Abstract
As more bacterial protein glycosylation systems are identified and characterized, a central question that arises is, what governs the prevalence of particular glycans associated with them? In addition, accumulating evidence shows that bacterial protein glycans can be subject to the phenomenon of microheterogeneity, in which variant glycan structures are found at specific attachment sites of a given glycoprotein. Although factors underlying microheterogeneity in reconstituted expression systems have been identified and modeled, those impacting natural systems largely remain enigmatic. On the basis of a sensitive and specific glycan serotyping system, microheterogeneity has been reported for the broad-spectrum, O-linked protein glycosylation system in species within the genus Neisseria. To elucidate the mechanisms involved, a genetic approach was used to identify a hypomorphic allele of pglA (encoding the PglA galactosyltransferase) as a significant contributor to simultaneous expression of multiple glycoforms. Moreover, this phenotype was mapped to a single amino acid polymorphism in PglA. Further analyses revealed that many pglA phase-off variants (containing out-of-frame configurations in simple nucleotide repeats within the open reading frame) were associated with disproportionally high levels of the N,N′-diacetylbacillosamine–Gal disaccharide glycoform generated by PglA. This phenotype is emblematic of nonstandard decoding involving programmed ribosomal frameshifting and/or programmed transcriptional realignment. Together, these findings provide new information regarding the mechanisms of neisserial protein glycan microheterogeneity and the anticipatory nature of contingency loci.
INTRODUCTION
Protein glycosylation systems based on both N- and O-linked modifications are well documented in eubacteria. In addition to the distinctions based on the nature of site attachment, bacterial protein glycosylation systems can be differentiated on the basis of whether they are dedicated to a single class of proteins or target a broad spectrum of protein substrates. These systems can also be classified as to the degree of glycan diversity observed within and between related species. For example, in the type IV pilus (Tfp) O-linked glycosylation in Pseudomonas aeruginosa, some strains utilize the endogenous O-antigen unit found in lipopolysaccharide (LPS; which in itself varies between strains), while others employ d-arabinofuranose moieties synthesized by an apparently dedicated biosynthetic machinery (11, 28). In the N-linked, broad-spectrum protein glycosylation system of Campylobacter jejuni, a conserved heptasaccharide has been presumed to be highly disseminated. Major disparities in protein glycan diversification have been observed for flagellum-associated, O-linked systems, with those in Campylobacter, Helicobacter, and Clostridium exhibiting high levels of intra- and interstrain variation, while the system in P. aeruginosa appears to be limited to two major glycoforms (4, 10, 13, 26). Why protein-associated glycans display high intra- and interstrain variation in some systems and not in others remains unknown. Many of these and other systems have only begun to be rigorously characterized, and thus, detailed studies of glycan diversity are still in their infancy.
The broad-spectrum O-linked protein glycosylation (pgl) system expressed by species within the genus Neisseria provides a unique model system in which to study the long-term evolutionary trends and dynamics of protein glycan diversification. The highly related species Neisseria gonorrhoeae (the agent of gonorrhea), Neisseria meningitidis (an agent of epidemic meningitis), and Neisseria lactamica (a commensal colonizing the oropharynx of young children) are of particular importance to human health and disease. The basic genetics and biochemistry of glycan biosynthesis, elaboration, and transfer to protein have begun to be well characterized in these species (7, 8, 27). The nascent glycan chain is first synthesized on the lipid carrier undecaprenyl-pyrophosphate on the inner leaflet of the cytoplasmic membrane and translocated into the periplasm. PglO, a protein-targeting oligosaccharyltransferase structurally related to the WaaL family of O-antigen ligases, then transfers the oligosaccharides en bloc to protein substrates (15). These compartmentalized processes are analogous to those seen for eukaryotic systems in which the transfer of a preassembled oligosaccharide from a dolichyl-pyrophosphate donor to the asparagine side chain of nascent proteins occurs within the endoplasmic reticulum.
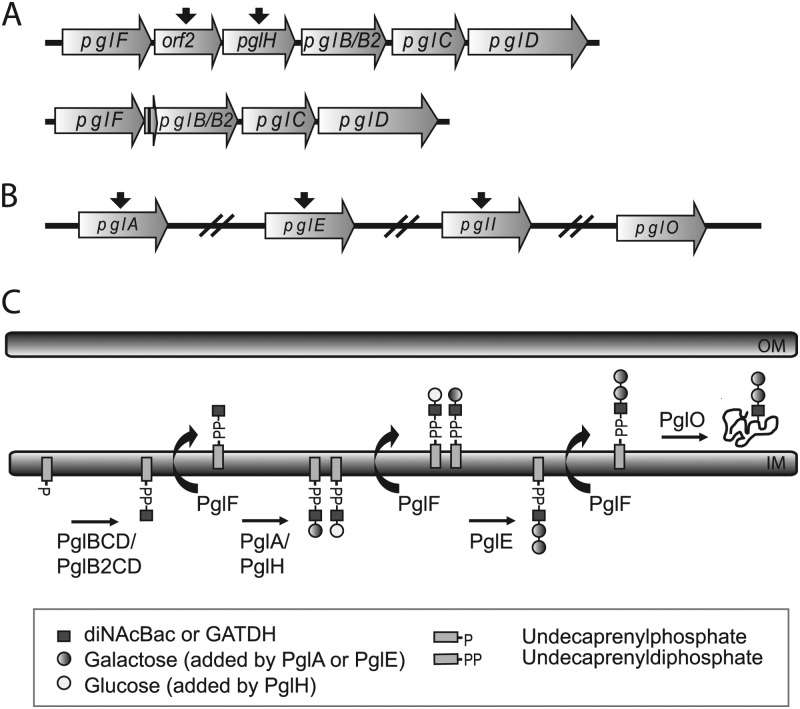
The neisserial pgl system is unique among broad-spectrum bacterial glycosylation systems in the extreme degree of glycan diversity observed within and between strains. Here, the glycan repertoire is governed by the pgl gene content and gene interactions as well as by phase-variable expression of individual components (Fig. 1). Several strains carry a conserved pgl core locus consisting of pglB, pglC, and pglD, whose products (PglB, PglC, and PglD) function in the synthesis of undecaprenyl diphosphate (UndPP)–N,N′-diacetylbacillosamine (diNAcBac) (15). Approximately one-half of N. meningitidis strains carry an alternative pglB allele, namely, pglB2, at this locus. pglB2 is associated with the synthesis of UndPP–glyceramido-acetamido trideoxyhexose (GATDH) rather than UndPP-diNAcBac (12). GATDH differs from diNAcBac by the presence of a glyceroyl moiety instead of an acetyl modification at C-4. Both of these can be further elaborated by the sequential action of two galactosyltransferases, termed PglA and PglE, to form di- and trisaccharide forms (2). The pglA and pglE genes are found in all strains of N. gonorrhoeae, N. meningitidis, and N. lactamica examined to date. The pglA and pglE alleles primarily exist in phase-variable forms due to the presence of simple nucleotide repeats within their open reading frames (ORFs) (22). As such repeats are hypermutable due to processes associated with DNA replication and metabolism, this leads to a high rate of indel mutations that in turn alter protein expression. Such genes are referred to as contingency loci, which underlie a bet-hedging strategy in which even small populations carry variants in altered indel configurations. Variations at contingency loci are generally perceived to be dictating on-off phenotypic switching.
