Abstract
Bacteria possess multiple mechanisms to survive exposure to various chemical stresses and antimicrobial compounds. In the enteric bacterium Escherichia coli, three homologous transcription factors—MarA, SoxS, and Rob—play a central role in coordinating this response. Three separate systems are known to regulate the expression and activities of MarA, SoxS, and Rob. However, a number of studies have shown that the three do not function in isolation but rather are coregulated through transcriptional cross talk. In this work, we systematically investigated the extent of transcriptional cross talk in the mar-sox-rob regulon. While the three transcription factors were found to have the potential to regulate each other's expression when ectopically expressed, the only significant interactions observed under physiological conditions were between mar and rob systems. MarA, SoxS, and Rob all activate the marRAB promoter, more so when they are induced by their respective inducers: salicylate, paraquat, and decanoate. None of the three proteins affects the soxS promoter, though unexpectedly, it was mildly repressed by decanoate by an unknown mechanism. SoxS is the only one of the three proteins to repress the rob promoter. Surprisingly, salicylate somewhat activates transcription of rob, while decanoate represses it a bit. Rob, in turn, activates not only its downstream promoters in response to salicylate but also the marRAB promoter. These results demonstrate that the mar and rob systems function together in response to salicylate.
INTRODUCTION
The enteric bacterium Escherichia coli can resist a broad spectrum of antimicrobial compounds by altering its metabolism and physiology (2, 26, 27, 56). These changes include expressing multidrug efflux pumps and superoxide dismutases, redirecting metabolic flux, and altering outer membrane porin composition (3, 5, 11, 26, 28, 48). Three homologous AraC/XylS-type transcription factors—MarA, SoxS, and Rob—play a central role in governing this coordinated response (35, 41). The three regulate a common set of genes known as the mar-sox-rob regulon. They do so by binding to the same degenerate operator site within the promoters of these genes (24, 25, 29, 30). Despite the overlapping nature of this regulon, MarA, SoxS, and Rob can differentially activate these promoters (31), enabling the cell to tune its response to specific chemical stresses and antimicrobial compounds.
Three separate systems are known to individually regulate the respective expression and activities of MarA, SoxS, and Rob in response to these different chemical signals. MarA is regulated at the transcriptional level by the MarR repressor (12). The genes for these two proteins are encoded within the marRAB operon. MarR regulates the transcription of this operon in response to phenolic compounds such as salicylate and 2,3-dihydroxybenzoate (1, 10, 13, 33, 36, 55). In addition, MarA can bind to and activate the marRAB promoter (32). An interesting feature of the marRAB operon is that its transcription is governed by both a negative-feedback loop involving MarR and a positive one involving MarA.
SoxS is also transcriptionally regulated, albeit in a different manner than MarA. SoxR, a [2Fe-2S] cluster containing a transcriptional regulator found in many bacterial species (19, 21, 47), positively regulates the expression of SoxS in response to redox-cycling compounds such as paraquat and plumbagin (19, 20, 45, 60). Oxidation of the [2Fe-2S] cluster by these redox-cycling compounds activates SoxR (16), which in turn activates soxS transcription (22, 23, 60). In addition, SoxR and SoxS both repress their own transcription (23, 46).
Unlike MarA and SoxS, Rob is regulated posttranslationally by a sequestration-dispersal mechanism (15). When Rob is inactive, it forms clusters within the cell. These clusters are thought to sequester Rob and prevent it from activating its target promoters. A number of diverse compounds, including 2,2′-dipyridyl, deoxycholate, and decanoate, activate Rob (49, 50). When these compounds activate Rob, it no longer aggregates within these clusters and thus is free to activate the transcription of its target genes.
Although MarA, SoxS, and Rob are regulated by distinct systems, they can also regulate each other's expression. Both MarA and SoxS are known to repress the rob promoter (37, 38, 54), and SoxS, Rob, and MarA are known to activate the marRAB promoter (32, 34, 40, 58). These results suggest that the mar-sox-rob regulon may be highly interconnected through transcriptional cross talk. In this work, we aimed to systematically study both self-regulation and cross-regulation, particularly using canonical inducers and deletions of chromosomal genes. While many of these interactions have been documented previously, an integrated model for the three is still lacking. Our goal in the present study was to develop such a model under a common set of experimental conditions.
MATERIALS AND METHODS
Media and growth conditions.
Luria-Bertani liquid medium (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl) and solid medium (15 g/liter agar) were used for routine bacterial culture and genetic manipulation (39). Unless otherwise indicated, experiments were conducted in MOPS (morpholinepropanesulfonic acid)-buffered glucose medium (MGC; 40 mM MOPS, 4 mM Tricine, 9.5 mM NH4Cl, 0.276 mM K2SO4, 5 × 10−4 mM CaCl2, 0.525 mM MgCl2, and 50 mM NaCl with micronutrients, pH 7.2) using the formulation described by Neidhardt and coworkers supplemented with 20 mM glucose and 0.2% Casamino Acids as carbon sources (43). All bacterial cultures were grown at 37°C except for strains containing plasmids pKD46, pINT-Ts, and pCP20, which were grown at 30°C. The following antibiotics were used at the indicated concentrations: ampicillin, 100 μg/ml; kanamycin, 20 μg/ml; and chloramphenicol, 20 μg/ml. Salicylate, paraquat (methyl viologen), or decanoate was added to the growth medium at 5 mM, 50 μM, and 5 mM concentrations, respectively.
Strain and plasmid construction.
Table 1 provides a list of all strains and plasmids used in this work. All strains except BL21(DE3) are isogenic derivatives of the sequenced E. coli K-12 strain MG1655. The generalized transducing phage P1vir was used in all genetic crosses according to standard methods (57). Targeted gene deletions and subsequent marker removal were made using the bacteriophage λ Red-recombinase method of Datsenko and Wanner (14). Site-specific integrations were made using the λInt/CRIM method of Haldimann and Wanner (18).
