated promoters tested (26), whereas HTH-3 appears to serve an accessory role for binding at certain promoters. Two central PRD domains are sites of PTS phosphorylation on conserved histidines that regulate Mga activity (E. R. Hondorp and K. S. McIver, submitted for publication). The C terminus of Mga contains a PTS enzyme IIB (EIIB)-like domain that is necessary for multimerization and transcriptional activity of the protein, yet apparently not for interacting with promoter DNA (9).
Based on studies primarily done in the serotype M6 strain JRS4, three categories of Mga-regulated promoters (categories A, B, and C) were proposed based on the number of binding sites and their position relative to the start of transcription (2). Category A promoters (Pemm and PscpA) were defined using DNase I footprinting; these promoters are composed of a single 45-bp binding site centered at −54 from the start of transcription overlapping the −35 hexamer (16). A category B promoter (PsclA and Psof) was defined by sequence alignment, electrophoretic mobility shift assay (EMSA) analysis, and in vitro transcription. These promoters have a single 45-bp binding site that is located further upstream (−168) from the start of transcription (2, 3). A category C promoter (Pmga), defined by DNase I footprinting, is composed of two 59-bp binding sites located far upstream (−100 and −181) from the start of transcription (18). Based on the positions of putative binding sites, category A appears to be the most common pattern among Mga-regulated promoters in sequenced GAS strains. Interestingly, sequence alignments of these binding sites exhibit very low sequence identity, making it difficult to determine how Mga interacts with its promoters. In this study, we dissect the protein-DNA interactions between Mga and a model category A promoter (Pemm) to understand how this process occurs and test whether these findings can be applied to other Mga-regulated promoter binding sites in GAS.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains and plasmids used in this study are shown in Table 1. GAS strain MGAS5005 (covS mutant) is a well-characterized M1T1 invasive strain and its genome has been sequenced (24). Escherichia coli TOP10 was used for site-directed mutagenesis, and E. coli DH5α was used for general cloning. E. coli C41(DE3) was used for protein expression (19). E. coli strains were grown in Luria-Bertani (LB) broth for plasmid construction and in ZYP autoinduction medium (23) for protein purification. GAS was cultured in Todd-Hewitt medium supplemented with 0.2% yeast extract (THY), and growth was assayed by absorbance measurement using a Klett-Summerson photoelectric colorimeter with the A filter. Antibiotics were used at the following concentrations: ampicillin at 100 μg ml−1 for E. coli; spectinomycin at 100 μg ml−1 for E. coli and GAS; and kanamycin at 50 μg ml−1 for E. coli.
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| GAS strains | ||
| MGAS5005 | M1T1, covS, clinical isolate from CNS, Ontario, Canada, in 1996 | 14 |
| KSM165-L.5005 | mga-inactivated derivative of MGAS5005 | 9 |
| E. coli strains | ||
| DH5α | hsdR17 recA1 gyrA endA1 relA1 | 7 |
| C41(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | 19 |
| TOP10 | F− mcrAΔ(mrr hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 deoR araD139Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| Plasmids | ||
| pCR-Blunt-II-TOPO | pUC ori f1 ori Kanr Ampr LacZα | Invitrogen |
| pMga1-His6 | M1 Mga-His6 under PT7 in pET21a; C-terminally tagged protein in E. coli | 9 |
| pKSM720 | Spectinomycin-resistant promoterless luciferase plasmid | 12 |
| pKSM210 | Pemm1-luciferase reporter plasmid in pKSM720 backbone | This study |
| pKSM211 | PscpA1-luciferase reporter plasmid in pKSM720 backbone | This study |
| pKSM212 | Pemm1-luciferase reporter with the C43A mutation | This study |
| pKSM213 | Pemm1-luciferase reporter with the A35C mutation | This study |
| pKSM214 | Pemm1-luciferase reporter with the G40A mutation | This study |
| pKSM215 | Pemm1-luciferase reporter with the G37A mutation | This study |
| pKSM216 | Pemm1-luciferase reporter with the C38A mutation | This study |
| pKSM217 | Pemm1-luciferase reporter with the T44C mutation | This study |
| pKSM218 | Pemm1-luciferase reporter with the T45C mutation | This study |
| pKSM219 | Pemm1-luciferase reporter with the C12A mutation | This study |
| pKSM220 | Pemm1-luciferase reporter with the C23A mutation | This study |
| pKSM221 | Pemm1-luciferase reporter with the C29A mutation | This study |
| pKSM222 | Pemm1-luciferase reporter with the T11C mutation | This study |
| pKSM232 | Pemm1-luciferase reporter with the C3A mutation | This study |
| pKSM240 | Pemm1-luciferase reporter with the G9A mutation | This study |
| pKSM241 | Pemm1-luciferase reporter with the G10A mutation | This study |
| pKSM242 | Pemm1-luciferase reporter with the T39C mutation | This study |
| pKSM243 | PscpA1-luciferase reporter with the C12A mutation | This study |
| pKSM244 | Pemm1-luciferase reporter with the C12/43A mutation | This study |
| pKSM245 | Psic1-luciferase reporter plasmid in pKSM720 backbone | This study |
| pKSM256 | PscpA1-luciferase reporter with the C43A mutation | This study |
| pKSM257 | PscpA1-luciferase reporter with the C12/43A mutation | This study |
| pKSM258 | Pemm1-luciferase reporter with the G41A mutation | This study |
| pKSM259 | Pemm1-luciferase reporter with the A13C mutation | This study |
| pKSM260 | Pemm1-luciferase reporter with the G18A mutation | This study |
| pKSM261 | Pemm1-luciferase reporter with the G19A mutation | This study |
| pKSM262 | Pemm1-luciferase reporter with the A33C mutation | This study |
| pKSM263 | Pemm1-luciferase reporter with the A34C mutation | This study |
| pKSM271 | Psic1-luciferase reporter with the G40A mutation | This study |
| pKSM272 | Psic1-luciferase reporter with the C12A mutation | This study |
| pKSM273 | Psic1-luciferase reporter with the C43A mutation | This study |
| pKSM274 | Psic1-luciferase reporter with the C12/43A mutation | This study |
| pKSM275 | PspcA1-luciferase reporter with the G40A mutation | This study |
| TOPO-Pemm | Pemm1 in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm C3A | Pemm1 with the C3A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm G9A | Pemm1 with the G9A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm G10A | Pemm1 with the G10A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm T11C | Pemm1 with the T11C mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm C12A | Pemm1 with the C12A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm A13C | Pemm1 with the A13C mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm G18A | Pemm1 with the G18A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm G19A | Pemm1 with the G19A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm C23A | Pemm1 with the C23A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm C29A | Pemm1 with the C29A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm A33C | Pemm1 with the A33C mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm A34C | Pemm1 with the A33C mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm A35C | Pemm1 with the A35C mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm G37A | Pemm1 with the G37A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm C38A | Pemm1 with the C38A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm T39C | Pemm1 with the T39C mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm G40A | Pemm1 with the G40A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm G41A | Pemm1 with the G41A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm C43A | Pemm1 with the C43A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm T44C | Pemm1 with the T44C mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm T45C | Pemm1 with the T45C mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-Pemm C12/43A | Pemm1 with the C12/43A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-PscpA | PspA1 in pCR-Blunt-II-TOPO | This study |
| TOPO-PscpA C12A | PspA1 with the C12A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-PscpA G40A | PspA1 with the G40A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-PscpA C43A | PspA1 with the C43A mutation in pCR-Blunt-II-TOPO | This study |
| TOPO-PscpA C12/43A | PspA1 with the C12/43A mutation in pCR-Blunt-II-TOPO | This study |
CNS, central nervous system.
DNA manipulations.
Plasmid DNA was isolated from E. coli using the Wizard Plus SV miniprep system (Promega). DNA fragments were gel purified from agarose using the QIAquick gel extraction kit (Qiagen) or the Wizard SV gel and PCR clean-up system (Promega). PCR for cloning and generating probes was performed using Taq DNA polymerase (New England BioLabs). PCR for site-directed mutagenesis was performed using Pfu Ultra HF DNA polymerase (Stratagene). DNA sequencing was performed either using the SequiTherm Excel II DNA sequencing kit (Epicentre, Inc.) or through Genewiz, Inc.
Construction of luciferase reporter plasmids.
Pemm was amplified from GAS strain MGAS5005 genomic DNA (gDNA) using the primers M1 SF370 Pemm L (L stands for left) and M1 SF370 Pemm R (R stands for right) (Table 2). PscpA was amplified from MGAS5005 gDNA using the primers M1 PscpA Bam L and M1 PscpA Xho R (Table 2). Psic was amplified from MGAS5005 gDNA using the primers Psic1 BglII and Psic1 XhoI (Table 2). The resulting Pemm and PscpA PCR products were end filled using T4 DNA polymerase (New England BioLabs [NEB]) and cloned by blunt ligation into pCR-Blunt-II-TOPO (Invitrogen) to produce TOPO-Pemm and TOPO-PscpA (Table 1). The resulting Psic PCR products were digested with BglII and XhoI, gel purified, and ligated into BglII-XhoI-digested pKSM720 (Table 1). Mutagenic oligonucleotide pairs (Table 3) were synthesized to introduce point mutations into the Pemm, PscpA, and Psic Mga binding sites using the QuikChange site-directed mutagenesis kit (Stratagene), and the resulting mutations were verified by DNA sequencing. Each insert was digested with BamHI and XhoI or with BglII and XhoI, gel purified, and ligated into BglII-XhoI-digested pKSM720 to produce the respective promoter-luciferase fusions (Table 1). Plasmids were verified by DNA sequencing and transformed into GAS MGAS5005 for luciferase assays.
Table 2.