Fig 1.
pgl genes and glycosylation pathway in Neisseria. (A) Polymorphisms at the pgl locus of Neisseria strains. The ancestral pgl locus consists of pglF, orf2, pglH, pglB-pglB2, pglC, and pglD, while other strains lost two genes, orf2 and pglH (see Table S1 in the supplemental material). The first 40 bp of orf2 and the last 100 bp of pglH remain in the deleted pglFBCD locus (short ORF between pglF and pglB/B2). The function of orf2 is not known at the present. Both ancestral and deleted pgl loci are found in combination with both pglB and pglB2 alleles. (B) Additional pgl genes, such as pglA, pglE, pglI, and pglO, are located elsewhere in the genome. Phase-variable pgl genes are indicated with a black arrow above the genes. (C) Current model of the broad-spectrum O-linked glycosylation pathway in Neisseria. OM, outer membrane; IM, inner membrane.
We recently identified a third glycosyltransferase gene, pglH, that encodes a glucosyltransferase that generates disaccharide products using UndPP-linked monosaccharide substrates (8). However, the resulting glycoform is incapable of being further elaborated by PglE. The pglH gene is also a contingency locus, while in some strains, the gene is inactivated due to a large deletion encompassing its 5′ end. Finally, the galactose and glucose residues in the di- and trisaccharide forms are capable of being O-acetylated by the activity of PglI, whose gene also exists in phase-variable and non-phase-variable forms (2). Solely on the basis of differences in gene content and polymorphisms, single strains can vary their glycan repertoire from as few as 4 to as many as 7 glycoforms. In the case of N. meningitidis, which can carry either the pglB or the pglB2 allele, at least 13 different glycoforms can be expressed within the species (8). These findings indicate that the PglB/PglB2-associated glycosyl-1-phosphate transferase, PglF flippase, and PglO oligosaccharide transferase (which all act downstream of the synthesis of the UndPP-linked saccharides) must each possess relaxed donor specificity.
The phenomenon in which a range of glycan structure variation occurs at a given glycosylation site on a given protein within a defined population of cells is termed microheterogeneity. Although this condition is observed across broad taxonomic boundaries, its biological significance and mechanistic basis remain poorly understood. In part, this situation reflects the technological difficulties in monitoring and quantitating glycan diversity at single sites. In the neisserial pgl system, microheterogeneity has been observed in a number of instances (7, 8). For example, microheterogeneity at the level of glycan O-acetylation has been documented at a number of occupancy sites. Moreover, some strains have been shown to simultaneously express mono- and disaccharides, di- and trisaccharides, and even all three glycoforms. A unique form of microheterogeneity occurs in strains that carry active alleles of both pglA and pglH, resulting in the simultaneous expression of a mixture of galactose- and glucose-containing oligosaccharides (8). Here, we investigated the molecular basis for glycan microheterogeneity and identified an important role for hypomorphic glycosyltransferase alleles. We also found evidence for the influence of recoding at these contingency loci, as some phase-off (frameshift-containing) pglA alleles were associated with considerable glycosyltransferase activity.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are described in Table 1 and were grown on conventional GC medium as described previously (14). Protein glycosylation mutations (pglA, pglEon) were introduced into various strain backgrounds using transformation, as previously described (1, 2). The following antibiotics were used for selection of transformants at the indicated concentrations: streptomycin, 750 μg/ml; erythromycin, 8 μg/ml; kanamycin, 50 μg/ml.
Table 1.
Strains used in this study
| Strain | Parental strain | Relevant genotype | Reference or source |
|---|---|---|---|
| N. gonorrhoeae N400 strains | |||
| KS100 (N400) | VD300 | recA6a | 25 |
| KS461 | KS100 | recA6 pglEon pglA::ermC-rpsL | This study |
| KS462 | KS100 | recA6 pglA::ermC-rpsL | This study |
| KS488 | KS462 | recA6 pglA5′ FA1090 | This study |
| KS489 | KS462 | recA6 pglA5′ N400 | This study |
| KS477 | KS462 | recA6 pglAST3787 | This study |
| KS478 | KS462 | recA6 pglAST640 | This study |
| KS483 | KS462 | recA6 pglAFA1090 | This study |
| KS484 | KS462 | recA6 pglAN400 | This study |
| KS742 | KS462 | recA6 pglAZ2491 | This study |
| KS743 | KS462 | recA6 pglAMC58 | This study |
| KS744 | KS462 | recA6 pglA8013 | This study |
| KS745 | KS462 | recA6 pglAFAM18 | This study |
| KS473 | KS462 | recA6 pglAGGGG | This study |
| KS474 | KS462 | recA6 pglAGGGG, E275A | This study |
| KS492 | KS462 | recA6 pglAG350A | This study |
| KS493 | KS461 | recA6 pglEon pglAG350A | This study |
| KS495 | KS462 | recA6 pglAE275A | This study |
| KS101 | VD300 | pilEind | 29 |
| KS122 | KS101 | pilEind pglA::kan | 27 |
| KS773 | KS122 | pilEind pglA::kan lct::pglAN400 | This study |
| KS771 | KS122 | pilEind pglA::kan lct::pglAFA1090 | This study |
| KS772 | KS122 | pilEind pglA::kan lct::pglAST3787 | This study |
| N. gonorrhoeae FA1090 strains | |||
| KS300 | FA1090 | recA6a | 25 |
| KS463 | KS300 | recA6 pglA::ermC-rpsL | This study |
| KS464 | KS300 | recA6 pglEoff pglA::ermC-rpsL | This study |
| KS465 | KS463 | recA6 pglAPRGSG | This study |
| KS466 | KS463 | recA6 pglAPRGSGDLEQPA | This study |
| KS467 | KS463 | recA6 pglAQPA, A275E | This study |
| KS468 | KS463 | recA6 pglAPRGSG, A275E | This study |
| KS469 | KS463 | recA6 pglAQPA | This study |
| KS470 | KS463 | recA6 pglAGSG, A275E | This study |
| KS471 | KS463 | recA6 pglAPRGSGDLEQPA, A275E | This study |
| KS472 | KS463 | recA6 pglAGSG | This study |
| KS475 | KS463 | recA6 pglAA146T | This study |
| KS476 | KS463 | recA6 pglAPRGSGDLEQPA, A275E, A146T | This study |
| KS479 | KS463 | recA6 pglAST3787 | This study |
| KS480 | KS463 | recA6 pglAST640 | This study |
| KS481 | KS463 | recA6 pglAN400 | This study |
| KS482 | KS463 | recA6 pglAFA1090 | This study |
| KS763 | KS463 | recA6 pglAZ2491 | This study |
| KS764 | KS463 | recA6 pglAMC58 | This study |
| KS765 | KS463 | recA6 pglA8013 | This study |
| KS766 | KS463 | recA6 pglAFAM18 | This study |
| KS486 | KS463 | recA6 pglA5′ N400 | This study |
| KS487 | KS463 | recA6 pglA5′ FA1090 | This study |
| KS490 | KS463 | recA6 pglAA350G | This study |
| KS491 | KS463 | recA6 pglAPRGSGDLEQPA, A275E, A146T, A350G | This study |
| KS494 | KS464 | recA6 pglEoff, pglAA350G | This study |
| KS496 | KS463 | recA6 pglAA275E | This study |
| KS769 | KS300 | recA6 pglA::kan | This study |
| KS422 | KS300 | recA6 pglH::ermC-rpsL | 8 |
| KS459 | KS422 | recA6 pglHH371R | 8 |
| KS423 | KS422 | recA6 pglA pglH | 8 |
| KS460 | KS423 | recA6 pglA::kan pglHH371R | 8 |
| KS770 | KS460 | recA6 pglAN400 pglHH371R | This study |
recA6 is an IPTG-inducible allele of recA.
Allelic exchange of pglA.