Table 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Genotype or relevant characteristicsa | Source or referenceb |
|---|---|---|
| Strains | ||
| MG1655 | F− λ− ilvG rph-1 | CGSC 7740 |
| BW25141 | F− λ− Δ(araB-araD)567 ΔlacZ4787(::rrnB3) Δ(phoB-phoR)580 galU95 ΔuidA3::pir+ recA1 ΔendA9::FRT rph-1 Δ(rhaB-rhaD)568 hsdR514 | CGSC 7635 |
| BL21(DE3) | F− ompT gal dcm lon hsdSB(rb− rm−) λ(DE3 [lacI lacUV5-T7 gene1 ind1 sam7 nin5]) | G. W. Ordal |
| JW5249 | F− λ− rph-1 Δ(araB-araD)567 ΔlacZ4787(::rrnB3) Δ(rhaB-rhaD)568 hsdR514 ΔmarA752::kan | CGSC 11269 |
| JW4023 | F− λ− rph-1 Δ(araB-araD)567 ΔlacZ4787(::rrnB3) Δ(rhaB-rhaD)568 hsdR514 ΔsoxS756::kan | CGSC 10891 |
| JTG1078 | F− λ− rph-1 rfb-50 INV(rrnD-rrnE)1 rpsL179 soxR105 zjc-2206::Tn10dKan | CGSC 7594 |
| CR700 | attλ::[kan marR′-yfp oriR6K] | 10 |
| CR715 | attλ::[kan micF′-yfp oriR6K] | 11 |
| CR719 | ΔmarRAB::FRTc | 11 |
| CR720 | ΔsoxRS::FRT | 11 |
| CR721 | Δrob::FRT | 11 |
| CR723 | ΔmarRAB::FRT ΔsoxRS::FRT | 11 |
| CR724 | ΔmarRAB::FRT Δrob::FRT | 11 |
| CR725 | ΔsoxRS::FRT Δrob::FRT | 11 |
| CR726 | ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT | 11 |
| CR765 | ΔmarRAB::FRT attλ::[kan micF′-yfp oriR6K] | 11 |
| CR766 | ΔsoxRS::FRT attλ::[kan micF′-yfp oriR6K] | 11 |
| CR777 | Δrob::FRT attλ::[kan micF′-yfp oriR6K] | 11 |
| CR779 | ΔmarRAB::FRT ΔsoxRS::FRT attλ::[kan micF′-yfp oriR6K] | 11 |
| CR782 | ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT attλ::[kan micF′-yfp oriR6K] | 11 |
| CR903 | JTG1078 ΔsoxS::cat (4275086–4275406) | |
| CR904 | ΔsoxS::cat soxR105 (Kans) | |
| CR905 | ΔmarRAB::FRT ΔsoxS::cat soxR105 | |
| CR906 | Δrob::FRT ΔsoxS::cat soxR105 | |
| CR907 | ΔmarRAB::FRT Δrob::FRT ΔsoxS::cat soxR105 | |
| CR908 | ΔmarA752::kan | |
| CR909 | ΔsoxS756::kan | |
| CR910 | ΔmarA752::FRT ΔsoxS756::FRT | |
| CR911 | ΔmarA752::FRT Δrob::FRT | |
| CR912 | ΔsoxS756::FRT Δrob::FRT | |
| CR913 | ΔmarA752::FRT ΔsoxS756::FRT Δrob::FRT | |
| CR914 | attλ::[kan soxS′-yfp oriR6K] | |
| CR915 | attλ::[kan rob′-yfp oriR6K] | |
| CR916 | attλ::[kan inaA′-yfp oriR6K] | |
| CR917 | ΔmarRAB::FRT Δrob::FRT ΔsoxS::cat soxR105 attλ::[kan marR′-yfp oriR6K] | |
| CR918 | ΔmarRAB::FRT Δrob::FRT ΔsoxS::cat soxR105 attλ::[kan soxS′-yfp oriR6K] | |
| CR919 | ΔmarRAB::FRT Δrob::FRT ΔsoxS::cat soxR105 attλ::[kan rob′-yfp oriR6K] | |
| CR920 | ΔmarRAB::FRT Δrob::FRT ΔsoxS::cat soxR105 attλ::[kan inaA′-yfp oriR6K] | |
| CR921 | ΔmarRAB::FRT attλ::[kan marR′-yfp oriR6K] | |
| CR922 | ΔmarRAB::FRT ΔsoxRS::FRT attλ::[kan marR′-yfp oriR6K] | |
| CR923 | ΔmarRAB::FRT Δrob::FRT attλ::[kan marR′-yfp oriR6K] | |
| CR924 | ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT attλ::[kan marR′-yfp oriR6K] | |
| CR925 | ΔsoxS::cat soxR105 attλ::[kan soxS′-yfp oriR6K] | |
| CR926 | ΔmarRAB::FRT ΔsoxS::cat soxR105 attλ::[kan soxS′-yfp oriR6K] | |
| CR927 | Δrob::FRT ΔsoxS::cat soxR105 attλ::[kan soxS′-yfp oriR6K] | |
| CR928 | ΔmarRAB::FRT Δrob::FRT ΔsoxS::cat soxR105 attλ::[kan soxS′-yfp oriR6K] | |
| CR929 | Δrob::FRT attλ::[kan rob′-yfp oriR6K] | |
| CR930 | ΔmarRAB::FRT Δrob::FRT attλ::[kan rob′-yfp oriR6K] | |
| CR931 | ΔsoxRS::FRT Δrob::FRT attλ::[kan rob′-yfp oriR6K] | |
| CR932 | ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT attλ::[kan rob′-yfp oriR6K] | |
| CR933 | ΔsoxS756::FRT attλ::[kan marR′-yfp oriR6K] | |
| CR934 | Δrob::FRT attλ::[kan marR′-yfp oriR6K] | |
| CR935 | ΔsoxS756::FRT Δrob::FRT attλ::[kan marR′-yfp oriR6K] | |
| CR936 | ΔmarA752::FRT ΔsoxS756::FRT Δrob::FRT attλ::[kan marR′-yfp oriR6K] | |
| CR937 | ΔmarA752::FRT attλ::[kan soxS′-yfp oriR6K] | |
| CR938 | Δrob::FRT attλ::[kan soxS′-yfp oriR6K] | |
| CR939 | ΔmarA752::FRT Δrob::FRT attλ::[kan soxS′-yfp oriR6K] | |
| CR940 | ΔmarA752::FRT ΔsoxS756::FRT Δrob::FRT attλ::[kan soxS′-yfp oriR6K] | |
| CR941 | ΔmarA752::FRT attλ::[kan rob′-yfp oriR6K] | |
| CR942 | ΔsoxS756::FRT attλ::[kan rob′-yfp oriR6K] | |
| CR943 | ΔmarA752::FRT ΔsoxS756::FRT attλ::[kan rob′-yfp oriR6K] | |
| CR944 | ΔmarA752::FRT ΔsoxS756::FRT Δrob::FRT attλ::[kan rob′-yfp oriR6K] | |
| CR945 | ΔsoxRS::FRT Δrob::FRT attλ::[kan inaA′-yfp oriR6K] | |
| CR946 | ΔmarRAB::FRT Δrob::FRT attλ::[kan inaA′-yfp oriR6K] | |
| CR947 | ΔmarRAB::FRT ΔsoxRS::FRT attλ::[kan inaA′-yfp oriR6K] | |
| CR948 | ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT attλ::[kan inaA′-yfp oriR6K] | |
| CR949 | ΔsoxRS::FRT Δrob::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR950 | ΔmarRAB::FRT Δrob::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR951 | ΔmarRAB::FRT ΔsoxRS::FRT attλ::[kan micF′-yfp oriR6K] | |
| CR952 | ΔmarRAB::FRT ΔsoxRS::FRT Δrob::FRT attλ::[kan micF′-yfp oriR6K] | |
| Plasmids | ||
| pKD46 | bla PBAD gam bet exo pSC101 ori(Ts) | 14 |
| pCP20 | bla cat cI857 λPR′-flp pSC101 ori(Ts) | 9 |
| pKD3 | bla rgnB FRT cat FRT oriR6K | 14 |
| pET28a | kan PT7/lacO-6 × His-MCS lacIq ColE1 | Novagen |
| pBAD30 | bla araC PBADp15A ori | 17 |
| pMarA | pBAD30::RBS-marA | 11 |
| pRob | pBAD30::RBS-rob | 11 |
| pSoxS | pBAD30::RBS-soxS | |
| pVenus | kan MCS yfp(venus) t0 attλ oriR6K | 53 |
| pVenus-soxS | kan MCS soxS′-yfp t0 attλ oriR6K | |
| pVenus-rob | kan MCS rob′-yfp t0 attλ oriR6K | |
| pVenus-inaA | kan MCS inaA′-yfp t0 attλ oriR6K | |
| pET28a-rob | kan PT7/lacO-6×His-rob lacIq ColE1 |
Except for BL21(DE3), all strains are isogenic derivatives of E. coli K-12 strain MG1655. Numbers in parentheses indicate deletion endpoints determined using the MG1655 genome sequence.