PCR primers used in this study
| Target | PCR primer | Sequence (5′–3′)a | Reference |
|---|---|---|---|
| Pemm | M1 SF370 Pemm L | GGATCCTCCACAACTTAGACAGC | This study |
| M1 SF370 Pemm R | CTCGAGCGTGTTATTTTTAGCCA | This study | |
| M1 Pemm Luc L | gggGGATCCTCCACAACTTAGACAGC | This study | |
| M1 Pemm Luc R | gggCTCGAGCGTGTTATTTTTAGCCA | This study | |
| M1 FPR Pemm L | CCCAGTCACGACGTTGTAAAA | This study | |
| M1 FPR Pemm R | CCCTCATTTTCAGGGTTTAACTCTAA | This study | |
| PscpA | M1 FPL PscpA L | AGTCCGTAATACGACTCACTTAAGGCCT | This study |
| M1 FPL PscpA R | GCAAACAGGGGTTATTTGCATATGATACA | This study | |
| M1 FPR PscpA L New | TAACGCCAGGGTTTTCCCAG | This study | |
| M1 FPR PscpA R New | CTTGCTTTTGTCATAATGATTAAATGT | This study | |
| M1 PscpA Bam L | gcGGATCCTATGTCTAAAAGAATGAG | This study | |
| M1 PscpA Xho R | gcCTCGAGGATGAGAGACTTTGTCTT | This study | |
| Psic | M1 Psic Luc BglII L | cacAGATCTCAGCAGTTGTAAAACGCAAAG | This study |
| M1 Psic Luc XhoI L | gggCTCGAGTAGTATTCTCTCCTTAATAAATT | This study | |
| M1 FP Psic L | CGCAAAGAAGAAAACTAAGCTATC | This study | |
| M1 FP Psic | TGCAGGAATTCCTCGAGTAGTAT | This study | |
| pKSM720 | 720 conf L | ACGACGTTGTAAAACGACGGC | This study |
| 720 conf R | AGCCTTATGCAGTTGCTCTCC | This study |
Nucleotides in clamp sequences are shown in lowercase type, and nucleotides in restriction sites are shown in bold italic type.
Table 3.
Mutagenic oligonucleotides used in this studya
| Target | Mutagenic oligonucleotide | Sequence (5′–3′)a | Reference |
|---|---|---|---|
| Pemm1-TOPO | Pemm1 C3A SDM L | TCAAAAACAGATTCATCATTAATAGAATTTAGGTCAAAAAGGTGGCAAAAG | This study |
| Pemm1 C3A SDM R | CTTTTGCCACCTTTTTGACCTAAATTCTATTAATGATGAATCTGTTTTTGA | This study | |
| Pemm1 G9A SDM L | ACTCAAAAACAGATTCATCATTAATAGCATTTAAGTCAAAAAGGTGGCAAAA | This study | |
| Pemm1 G9A SDM R | TTTTGCCACCTTTTTGACTTAAATGCTATTAATGATGAATCTGTTTTTGAGT | This study | |
| Pemm1 G10A SDM L | CAGATTCATCATTAATAGCATTTAGATCAAAAAGGTGGCAAAAGCTAAAAA | This study | |
| Pemm1 G10A SDM R | TTTTTAGCTTTTGCCACCTTTTTGATCTAAATGCTATTAATGATGAATCTG | This study | |
| Pemm1 T11C SDM L | GATTCATCATTAATAGCATTTAGGCCAAAAAGGTGGCAAAAGCTAAAAA | This study | |
| Pemm1 T11C SDM R | TTTTTAGCTTTTGCCACCTTTTTGGCCTAAATGCTATTAATGATGAATC | This study | |
| Pemm1 C12A SDM L | GATTCATCATTAATAGCATTTAGGTAAAAAAGGTGGCAAAAGCTAAAAAAG | This study | |
| Pemm1 C12A SDM R | CTTTTTTAGCTTTTGCCACCTTTTTTACCTAAATGCTATTAATGATGAATC | This study | |
| Pemm1 A13C SDM L | TTCATCATTAATAGCATTTAGGTCCAAAAGGTGGCAAAAGCTAAAAAAG | This study | |
| Pemm1 A13C SDM R | CTTTTTTAGCTTTTGCCACCTTTTGGACCTAAATGCTATTAATGATGAA | This study | |
| Pemm1 G18A SDM L | ACAGATTCATCATTAATAGCATTTAGGTCAAAAAAGTGGCAAAAGCTAAAAAA | This study | |
| Pemm1 G18A SDM R | TTTTTTAGCTTTTGCCACTTTTTTGACCTAAATGCTATTAATGATGAATCTGT | This study | |
| Pemm1 G19A SDM L | TTAATAGCATTTAGGTCAAAAAGATGGCAAAAGCTAAAAAAGCTGG | This study | |
| Pemm1 G19A SDM R | CCAGCTTTTTTAGCTTTTGCCATCTTTTTGACCTAAATGCTATTAA | This study | |
| Pemm1 C23A SDM L | GCATTTAGGTCAAAAAGGTGGAAAAAGCTAAAAAAGCTGGTCT | This study | |
| Pemm1 C23A SDM R | AGACCAGCTTTTTTAGCTTTTTCCACCTTTTTGACCTAAATGC | This study | |
| Pemm1 C29A SDM L | GGTCAAAAAGGTGGCAAAAGATAAAAAAGCTGGTCTTTACC | This study | |
| Pemm1 C29A SDM R | GGTAAAGACCAGCTTTTTTATCTTTTGCCACCTTTTTGACC | This study | |
| Pemm1 A33C SDM L | TAGGTCAAAAAGGTGGCAAAAGCTAACAAAGCTGGTCTTTAC | This study | |
| Pemm1 A33C SDM R | GTAAAGACCAGCTTTGTTAGCTTTTGCCACCTTTTTGACCTA | This study | |
| Pemm1 A34C SDM L | AGGTCAAAAAGGTGGCAAAAGCTAAACAAGCTGGTCTTTAC | This study | |
| Pemm1 A34C SDM R | GTAAAGACCAGCTTGTTTAGCTTTTGCCACCTTTTTGACCT | This study | |
| Pemm1 A35C SDM L | AGGTCAAAAAGGTGGCAAAAGCTAAAACAGCTGGTCTTTACC | This study | |
| Pemm1 A35C SDM R | GGTAAAGACCAGCTGTTTTAGCTTTTGCCACCTTTTTGACCT | This study | |
| Pemm1 G37A SDM L | TCAAAAAGGTGGCAAAAGCTAAAAAAACTGGTCTTTACCTTTTGG | This study | |
| Pemm1 G37A SDM R | CCAAAAGGTAAAGACCAGTTTTTTTAGCTTTTGCCACCTTTTTGA | This study | |
| Pemm1 C38A SDM L | GGTGGCAAAAGCTAAAAAAGATGGTCTTTACCTTTTGGCTT | This study | |
| Pemm1 C38A SDM R | AAGCCAAAAGGTAAAGACCATCTTTTTTAGCTTTTGCCACC | This study | |
| Pemm1 T39C SDM L | GTGGCAAAAGCTAAAAAAGCCGGTCTTTACCTTTTGGCTTT | This study | |
| Pemm1 T39C SDM R | AAAGCCAAAAGGTAAAGACCGGCTTTTTTAGCTTTTGCCAC | This study | |
| Pemm1 G40A SDM L | GGTGGCAAAAGCTAAAAAAGCTAGTCTTTACCTTTTGGCTTTTAT | This study | |
| Pemm1 G40A SDM R | ATAAAAGCCAAAAGGTAAAGACTAGCTTTTTTAGCTTTTGCCACC | This study | |
| Pemm1 G41A SDM L | GGTGGCAAAAGCTAAAAAAGCTGATCTTTACCTTTTGGCTTTTATTA | This study | |
| Pemm1 G41A SDM R | TAATAAAAGCCAAAAGGTAAAGATCAGCTTTTTTAGCTTTTGCCACC | This study | |
| Pemm1 C43A SDM L | GTGGCAAAAGCTAAAAAAGCTGGTATTTACCTTTTGGCTTTTATTATTT | This study | |
| Pemm1 C43A SDM R | AAATAATAAAAGCCAAAAGGTAAATACCAGCTTTTTTAGCTTTTGCCAC | This study | |
| Pemm1 T44C SDM L | GGCAAAAGCTAAAAAAGCTGGTCCTTACCTTTTGGCTTTTATTATTT | This study | |
| Pemm1 T44C SDM R | AAATAATAAAAGCCAAAAGGTAAGGACCAGCTTTTTTAGCTTTTGCC | This study | |
| Pemm1 T45C SDM L | GGCAAAAGCTAAAAAAGCTGGTCTCTACCTTTTGGCTTTTATTATTTAC | This study | |
| Pemm1 T45C SDM R | AATAATAAAAGCCAAAAGGTAGAGACCAGCTTTTTTAGCTTTTGCC | This study | |
| PscpA1-TOPO | PscpA1 C12A SDM L | TCTAAAAGAATGTGGATAAGGAGGTAACAAACTAAGCAACTCTTAA | This study |
| PscpA1 C12A SDM R | TTTAAGAGTTGCTTAGTTTGTTACCTCCTTATCCTCATTCTTTTAGA | This study | |
| PscpA1 G40A SDM L | CTAAGCAACTCTTAAAAAGCTAACCTTTACTAATAATCATC | This study | |
| PscpA1 G40A SDM R | GATGATTATTAGTAAAGGTTAGCTTTTTAAGAGTTGCTTAG | This study | |
| PscpA1 C43A SDM L | ACAAACTAAGCAACTCTTAAAAAGCTGACATTTACTAATAATCATCTTTGTTTTATAAT | This study | |
| PscpA1 C43A SDM R | ATTATAAAACAAAGATGATTATTAGTAAATGTCAGCTTTTTAAGAGTTGCTTAGTTTGT | This study | |
| pKSM245 | Psic1 C12A SDM L | AATGAGGTTAAGGAGAGGTAACAAACTAAACAACTC | This study |
| Psic1 C12A SDM R | GAGTTGTTTAGTTTGTTACCTCTCCTTAACCTCATT | This study | |
| Psic1 G40A SDM L | TAAACAACTCTTAAAAAGCTAACCTTTACTAATAATCGTC | This study | |
| Psic1 G40A SDM R | GACGATTATTAGTAAAGGTTAGCTTTTTAAGAGTTGTTTA | This study | |
| Psic1 C43A SDM L | CAACTCTTAAAAAGCTGACATTTACTAATAATCGTCTTTG | This study | |
| Psic1 C43A SDM R | CAAAGACGATTATTAGTAAATGTCAGCTTTTTAAGAGTTG | This study |
Mutated nucleotides are shown in bold underlined type.