The introduction of the different pglA alleles into N400 was performed through a two-step mutagenesis strategy that allowed gene replacement without introducing any selectable marker into the final strain. The method uses a two-gene cassette containing both a selectable marker (ermC′) and a counterselectable marker (rpsL+) (18). First, N400 was transformed with pFLOB-pglA::ermC′-rpsL+ (3), and erythromycin-resistant gonococci were selected. N400 is naturally streptomycin resistant, but the introduction of the rpsL+ allele made the intermediate strain, N400 pglA::ermC′-rpsL+, streptomycin sensitive. The next step was transformation with genomic DNA from different N. gonorrhoeae, N. meningitidis, and N. lactamica strains, where homologous recombination replaced the ermC′-rpsL+ cassette with pglA from the genomic DNA and the final strain could be selected on streptomycin plates. For the derived strains, DNA sequencing was used to confirm the absence of other mutations.
Construction of pglA mutant strains.
First, the pCRII-pglAN400 and pCRII-pglAFA1090 plasmids were constructed by PCR amplifying the whole pglA gene and surrounding sequences (3,734 bp) from N. gonorrhoeae strains N400 and FA1090 (pglAN400 and pglAFA1090, respectively; primers pglA-F-EcoRI [5′-GGAATTCACTAACTGGAATTAACAAATACGGC-3′, where the underlining indicates the restriction site] and pglA-R-HindIII [5′-CCCAAGCTTAGCAATGCTGAAACAAGCCC-3′]) and inserting the PCR products into the pCRII-TOPO vector (Invitrogen). To exchange the upstream sequences between N400 and FA1090 pglA, the pCRII-pglAN400, and pCRII-pglAFA1090 plasmids were cut with BamHI and MluI and the 5′ sequence fragments were exchanged and ligated with the other pCRII-pglA plasmid to generate pCRII-5′N400-pglAFA1090 and pCRII-5′FA1090-pglAN400. Since there are very few nucleotides different between N400 and FA1090 in this region, we could use the available restriction enzymes BamHI and MluI, whose fragments include all nucleotides different between the N400 and FA1090 pglA 5′ sequences.
To introduce the different mutations into the endogenous site of pglAN400 and pglAFA1090, the pCRII-pglA plasmids were mutated by using specific primers (see Table S2 in the supplemental material) containing the mutation of interest and a QuikChange XL site-directed mutagenesis kit (Agilent Technologies, Inc., 2009), as described in the manual. The mutation was then introduced into the N400 pglA::ermC-rpsL (KS462) and FA1090 pglA::ermC-rpsL (KS463) strains by homologous recombination that replaced the ermC′-rpsL+ cassette with pglA containing the different mutations of interest.
Complementation analysis.
The coding regions of the pglAFA1090, pglAN400, and pglAST3787 genes were amplified from genomic DNA from the respective strains with primers specific for all strains (primers BP114 [5′-CCTTAATTAAATGAAAATCGTTTTTATCAC-3′] and BP115 [5′-AGCTTTGTTTAAACTTACGCCTTCAAAATATCGCG-3′]) containing PacI and PmeI restriction sites and subcloned into plasmid pGCC4 that had been digested with PacI and PmeI. The pglA genes from FA1090, N400, and ST3787 were then inserted at an intergenic chromosomal site located between the lctP and aspC genes and linked to an erythromycin resistance cassette (20, 21) in the pGCC4::pglA plasmid. To do complementation analysis, this plasmid was then transformed into strain N400 derivative 4/3/1 pglA (KS122) gonococci and selected on plates containing erythromycin.
SDS-PAGE and immunoblotting.
Procedures for SDS-PAGE and immunoblotting have been described previously (14). Whole-cell lysates were prepared from equivalent numbers of cells by heating cell suspensions to 65°C for 10 min in SDS-sample loading buffer. Immunoreactive proteins were detected by immunoblotting using the glycan-specific rabbit antibodies pDAb2 (which recognizes diNAcBac-Glc) (8), npg1 (which recognizes diNAcBac), npg2 (which recognizes diNAcBac-Gal), and npg3 (which recognizes diNAcBac-Gal-Gal) (7) and an alkaline phosphatase-coupled goat anti-rabbit secondary antibody (Sigma).
RESULTS
Microheterogeneity is associated with pglAFA1090 and pglAN400 alleles.
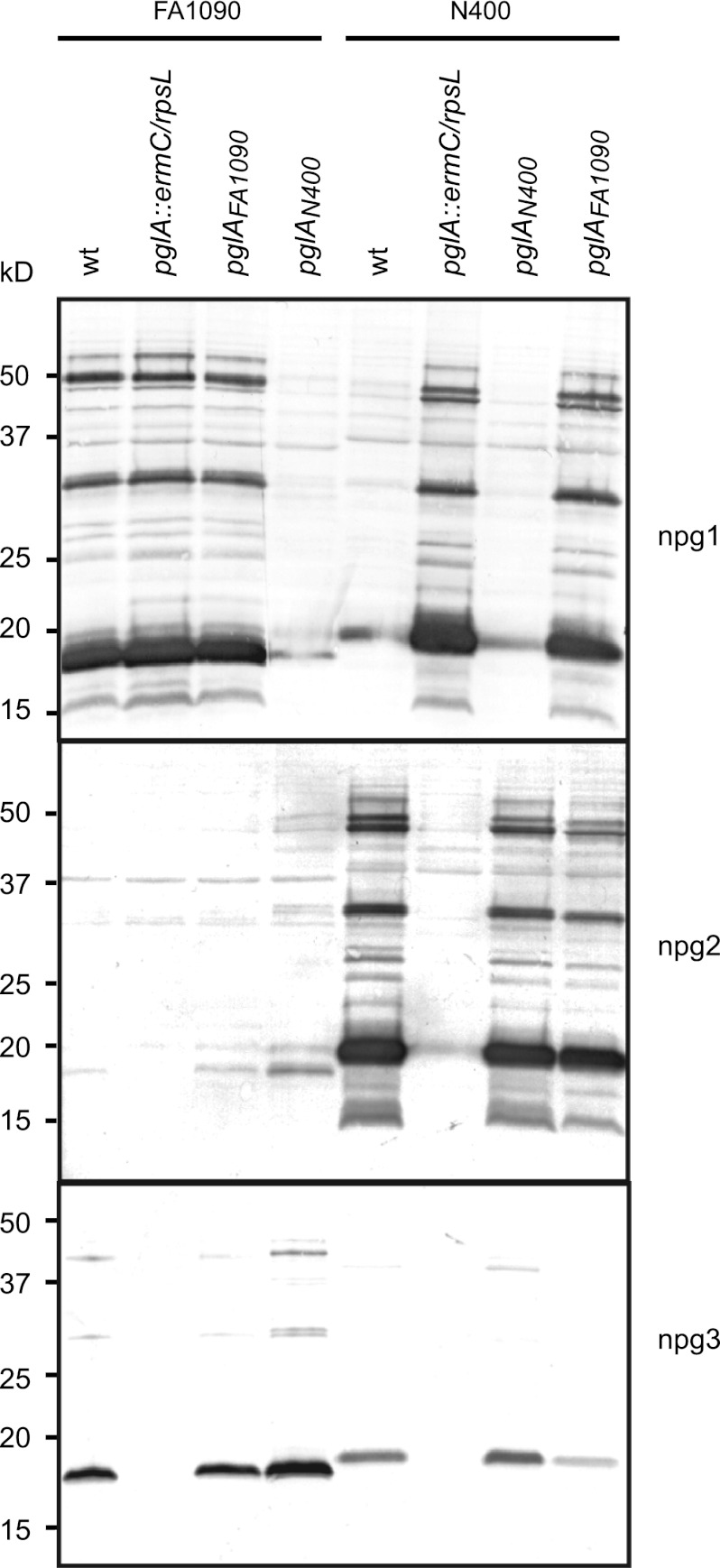
Our previous studies of glycosylation revealed a high degree of glycoform variation at both the intra- and interstrain level in Neisseria strains (7, 8). Although microheterogeneity was seen in many backgrounds, we were particularly struck by the unique pattern of microheterogeneity in gonococcal strain FA1090, which simultaneously expressed high levels of the diNAcBac monosaccharide glycoform, no disaccharide glycoform, and moderate levels of trisaccharide. This observation was made through the use of the highly specific monoclonal antibodies (MAbs) npg1, npg2, and npg3, which recognize diNAcBac- and diNAcBac-based di- and trisaccharide-associated epitopes, respectively. This result was unanticipated, as FA1090 carries phase-on alleles of pglA and pglE and is therefore expected to express primarily the diNAcBac trisaccharide glycoform. For example, strain N400 pglEon carries the same functional genotypic background as FA1090 but nevertheless largely synthesizes diNAcBac-Gal-Gal trisaccharide (7, 8). To examine this phenomenon in more detail, glycoform expression in N. gonorrhoeae strains FA1090 and N400 and genetically defined derivatives was monitored using the glycan epitope-specific MAbs. As shown earlier, FA1090 expressed high levels of the diNAcBac glycoform and relatively reduced levels of the trisaccharide glycoforms (Fig. 2, lane wt). Replacing the wild-type pglA allele with a pglA-null allele in this background abolished trisaccharide glycoform expression but had no discernible effect on diNAcBac expression. Reintroduction of the pglAFA1090 gene into the null mutant restored the parental phenotype, while reintroduction of the pglAN400 gene increased expression of both the di- and trisaccharide forms, which came at the expense of diNAcBac expression, which was nearly abolished. To confirm the inference that the microheterogeneity in FA1090 is related to its pglA allele, analogous gene disruption and transfers were carried out in N400. As this background has pglAon and pglEoff alleles, it predominantly expresses the disaccharide glycoform, and when a pglA disruption was introduced, a major shift to expression of the diNAcBac glycoform was observed. While this alteration was corrected by introduction of the pglAN400 gene, the pglAFA1090 gene led to a mixture of the diNAcBac and disaccharide glycoforms (Fig. 2). Together, these findings showed that the differences in microheterogeneity could be primarily attributed to pglA allele status and, accordingly, that these phenotypes were the result of altered pglA-associated activity.
