Abstract
A sequence encoding a putative extracellular endoglucanase (sso1354) was identified in the complete genome sequence of Sulfolobus solfataricus. The encoded protein shares signature motifs with members of glycoside hydrolases family 12. After an unsuccessful first attempt at cloning the full-length coding sequences in Escherichia coli, an active but unstable recombinant enzyme lacking a 27-residue N-terminal sequence was generated. This 27-amino-acid sequence shows significant similarity with corresponding regions in the sugar binding proteins AraS, GlcS, and TreS of S. solfataricus that are responsible for anchoring them to the plasma membrane. A strategy based on an effective vector/host genetic system for Sulfolobus and on expression control by the promoter of the S. solfataricus gene which encodes the glucose binding protein allowed production of the enzyme in sufficient quantities for study. In fact, the enzyme expressed in S. solfataricus was stable and highly thermoresistant and showed optimal activity at low pH and high temperature. The protein was detected mainly in the plasma membrane fraction, confirming the structural similarity to the sugar binding proteins. The results of the protein expression in the two different hosts showed that the SSO1354 enzyme is endowed with an endo-β-1-4-glucanase activity and specifically hydrolyzes cellulose. Moreover, it also shows significant but distinguishable specificity toward several other sugar polymers, such as lichenan, xylan, debranched arabinan, pachyman, and curdlan.
INTRODUCTION
Plant cells are enclosed by primary cell walls built largely from three classes of complex carbohydrates organized in interdependent networks: pectins, cellulose, and cross-linking hemicellulose (37). Cellulose is the most widespread natural and renewable carbon source on earth, constituting the main component of the plant cell wall and also occurring in some algae, fungi, bacteria, and animals (51). Cellulose molecules are linear polymers of 500 to 15,000 glucose units linked by β-1-4 diether bonds, and extended carbohydrate chains tightly associate by Van der Waals and hydrogen bonds to form compact microfibrils (32). This complex crystalline organization together with the heterogeneous polysaccharide network renders plant material particularly recalcitrant to deconstruction and poorly susceptible to both chemical and enzymatic hydrolysis (9, 41, 42).
Complete breakdown of cellulose requires the synergistic hydrolytic action of three main enzyme classes: (i) endo-β-1-4-glucanases, (ii) exoglucanases, and (iii) β-glucosidases. Endo-β-1-4-glucanases (EC 3.2.1.4) comprise cellulases and act directly on polymer and/or on the shorter (poly)-oligosaccharides produced. They randomly hydrolyze the 1,4-β bonds in more exposed chains of less-structured and/or amorphous regions and produce oligosaccharides of various lengths. Exoglucanases include glucan 1,4-β-glucosidases (cellulodextrinases) (EC 3.2.1.74) and cellulose 1,4-β-cellobiosidases (cellobiosidases); (EC 3.2.1.91). Cellulodextrinases and cellobiosidases act progressively on reducing and nonreducing ends of chains even in microcrystalline (structured) cellulose and release cellobiose and glucose, respectively. β-d-Glucoside glucohydrolases (β-glucosidases) (EC 3.2.1.21) are involved in the final step of cellulose hydrolysis, acting on the products of the reactions described above. In fact, they catalyze the hydrolysis of soluble cellulodextrins (oligosaccharides composed of three to seven glucose molecules) and cellobiose produced by both endo- and exo-glucanases to glucose (22, 29, 36, 56).
The biotechnological potential of cellulases is currently being exploited in industrial processes, e.g., in the paper, textile, and detergent industries (6, 35, 57). However, the paucity of enzymes that efficiently hydrolyze cellulose under the harsh conditions required by the specific processes represents a serious limit in the development of industrial-scale biomass breakdown (26).