All strains and plasmids are from this work unless otherwise noted. CGSC, E. coli Genetic Stock Center, Yale University.
FRT, FLP recombination target.
The soxS deletion cassette was generated using the plasmid templates pKD3 and standardized priming sites (14). The ΔsoxS deletion cassette was generated by PCR using the primer pairs TGA ATT AAC GAA CTG AAC ACT GAA AAG AGG CAG ATT TAT GTG TAG GCT GGA GCT GCT TCG and AAT TAC CCG CGC GGG AGT TAA CGC GCG GGC AAT AAA ATT ACA TAT GAA TAT CCT CCT TAG. All cassettes were transformed into MG1655 cells expressing bacteriophage λ Red-recombinase from the pKD46 helper plasmid. Deletions were verified by PCR using primers in the antibiotic resistance marker and sites adjacent on the host chromosome. All deletions were subsequently transduced into a clean MG1655 or parental background prior to antibiotic cassette removal using the FLP-recombinase-expressing pCP20 helper plasmid.
Single-copy transcriptional and translational fusions were constructed in trans using the pVenus integration vector (53). Transcriptional fusions to the soxS, rob, and inaA promoters were made by PCR amplifying the promoter regions of the soxS, rob, and inaA genes using primers ATA GGT ACC TTC TCG CCA TTG GGA CGA AA and ATA GAA TTC AAG ATC CTG AAT AAT TTT CTG ATG G, ATA GGT ACC CTG AGC TTT GCC GTT TTT AA and ATA GAA TTC AAG GTC GCG AAT AAT GCC G, and ATA GGT ACC CAAT GCT TTT CAG CGT TAA C and ATA GAA TTC AAA TTC GTC GTA CTT TGC TG, respectively (the underlined italic sequences represent restriction sites). Following amplification, the PCR products were digested with KpnI and EcoRI restriction endonucleases and ligated into the corresponding restriction sites of pVenus to produce pVenus-soxS, pVenus-rob, and pVenus-inaA. The pVenus derivatives described above were then integrated into the phage λ attachment site in MG1655 cells expressing λInt from the pINT-Ts helper plasmid. Single-copy integrations were verified by PCR using primers described by Haldimann and Wanner (18). The resulting single-copy fusions were transduced back into a clean MG1655 background.
The overexpression vectors for soxS were constructed using the medium-copy, arabinose-inducible expression vector pBAD30. The soxS-coding region was amplified by PCR using primers ATA GAA TTC TTT ATA AGG AGG AAA AAC ATA TGT CCC ATC AGA AAA TTA TTC AG and ATA TCT AGA TTA CAG GCG GTG GCG ATA. The resulting soxS PCR fragment was treated with EcoRI and XbaI. The digested fragments were then ligated into the corresponding restriction sites of the pBAD30 multiple-cloning site to produce pSoxS. The construct encodes a strong ribosome binding site upstream of soxS common to pMarA and pRob to ensure high-level expression (11).
The 6×His-Rob overexpression vector pET28a-rob was made by amplifying the rob-coding region by PCR using primers ATA GAG CTC TTT ATA AGG AGG AAA AAC ATA TGG ATC AGG CCG GCA TTA T and ATA GGT ACC TTA ACG ACG GAT CGG AAT CA, followed by digestion with NdeI and SacI. The digested rob fragment was then ligated into the corresponding restriction sites of pET28a, creating an in-frame 6×His-rob-coding region and producing pET28a-rob.
Fluorescence-based promoter activity assays.
Cells were grown overnight in MGC to saturation and subcultured 1:200 in fresh medium. For experiments, 0.45 ml was dispensed to individual wells of 96-well, deep, square-well microtiter plates (82006-448; VWR). Plates were sealed with Breath-Easy membranes (Diversified Biotech) to reduce evaporation and placed on a high-speed, microplate shaker (VWR) at 1,000 rpm and 37°C. Cultures were grown to mid-logarithmic phase (optical density [OD], ∼0.5) and induced with 100 μl medium containing inducer, bringing the final culture volume to 0.55 ml. Negative-control samples were treated with fresh medium without inducers. Growth after induction was continued at 37°C and 1,000 rpm for 1 h prior to fluorescence and optical density measurements, unless noted otherwise.
To measure fluorescence and optical density, 250 μl of culture was transferred from the deep-well plates to black, clear-bottomed Costar 96-well microtiter plates. Fluorescence (excitation and emission, 515 and 530 nm, respectively) and optical density (600 nm) were measured using a Tecan Safire2 microplate reader. Fluorescence measurements are reported as the relative fluorescence normalized to the optical density of the sample to correct for variation in cell density. All experimental data presented are the average and standard deviation of four replicate samples.
Purification of Rob.
Rob purification was performed using Ni2+-affinity chromatography using an Akta Prime fast-performance liquid chromatograph (GE Health Sciences) under native conditions. Rob was expressed with an N-terminal 6×His tag from pET28a in strain BL21(DE3). Cells were grown in 2 liters of LB medium at 37°C and with shaking at 250 rpm to an OD of 0.7, followed by induction with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cultures were grown for an additional 3 h. Cells were then pelleted, washed once in phosphate-buffered saline (PBS; 10 mM Na2HPO4, 2 mM KH2PO4, 138 mM NaCl, 2.7 mM KCl, pH 7.4), and repelleted. The cell pellet was then frozen at −80°C before any further steps.