Fig 2.
Microheterogeneity is associated with the pglA alleles from FA1090 and N400. Immunoblotting of whole-cell lysates from N. gonorrhoeae strains FA1090 and N400 and mutant derivatives with glycan-specific monoclonal antibodies (npg1, npg2, and npg3). Note that N400 has pglEoff and FA1090 has pglEon and that the glycosylation patterns change according to pglE genotype and the presence of the pglA allele. Strains used were KS300, KS463, KS482, KS481, KS100, KS462, KS484, and KS483. wt, wild type.
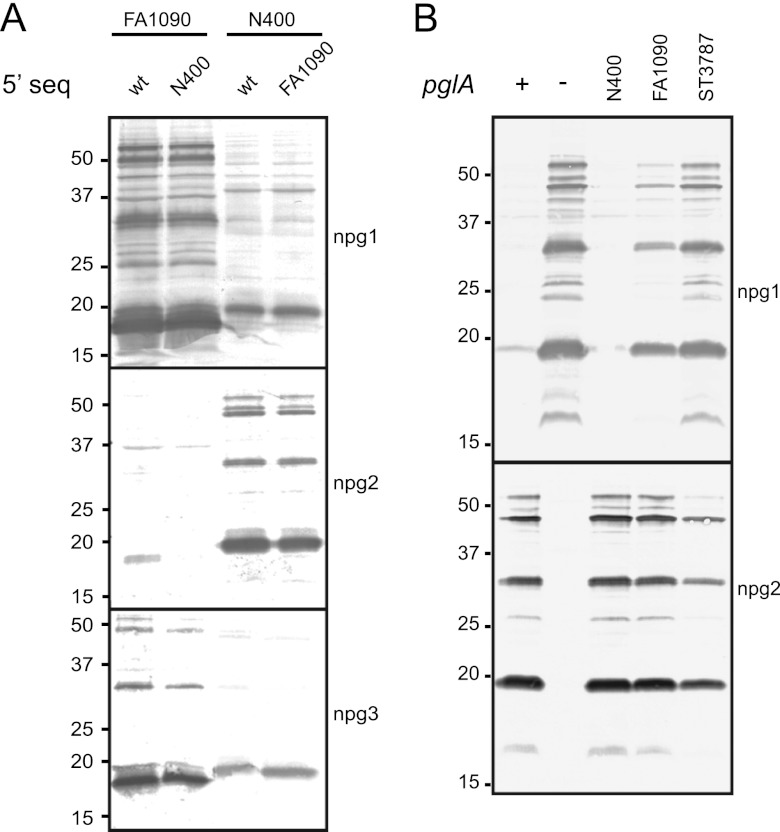
The glycosylation microheterogeneity phenotype is linked to the pglAFA1090 ORF.
Having established that the distinctive glycosylation phenotypes were linked to pglA, we wanted to further elucidate whether this was due to differences in the pglA ORF or features mapping 5′ to the ORF. Therefore, we exchanged the upstream sequences between the N400 and FA1090 pglA alleles and examined the patterns of glycoform expression. Neither fusion of the pglAN400 5′ sequence to the pglAFA1090-coding region nor the reciprocal exchange influenced glycosylation expression (Fig. 3A), proving that the glycosylation phenotype is not linked to sequences 5′ of the pglA ORF. To confirm that the outcomes seen in strains carrying different pglA loci were ascribed to the coding region of pglA and were not indirect effects associated with strain construction, recombinants were created in which the coding region of pglA alone was expressed from an ectopic site in a pglA-null background. In this experiment, we included the pglA allele from N. lactamica ST3878, which we previously observed has a glycosylation pattern similar to that of FA1090 (7). Interestingly, this allele is annotated as being in an out-of-frame configuration, and we confirmed this by resequencing. The coding regions of pglA from strains FA1090, N400, and ST3787 were placed downstream of a derepressible promoter and operator and expressed in the N400 derivative 4/3/1 pglA from an ectopic site. Using immunoblotting and glycan-specific monoclonal antibodies, we confirmed that the coding regions of pglA were both necessary and sufficient to obtain the same glycosylation phenotypes observed when exchanging the whole pglA allele into the endogenous site. Since pglE is phase off in this background, only mono- and disaccharide reactivities are shown in Fig. 3. The complementation studies hence showed that the glycosylation phenotype is coupled to the pglA ORF from N400, FA1090, and ST3787 (Fig. 3B).
Fig 3.
Glycosylation microheterogeneity phenotype is linked to the pglAFA1090 ORF. (A) The glycosylation phenotype is not linked to different 5′ sequences of pglA. Immunoblotting of whole-cell lysates from N. gonorrhoeae strains FA1090 and N400 and mutant strains where only the 5′ noncoding sequences are exchanged using glycan-specific monoclonal antibodies (npg1, npg2 and npg3). Strains used were KS487, KS486, KS489, and KS488. (B) Complementation studies show that the glycosylation phenotype is coupled to the pglA ORF. Variant alleles of pglA were IPTG (isopropyl-β-d-thiogalactopyranoside) induced and expressed ectopically in the strain N400 derivative 4/3/1 pglA, and expression of mono- and disaccharides was monitored by immunoblotting whole-cell lysates with the glycan-specific monoclonal antibodies npg1 and npg2. The immunoblot demonstrates that microheterogeneity is associated with the respective pglA ORFs and is not dependent on endogenous expression. Lane −, pglA::kan; lane +, N400 wild type (pglA in the endogenous site). The last three lanes have the respective pglA ORFs expressed ectopically. Strains used were KS101, KS122, KS773, KS771, and KS772.