Over the last few years, the growing challenge has been the setup of industrial-scale conversion of cellulosic biomass into fermentable sugars, which can in turn be used as raw material for the production of biofuels and other biobased high-value-added products (2, 10, 24, 26, 35, 47, 50, 55). The main bottleneck in the utilization of lignocellulosic plant material as a renewable energy source alternative to fossil fuels lies in the recalcitrance of this biomaterial to hydrolysis (15, 41). The traditional, full chemical process (first developed in late 19th century) uses concentrated acid solutions with heat and pressure to break down the cellulose structure and hydrolyze the polymer. The high energy demand and waste production (coming from the harsh process parameters), together with the production of degradation compounds that can be toxic for a further fermentation step, shaped the research in developing a more sustainable mixed chemical-physical/biological process (3, 23, 40, 49). Nowadays, a first pretreatment step usually uses dilute acids and/or steam to weaken the crystalline structure of cellulose, followed by an enzymatic hydrolysis step where a cellulolytic enzyme mixture is added to convert the cellulose chains to glucose (1, 3, 12, 15, 19, 36). The use of enzymes enables a reduction in the utilization of harsh chemicals and/or heat needed to produce fermentable sugars, raising the environmental feasibility of the process.
However, biotechnological bioconversion processes still appear inefficient since they rely on the low activity and scarce resistance of the currently available hydrolytic enzymes and/or intact microorganisms (26, 46, 55, 60).
In this respect, enzymes from (hyper)thermophilic organisms represent a key opportunity to overcome these obstacles, since they show resistance to extreme physical-chemical conditions, prohibitive for their counterparts from mesophilic organisms (13, 53). In fact, these enzymes can work efficiently at high temperatures and low pH, required for the pretreatment of lignocellulose biomasses (52). They can also directly participate in processes at temperatures up to 90 to 100°C that per se accelerate transfer rates by increasing substrate accessibility and product solubility. Sulfolobus solfataricus is a thermoacidophile that lives in acidic volcanic hot springs and grows optimally up to 87°C and pH 2 to 4. It has been shown to possess several glycoside hydrolases belonging to the different classes mentioned above. Among others, it produces enzymes with carbohydrate depolymerizing activities, such as endoglucanases and xylanases, as well as β-glucosidases/xylosidases involved in the degradation of plant-derived complex polysaccharides (18, 20, 31, 38). The genome of S. solfataricus has been sequenced, and three open reading frames (sso1354, sso1949, and sso2534) coding for putative extracellular endoglucanases have been identified (48). These enzymes belong to a family 12 of glycoside hydrolases, clan C. So far, SSO2534 and SSO1949 have been characterized, as has the sso1949 gene expressed in Escherichia coli (31, 38).
In this article, we describe the cloning and expression of the sso1354 gene in different hosts—E. coli and S. solfataricus itself—and the characterization of the recombinant endo-β-glucanase produced. Heterologous expression in the two hosts allowed elucidation of the modular structure of the enzyme and its cell localization as a cell-bound protein. Comparative characterization also highlighted interesting differences in the properties of the recombinant enzymes produced by the two sources, with the version of the enzyme isolated from the natural host showing exceptional thermostability and activity on a variety of soluble substrates.
MATERIALS AND METHODS
Strains, enzymes, reagents, and devices.
The reagents used for preparation of buffers and growth media of Sulfolobus solfataricus were supplied by Sigma-Aldrich; the yeast extract and Casamino Acids were supplied by Becton, Dickinson (BD). The reagents for polyacrylamide gel electrophoresis were supplied by Bio-Rad. The restriction enzymes, modification enzymes (alkaline phosphatase, T4 DNA ligase, and T4 DNA polymerase), and molecular weight markers for nucleic acids were supplied by New England BioLabs (NEB). The Phusion DNA polymerase was supplied by Finnzymes. The oligonucleotides were synthesized by PRIMM s.r.l.; the radioactive material was supplied by Perkin Elmer. The pET 28c vector was supplied by Novagen. The Escherichia coli MOS-blue strain used for cloning was supplied by Amersham Pharmacia biotech; E. coli BL21-CodonPlus(DE3) RIL cells were supplied by Stratagene. The strain S. solfataricus GθW, a spontaneous derivative mutant of the Gθ strain lacking the β-galactosidase activity (11), was used as the homologous expression host.
sso1354 gene expression in E. coli. (i) Plasmid constructions.