Luciferase assay.
Luciferase assays were performed as described previously (12). Briefly, strain MGAS5005 containing each luciferase plasmid was grown in THY broth with spectinomycin at 37°C to early logarithmic phase (20 Klett units), and 500-μl samples were taken every 15 Klett units into stationary phase to assess activity across growth. At least three replicates were sampled at mid-logarithmic phase (80 Klett units) to determine the percent luciferase activity of each point mutation compared to the wild type. Sample pellets were stored at −20°C overnight and assayed the following day using the luciferase assay system (Promega). All samples were normalized to cell units according to the equation 4.5 = (x ml)(65 Klett units/2), where x is the volume of sample. The luciferase assay was read using a Centro XS3 LB 960 luminometer (Berthold Technologies), into which 50 μl of Luciferin-D reagent was directly injected.
Expression and purification of Mga1-His6 proteins from E. coli.
Mga1 protein with a six-histidine tag (Mga1-His6) was purified as follows. E. coli C41(DE3) containing pMga1-His6 (Table 1) was grown in ZYP autoinduction medium for ∼62 h at 37°C, and the cells were harvested by centrifugation at 6,000 rpm at 4°C. The pellet was resuspended in nickel-nitrilotriacetic acid (NiNTA) lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]), incubated on ice with lysozyme for 30 min, followed by sonication on ice using a Branson sonifier 450 with a tapered microtip (setting 6, 50% duty cycle) pulsing 6 times for 30 s each time with 1-min breaks between pulses. The lysate was loaded onto a 750-μl NiNTA agarose column (Qiagen), washed with 20, 50, 70, and 90 mM imidazole NiNTA wash buffer, and eluted with 250 mM imidazole NiNTA elution buffer. Proteins were separated on a 10% SDS-polyacrylamide gel and detected by Coomassie blue staining. Fractions were dialyzed overnight at 4°C into 50 mM HEPES citrate (pH 7.5) with 50 mM EDTA. EDTA was washed out with 50 mM HEPES citrate (pH 7.5), and protein samples were determined to be EDTA free by 4-(2-pyridylazo) resorcinol (PAR) analysis (9). Protein concentration was analyzed by absorbance at 280 nm with the extinction coefficient of ε280 of 59,650 M−1 cm−1 and Coomassie blue staining.
EMSA.
Electrophoretic mobility shift assays (EMSAs) were performed as described previously (16). Briefly, 49-bp DNA probes were generated by annealing oligonucleotide pairs (Table 4) representing wild-type Pemm1, PscpA1, Psic1, and respective point mutations. Each gel-purified oligonucleotide pair (12.5 μM) was mixed with 10 mM Tris-HCl (pH 8.0) and 5 mM MgCl2, heated to 85°C for 5 min, and allowed to anneal by slowly cooling to room temperature. Annealed oligonucleotides were end labeled with [γ-32P]ATP using T4 polynucleotide kinase (NEB). Mga1-His6 (2.5 μM) was incubated with 0.1 nM each probe in band shift buffer [20 mM HEPES (pH 7.5), 1 mM EDTA, 0.6 mM dithiothreitol [DTT], 60 mM KCl, 5 mM MgCl2, and 50 ng/μl poly(dI-dC)] for 20 min at room temperature. Loading dye (5% Ficoll, 0.1% bromophenol blue) (1/5 volume) was added, and each sample was separated on a 5% polyacrylamide gel at 140 V. The gels were then dried for 1 h at 80°C, exposed to a phosphorimager plate, and scanned using a FLA-1500 phosphorimager (GE Healthcare).
Table 4.
Oligonucleotides for binding site studies
| Binding oligonucleotide | Sequence (5′–3′)a | Reference |
|---|---|---|
| Pemm1 MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGCAAAAGCTAAAAAAGCTGGTCTTTACC | 1 |
| TCGTAAATCCAGTTTTTCCACCGTTTTCGATTTTTTCGACCAGAAATGG | ||
| Pemm1 G9A MBS 49-mer | AGCATTTAAGTCAAAAAGGTGGCAAAAGCTAAAAAAGCTGGTCTTTACC | This study |
| TCGTAAATTCAGTTTTTCCACCGTTTTCGATTTTTTCGACCAGAAATGG | ||
| Pemm1 G10A MBS 49-mer | AGCATTTAGATCAAAAAGGTGGCAAAAGCTAAAAAAGCTGGTCTTTACC | This study |
| TCGTAAATCTAGTTTTTCCACCGTTTTCGATTTTTTCGACCAGAAATGG | ||
| Pemm1 T11C MBS 49-mer | AGCATTTAGGCCAAAAAGGTGGCAAAAGCTAAAAAAGCTGGTCTTTACC | This study |
| TCGTAAATCCGGTTTTTCCACCGTTTTCGATTTTTTCGACCAGAAATGG | ||
| Pemm1 C12A MBS 49-mer | AGCATTTAGGTAAAAAAGGTGGCAAAAGCTAAAAAAGCTGGTCTTTACC | This study |
| TCGTAAATCCATTTTTTCCACCGTTTTCGATTTTTTCGACCAGAAATGG | ||
| Pemm1 A13C MBS 49-mer | AGCATTTAGGTCCAAAAGGTGGCAAAAGCTAAAAAAGCTGGTCTTTACC | This study |
| TCGTAAATCCAGGTTTTCCACCGTTTTCGATTTTTTCGACCAGAAATGG | ||
| Pemm1 G18A MBS 49-mer | AGCATTTAGGTCAAAAAAGTGGCAAAAGCTAAAAAAGCTGGTCTTTACC | This study |
| TCGTAAATCCAGTTTTTTCACCGTTTTCGATTTTTTCGACCAGAAATGG | ||
| Pemm1 G19A MBS 49-mer | AGCATTTAGGTCAAAAAGATGGCAAAAGCTAAAAAAGCTGGTCTTTACC | This study |
| TCGTAAATCCAGTTTTTCTACCGTTTTCGATTTTTTCGACCAGAAATGG | ||
| Pemm1 C23A MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGAAAAAGCTAAAAAAGCTGGTCTTTACC | This study |
| TCGTAAATCCAGTTTTTCCACCTTTTTCGATTTTTTCGACCAGAAATGG | ||
| Pemm1 C29A MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGCAAAAGATAAAAAAGCTGGTCTTTACC | This study |
| TCGTAAATCCAGTTTTTCCACCGTTTTCTATTTTTTCGACCAGAAATGG | ||
| Pemm1 A33C MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGCAAAAGCTAACAAAGCTGGTCTTTACC | This study |
| TCGTAAATCCAGTTTTTCCACCGTTTTCGATTGTTTCGACCAGAAATGG | ||
| Pemm1 A34C MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGCAAAAGCTAAACAAGCTGGTCTTTACC | This study |
| TCGTAAATCCAGTTTTTCCACCGTTTTCGATTTGTTCGACCAGAAATGG | ||
| Pemm1 A35C MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGCAAAAGCTAAAACAGCTGGTCTTTACC | This study |
| TCGTAAATCCAGTTTTTCCACCGTTTTCGATTTTGTCGACCAGAAATGG | ||
| Pemm1 G37A MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGCAAAAGCTAAAAAAACTGGTCTTTACC | This study |
| TCGTAAATCCAGTTTTTCCACCGTTTTCGATTTTTTTGACCAGAAATGG | ||
| Pemm1 C38A MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGCAAAAGCTAAAAAAGATGGTCTTTACC | This study |
| TCGTAAATCCAGTTTTTCCACCGTTTTCGATTTTTTCTACCAGAAATGG | ||
| Pemm1 T39C MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGCAAAAGCTAAAAAAGCCGGTCTTTACC | This study |
| TCGTAAATCCAGTTTTTCCACCGTTTTCGATTTTTTCGGCCAGAAATGG | ||
| Pemm1 G40A MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGCAAAAGCTAAAAAAGCTAGTCTTTACC | This study |
| TCGTAAATCCAGTTTTTCCACCGTTTTCGATTTTTTCGATCAGAAATGG | ||
| Pemm1 G41A MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGCAAAAGCTAAAAAAGCTGATCTTTACC | This study |
| TCGTAAATCCAGTTTTTCCACCGTTTTCGATTTTTTCGACTAGAAATGG | ||
| Pemm1 C43A MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGCAAAAGCTAAAAAAGCTGGTATTTACC | This study |
| TCGTAAATCCAGTTTTTCCACCGTTTTCGATTTTTTCGACCATAAATGG | ||
| Pemm1 T44C MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGCAAAAGCTAAAAAAGCTGGTCCTTACC | This study |
| TCGTAAATCCAGTTTTTCCACCGTTTTCGATTTTTTCGACCAGGAATGG | ||
| Pemm1 T45C MBS 49-mer | AGCATTTAGGTCAAAAAGGTGGCAAAAGCTAAAAAAGCTGGTCTCTACC | This study |
| TCGTAAATCCAGTTTTTCCACCGTTTTCGATTTTTTCGACCAGAGATGG | ||
| Pemm1 C12/43A MBS 49-mer | AGCATTTAGGTAAAAAAGGTGGCAAAAGCTAAAAAAGCTGGTATTTACC | This study |
| TCGTAAATCCATTTTTTCCACCGTTTTCGATTTTTTCGACCATAAATGG | ||
| PscpA1 MBS 49-mer | GGATAAGGAGGTCACAAACTAAGCAACTCTTAAAAAGCTGACCTTTACT | This study |
| CCTATTCCTCCAGTGTTTGATTCGTTGAGAATTTTTCGACTGGAAATGA | ||
| PscpA1 C12A MBS 49-mer | GGATAAGGAGGTAACAAACTAAGCAACTCTTAAAAAGCTGACCTTTACT | This study |
| CCTATTCCTCCATTGTTTGATTCGTTGAGAATTTTTCGACTGGAAATGA | ||
| PscpA1 G40A MBS 49-mer | GGATAAGGAGGTCACAAACTAAGCAACTCTTAAAAAGCTAACCTTTACT | This study |
| CCTATTCCTCCAGTGTTTGATTCGTTGAGAATTTTTCGATTGGAAATGA | ||
| PscpA1 C43A MBS 49-mer | GGATAAGGAGGTCACAAACTAAGCAACTCTTAAAAAGCTGACATTTACT | This study |
| CCTATTCCTCCAGTGTTTGATTCGTTGAGAATTTTTCGACTGTAAATGA | ||
| PscpA1 C12/43A MBS 49-mer | GGATAAGGAGGTAACAAACTAAGCAACTCTTAAAAAGCTGACATTTACT | This study |
| CCTATTCCTCCATTGTTTGATTCGTTGAGAATTTTTCGACTGTAAATGA | ||
| Psic1 MBS 49-mer | GTAAGGAGAGGTCACAAACTAAACAACTCTTAAAAAGCTGACCTTTACT | This study |
| CATTCCTCTCCAGTGTTTGATTTGTTGAGAATTTTTCGACTGGAAATGA | ||
| Psic1 C12A MBS 49-mer | GTAAGGAGAGGTAACAAACTAAACAACTCTTAAAAAGCTGACCTTTACT | This study |
| CATTCCTCTCCATTGTTTGATTTGTTGAGAATTTTTCGACTGGAAATGA | ||
| Psic1 G40A MBS 49-mer | GTAAGGAGAGGTCACAAACTAAACAACTCTTAAAAAGCTAACCTTTACT | This study |
| CATTCCTCTCCAGTGTTTGATTTGTTGAGAATTTTTCGATTGGAAATGA | ||
| Psic1 C43A MBS 49-mer | GTAAGGAGAGGTCACAAACTAAACAACTCTTAAAAAGCTGACATTTACT | This study |
| CATTCCTCTCCAGTGTTTGATTTGTTGAGAATTTTTCGACTGTAAATGA | ||
| Psic1 C12/43A MBS 49-mer | GTAAGGAGAGGTAACAAACTAAACAACTCTTAAAAAGCTGACATTTACT | This study |
| CATTCCTCTCCATTGTTTGATTTGTTGAGAATTTTTCGACTGTAAATGA | ||
| MBS Random 49-mer | TTTAGAAACAAAGGCATCAGTCGACCTGAAGCTATTTAGAAAAAGGGTC | This study |
| AAATCTTTGTTTCCGTAGTCACGTGGACTTCGATAAATCTTTTTCCCAG |
Mutated nucleotides are shown in bold underlined type.