Cell pellets were thawed and resuspended in 5 ml of lysis buffer (50 mM Tris-HCl, 500 mM NaCl, 10 mM imidazole, 0.1% Triton X-100, pH 7.4) per gram of cells. Resuspended cells were then disrupted by sonication (8 10-s pulses). Extracts were then clarified by centrifugation at 30,000 × g for 1 h, followed by filtration through a 0.45-μm-pore-size membrane. Clarified extracts were then loaded at 1 ml/min onto a 5 ml HiTrap HP (Ni2+-charged) column preequilibrated with wash buffer (50 mM Tris-HCl, 500 mM NaCl, 20 mM imidazole, pH 7.4). The column was then washed with 10 bed volumes of wash buffer, followed by elution with wash buffer containing 150 mM imidazole. Fractions containing >95% pure Rob (determined by SDS-PAGE) were collected, concentrated 5 times with a 10,000-molecular-weight-cutoff concentrator cassette (Amicon), and dialyzed against Tris-buffered saline (TBS; 50 mM Tris-HCl, 500 mM NaCl, pH 7.4). Final proteins were determined using the bicinchoninic acid assay method using bovine serum albumin standards after trichloroacetic acid (TCA) precipitation.
ITC.
All isothermal titration calorimetry (ITC) experiments were conducted using a MicroCal VP-ITC titration calorimeter preincubated to 25°C for at least 1 h prior to the start of experiments. Rob protein solution was brought to a final concentration of 10 μM in TBS (500 mM NaCl), and the pH was measured (typically, it was between 7.2 and 7.4) using a Perkin-Elmer pH meter. Ligand solutions were prepared fresh in TBS, the pH of the solution was adjusted to that of the Rob solution, and the final concentration was brought to 10 mM. The 1.4-ml sample well was loaded with a blunt-end needle attached to a 5-ml Hamilton pipette, making sure to introduce no air bubbles into the sample cell. Likewise, the injection syringe was filled and expelled with the 10 mM ligand solution twice, prior to finally being filled and made free of any air bubbles. The experimental parameters used with the VP-ITC system were 28 10-μl injections at 5-min intervals, a 300 rpm stirring speed, and a reference power of 1 μcal/s.
RESULTS
Regulation of mar-sox-rob gene expression by ectopically expressed MarA, SoxS, and Rob.
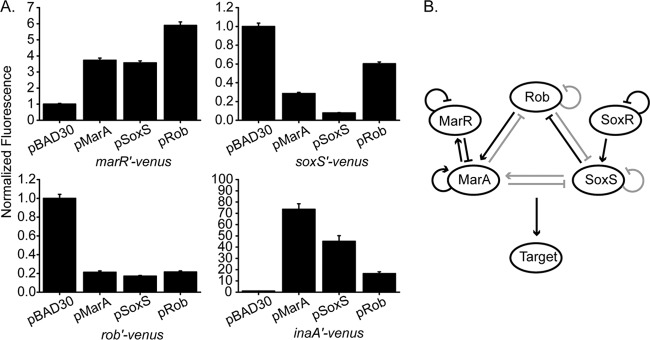
Both SoxS and Rob are known to activate the marRAB promoter (32, 34, 40), and both MarA and SoxS are known to repress the rob promoter (37, 38, 54). These interactions, along with other data, suggest that the mar-sox-rob regulon forms an integrated regulatory circuit. As a first step toward understanding this integrated regulation, we expressed MarA, SoxS, and Rob individually from an arabinose-inducible promoter in a marRAB soxS rob genetic background containing a constitutively active mutant of SoxR (soxR105) (44). Note that the soxS promoter is inactive in the absence of soxR or an inducer (22, 23, 60). Use of the soxR105 allele allowed us to examine the effects of MarA, SoxS, and Rob on the soxS promoter without needing to add an inducer for SoxR. As indirect measures of gene expression from the marRAB, soxS, and rob promoters, single-copy transcriptional fusions to the fast-folding yellow fluorescent protein (YFP) variant venus were employed (42). In addition, we included the downstream inaA promoter, which is known to be activated by all three transcription factors, as a positive control in these experiments.
As shown in Fig. 1A, MarA, SoxS, and Rob all regulate the marRAB, soxS, and rob promoters in a sign-consistent manner, albeit with various intensities. In particular, MarA, SoxS, and Rob are all activators of the marRAB promoter, consistent with previous studies. Likewise, MarA, SoxS, and Rob are all repressors of the soxS and rob promoters. These results confirm the results of previous experiments, except that it had not previously been shown that overexpressed Rob represses the rob promoter and that overexpressed MarA and Rob repress the soxS promoter. Nonetheless, these results were expected, as all three regulators bind the same sites by a common mechanism (24, 25, 29, 30).
Fig 1.
Regulation of mar-sox-rob gene expression by MarA, SoxS, and Rob. (A) Strains contain plasmids pBAD30, pMarA, pSoxS, and pRob and single-copy, yfp transcriptional fusions. marRAB, soxS, and rob were deleted from cells, and cells contained a constitutively active mutant of SoxR (soxR105). Cells were grown in LB–0.2% arabinose medium for 4 h prior to fluorescence and optical density measurements. Fluorescence values have been divided by the optical density and then normalized to the value for the empty-plasmid (pBAD30) negative control. (B) mar-sox-rob regulatory network inferred from the data in panel A. Dark lines, interactions found to be significant under physiological conditions; gray lines, interactions found to be significant only when regulators are overexpressed. Strains used were CR917 (marRAB promoter), CR918 (soxS promoter), CR219 (rob promoter), and CR920 (inaA promoter) harboring pBAD30, pMarA, pSoxS, and pRob, respectively.
Role of autoregulation on inducible expression.
The preceding experiments suggest that the mar-sox-rob regulon may be highly interconnected through transcriptional cross talk (Fig. 1). However, these results were obtained from experiments where the regulators were ectopically expressed. One question, then, is whether the same results hold when MarA, SoxS, or Rob are induced by salicylate, paraquat, or decanoate, respectively, as opposed to being overexpressed.
MarA and SoxS are known to positively and negatively regulate their own respective expression (23, 32, 46). In addition, we found that Rob is also capable of repressing its own expression when ectopically expressed (Fig. 1). We first tested whether these three regulators affect their own expression when independently induced. To control for cross talk, these experiments were performed in a genetic background where only one system was present. For example, the mar experiments were performed in a soxS rob genetic background.