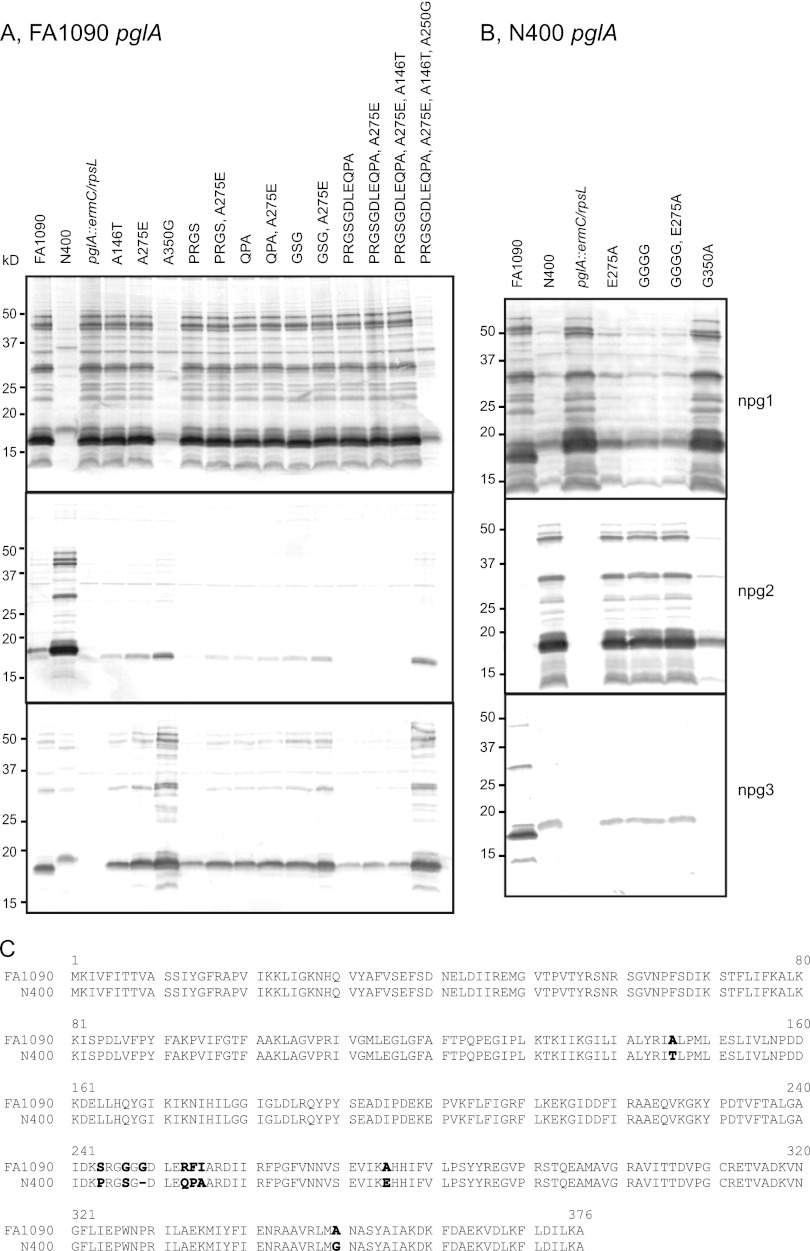
Identification of a single amino acid responsible for the hypomorphic character of the pglAFA1090 allele.
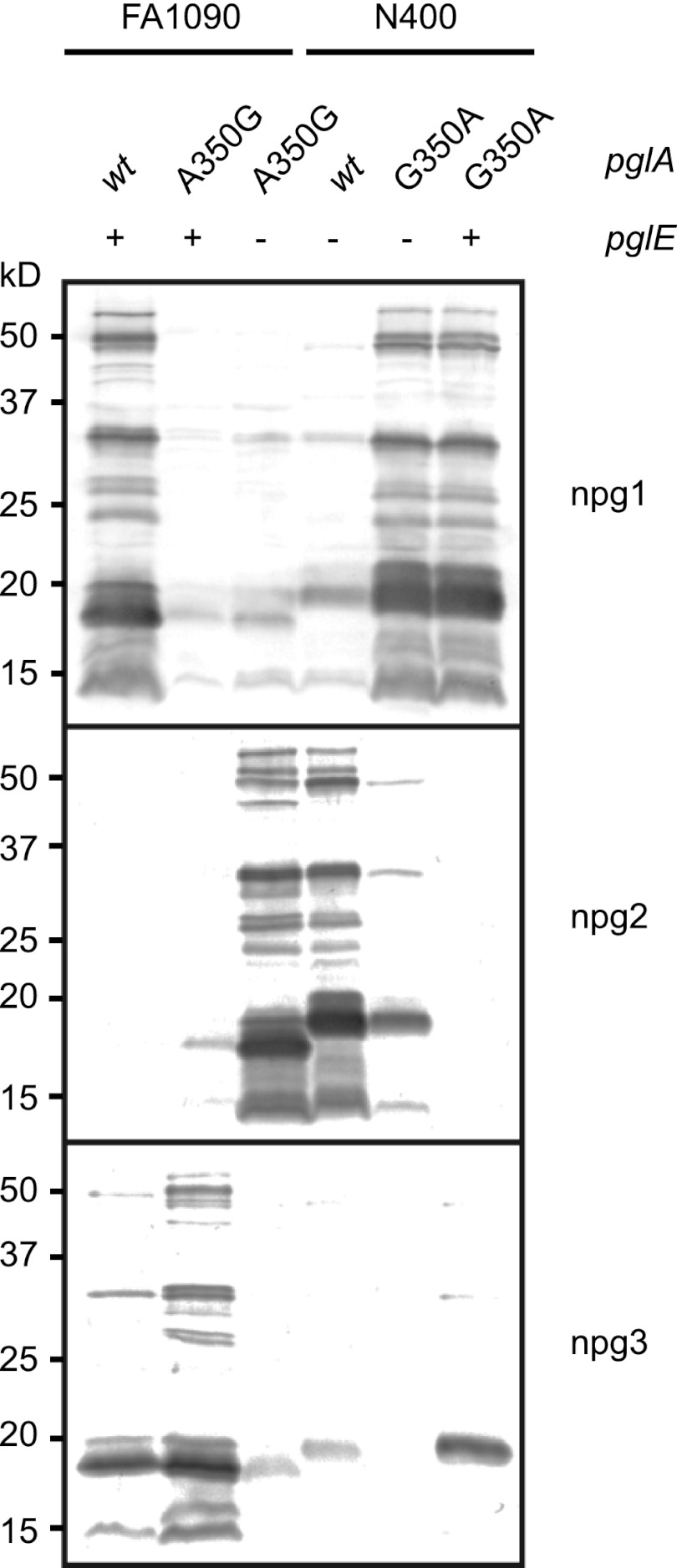
Having ascertained the importance of the pglA-coding region, we next wanted to examine which features of PglA were responsible for these differences. All amino acids distinct to PglAN400 were introduced into PglAFA1090 by site-directed mutagenesis in the FA1090 background (Fig. 4A). As anticipated, the simultaneous replacement of all polymorphisms unique to pglAN400 in the pglAFA1090 allele restored full activity. Of all of these, however, the change from an alanine to a glycine at residue 350 was both necessary and sufficient for the phenotype. Some of the distinguishing amino acids were also examined by mutagenesis of PglAN400 in the N400 background (Fig. 4B). Note that this was done in different pglE backgrounds, so that the glycosylation patterns cannot be directly compared. When a reciprocal G350A mutation was introduced into PglAN400, this resulted in a glycosylation pattern identical to that seen when inserting the whole pglAFA1090 allele into N400.
Fig 4.