DNase I footprint analysis.
Strand-specific labeled probes were generated by PCR amplifying with one end-labeled primer and one cold primer. Each PCR product was run on a 5% polyacrylamide gel, extracted by the crush and soak method, and purified using the QIAquick PCR purification kit (Qiagen). Binding reactions were set up as described above for EMSA except after equilibrium was reached, 1 μl Turbo DNase I (Ambion) was added to each reaction mixture for 90 s. The reaction product was precipitated with 150 μl of DNase I stop buffer (570 mM ammonium acetate [NH4OAc], 50 μg/ml tRNA, 80% [vol/vol] ethanol). The reaction products were washed twice with 70% ethanol, dried under vacuum, and resuspended in 5 μl DNase I gel loading dye (80% formamide, 1× Tris-borate-EDTA [TBE], 0.1% xylene cyanol, 0.1% bromophenol blue). The reaction products were separated on a 6% sequencing gel alongside a Sanger sequencing ladder produced using SequiTherm Excel II DNA sequencing kit (Epicentre, Inc.). The gels were dried for 1 h at 80°C, exposed to a phosphorimager plate, and scanned using a FLA-1500 or FLA-5000 phosphorimager (GE Healthcare).
Methylation protection and interference assays.
The binding reactions used were based on EMSA that resulted in a 50% shift of the probe with the following modifications. For the interference assay, labeled probes were methylated prior to incubation as follows: ∼300,000 cpm of each probe was incubated with 100 μl of 2× DMS buffer (120 mM NaCl, 20 mM Tris-HCl [pH 8.0], 20 mM MgCl2, and 2 mM EDTA) and distilled H2O (dH2O) to a volume of 200 μl. One microliter of dimethyl sulfate (DMS) was added to either the Pemm-R probe or the Pemm-L probe for approximately 75 s at room temperature to obtain approximately one methylation site per probe (see Fig. 2). The PscpA-L and PscpA-R probes were treated in the same manner (see Fig. 6). The reaction was stopped by the addition of 50 μl ice-cold DMS stop buffer (1.5 M sodium acetate [pH 7.0], 1 M 2-mercaptoethanol) followed by ethanol precipitation. For the protection assay, after the binding reaction had been performed, 20 μl of 0.01% DMS was added to the reaction mixture and incubated for 2 min. Then, 1/10 volume of 250 mM dithiothreitol was added, and the reaction products were then separated on a 5% polyacrylamide gel and subsequently exposed to film. Shifted probe and unbound probe were excised from the gel and extracted using the crush and soak method as described above, followed by PCR purification. To reveal the modified A and G nucleotides, the probes were dried and resuspended in 30 μl of 10 mM sodium phosphate (pH 6.8) and 1 mM EDTA. The probes were incubated for 15 min at 92°C to denature the protein, followed by the addition of 3 μl of 1 M NaOH for 30 min and finally ethanol precipitation (320 μl of 500 mM NaCl, 50 μg/ml tRNA, and 900 μl ethanol). The precipitated probes were washed once with 70% ethanol, dried, and resuspended in DNase I loading dye, prior to separation on a 6% sequencing gel alongside a Maxam-Gilbert sequencing ladder (15). The gels were dried for 1 h at 80°C, exposed overnight to a phosphorimager plate, and scanned using a Fuji FLA-5000 phosphorimager (GE Healthcare).
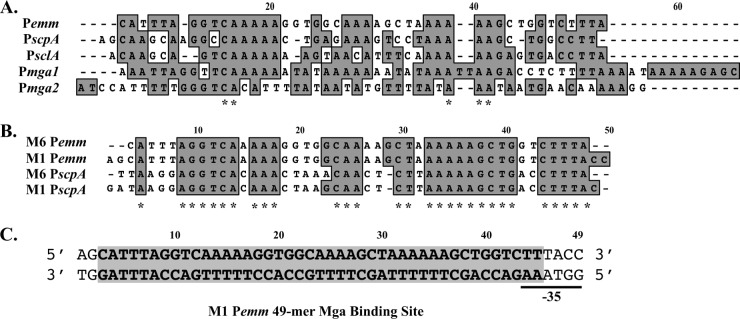
Fig 2.
Conservation of nucleotides in known Mga binding sites. (A) ClustalW nucleotide alignment of category A, B, and C Mga binding sites identified by DNase I footprinting (Pemm, PscpA, and Pmga) or overlapping EMSA (PsclA) in the serotype M6 strain JRS4 (2). A conserved nucleotide in all promoters is indicated by an asterisk below the sequence alignment. Gaps introduced to maximize sequence alignment are indicated by dashes. (B) ClustalW nucleotide alignment of category A Pemm and PscpA Mga binding sites from M6 JRS4 and M1T1 MGAS5005 GAS. A conserved nucleotide in all promoters is indicated by an asterisk below the sequence alignment. (C) Sequence of Pemm1 49-mer double-stranded oligonucleotide probe encompassing the 45-bp Mga binding site (9) used in this study (Table 4). Nucleotides are numbered from 5′ to 3′.
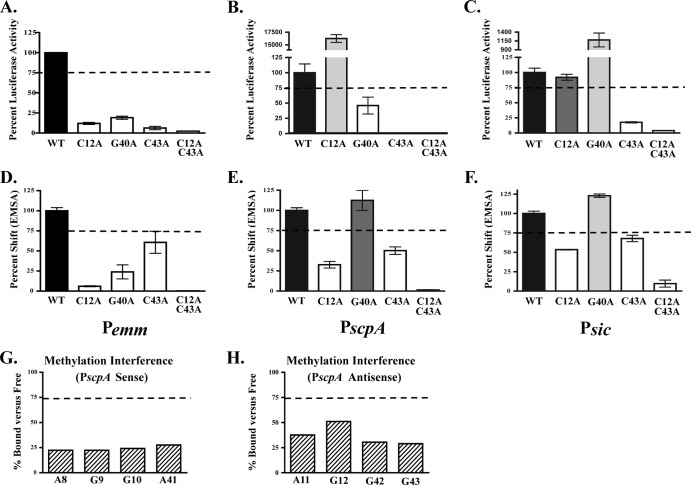
Fig 6.
Role of functional Pemm nucleotides conserved in PscpA and Psic. Three nucleotides conserved in Pemm, PscpA, and Psic (C12, G40, and C43) and found to be important for both activity and DNA binding in Pemm1 were chosen for analysis. (A to F) Single mutations (C12A, G40A, and C43A) and the C12A C43A double mutation in Pemm (A), PscpA (B), and Psic (C) were assayed for promoter activity by luciferase reporter assay (A to C) and DNA binding by EMSA (D to F) for each promoter. Quantification of RLU is shown as percent luciferase activity compared to the respective wild-type promoter. Mutants showing less than 75% wild-type activity (light gray bars), 75 to 100% of wild-type activity (dark gray bars), and greater than wild-type activity (white bars) are indicated. Quantification of EMSA Pemm (D), PscpA (E), and Psic (F) is shown as percent shifted by Mga1-His6 compared to the respective wild type. Mutants that shift less than the wild type (light gray bars), comparable to the wild type (dark gray bars), and greater than the wild type (white bars) are indicated. (G and H) Quantitation of methylation interference assays on the sense (G) and antisense (H) strands of PscpA. Nucleotides exhibiting a reduction in percentage bound versus free presented from 5′ to 3′. The values are averages of two independent experiments. The broken line in all panels denotes 75% of either wild-type binding or luciferase activity.
Uracil and missing thymine interference assay.