Consistent with previous studies, marA was found to increase the activation of the marRAB promoter in response to salicylate and soxS was found to decrease the activation of the soxS promoter in response to paraquat (Fig. 2A and B). However, autoregulation by SoxS had only a small effect, contrary to previous reports (46). Consistent with our overexpression experiments, rob was found to decrease the activation of the rob promoter (Fig. 2C). The effect, however, was minor. This is consistent with a model where Rob is regulated primarily at the posttranslational level. We also found that decanoate represses the rob promoter and that this repression is independent of rob. Collectively, these results indicate that autoregulation plays a significant role in inducible expression only in the case of the mar system.
Fig 2.
Effect of autoregulation on mar-sox-rob regulon activation in the absence of genetic cross talk. Activation of the marRAB (A), soxS (B), and rob (C) promoters during induction with 5 mM salicylate, 50 μM paraquat, and 5 mM decanoate, respectively. Strains used were CR935 and CR936 (A), CR939 and CR940 (B), and CR934 and CR935 (C). A.U., absorbance units.
Effect of inducible transcriptional cross talk on mar-sox-rob gene expression.
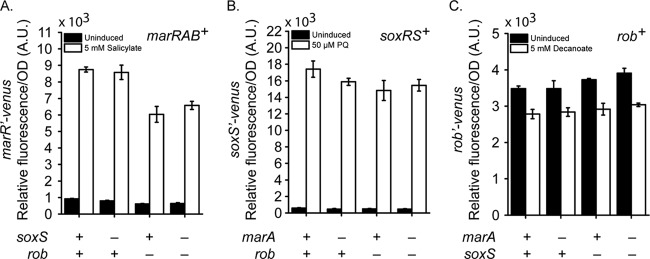
We next investigated cross talk and the ability of these regulators to activate each other's expression when induced. To isolate the affects of individual systems, the marRAB promoter experiments were performed in a marRAB genetic background where the soxRS and/or rob operon was deleted, the soxS promoter experiments were performed in a soxS soxR105 genetic background where the marRAB and/or rob operon was deleted, and the rob promoter experiments were performed in a rob genetic background where the marRAB and/or soxRS operons were deleted. Once again, we employed the soxR105 allele in these experiments to artificially induce the soxS promoter.
In confirmation of previous reports (32), we found that paraquat activates the marRAB promoter in a soxRS-dependent manner (Fig. 3A). Decanoate was also found to activate the marRAB promoter in a rob-dependent manner, though weak activation was also observed in the absence of rob. Rob was also found to increase marRAB promoter activity independently of inducer, in confirmation of previous reports (34). These results indicate that not all Rob is in the inactive form. Likely, only a fraction aggregates within clusters, leaving some of it in the free and active form even in the absence of its cognate inducer, decanoate. In the case of the soxS promoter (Fig. 3B), salicylate was found to have no effect and decanoate was found to repress it independently of rob. In the case of the rob promoter (Fig. 3C), we found that paraquat represses it in a soxRS-dependent manner, in confirmation of previous reports (38). Salicylate, on the other hand, was found to activate the rob promoter independently of the marRAB operon. This activation is enhanced in the absence of the marRAB operon, consistent with MarA being a repressor of the rob promoter.
Fig 3.
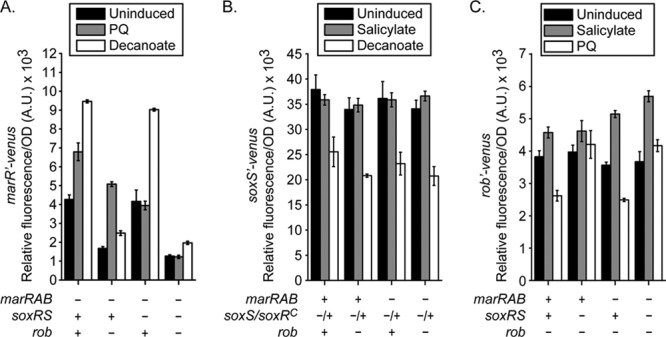
Effect of inducible cross talk on mar-sox-rob gene expression. (A) Activation of the marRAB promoter in response to paraquat (PQ) and decanoate in the presence or absence of SoxS and Rob. (B) Activation of the soxS promoter in response to salicylate and decanoate in the presence or absence of MarA and Rob. (C) Activation of the rob promoter in response to salicylate and paraquat in the presence or absence of MarA and SoxS. Salicylate, paraquat, and decanoate were used at concentrations of 5 mM, 50 μM, and 5 mM, respectively. Strains used were CR921 to CR924 (A), CR925 to CR928 (B), and CR929 to CR932 (C).
The results from these and the preceding experiments indicate that transcriptional cross talk is less extensive than suggested by the overexpression experiment (Fig. 1). They also indicate that cross talk may occur independently of MarA, SoxS, and Rob, as indicated by the repression of the soxS promoter by decanoate and the activation of the rob promoter by salicylate. How this occurs is not known, though in the former case, decanoate may inhibit the activity of SoxR (note that these experiments were performed using a constitutively active variant of SoxR [soxR105]).
Effect of transcriptional cross talk on native regulation and downstream gene expression.
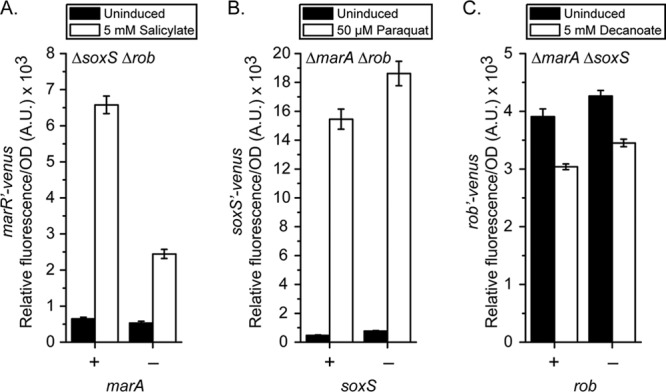
Our preceding results demonstrate that while cross talk is less extensive than that inferred from overexpression experiments, it is still present nonetheless. This would suggest that cross talk may amplify or attenuate the response of a given system to its cognate inducer. Our results also suggest that an inducer may act through noncognate genes. For example, decanoate represses the soxS promoter independently of Rob and salicylate activates the rob promoter independently of MarA. To determine whether such mechanisms are present, we measured the response of the intact mar-sox-rob regulon and downstream promoters when individual systems were selectively deleted.