Mutagenesis identifies one PglA amino acid responsible for diminished galactosyltransferase activity. Expression of different glycans was monitored by immunoblotting with the glycan-specific antibodies npg1, npg2, and npg3. (A) Various amino acid substitutions were introduced into PglAFA1090, and the following glycosylation patterns were examined. Mutating only one amino acid, A350G, was sufficient to obtain the same glycosylation pattern obtained when pglAN400 was introduced into FA1090 (sixth lane). All amino acids mutated are shown for each lane. Strains used were KS300, KS100, KS462, KS475, KS496, KS490, KS465, KS468, KS469, KS467, KS472, KS470, KS466, KS471, KS476, and KS491. (B) An assortment of amino acid mutations was introduced into PglAN400, and the glycosylation patterns that followed were examined. The analogous G350A mutation in PglAN400 changed the glycosylation pattern, which was similar to that of pglAFA1090 expressed in N400 (last lane). All amino acids mutated are shown for each respective lane. Strains used were KS300, KS100, KS463, KS495, KS473, KS474, and KS492. (C) An alignment of PglA protein sequences from FA1090 and N400, where all dissimilar amino acids are shown in bold.
Next, we wanted to alter amino acid 350 in the context of the respective pglE backgrounds so that the glycosylation patterns could be directly compared. By introducing pglAA350G in a FA1090 pglEoff background, the glycosylation pattern changed accordingly and was identical to that of N400 wild type (Fig. 5; compare the third and the fourth lanes from the left). Furthermore, by introducing pglAG350A in an N400 pglEon background, the glycosylation pattern was identical to that of FA1090 wild type (Fig. 5, first and sixth lanes). In conclusion, the differences between N400 and FA1090 could be mapped to amino acid 350 of PglA. It is worth noting that only PglA from FA1090 has alanine at amino acid 350, while all other strains have glycine in this position (see Fig. S1 in the supplemental material).
Fig 5.
Validation of the roles of amino acid 350 substitutions in common pglE backgrounds. Glycosylation patterns were monitored by immunoblotting with the glycan-specific antibodies npg1, npg2, and npg3. By introducing the A350G PglA mutation in the FA1090 pglEoff background, the obtained glycosylation was identical to that of N400 wild type. Similarly, by introducing the G350A PglA mutation in the N400 pglEon background, the achieved glycosylation was identical to that of FA1090 wild type. −, pglE from strain N400, which is pglEoff; +, pglE from strain FA1090, which is pglEon. Strains used were KS300, KS490, KS494, KS100, KS492, and KS493.
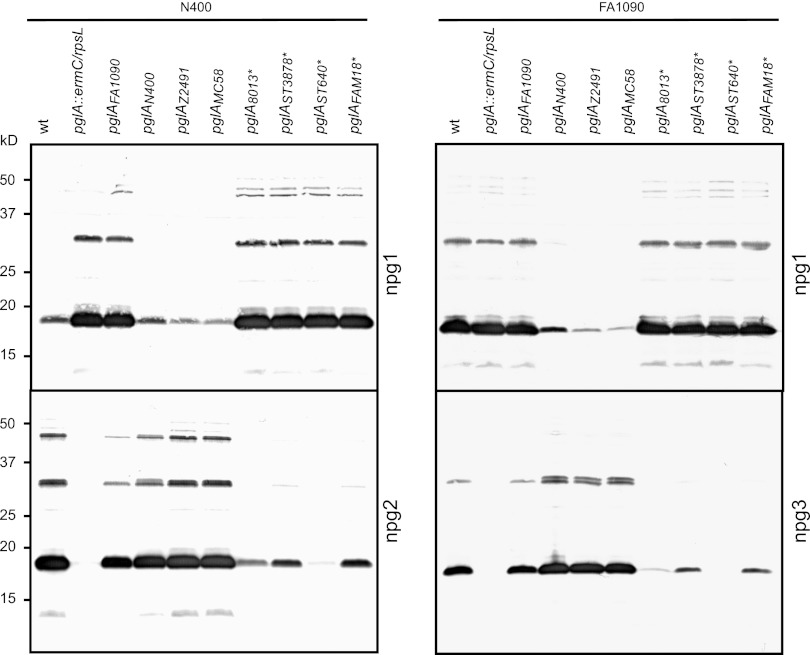
Distinct pglA alleles are associated with variable levels of microheterogeneity.
Having demonstrated the importance of PglA in the distinct FA1090 and N400 glycosylation patterns, we wanted to examine the influence of other pglA alleles on microheterogeneity. Therefore, several pglA alleles from N. gonorrhoeae, N. meningitidis, and N. lactamica strains were exchanged into the N400 and FA1090 backgrounds using the allelic exchange system. Immunoblotting with the glycan-specific monoclonal antibodies revealed that the different pglA alleles accounted for distinct glycosylation patterns (Fig. 6). The pglA alleles from strains FA1090, N400, Z2491, and MC58 are all in a phase-on configuration, while the pglA alleles from strains 8013, ST3787, ST640, and FAM18 are in a phase-off configuration but the strains nevertheless express detectable levels of the disaccharide glycoform (in the N400 background, as pglE is off) or trisaccharide glycoform (in the FA1090 background, as pglE is on). Interestingly, the phase-off pglA alleles were associated with considerable levels of PglA activity. Furthermore, the levels of activity varied to quite a degree between strains carrying the different pglAoff alleles. To investigate the levels of glycoform expression in these strains in more detail, we examined immunoblot signal intensity versus that for standards consisting of admixtures of samples from pglAN400 and pglA-null backgrounds. In this way, the strain carrying the pglAFA1090 allele had about 30% of the npg2-reactive glycoform expression compared to N400, while the strains carrying pglAST3787 and pglAFAM18 had about 4% of the npg2-reactive glycoform (Fig. 7). The strain carrying pglA8013 had about 1 to 2% of the npg2-reactive glycoform and the strain carrying pglAST640 had about 0.5% of the npg2-reactive glycoform expression compared with the standard. Even pglAST640 (which had the lowest npg2 reactivity level) had reactivity above the background reactivity seen for the pglA-null mutant strain.
Fig 6.
Distinct pglA alleles are associated with variable levels of microheterogeneity. Immunoblotting of whole-cell lysates from mutant strains of FA1090 and N400, carrying pglA alleles from different N. gonorrhoeae, N. meningitidis, and N. lactamica strains, with glycan-specific monoclonal antibodies (npg1, npg2, and npg3). Note that the pglA genes from N400, FA1090, Z2491, and MC58 are in the phase-on configuration, while those from 8013, ST3787, ST640, and FAM18 are in the phase-off configuration and are marked with asterisks (the alignment is shown in Fig. S1 in the supplemental material). N400-derived strains were KS100, KS462, KS483, KS484, KS742, KS743, KS744, KS477, KS478, and KS745. FA1090-derived strains were KS300, KS463, KS482, KS481, KS763, KS764, KS765, KS479, KS480, and KS766.
Fig 7.