The primers were end labeled, and then one labeled primer and one cold primer were used in a PCR that had a 1:20 dUTP/dTTP ratio, so that on average, one thymine was replaced with uracil per binding site. For the missing thymine interference assay, the probes were digested with uracil deglycosolase (NEB) for 1 h at 37°C, followed by PCR purification and incubation in the binding reaction mixture. The binding reaction mixtures were based on EMSA results found to shift 50% of the probe, run on a 5% polyacrylamide gel, and exposed to film. The bound and unbound fractions were extracted by the crush and soak method as described above. Probes from the uracil interference assay were then digested with uracil deglycosylase. The resulting probes were dried and resuspended in 50 μl of 1 M piperidine to generate strand breaks, incubated at 90°C for 30 min, and then placed on ice. One hundred twenty microliters of n-butanol and 50 μl of 1% SDS were added, and the upper phase was extracted. This was repeated with 50 μl n-butanol, and then the probes were dried. The probes were resuspended in 50 μl dH2O, redried, and then resuspended in 10 μl DNase I gel loading dye. The reaction products were separated on a 6% sequencing gel run at 1,700 V for 1.5 h, dried for 1 h at 80°C, then exposed overnight to a phosphorimager plate, and scanned using a FUJI FLA-5000 phosphorimager (GE Healthcare).
RESULTS
Characterization of MBSs in the M1T1 MGAS5005.
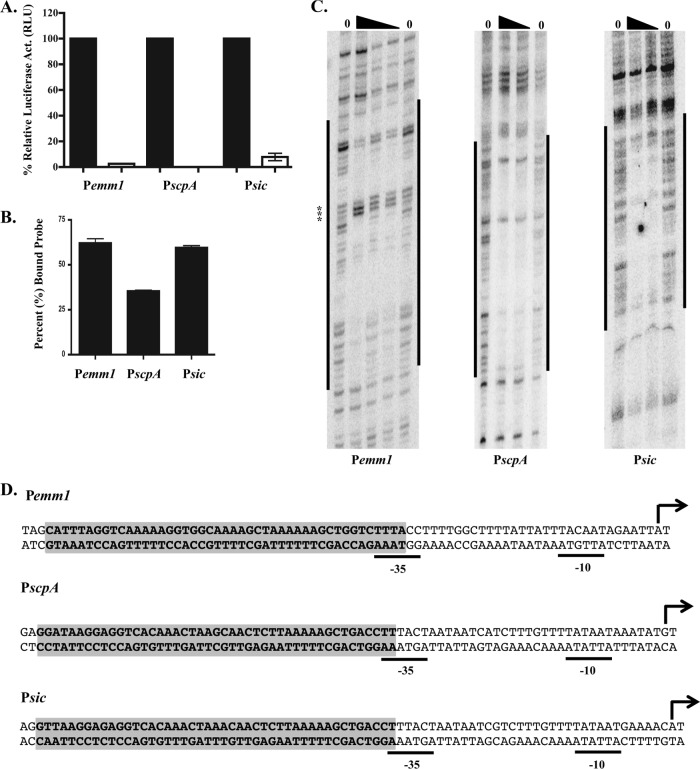
Published biochemical analyses of Mga binding sites (MBSs) (EMSA, DNase I footprinting) have focused on a single serotype M6 GAS strain JRS4 (16, 18). To determine whether Mga-promoter interactions were conserved in other GAS serotypes, Mga binding sites for Pemm and PscpA, two category A promoters (2), were characterized in the invasive M1T1 strain MGAS5005. The Mga-regulated sic gene is found exclusively in M1 GAS and possesses a predicted category A promoter (Psic) based on sequence alignment with M6 sequences; therefore, direct DNA binding studies on Psic were also performed. Each promoter was amplified from the MGAS5005 genome and was cloned in front of a promoterless firefly luciferase (luc) gene in the reporter plasmid pKSM720 (12) for analysis in wild-type MGAS5005 and in the isogenic mga-inactivated strain KSM165L.5005 (Fig. 1A). Luciferase activity was assessed at mid-logarithmic phase (80 Klett units) at a point associated with maximal Mga activity. The Pemm-luc promoter showed the highest luciferase activity (1.6 × 105 relative luciferase units [RLU]), Psic-luc showed intermediate activity (8.7 × 103 RLU), and PscpA-luc exhibited the lowest activity (2.7 × 102 RLU) at this time point (data not shown). All three promoters showed significantly reduced luciferase activity in the mga-inactivated KSM165L.5005 compared to the wild type (Fig. 1A), confirming the Mga-dependent transcriptional activation of Pemm, PscpA, and Psic in the M1T1 background.
Fig 1.
Determination of M1T1 Pemm1, PscpA, and Psic Mga binding sites. (A) Relative Mga-regulated promoter activity determined by luciferase assay at mid-logarithmic phase for Pemm-luc, PscpA-luc, and Psic-luc reporter plasmids in wild-type M1T1 MGAS5005 GAS (black bars) and isogenic mga-inactivated strain KSM165L-5005 (white bars). Data are expressed as percentages of relative luciferase activity (relative light units [RLU]) in the wild-type background (100%) compared to the mga mutant. (B) Relative Mga-binding activity for Pemm, PscpA, and Psic determined by EMSA. Mga1-His6 (2.5 μM) was incubated with a 0.1 nM concentration of each double-stranded oligonucleotide probe, and densitometry was used to quantify the percentage of probe shifted versus total probe added based on a saturating shift. (C) DNase I footprint analysis of Mga1-His6 to antisense strand of Pemm, PscpA, and Psic from strain MGAS5005. Labeled probe was incubated with increasing amounts of Mga1-His6 (0, 0.7, 1.5, and 2.5 μM [the amount of Mga1-His6 indicated by the height of the black triangle above the lane]). Protected regions are indicated by vertical black bars to the sides of the gels. Hypersensitive residues are marked by asterisks. The positions were determined by DNA sequencing ladders. (D) Locations of DNase I footprints of both strands (bold nucleotides) within M1T1 Pemm1, PscpA, and Psic. The numbering of the predicted starts of transcription (+1 [black arrow]) and −10/−35 hexamers are based on those experimentally determined for M6 GAS.
EMSAs using 0.1 nM double-stranded 49-mer oligonucleotide probes of each promoter binding site and various amounts of purified Mga1-His6 protein found that Mga bound to each with maximal binding at 2.5 μM protein (data not shown). At this level of protein, Mga shifted 62% of the Pemm1-MBS 49-mer probe, 60% of the Psic1-MBS 49-mer probe, and 35% of the PscpA1-MBS 49-mer probe (Fig. 1B). To delineate the nucleotides bound by Mga, DNase I footprint assays were performed on both strands of each of the three MGAS5005 promoters using increasing amounts of purified Mga1-His6 (Fig. 1C, antisense; data not shown, sense). In each case, Mga protected a 45-bp region of DNA immediately upstream of the −35 region (Fig. 1D) that correlated exactly with the binding sites predicted by sequence alignments to the established M6 category A binding sites (Fig. 2A).
Comparison of Mga binding sites between different promoter categories.
The goal of this study was to identify the nucleotides within a Mga binding site that are important for interacting with Mga, resulting in functional activation of transcription. A sequence alignment using a modified ClustalW of the published Mga binding sites (Pemm6, PscpA, PsclA, Pmga1, and Pmga2) from M6 JRS4 (16, 18) with the M1T1 MGAS5005 sites (Pemm1, PscpA, and Psic) representing all three categories of Mga-regulated promoters, exhibits only 13.4% nucleotide identity (Fig. 2B). This variability across the different types of binding sites has made it difficult to define a “core DNA-binding sequence.” However, Mga binding sites from comparable promoters found in other GAS serotypes exhibit much higher nucleotide similarity, as seen with Pemm and PscpA from M1T1 and M6 GAS, which shows a nucleotide identity of 49.1% (Fig. 2B, asterisks). Because Pemm is conserved in many GAS serotypes, is strongly regulated by Mga, and shows one of the highest transcript levels of any GAS gene in vivo (6), the 45-bp M1T1 Pemm1 from strain MGAS5005 was chosen as the paradigm Mga binding site for the studies described here (Fig. 2C, shaded region). Conserved nucleotides found to be important for Mga binding and activation in Pemm1 were then tested in other category A Mga-regulated promoters (PscpA and Psic).
Biochemical analysis of the Pemm1 Mga binding site.
Biochemical assays were performed to assess the role of each thymine, adenine, and guanine of the Pemm binding site for Mga interaction. The methyl group of thymine, nitrogen-3 of adenine and nitrogen-7 of guanine have all been identified as points of contact between protein and DNA (11, 28). Therefore, biochemical assays that specifically disrupt these potential sites of Mga interaction were chosen. In each assay, Mga1-His6 was incubated with a randomly modified 226-bp M1 Pemm PCR probe so that 50% of the probe was bound, and separated by EMSA. Strand scissions were then induced in the bound and free DNA fractions to reveal the modified nucleotides, followed by separation on a 6% acrylamide sequencing gel. Nucleotides important for DNA binding are those found in the free DNA lane but are diminished or missing in the bound DNA lane.
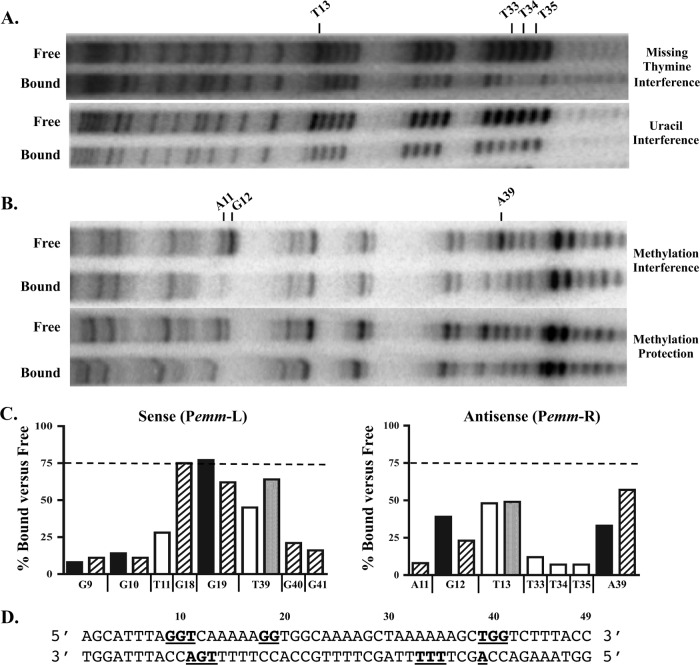
Uracil interference assays were used to target the thymines in the binding site by randomly replacing them with uracil during the PCR amplification of the probe using a dTTP/dUTP ratio that gave one substitution per binding site (Fig. 3A, bottom gel). Nucleotides in the binding site were numbered 5′ to 3′ using the Pemm1 49-mer Mga binding probe as a reference (Fig. 2C). On the sense strand, thymine 39 (T39) was reduced (64% of free) in the bound fraction, while on the antisense strand, T13 was also diminished (49%) in the bound fraction (Fig. 3A, bottom gel, and C).