In the case of salicylate and the marRAB promoter (Fig. 4A), only rob was found to have an effect. While Rob is known to increase marRAB promoter activity in response to salicylate (34), our results indicate that this increase is not solely due to the basal activity of Rob, as previously proposed, but is also due to the fact that Rob is being activated by salicylate, as discussed below. We also note that salicylate has previously been shown to activate the marRAB promoter independently of mar, sox, and rob (12, 34, 51). In the case of paraquat and the soxS promoter (Fig. 4B), both marA and rob were found to have little or no effect. Similarly, in the case of decanoate and the rob promoter (Fig. 4C), marA and soxS were found to have little or no effect. In fact, our preceding results show that this repression of rob promoter activity by decanoate is also independent of Rob itself (Fig. 3C).
Fig 4.
Effect of cross talk on activation of natively encoded systems observed through monitoring of the transcriptional responses of the marRAB (A), soxS (B), and rob (C) promoters. Each system was examined in the absence of one or both systems capable of cross regulation. Strains used were CR700 and CR933 to CR935 (A), CR914 and CR937 to CR939 (B), and CR915 and CR941 to CR943 (C).
We also investigated how cross talk affects the expression of downstream genes (Fig. 5). Here, we tested the inaA and micF promoters, two known targets of MarA, SoxS, and Rob. In the case of salicylate, activation of the inaA and micF promoters is reduced roughly 2-fold in the absence of the soxRS and rob operons. Salicylate can also induce these two promoters through Rob independently of MarA. While this Rob-dependent activation by salicylate is relatively minor in the case of the inaA promoter, it yields a 2-fold increase in activity in the case of the micF promoter. In the case of paraquat, we found that the activation of the inaA and micF promoters requires the soxRS operon and that the degree of activation was somewhat reduced in the absence of the marRAB and rob operons, consistent with previous observations in the case of the inaA promoter (52). Our results show that this reduction in activity can be attributed to the loss of Rob in the case of the micF promoter; however, they do not explain why inaA promoter activity is reduced. In the case of decanoate, we found that activation of the inaA and micF promoters requires Rob and that the marRAB and soxRS operons have little or no effect.
Fig 5.
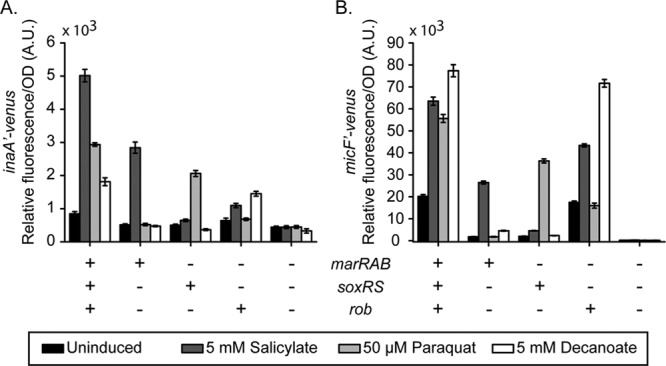
Maximal activation of the downstream mar-sox-rob regulon in response to canonical inducers requires a fully intact mar-sox-rob network. Levels of transcriptional activity of two downstream promoters, the inaA (A) and micF (B) promoters, during exposure to the canonical mar-sox-rob inducers salicylate, paraquat, and decanoate. Strains used were CR916 and CR945 to CR948 (A) and CR715 and CR949 to CR952 (B).
Both the inaA and micF promoters are active in the absence of salicylate, paraquat, and decanoate. In the case of the micF promoter, this basal activity can be attributed to Rob. Upon loss of Rob, this promoter is effectively in the off state. These results suggest that Rob may play an important role in setting the basal activity of some downstream promoters. They also demonstrate that these regulators differentially regulate downstream promoters, consistent with the findings of previous studies (31).
Collectively, these results (Fig. 4 and 5) suggest a further reduced model for cross talk within the mar-sox-rob regulon. In particular, cross talk is significant only in the case of salicylate due to its ability to activate the mar and rob systems in parallel (Fig. 1B). Aside from cross talk, we also found that Rob sets the basal level of expression for the micF promoter and that this basal activity can affect how strongly this promoter is activated by paraquat through the soxRS system.
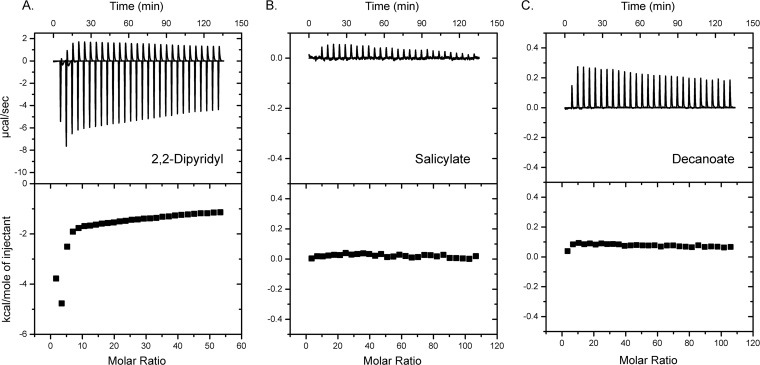
Rob responds to salicylate by an indirect mechanism.
Rob alone is capable of activating the inaA and micF promoters in response to salicylate (Fig. 5). Although several compounds such as decanoate, dipyridyl, and deoxycholate have been observed to bind and activate Rob, salicylate is not known to bind and activate it, to the best of our knowledge (49–51). To test for binding, we employed isothermal titration calorimetry (ITC) using purified Rob protein titrated with a salicylate solution (Fig. 6). As controls, we also tested whether Rob binds to 2,2-dipyridyl and decanoate. Of the three compounds tested, we observed an appreciable enthalpic change only with 2,2-dipyridyl. No significant enthalpic changes were observed with salicylate or decanoate, even though the latter is known to bind Rob. While the results are not definitive, they nonetheless suggest that Rob does not directly bind salicylate and that instead salicylate may regulate Rob at the transcriptional level.
Fig 6.
Rob does not directly bind salicylate. Measurements were made using a VP-ITC (MicroCal) calorimeter with purified Rob (10 μM) and 5 mM 2,2-dipyridyl (A), 5 mM salicylate (B), and 10 mM decanoate (C). Data were collected and analyzed using the Origin-based MicroCal software suite. We also tested whether a buffer-only control with 5 mM 2,2-dipyridyl would also yield an enthalpic change due to metal chelation. While the buffer-only control yielded an enthalpic change, it was appreciably less than that with Rob present (data not shown).
DISCUSSION
The goal of this study was to investigate transcriptional cross talk within the mar-sox-rob regulon. While many interactions between these systems have been identified in the past, a systematic investigation under a common set of experimental conditions has been lacking. We found that MarA, SoxS, and Rob all have the potential to regulate each other's expression in a sign-consistent manner, suggesting that the three form a fully connected network. However, this fully connected network is not realized under the conditions tested (Fig. 1B). Only in response to salicylate did we observe any significant cross talk.