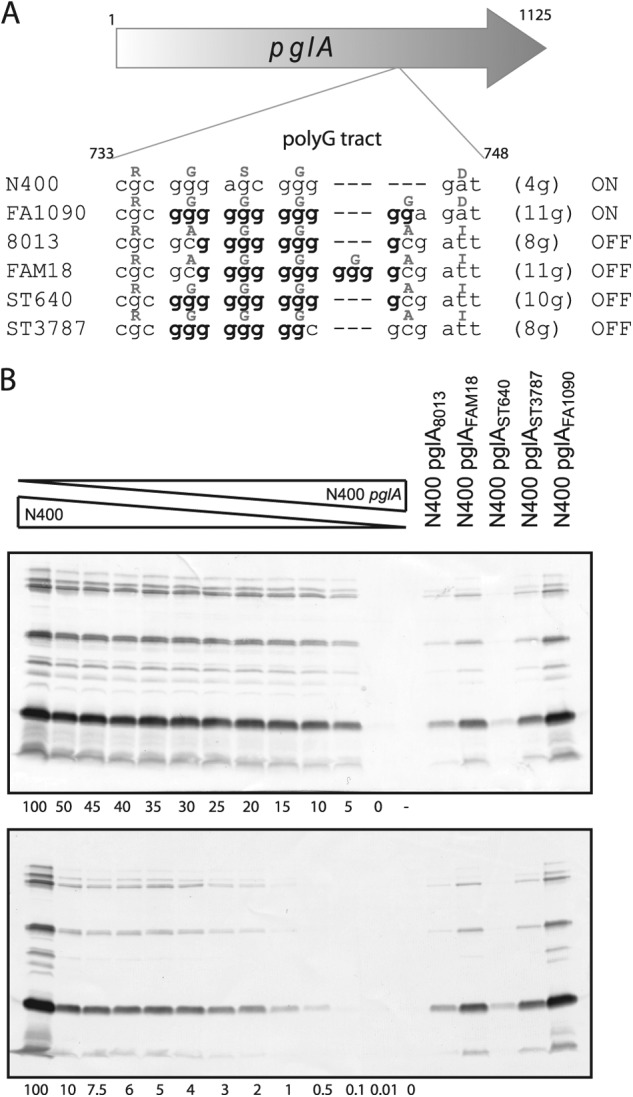
Quantification of the variable levels of microheterogeneity. (A) The poly(G) tracts within the pglA ORFs from N. gonorrhoeae strains (N400, FA1090), N. meningitidis strains (8013, FAM18), and N. lactamica strains (ST640, ST3787) are shown with the corresponding amino acids above (gray). The nucleotide numbering is according to pglAN400. (B) Immunoblotting of whole-cell lysates from mutant strains of N400 carrying pglA alleles from different N. gonorrhoeae, N. meningitidis, and N. lactamica strains with the npg2 glycan-specific monoclonal antibody. The amount of disaccharide glycan in N400 carrying different pglA alleles was compared to the level of disaccharide glycan in admixtures of N400 wild type (KS100) and N400 pglA::ermC-rpsL (KS462). Other strains used were KS744, KS745, KS478, KS477, and KS483. Note that the pglA genes from 8013, ST3787, ST640, and FAM18 are in the phase-off configuration.
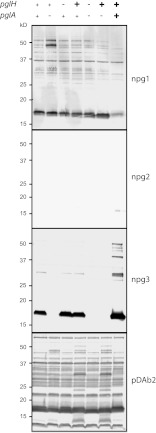
Interactions of pglH and pglA alleles and their contributions to microheterogeneity.
PglH acts as a glucosyltransferase that further elaborates both UndPP-linked GATDH and diNAcBac to glucose-containing disaccharides (8, 15). During the earlier study, we observed a high degree of pglH polymorphisms and found that the pglHFA1090 allele is grossly hypomorphic and mapped the reduced glucosyltransferase activity trait to a unique histidine at residue 371. Interestingly, the hypomorphic pglA and pglH alleles thus exist in the same FA1090 background. Therefore, we wanted to investigate how these hypomorphic glycosyltransferases might influence microheterogeneity when they are expressed together. Subsequently, FA1090-manipulated strains that express the active and/or the less active pglA and pglH alleles were monitored by immunoblotting with glycan-specific antibodies (Fig. 8). By introducing the most active allele of pglH (pglHFA1090 H371R), increased PglH-based disaccharide was detected in backgrounds without PglA or with less active PglA (pglAFA1090). By introducing the most active allele of pglA (pglAN400), PglH-based disaccharide levels were decreased, while a compensatory increase in trisaccharide glycoform (based on PglA and PglE) was detected. Together, these results show how allelic differences and interactions can dramatically modulate glycoform expression.
Fig 8.
Polymorphic alleles of pglA and pglH influence microheterogeneity. Immunoblotting of whole-cell lysates from mutant strains of FA1090 carrying different pglA and pglH alleles with the glycan-specific monoclonal (npg1, npg2, and npg3) and polyclonal (pDAb2, which recognizes diNAcBac-Glc) antibodies. +, wild-type pglA and pglH alleles; −, null alleles pglA::kan and pglH::ermC-rpsL; +, hypermorphic alleles pglHFA1090 H371R and pglAN400. Strains with the most active allele of pglH had increased diNAcBac-Glc in backgrounds without PglA or with less active PglA (compare the fourth and sixth lanes with the second lane). Strains with most active allele of pglA had less diNAcbac-Glc, and instead, more trisaccharide was detected (seventh lane). Strains used were KS300, KS769, KS422, KS459, KS423, KS460, and KS770.
DISCUSSION
Despite its ubiquitous nature, the mechanisms underlying glycosylation microheterogeneity remain poorly understood. Likewise, the biological significance of these phenomena remains largely unexplored. In systems in which oligosaccharides are transferred en bloc to proteins, microheterogeneity may be explained by the network of rapid, sequential reactions taking place in a cellular compartment distinct from that in which substrate transfer occurs. In the absence of any quality control surveillance, simple enzyme kinetics may dictate the degree of oligosaccharide maturation prior to membrane translocation and transfer to substrate. This appears to be the situation operating in the neisserial O-linked pgl system since it has evolved to tolerate alternate UndPP-linked glycoforms as part of its phase and antigenic variation program. It follows, then, that the relative activities of glycosyltransferases required for synthesis of UndPP-linked oligosaccharides should be a major factor in microheterogeneity. Our data here are entirely congruent with this model.
Based on the extreme specificities and sensitivities of the glycoform-specific MAbs used here, we observed a number of insightful examples of microheterogeneity. In the cases of low-level glycoform expression, an important distinction to be made is whether the signals detected represent those from a small number of cells in which phase expression has been altered or a small amount of expression within each individual cell. For example, gonococcal strain N400 (which carries pglE in an off configuration) expresses low levels of diNAcBac and trisaccharide, although it is predicted, on the basis of its genotype, to express solely the disaccharide glycoform. Here, the low levels of trisaccharide are likely attributable to a small number of pglE phase-on variants in the cell population, while the low level of diNAcBac expression must be due to incomplete modification in an otherwise wild-type, PglA-expressing cell since the pglAN400 allele is not subject to phase variation (as it lacks the extended nucleotide repeat motif) (Fig. 7A; see Table S1 in the supplemental material). Similar concerns also apply to those instances where di- and trisaccharides are detected in strains carrying phase-off pglA alleles (such as those from strains 8013, ST3787, ST640, and FAM18). On the basis of the number of guanine residues spanning the simple repeat region (varying from 8 to 11) in these alleles, the frequencies with which phase-on variants are predicted to arise are orders of magnitude below those required to account for the glycoform levels observed (5). This is especially evident in the case of the pglAST3787 and pglAFAM18 alleles, which were associated with levels of disaccharide glycoform expression that were only 20- to 25-fold reduced from the level seen in the pglAN400 background. We propose that these alleles are examples of what have been termed “expressed pseudogenes” (16) or “pseudopseudogenes” (6), in which significant levels of gene product are expressed from out-of-frame alleles. Specifically, the poly(G) stretches contained within these pglA alleles are sequence patterns for nonstandard decoding at the levels of both programmed ribosomal frameshifting and programmed transcriptional realignment (23). Delineation of these two mechanisms cannot be achieved even at the protein level, as their protein products are identical. Additionally, we documented clear differences in the levels of microheterogeneity among the out-of-frame alleles. This could be due to context-specific, cis-acting effects or to other polymorphisms in the pglA ORFs. Whatever the situation, our findings indicate that in this system, the contingency locus is not simply a mechanism for on-off expression but, rather, operates to modulate glycoform expression across a broad spectrum from low to high levels. These findings are clearly evocative of findings made in the gonococcal lipooligosaccharide expression system, in which out-of-frame glycosyltransferase alleles are associated with considerable activity (9, 24).