Fig 3.
Biochemical analyses of Mga binding to Pemm. A 226-bp Pemm1 probe was subjected to various chemical mutagenesis strategies to obtain one mutation per binding site. The resulting probes were assayed by EMSA using Mga1-His6 such that 50% of the probe was bound, followed by excision of bound and free probe. (A and B) Missing thymine and uracil interference assays (thymines) (A) and methylation protection and interference assays (adenines and guanines) (B) were performed on each fraction to identify those nucleotides important for DNA binding (reduced or missing in bound). The antisense strand for each experiment is shown from 5′ to 3′ with the nucleotides identified indicated above the gels. (C) Quantitation of Pemm nucleotides exhibiting a reduction in the percentage bound versus free for each biochemical analysis presented from 5′ to 3′. Methylation protection (black bars), methylation interference (hatched bars), uracil interference (light gray bars), and missing thymine interference (white bars) are shown. The values are averages of two experiments. The broken line denotes 75% of wild-type binding. (D) Schematic diagram showing the locations of all nucleotides within the Pemm1 49-mer identified as important for DNA binding (bold and underlined).
In missing thymine interference assays, incorporated uracils were cleaved by a uracil DNA deglycosylase, leaving only the sugar phosphate backbone prior to incubation with protein (Fig. 3A, top gel). On the sense strand, T11 (28%) and T39 (45%) were identified as being important for binding (Fig. 3C), and on the antisense strand, T13 (48%), T33 (12%), T34 (7%), and T35 (7%) were also reduced in the bound fraction (Fig. 3A, top gel, and C).
Methylation protection assays were performed to identify those guanines or adenines protected from methylation by Mga binding (Fig. 3B, top gel). Guanines are methylated on the nitrogen-7 position in the major groove of the DNA helix, while adenines are methylated on the nitrogen-3 located in the minor groove. Guanines G9 (8%), G10 (14%), and G19 (77%) were identified on the sense strand (Fig. 3C), and G12 (39%) and A39 (57%) were identified on the antisense strand (Fig. 3B, top gel, and C). Methylation interference assays were performed to determine at which guanines and adenines would prior methylation prevent Mga binding (Fig. 3B, bottom gel). Nucleotides G9 (11%), G10 (11%), G18 (75%), G19 (62%), G40 (21%), and G41 (16%) were identified on the sense strand (data not shown), and A11 (8%), G12 (23%), and A39 (33%) were identified on the antisense strand (Fig. 3B, bottom gel, and C). A summary of all the biochemical results is provided using the Pemm1 sequence (Fig. 3D).
In vivo analysis of Pemm1 binding site mutants.
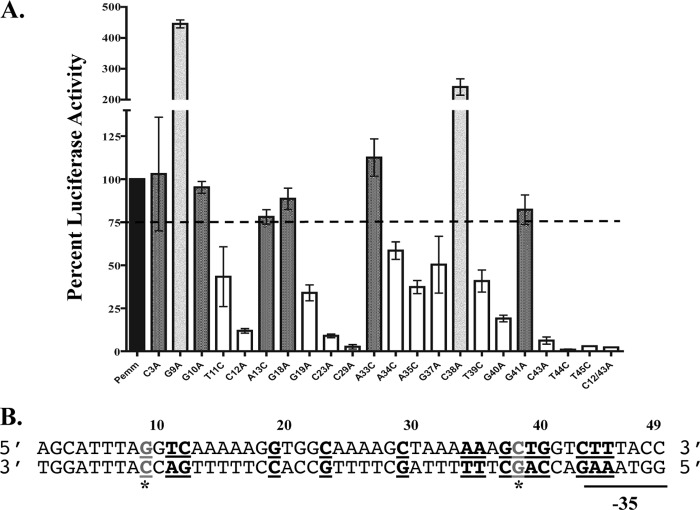
Luciferase assays were performed to study the effect on transcriptional activity of a Pemm1-luc reporter by directed mutagenesis of selected conserved nucleotides based on the alignment of the M1 and M6 Pemm and PscpA Mga binding site (Fig. 2B). In addition, mutations were introduced into all cytosine nucleotides on the sense strand (C3, C12, C23, C29, C38, and C42) as well as any nucleotides identified as important for binding in the biochemical assays above yet not already targeted. Pemm-luc plasmids containing each mutant promoter, a wild-type Pemm-luc plasmid, and a promoterless luc control plasmid were transformed into wild-type strain MGAS5005. Samples were taken at mid-logarithmic phase (80 Klett units), a time of maximal Mga-regulated expression, in order to quantify activity. The wild-type Pemm promoter was set at 100% relative luciferase activity, and the activity of each mutated promoter was calculated as a percentage of the activity of the wild type (Fig. 4A). Strains with the C3A, G10A, A13C, G18A, A33C, and G41A mutations had expression greater than 75% and were considered to have wild-type activity (Fig. 4A, dark gray bars). Strains with the single mutations T11C, C12A, G19A, C23A, C29A, A34C, A35C, G37A, T39C, G40A, C43A, T44C, and T45C and the double mutation C12/43A (C-to-A mutations at positions 12 and 43) had a significant decrease (less than 75%) in luciferase activity (Fig. 4A, white bars). Strains with two different mutations, G9A and C38A, had increased luciferase activity, which increased transcriptional activity to 445% and 241% of the wild type, respectively (Fig. 4A, light gray bars). These two mutated plasmids were also transformed into the mga-inactivated KSM165L-5005 strain. Luciferase assays with this strain showed that these mutations caused the same amount of activity as the wild-type Pemm promoter in the absence of Mga and that the increase in transcriptional activation with each Pemm1 mutant is Mga dependent (data not shown). A summary of all the in vivo reporter results is provided using the Pemm1 sequence (Fig. 4B).
Fig 4.
Luciferase promoter reporter assays of Pemm site-directed mutations in vivo. The 226-bp Pemm1 probe for the wild-type strain and mutant strains with each point mutation was cloned into the firefly luciferase reporter pKSM720 and transformed into M1T1 MGAS5005 GAS for in vivo analysis. (A) Quantification of the relative luciferase activity (RLU) of each Pemm point mutation was compared to that of the wild type and shown as percent luciferase activity. Mutants showing less than 75% wild-type activity (white), 75 to 100% of wild-type activity (dark gray), and greater than wild-type activity (light gray) are indicated. The broken line denotes 75% of wild-type luciferase activity. (B) Schematic diagram showing the locations of all nucleotides within the Pemm1 49-mer identified as important for Pemm activity (bold and underlined). The nucleotides leading to increased promoter activity (G9A and C38A) are shown as gray underlined letters with an asterisk.
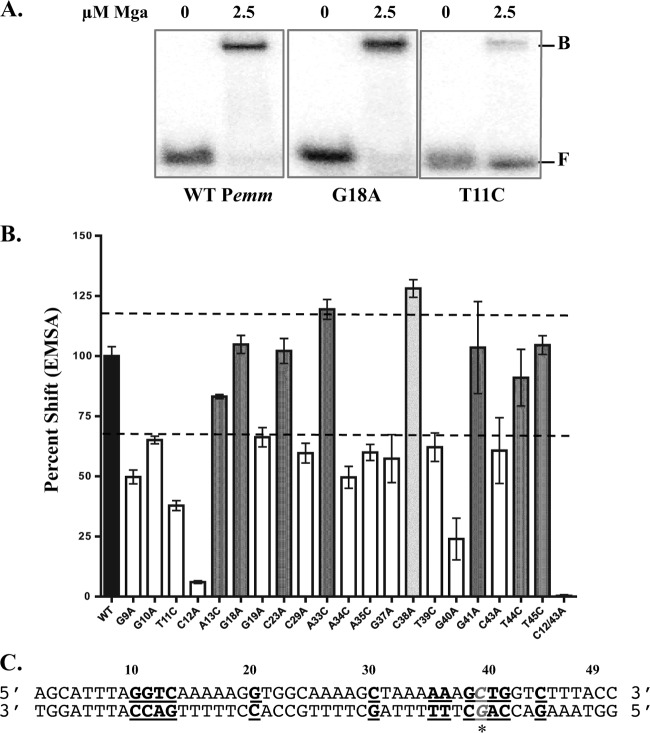
EMSA analysis of Mga binding to Pemm mutants.
EMSA analysis was performed in order to determine the effect on Mga binding of the nucleotides identified by either the biochemical binding assays or luciferase reporter assays. In each assay, 2.5 μM Mga1-His6 was incubated with either 0.1 nM concentration of the Pemm1 MBS 49-mer probe or a mutated probe at the ratio of protein to probe previously determined to be saturating with the probe (Fig. 5A and data not shown). All mutant Pemm1 probes were constructed so that guanines and cytosines were mutated to adenines, whereas the adenines and thymines were mutated to cytosine. Following EMSA, densitometry was performed to measure the amount of total probe bound, and each mutated probe was then compared to the wild type to calculate the percentage shift (Fig. 5B). Since the EMSA was saturating for the wild type, this was set at 100%. Mga shifted wild-type amounts (>75%) of the Pemm1 mutants A13C, G18A, C23A, A33C, G41A, T44C, and T45C MBS 49-mer probes (Fig. 5B, dark gray bars). Mga shifted significantly less (<75%) of the G9A, G10A, T11C, C12A, G19A, C29A, A34C, A35C, G37A, T39C, G40A, and C43A Pemm1 mutants and the double mutant C12/43A MBS 49-mers (Fig. 5B, white bars). The Pemm1 C38A MBS 49-mer was found to have a wild-type shift when incubated with 2.5 μM protein/0.1 nM probe (data not shown); however, when incubated with 1.25 μM protein/0.1 nM probe, Pemm1 C38A MBS 49-mer bound 128% of the probe compared to the wild type (Fig. 5B, light gray bars). A summary of all the DNA-binding results is provided using the Pemm sequence (Fig. 5C).