One possibility is that cross talk between the mar, sox, and rob systems becomes significant only when two or more of these systems are activated. In particular, we do not expect SoxS, for example, to activate the marRAB promoter when MarR is still repressing it. Similar arguments can also be extended to the other two systems. In fact, previous studies have shown that when cells are exposed to multiple antibiotics, the effects can be nonlinear (5, 6, 8, 61). A similar process may occur with the mar-sox-rob regulon. We tested this hypothesis by measuring the response to different pairs of inducers (data not shown). Our data indicate that activation of the marRAB promoter by salicylate and decanoate is simply additive. In the case of the soxS promoter, we observed that salicylate enhances the response to paraquat, though this effect cannot be explained by transcription cross talk between the mar and sox systems. The two most likely work synergistically on SoxR. Otherwise, no significant interactions were observed with two inducers. These results provide further evidence that cross talk plays only a minor role within the mar-sox-rob regulon, aside from the interactions observed between the mar and rob systems. This conclusion is further supported by our results where we observed limited transcriptional cross talk when one system was constitutively active and the other two were selectively induced.
A novel finding of this study is that salicylate is capable of activating the marRAB, inaA, and micF promoters through Rob. In the case of the marRAB promoter, Rob has previously been shown to contribute to its activation by salicylate (34). Moreover, salicylate is known to induce the marRAB promoter independently of mar, sox, and rob (12, 34, 51). Our contribution was to show that salicylate activates Rob and that this activation contributes not only to the activation of the marRAB promoter by salicylate but also to the activation of the downstream inaA and micF promoters. These results would suggest that the marRAB operon does not form an independent regulatory system in its own right but, rather, forms a regulatory system also involving Rob. Interestingly, this regulatory network adopts different topologies depending on the inducer. In the case of salicylate, Rob functions in a feed-forward loop with MarA, where it activates both the marRAB promoter and downstream ones. In response to decanoate, however, Rob functions autonomously. Such a regulatory structure would enable decanoate to activate a subset of the genes activated by salicylate. Consistent with this model, Warner and Levy (58) found that cationic antimicrobial peptides activate the marRAB operon through Rob alone.
One open question is how salicylate activates Rob. The mechanism appears to be indirect, as salicylate does not bind to Rob in vitro, as determined using isothermal calorimetry. One possibility is that the binding of salicylate to Rob does not yield any appreciable enthalpic change. Rather, binding could yield an entropic change, possibly by disordering the protein (7, 59). Such a binding mechanism would not be detected using isothermal calorimetry. In fact, we were unable to detect the binding of decanoate to Rob using this method. An alternate possibility is that salicylate increases the expression of rob. Our data (Fig. 3C) show that salicylate activates the rob promoter independently of the marRAB operon. If anything, marA seems to attenuate this response. Whether this increase in promoter activity is sufficient to activate Rob is unknown. What is clear is that not all Rob is in the inactive form (Fig. 3A). This would imply that Rob can also be controlled at the transcriptional level, as increased expression of rob would also increase the concentration of Rob in the free and active form.
We also do not know how salicylate is able to activate the rob promoter. What is known is that salicylate also activates the marRAB promoter independently of MarR, MarA, SoxS, and Rob (32, 34). EmrR, a transcription factor also responsive to salicylate, is also known not to be involved (34). In addition, we found that decanoate represses the soxS and rob promoters and induces the marRAB promoter independently of Rob. Again, the mechanisms are unknown.
One limitation of our experimental analysis is that we did not control for the action of downstream genes. In particular, the mar-sox-rob regulon includes a number of genes that encode efflux transporters and other detoxifying systems (3, 4, 35, 48). In our experiments, where we selectively deleted different systems, it is possible that we were affecting the ability of the cells to adapt to these chemical stresses. If such a process were occurring, the various mutants used in this study would become hypersensitive to the compounds tested. However, we did not observe such an effect.
In conclusion, we have systematically mapped the interactions between the marRAB, soxRS, and rob operons under a common set of experimental conditions. The main contribution of this work was to show that transcriptional cross talk is limited under physiological conditions, even though multiple studies have previously suggested otherwise. Only the marRAB operon was found to be subject to appreciable cross talk through its interactions with Rob. Moreover, our results suggest that the marRAB and rob operons function together in a conditional manner and that the two systems should not be viewed as autonomous systems but rather as an integrated network in their own right.
ACKNOWLEDGMENTS
We thank J. M. Slauch for helpful discussions and advice regarding this work.
This work was supported in part by grants from the National Science Foundation and the National Institutes of Health. C.V.R. is affiliated with the Energy Biosciences Institute.