Our observations on protein glycan microheterogeneity in gonococcal strain FA1090 are particularly noteworthy. This strain carries hypomorphic alleles of both pglA and pglH, and both genes are phase variable, endowing it with particularly unique forms of microheterogeneity. It remains unclear, however, how and why these genotypes have arisen. It is interesting that this strain has been and is continuing to be used in experimental infection of human male volunteers (17). As such, it should be possible to assess how glycoform variation and microheterogeneity might occur in vivo in this particular strain.
One important aspect of this work relating to both PglAFA1090 and PglHFA1090 centers on how single-residue substitutions can have such profound effects on glycosyltransferase activity. It is remarkable that the single substitutions in both proteins are localized to the C terminus and outside the conserved carbohydrate-active enzyme (CAZy) family 1 glycosyltransferase domain (see Fig. S2 in the supplemental material). A trivial explanation would be that these alterations simply reduce steady-state levels of the enzymes. We currently lack the reagents necessary to monitor PglA levels but have previously shown that the single alteration at residue 371 of PglH does not significantly alter protein expression levels (8). An alternative explanation stems from observation made for the WaaJ glycosyltransferase involved in the synthesis of the outer core region of Escherichia coli LPS (19). This work reported that the catalytic activity of WaaJ was severely compromised by limited deletions at the very C terminus. This region also maps outside the major catalytic domain but is rich in hydrophobic and positively charged amino acids that were implicated in membrane association. These features are also observed within the C-terminal tail of PglA and PglH. As the acceptors in all these cases are nascent, membrane-associated oligosaccharides, this property might be beneficial for activity, as previously suggested (19). Thus, the single-residue substitutions in the defective PglA and PglH forms might result in diminished membrane targeting. Albeit plausible, more work is needed to firmly address these possibilities.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by grants 166931, 183613, and 183814 from the Research Council of Norway and by funds from the Department of Molecular Biosciences and Center for Molecular Biology and Neuroscience of the University of Oslo.
We thank Dan Stein, Warren Wakarchuk, and Chris Whitfield for helpful discussions.
Footnotes
Published ahead of print 13 July 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Aas FE, et al. 2006. Neisseria gonorrhoeae type IV pili undergo multisite, hierarchical modifications with phosphoethanolamine and phosphocholine requiring an enzyme structurally related to lipopolysaccharide phosphoethanolamine transferases. J. Biol. Chem. 281:27712–27723 [DOI] [PubMed] [Google Scholar]
- 2. Aas FE, Vik A, Vedde J, Koomey M, Egge-Jacobsen W. 2007. Neisseria gonorrhoeae O-linked pilin glycosylation: functional analyses define both the biosynthetic pathway and glycan structure. Mol. Microbiol. 65:607–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anonsen JH, et al. 2012. Novel protein substrates of the phospho-form modification system in Neisseria gonorrhoeae and their connection to O-linked protein glycosylation. Infect. Immun. 80:22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Asakura H, et al. 2010. Helicobacter pylori HP0518 affects flagellin glycosylation to alter bacterial motility. Mol. Microbiol. 78:1130–1144 [DOI] [PubMed] [Google Scholar]
- 5. Banerjee A, et al. 2002. Implications of phase variation of a gene (pgtA) encoding a pilin galactosyl transferase in gonococcal pathogenesis. J. Exp. Med. 196:147–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baranov PV, Hammer AW, Zhou J, Gesteland RF, Atkins JF. 2005. Transcriptional slippage in bacteria: distribution in sequenced genomes and utilization in IS element gene expression. Genome Biol. 6:R25 doi: 10.1186/gb-2005-6-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Børud B, et al. 2010. Genetic, structural, and antigenic analyses of glycan diversity in the O-linked protein glycosylation systems of human Neisseria species. J. Bacteriol. 192:2816–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Børud B, et al. 2011. Genetic and molecular analyses reveal an evolutionary trajectory for glycan synthesis in a bacterial protein glycosylation system. Proc. Natl. Acad. Sci. U. S. A. 108:9643–9648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burch CL, Danaher RJ, Stein DC. 1997. Antigenic variation in Neisseria gonorrhoeae: production of multiple lipooligosaccharides. J. Bacteriol. 179:982–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carter AT, et al. 2009. Independent evolution of neurotoxin and flagellar genetic loci in proteolytic Clostridium botulinum. BMC Genomics 10:115 doi: 10.1186/1471-2164-10-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Castric P, Cassels FJ, Carlson RW. 2001. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J. Biol. Chem. 276:26479–26485 [DOI] [PubMed] [Google Scholar]
- 12. Chamot-Rooke J, et al. 2007. Alternative Neisseria spp. type IV pilin glycosylation with a glyceramido acetamido trideoxyhexose residue. Proc. Natl. Acad. Sci. U. S. A. 104:14783–14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Champion OL, et al. 2005. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc. Natl. Acad. Sci. U. S. A. 102:16043–16048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Freitag NE, Seifert HS, Koomey M. 1995. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol. Microbiol. 16:575–586 [DOI] [PubMed] [Google Scholar]
- 15. Hartley MD, et al. 2011. Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N′-diacetylbacillosamine. Biochemistry 50:4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hirotsune S, et al. 2003. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 423:91–96 [DOI] [PubMed] [Google Scholar]
- 17. Hobbs MM, et al. 2011. Experimental gonococcal infection in male volunteers: cumulative experience with Neisseria gonorrhoeae strains FA1090 and MS11mkC. Front. Microbiol. 2:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnston DM, Cannon JG. 1999. Construction of mutant strains of Neisseria gonorrhoeae lacking new antibiotic resistance markers using a two gene cassette with positive and negative selection. Gene 236:179–184 [DOI] [PubMed] [Google Scholar]
- 19. Leipold MD, Kaniuk NA, Whitfield C. 2007. The C-terminal domain of the Escherichia coli WaaJ glycosyltransferase is important for catalytic activity and membrane association. J. Biol. Chem. 282:1257–1264 [DOI] [PubMed] [Google Scholar]
- 20. Mehr IJ, Long CD, Serkin CD, Seifert HS. 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154:523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mehr IJ, Seifert HS. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol. Microbiol. 30:697–710 [DOI] [PubMed] [Google Scholar]
- 22. Power PM, et al. 2003. Genetic characterization of pilin glycosylation and phase variation in Neisseria meningitidis. Mol. Microbiol. 49:833–847 [DOI] [PubMed] [Google Scholar]
- 23. Sharma V, et al. 2011. A pilot study of bacterial genes with disrupted ORFs reveals a surprising profusion of protein sequence recoding mediated by ribosomal frameshifting and transcriptional realignment. Mol. Biol. Evol. 28:3195–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tong Y, et al. 2002. Neisseria gonorrhoeae strain PID2 simultaneously expresses six chemically related lipooligosaccharide structures. Glycobiology 12:523–533 [DOI] [PubMed] [Google Scholar]
- 25. Tønjum T, Freitag NE, Namork E, Koomey M. 1995. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol. Microbiol. 16:451–464 [DOI] [PubMed] [Google Scholar]
- 26. Twine SM, et al. 2009. Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 191:7050–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vik A, et al. 2009. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 106:4447–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Voisin S, et al. 2007. Glycosylation of Pseudomonas aeruginosa strain Pa5196 type IV pilins with Mycobacterium-like alpha-1,5-linked d-Araf oligosaccharides. J. Bacteriol. 189:151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wolfgang M, van Putten JP, Hayes SF, Dorward D, Koomey M. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 19:6408–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.