Fig 5.
DNA-binding activity of Pemm site-directed mutations. EMSA analysis of strains with Pemm1 49-mer point mutations compared to the wild-type strain was performed to identify nucleotides important for Mga binding. Each probe (0.1 nM) was incubated with 2.5 μM Mga1-His6 (except the C38A 49-mer was incubated with 1.25 μM Mga1-His6). (A) Representative EMSA results for Pemm1 wild-type (WT), G18A, and T11C 49-mer probes with Mga1-His6 (2.5 μM) or without Mga1-His6 (0) are shown. The positions of free (F) and bound (B) bands are indicated to the right of the gel. (B) Quantification of EMSA results for each Pemm1 49-mer point mutant (shown below the bars) as determined by densitometry and shown as percent shifted by Mga1-His6 compared to wild-type Pemm1. Mutants that shift less than the wild type (white), comparable to the wild type (dark gray), and greater than the wild type (light gray) are indicated. The broken line denotes 70% and 120% of wild-type binding. (C) Schematic diagram showing the locations of all nucleotides within the Pemm1 49-mer identified as important for DNA binding (bold and underlined). The C38A mutation leading to increased binding is shown in gray italic underlined letters with an asterisk.
Conservation of critical Pemm1 nucleotides in other category A Mga-regulated promoters.
A goal of this study was to use our in-depth analysis of Pemm1 to determine whether these results could be used to predict important nucleotides in other category A binding sites. To test this, directed mutations were subsequently made in PscpA (C5a peptidase gene promoter) and Psic (secreted inhibitor of complement gene promoter) M1T1 Mga binding sites. Three conserved nucleotides were chosen for analysis, C12A, G40A, C43A, and a double mutation C12/43A, that had exhibited both binding and activation defects in Pemm, and were located at either end of the binding site. Luciferase reporter assays using wild-type and mutant PscpA-luc and Psic-luc alleles were performed as described above (Fig. 6A to C). The C12A mutation showed widely variable impacts in the various promoters, with 12% of wild-type activity in Pemm1, yet 16,265% of wild-type activity in PscpA and wild-type levels in Psic. The G40A mutation had decreased luciferase expression in PscpA similar to Pemm, but dramatically increased expression (1,179% of wild type) in Psic. Only the C43A single mutation and the C12/43A double mutation resulted in a comparable decrease in promoter activity from all three promoters compared to their wild-type allele.
EMSA analysis was performed on the same mutations introduced into a PscpA MBS 49-mer and a Psic MBS 49-mer (Fig. 6D to F). The strain with the C12A mutation shifted less than the wild type did for all three binding sites, despite the fact that normal (Psic) and even increased (PscpA) expression was observed in the cognate luciferase reporter assays. The G40A mutation resulted in normal wild-type binding in PscpA that did not correlate with luciferase results. However, the Psic G40A mutant showed 123% of wild-type binding that mirrored the increased Psic G40A luciferase expression. Finally, the C43A and C12/43A probes had a decrease in the amount of protein shifted for all three promoters that correlated directly with reduced luciferase activity. Overall, C43 appears to play a conserved role in both binding and transcriptional activation in all category A Mga-regulated promoters tested. In contrast, C12 and G40 impacted Mga activation and binding differently between the three.
Given the observed variability in the importance of Pemm1 residues conserved in other category A promoters for Mga binding, we performed a methylation interference assay on PscpA as previously described for Pemm1. On the sense strand, the nucleotides A8 (64%), G9 (22%), G10 (24%), and A41 (28%) were identified as important for Mga4-His6 binding (Fig. 6G), whereas on the antisense strand, A11 (38%), G12 (51%), G42 (30%), and G43 (29%) were critical (Fig. 6H). G9 and G10 (sense) and A11 and G12 (antisense) were identified in both Pemm1 and PscpA, suggesting that they play comparable roles. However, the identified A8 (sense) and G42 (antisense) in PscpA are irrelevant thymines (T8 and T42) in Pemm1. Nucleotides at position 41 were identified as important for binding in both Pemm1 (G41) and PscpA (A41) but were different residues. While G40 was important in Pemm1 (Fig. 6D), it was not identified by methylation interference in PscpA and gave an opposite EMSA result when mutated (Fig. 6E). Finally, G43 on the sense strand was identified in PscpA, but not Pemm; however, the cognate sense strand C43A mutation resulted in decreased binding in both promoters. These data further support the conclusion that while Mga does utilize conserved residues for binding at different category A promoters, overall binding occurs in a promoter-specific context.
DISCUSSION
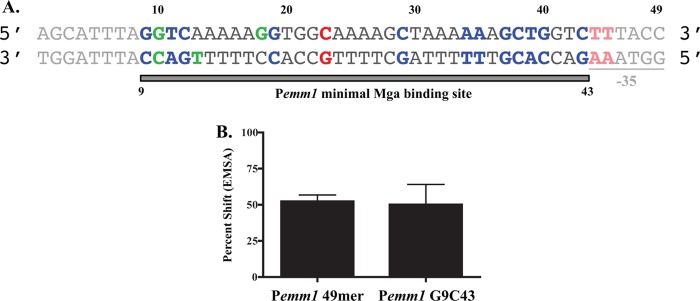
This study identified 34 separate nucleotides within the 45-bp Pemm1 Mga binding site established by DNase I footprinting (Fig. 1) that contribute to either DNA binding, transcriptional activation, or both (Fig. 7A, colored nucleotides). Some nucleotides (C23, T44, and T45) and their complementary antisense bases contribute only to Mga-dependent transcriptional activation (Fig. 7A, red nucleotides). Nucleotides G10 and G18 (sense strand) and C10 and T13 (antisense strand) show a contribution to binding by at least one biochemical method yet have only minor effects on transcriptional activation (Fig. 7A, green nucleotides). The nucleotides G9, T11, C12, G19, C29, A34, A35, G37, C38, T39, G40, and C43 (sense strand), along with their complementary bases (antisense strand), had effects on both binding and transcriptional activation (Fig. 7A, blue nucleotides). Therefore, the most common phenotype reflected in this study showed mutations that both reduced binding and activity. Overall, we propose that the minimal nucleotides within Pemm1 critical for proper interaction with Mga should encompass the bases required for both binding and activation (Fig. 7A, gray bar), resulting in a smaller 35-bp binding region from G9 to C43. In support of this hypothesis, EMSA analyses comparing this minimal Pemm1 G9C43 35-mer probe to the larger Pemm1 49-mer probe using Mga1-His6 revealed that they had essentially identical binding profiles (Fig. 7B). Therefore, Mga requires only a 35-bp binding site for interaction with Mga-regulated Pemm1 promoter sequences.
Fig 7.
Summary of Pemm nucleotides important for Mga DNA binding and activity. (A) Schematic diagram summarizing nucleotides important for Mga-dependent activation (red), Mga binding (green), or both (blue) identified in this study. Based on these results, a proposed minimal Pemm1 Mga binding site of 35 bp from C9 to C43 is indicated by bar below and bold sequences, with nucleotides not essential for binding and activation in faded font. (B) Quantification of EMSA comparing Mga1-His6 binding to Pemm1 49-mer and Pemm1 G9C43 probes. Data are presented as a percent shift of the total probe.
The nucleotides identified within Pemm1 necessary for both binding and activation are biased toward guanines and cytosines (66.7%) compared to the overall G+C content (37.5%) found within the initial 45-bp binding site. Interestingly, most of the bases identified to be not required for Mga binding or activation are found as runs of 4 to 6 adenines (sense strand) that could be functioning to orient Mga to the DNA, as spacer regions between the points of direct contact, or introducing curvature to the DNA (8). The methylation protection and interference assays (Fig. 3) predominately identified guanine residues located within the major groove of the DNA helix as being important for Mga binding. Specifically, direct interactions are suggested to occur in the major groove at G9, G10, G18, G19, G40, and G41 (sense strand) and G12 (antisense strand). The predominant DNA-binding domain of Mga (wHTH-4) is a winged helix-turn-helix domain that would be expected to use its recognition helix to contact nucleotides in the major groove (17, 22). Furthermore, the charged residues within the Mga recognition helix are lysine (positions 5 and 9) and arginine (position 6), which have been shown in other wHTH proteins to form hydrogen bonds primarily with guanines at N7 or O6 in the major groove (17). This corresponds with our guanine methylation assays, since they target N7 in the major groove. Some minor groove interactions were identified at A11 and A39 on the antisense strand; however, these can result from DNA interactions with the C-terminal β-strand “wing” of the wHTH domain (4, 28). Nucleotides G18 and G19 are subject to hypercleavage by DNase I footprinting upon Mga binding (Fig. 1C, Pemm1, asterisks) and may indicate a location of DNA bending. It is possible that methylation of G18 and G19 may actually prevent this flexibility and indirectly lead to the observed reduction in Mga binding and activation. Interestingly, the methylation interference assay performed on PscpA did not show a potential bend, which may suggest that the flexibility of the DNA affects Mga's ability to activate transcription.
Most of the critical nucleotides in Pemm are found clustered at the 5′ and 3′ ends of the binding site with a few dispersed between the ends (Fig. 7A). Combined with the large size of the DNase I-protected region (45 bp), this suggests that Mga might interact with DNA as a dimer despite the lack of any apparent dyad symmetry. Recently, we were able to show that Mga can form dimers in solution and that this self-interaction occurs in vivo (9). Interestingly, although the dimerization of the protein is necessary for transcriptional activation, it does not change the affinity with which Mga interacts with DNA promoter targets. A newly available crystal structure for the Enterococcus faecalis Mga-like regulator EF_3013 (Protein Data Bank [PDB] accession no. 3SQN) showed that this apparent orthologue also formed a homodimer in the absence of bound DNA and possessed an amino-terminal wHTH DNA-binding domain in each monomer comparable to that of Mga. Using the PyMol molecular visualization system (www.pymol.org), the wHTH recognition helices in each dimer were estimated to be approximately 95 Å to 100 Å apart (data not shown), corresponding to about 30 nucleotides (3.4 Å × 30 = 102 Å). Although this is slightly smaller than the 35 bp predicted for the minimal Mga binding site (Fig. 7A), it is based on an orthologous protein, and it does support the hypothesis that Mga and related regulators might interact with target DNA at two distinct sites within the binding region. Further studies will be necessary to confirm the stoichiometry of Mga molecules in this interaction and to determine whether Mga can bind independently at these two potential “half sites” in Pemm1.