The whole sso1354 coding sequence was amplified by PCR with the primers 1354-Nhe (TAAAGTAGGATAATGGCTAGCAATAAATTATATATTG) and 1354-Sal (CAACTTTTCTGAAACTCTCCTCTAAACAGTCGACGAC). The S. solfataricus P2 genome was used as the template for amplification. After amplification, the fragment was purified (with the Stratagene DNA purification kit), digested with the NheI and SalI enzymes, and cloned into pET-28c (previously made compatible with the NheI and SalI enzymes [recognition sites are indicated in bold letters in the oligonucleotide sequences]). This construct allowed expression of the protein plus an N-terminal 6× His tag. A DNA sequence was PCR amplified to obtain an N-terminally truncated and His-tagged SSO1354 version, namely, a protein lacking the hydrophobic region (amino acids 1 to 27). The forward and the reverse primers were 1354-NheI (GATGCTAGCCAGTCTCTCAGCGTTAAACCCGTAACTA) and 1354-HindIII (GGTCTTAGAAGCTTATATTGTTTAGAGGAGAG), respectively. The DNA amplicon was cut with NheI and HindIII and cloned into the vector pET-28c (the restriction sites are indicated as bold letters, and the three nucleotides corresponding to codon 28 of the SSO1354 coding sequence are underlined).
(ii) Expression and purification of the SSO1354 protein.
The expression plasmids were used to transform E. coli BL21-CodonPlus(DE3) RIL cells. For expression, cells were grown overnight in 10 ml of LB medium with 50 μg/ml kanamycin and 50 μg/ml chloramphenicol at 37°C. After scale-up to a 1-liter culture, the growth was continued up to an A600 of about 0.6 to 0.8, and isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.4 mM. The BL21 cells were incubated for a further 16 h at 22°C. Cells were harvested by centrifugation, resuspended in 50 ml of buffer (100 mM Tris-HCl, pH 7.5, 300 mM NaCl, 2 mM 2-mercaptoethanol, 0.7 mM phenylmethylsulfonyl fluoride [PMSF], and 5% [vol/vol] glycerol), and disrupted by sonication for 5 cycles of 1 min each (B. Braun sonicator). The recombinant endoglucanase versions carrying the 6× His tag were then purified by nickel affinity chromatography. Both intact and N-terminus-truncated SSO1354 enzymes were eluted at an 250 mM imidazole concentration from a HIS-Select spin column (Sigma) and dialyzed against Tris-HCl buffer (pH 7.5).
sso1354 gene expression in S. solfataricus. (i) Plasmid constructions.
A Sulfolobus expression vector was constructed, inserting the sso1354 His-tagged coding sequence fused to the putative promoter of the glucose binding protein (glcS) gene into the pMMSV Sulfolobus/E. coli shuttle vector (7). As a first step, an expression cassette containing the glcS promoter and the sso1354 coding sequence was constructed. The glcS promoter sequence (414 bp) was amplified using the primers glcS-Fw (CCCAATAACTACTCGAGTTACTGACAACTC) and glcS-Rv (CTTCCTTTTCATGACAATTTATGGTAACC). The sso1354 gene previously cloned in the pGEM-T Easy vector was amplified in order to give a 5′ blunt extremity starting at the second codon; the primers were 1354-Fw (AATAAATTATATATTGTGCTTCCGG) and sp6 (ATTTAGGTGACACTATAG, target sequence on the pGEM-T Easy vector). The glcS amplification primers were designed to insert a XhoI site (bold letters) at the 5′ end and to amplify the promoter sequence plus the first four codons of the glcS gene. The S. solfataricus P2 genome was used as the template for amplification of the glcS sequence. After amplifications and digestion with NsiI and with XhoI of the sso1354 and glcS amplicons, the fragments were purified (with the Stratagene DNA purification kit). The vector pGEM-T Easy was digested with XhoI and NsiI and ligated to the two fragments, and the correct vector pGEMTglcS1354 was selected. The ApaI/NsiI fragment encompassing the expression cassette was inserted into the pMSSV shuttle vector previously made end compatible with ApaI and PstI and dephosphorylated, producing the pMSSVglcS1354 expression vector.