Footnotes
Published ahead of print 29 June 2012
REFERENCES
- 1. Alekshun MN, Levy SB. 1999. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 181:4669–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alekshun MN, Levy SB. 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 128:1037–1050 [DOI] [PubMed] [Google Scholar]
- 3. Barbosa TM, Levy SB. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bennik MH, Pomposiello PJ, Thorne DF, Demple B. 2000. Defining a rob regulon in Escherichia coli by using transposon mutagenesis. J. Bacteriol. 182:3794–3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bollenbach T, Kishony R. 2011. Resolution of gene regulatory conflicts caused by combinations of antibiotics. Mol. Cell 42:413–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bollenbach T, Quan S, Chait R, Kishony R. 2009. Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell 139:707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bray D, Williams D. 2008. How the “melting” and “freezing” of protein molecules may be used in cell signaling. ACS Chem. Biol. 3:89–91 [DOI] [PubMed] [Google Scholar]
- 8. Chait R, Craney A, Kishony R. 2007. Antibiotic interactions that select against resistance. Nature 446:668–671 [DOI] [PubMed] [Google Scholar]
- 9. Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 10. Chubiz LM, Rao CV. 2010. Aromatic acid metabolites of Escherichia coli K-12 can induce the marRAB operon. J. Bacteriol. 192:4786–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chubiz LM, Rao CV. 2011. Role of the mar-sox-rob regulon in regulating outer membrane porin expression. J. Bacteriol. 193:2252–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cohen SP, Hachler H, Levy SB. 1993. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cohen SP, Levy SB, Foulds J, Rosner JL. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 175:7856–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Griffith KL, Fitzpatrick MM, Keen EF, III, Wolf RE., Jr 2009. Two functions of the C-terminal domain of Escherichia coli Rob: mediating “sequestration-dispersal” as a novel off-on switch for regulating Rob's activity as a transcription activator and preventing degradation of Rob by Lon protease. J. Mol. Biol. 388:415–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gu M, Imlay JA. 2011. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol. Microbiol. 79:1136–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haldimann A, Wanner BL. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hidalgo E, Bollinger JM, Jr, Bradley TM, Walsh CT, Demple B. 1995. Binuclear [2Fe-2S] clusters in the Escherichia coli SoxR protein and role of the metal centers in transcription. J. Biol. Chem. 270:20908–20914 [DOI] [PubMed] [Google Scholar]
- 20. Hidalgo E, Demple B. 1996. Activation of SoxR-dependent transcription in vitro by noncatalytic or NifS-mediated assembly of [2Fe-2S] clusters into apo-SoxR. J. Biol. Chem. 271:7269–7272 [DOI] [PubMed] [Google Scholar]
- 21. Hidalgo E, Demple B. 1994. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 13:138–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hidalgo E, Demple B. 1997. Spacing of promoter elements regulates the basal expression of the soxS gene and converts SoxR from a transcriptional activator into a repressor. EMBO J. 16:1056–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hidalgo E, Leautaud V, Demple B. 1998. The redox-regulated SoxR protein acts from a single DNA site as a repressor and an allosteric activator. EMBO J. 17:2629–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jair KW, et al. 1995. Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J. Bacteriol. 177:7100–7104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jair KW, et al. 1996. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J. Bacteriol. 178:2507–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 27. Lee HH, Collins JJ. 2012. Microbial environments confound antibiotic efficacy. Nat. Chem. Biol. 8:6–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee HH, Molla MN, Cantor CR, Collins JJ. 2010. Bacterial charity work leads to population-wide resistance. Nature 467:82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z, Demple B. 1994. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J. Biol. Chem. 269:18371–18377 [PubMed] [Google Scholar]
- 30. Martin RG, Gillette WK, Rhee S, Rosner JL. 1999. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 34:431–441 [DOI] [PubMed] [Google Scholar]
- 31. Martin RG, Gillette WK, Rosner JL. 2000. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol. Microbiol. 35:623–634 [DOI] [PubMed] [Google Scholar]
- 32. Martin RG, Jair KW, Wolf RE, Jr, Rosner JL. 1996. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J. Bacteriol. 178:2216–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martin RG, Rosner JL. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. U. S. A. 92:5456–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin RG, Rosner JL. 1997. Fis, an accessorial factor for transcriptional activation of the mar (multiple antibiotic resistance) promoter of Escherichia coli in the presence of the activator MarA, SoxS, or Rob. J. Bacteriol. 179:7410–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martin RG, Rosner JL. 2002. Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol. Microbiol. 44:1611–1624 [DOI] [PubMed] [Google Scholar]
- 36. Martin RG, Rosner JL. 2004. Transcriptional and translational regulation of the marRAB multiple antibiotic resistance operon in Escherichia coli. Mol. Microbiol. 53:183–191 [DOI] [PubMed] [Google Scholar]
- 37. McMurry LM, Levy SB. 2010. Evidence that regulatory protein MarA of Escherichia coli represses rob by steric hindrance. J. Bacteriol. 192:3977–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Michan C, Manchado M, Pueyo C. 2002. SoxRS down-regulation of rob transcription. J. Bacteriol. 184:4733–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related organisms. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 40. Miller PF, Gambino LF, Sulavik MC, Gracheck SJ. 1994. Genetic relationship between soxRS and mar loci in promoting multiple antibiotic resistance in Escherichia coli. Antimicrob. Agents Chemother. 38:1773–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miller PF, Sulavik MC. 1996. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol. Microbiol. 21:441–448 [DOI] [PubMed] [Google Scholar]
- 42. Nagai T, et al. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87–90 [DOI] [PubMed] [Google Scholar]
- 43. Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nunoshiba T, Demple B. 1994. A cluster of constitutive mutations affecting the C-terminus of the redox-sensitive SoxR transcriptional activator. Nucleic Acids Res. 22:2958–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nunoshiba T, Hidalgo E, Amabile Cuevas CF, Demple B. 1992. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 174:6054–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nunoshiba T, Hidalgo E, Li Z, Demple B. 1993. Negative autoregulation by the Escherichia coli SoxS protein: a dampening mechanism for the soxRS redox stress response. J. Bacteriol. 175:7492–7494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park W, Pena-Llopis S, Lee Y, Demple B. 2006. Regulation of superoxide stress in Pseudomonas putida KT2440 is different from the SoxR paradigm in Escherichia coli. Biochem. Biophys. Res. Commun. 341:51–56 [DOI] [PubMed] [Google Scholar]
- 48. Pomposiello PJ, Bennik MH, Demple B. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48:1609–1619 [DOI] [PubMed] [Google Scholar]
- 50. Rosner JL, Dangi B, Gronenborn AM, Martin RG. 2002. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J. Bacteriol. 184:1407–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rosner JL, Martin RG. 2009. An excretory function for the Escherichia coli outer membrane pore TolC: upregulation of marA and soxS transcription and Rob activity due to metabolites accumulated in tolC mutants. J. Bacteriol. 191:5283–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rosner JL, Slonczewski JL. 1994. Dual regulation of inaA by the multiple antibiotic resistance (mar) and superoxide (soxRS) stress response systems of Escherichia coli. J. Bacteriol. 176:6262–6269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Saini S, Pearl JA, Rao CV. 2009. Role of FimW, FimY, and FimZ in regulating the expression of type i fimbriae in Salmonella enterica serovar Typhimurium. J. Bacteriol. 191:3003–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schneiders T, Levy SB. 2006. MarA-mediated transcriptional repression of the rob promoter. J. Biol. Chem. 281:10049–10055 [DOI] [PubMed] [Google Scholar]
- 55. Seoane AS, Levy SB. 1995. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 177:3414–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tamae C, et al. 2008. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J. Bacteriol. 190:5981–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thomason LC, Costantino N, Court DL. 2007. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. Chapter 1:Unit 1.17 [DOI] [PubMed] [Google Scholar]
- 58. Warner DM, Levy SB. 2010. Different effects of transcriptional regulators MarA, SoxS and Rob on susceptibility of Escherichia coli to cationic antimicrobial peptides (CAMPs): Rob-dependent CAMP induction of the marRAB operon. Microbiology 156:570–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Williams DH, Stephens E, O'Brien DP, Zhou M. 2004. Understanding noncovalent interactions: ligand binding energy and catalytic efficiency from ligand-induced reductions in motion within receptors and enzymes. Angew. Chem. Int. Ed. Engl. 43:6596–6616 [DOI] [PubMed] [Google Scholar]
- 60. Wu J, Weiss B. 1992. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J. Bacteriol. 174:3915–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yeh P, Tschumi AI, Kishony R. 2006. Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 38:489–494 [DOI] [PubMed] [Google Scholar]