Pemm possesses a single category A Mga binding site that is centered at −54 from the start of transcription and overlapping the −35 hexamer (2), suggesting that Mga is positioned to interact directly with RNA polymerase to activate transcription. If Mga acts as a class I transcription factor, it would be expected to interact with the carboxy-terminal domains of the α subunits of RNA polymerase (RNAP) to stabilize its binding at the Pemm promoter (10). Alternatively, Mga may function as a class II transcription factor, which would involve interaction with domain 4 of σ factor in the holoenzyme to accomplish the same result. In either case, one would predict that Mga binding would correlate directly with Mga-dependent transcriptional regulation. As discussed above, the majority of mutations (24/34) demonstrated both reduced Mga binding in vitro and reduced activation in vivo (Fig. 4, 5, and 7A). This supports a model whereby less Mga bound to a promoter leads to less transcriptional activation of that promoter. Even when the mutation led to an increase in Mga binding to Pemm (C38 mutant), the resulting Pemm-luc activity was also increased over that of the wild type. However, G9 mutants presented with decreased Mga binding yet showed an increase in Mga-dependent transcriptional activation (Fig. 4 and 5). It is possible that while Mga has less affinity for this mutation, it may still be positioned to interact with RNA polymerase, and the lower binding affinity may lead to enhanced promoter clearance, leading to an increase in activity. Regardless, this suggests that the exact role of G9 in Mga binding and activation is more complex and will require further investigation. We are currently exploring the effects of these mutations using an in vitro transcription assay in order to better understand this interplay between Mga binding, potential RNAP interaction, and initiation of Mga-regulated transcription.
Interestingly, there does not always appear to be a direct correlation between the amount of DNA bound versus the amount of transcriptional activation. For example, the binding site with the C12A mutation shifted 6.03% and had 11.2% of wild-type RLU, while the binding site with the C23A mutation shifted 59.65% and had 2.62% of wild-type RLU. The location of the mutation could potentially change the orientation of one dimer to another, affecting how Mga interacts with RNAP and influences transcription. In this case, the effect on transcription would be cumulative with the effect on binding. The T44C and T45C mutations both decreased transcription levels in vivo without altering Mga binding (Fig. 4 and 5), which was predicted, as both residues are part of the Pemm −35 hexamer recognized by RNA polymerase. Since these nucleotides are also outside the 35-bp minimal Mga binding site (Fig. 7A), it suggests that they are protected from DNase I digestion but do not contribute to direct protein-DNA contacts. The C23A mutation also showed a decrease in transcription but no effect on binding ability. As with the G9 mutation discussed above, future studies will focus on how much and where Mga binds DNA contributes to transcriptional activation. Combination mutants of the up transcriptional mutations G9A and C38A with a strong down mutation such as C12A or C23A could also be used to dissect how different mutations combine to affect both binding and transcriptional activity and whether one mutation can compensate for another.
The M1T1 Pemm1 binding site was chosen for analysis here to be a possible paradigm for how Mga interacts with DNA at other similar Mga-regulated promoters. To test this possibility, we made directed mutations in the category A binding sites for PscpA and Psic based on conserved nucleotides found to be essential for Mga-Pemm1 interactions (Fig. 6). Interestingly, the phenotypes varied considerably between PscpA, Psic, and Pemm for both Mga binding in vitro and promoter activation in vivo. A C43A mutation and a C12/43A double mutation had the same effect at each of the three category A promoters, suggesting that Mga may interact at this nucleotide in a conserved manner at each target. However, this was not the case for the other two conserved nucleotides. A C12A mutation resulted in a decrease in binding at all three promoters, but in vivo activity varied considerably (Fig. 6). A G40A mutation had the greatest variation between promoters with wild-type binding and reduced activation in PscpA compared to increased binding and activation in Psic. Methylation interference assays performed on the PscpA binding site further demonstrate that Mga interactions with its promoters are only partially conserved. Of the 7 nucleotides identified in PscpA, 3 were unique to this promoter. Interestingly, PscpA has an inverted trinucleotide repeat of GGT. This pattern is only partially conserved in Pemm1; the repeat is present at the 5′ end on the binding site, but the sequence differs at the 3′ end. Combined with the results of the DNase I footprint assay, the results of the methylation interference assay indicate that Pemm1 has a bend, while there is no bend in PscpA. It can be said that all of the conserved Pemm1 nucleotides did have some importance for Mga interactions. However, these results show that Pemm1 serves only as a general model for identifying important Mga contacts in other category A promoters. As Mga appears to interact differently with each of its promoters, detailed analysis of these interactions would need to be determined for each individual promoter.
ACKNOWLEDGMENTS
We thank Vincent Lee and members of the McIver lab for critically reviewing the manuscript.
L.L.H. was supported in part by an NIH/NIGMS T32 training grant in Cell and Molecular Biology (GM080201; principal investigator [PI], L. Pick). This work was supported by an NIH/NIAID R01 (AI47928) award to K.S.M.
Footnotes
Published ahead of print 6 July 2012
REFERENCES
- 1. Almengor AC, Kinkel TL, Day SJ, McIver KS. 2007. The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator Mga in the group A streptococcus. J. Bacteriol. 189:8405–8416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Almengor AC, McIver KS. 2004. Transcriptional activation of sclA by Mga requires a distal binding site in Streptococcus pyogenes. J. Bacteriol. 186:7847–7857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Almengor AC, Walters MS, McIver KS. 2006. Mga is sufficient to activate transcription in vitro of sof-sfbX and other Mga-regulated virulence genes in the group A streptococcus. J. Bacteriol. 188:2038–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29:231–262 [DOI] [PubMed] [Google Scholar]
- 5. Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5:685–694 [DOI] [PubMed] [Google Scholar]
- 6. Graham MR, et al. 2006. Analysis of the transcriptome of group A streptococcus in mouse soft tissue infection. Am. J. Pathol. 169:927–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hanahan D, Meselson M. 1983. Plasmid screening at high colony density. Methods Enzymol. 100:333–342 [DOI] [PubMed] [Google Scholar]
- 8. Haran TE, Mohanty U. 2009. The unique structure of A-tracts and intrinsic DNA bending. Q. Rev. Biophys. 42:41–81 [DOI] [PubMed] [Google Scholar]
- 8a. Hondorp ER, McIver KS. 2007. The Mga virulence regulon: infection where the grass is greener. Mol. Microbiol. 66:1056–1065 [DOI] [PubMed] [Google Scholar]
- 9. Hondorp ER, et al. 2012. Characterization of the group A streptococcus Mga virulence regulator reveals a role for the C-terminal region in oligomerization and transcriptional activation. Mol. Microbiol. 83:953–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ishihama A. 1993. Protein-protein communication within the transcription apparatus. J. Bacteriol. 175:2483–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ivarie R. 1987. Thymine methyls and DNA-protein interactions. Nucleic Acids Res. 15:9975–9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kinkel TL, McIver KS. 2008. CcpA-mediated repression of streptolysin S expression and virulence in the group A streptococcus. Infect. Immun. 76:3451–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kreikemeyer B, McIver KS, Podbielski A. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224–232 [DOI] [PubMed] [Google Scholar]
- 14. Lukomski S, et al. 2000. Identification and characterization of the scl gene encoding a group A streptococcus extracellular protein virulence factor with similarity to human collagen. Infect. Immun. 68:6542–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maxam AM, Gilbert W. 1977. New method for sequencing DNA. Proc. Natl. Acad. Sci. U. S. A. 74:560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McIver KS, Heath AS, Green BD, Scott JR. 1995. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. J. Bacteriol. 177:6619–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McIver KS, Myles RL. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol. Microbiol. 43:1591–1602 [DOI] [PubMed] [Google Scholar]
- 18. McIver KS, Thurman AS, Scott JR. 1999. Regulation of mga transcription in the group A streptococcus: specific binding of Mga within its own promoter and evidence for a negative regulator. J. Bacteriol. 181:5373–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289–298 [DOI] [PubMed] [Google Scholar]
- 20. Musser JM, DeLeo FR. 2005. Toward a genome-wide systems biology analysis of host-pathogen interactions in group A streptococcus. Am. J. Pathol. 167:1461–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pabo CO, Sauer RT. 1984. Protein-DNA recognition. Annu. Rev. Biochem. 53:293–321 [DOI] [PubMed] [Google Scholar]
- 22. Ribardo DA, McIver KS. 2006. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Mol. Microbiol. 62:491–508 [DOI] [PubMed] [Google Scholar]
- 23. Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207–234 [DOI] [PubMed] [Google Scholar]
- 24. Sumby P, et al. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192:771–782 [DOI] [PubMed] [Google Scholar]
- 25. Tart AH, Walker MJ, Musser JM. 2007. New understanding of the group A streptococcus pathogenesis cycle. Trends Microbiol. 15:318–325 [DOI] [PubMed] [Google Scholar]
- 26. Vahling CA. 2006. Functional domains in the multigene regulator of the group A Streptococcus. Ph.D. dissertation. University of Texas Southwestern Medical Center, Dallas [Google Scholar]
- 27. Vahling CM, McIver KS. 2006. Domains required for transcriptional activation show conservation in the Mga family of virulence gene regulators. J. Bacteriol. 188:863–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xiong Y, Sundaralingam M. 2001. Protein–nucleic acid interaction: major groove recognition determinants. Encyclopedia of Life Sciences. John Wiley & Sons Ltd, Chichester, United Kingdom: http://www.els.net [Google Scholar]