(ii) Expression and purification of the SSO1354 enzyme from conditioned medium and cell membrane. (a) Purification from medium.
S. solfataricus GθW cells infected with the SSV2 virus were grown in in Brock's basal medium containing 0.1% yeast extract and 0.1% Casamino Acids (mediumYCA) and transformed by electroporation with 50 ng of the pMSSVglcS1354 vector (following the procedure described by Aucelli et al., [7]). Transformed cells were scaled up in Brock's basal medium containing 1g/liter glucose and 1g/liter tryptone up to 50 ml. After a scale-up to 500 ml in the same medium, the cells were grown up to an optical density at 600 nm (OD600) of 1.2 and harvested by centrifugation. The proteins of the supernatant were precipitated with ammonium sulfate (90% saturation); the pellet was resuspended in Tris-HCl buffer (25 mM, pH 7.5), and the protein sample was extensively dialyzed against the same buffer. After dialysis, the sample was loaded onto a Resource Q anion exchange column (Amersham). After elution (the SSO1354 protein was eluted at 500 mM NaCl), the recombinant enzyme carrying the 6× His tag was purified by nickel affinity chromatography (HIS-Select spin columns; Sigma). The active fractions (around a 250 mM imidazole concentration) were pooled and dialyzed against Tris-HCl buffer, pH 7.5.
(b) CsCl gradient fractionation.
The supernatant from 500-ml cultures of S. solfataricus GθW cells transformed with the pMSSVglcS1354 vector was filtered through a 0.22-μm Seritop filter (Millipore). The filtrate was concentrated by ultrafiltration through a membrane (Amicon; Millipore) with a cutoff of 30 kDa, until the retained volume was ∼15 ml. Separation of macromolecular complexes was performed by centrifugation in a CsCl gradient (0.45 g/ml) for 48 h (SW60-Ti rotor, 250,000 × g, 4°C; Beckman Optima LE-80K ultracentrifuge), and the opaque bands obtained were isolated and dialyzed extensively against 5 mM Tris-HCl buffer (pH 7) at 4°C.
(c) Purification from cell membrane.
pMSSVglcS1354-transformed cells from 500 ml Brock's basal medium supplemented with 0.1% (wt/vol) glucose–0.1% (wt/vol) tryptone (tryptone-glucose medium), respectively, were harvested in the middle-log and stationary phases, namely, at about OD600s of 0.5 and 1.2. Cell pellets were suspended in 10 ml 50 mM Tris-HCl (pH 7.0) and ground in a mortar with sand for 1 h. After centrifugation at 2,000 × g for 10 min in order to remove sand and unbroken cells, the supernatants were ultracentrifuged at 55,000 × g for 30 min. The clear crude extracts were stored at 4°C, whereas the pellets, containing membrane fragments, were suspended in 25 ml 50 mM Tris-HCl (pH 7.0) and ultracentrifuged again. The clear pellets were resuspended in 10 ml of the same buffer containing 0.5% Triton X-100 and incubated overnight at 70°C.
After incubation, the suspensions were ultracentrifuged as described above. The pellets were discarded, and the supernatants were extensively dialyzed against 25 mM Tris-HCl (pH 7.5). The membrane extracts were loaded onto a nickel affinity gel (1.3 ml), and the specific enzyme was eluted with increasing amounts of imidazole (10 to 500 mM) in the same buffer.
N-terminal protein sequence.
Five or six sequencing steps were performed on all recombinant SSO1354 protein versions electroblotted onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad). A Perkin-Elmer Applied Biosystems 477A pulsed-liquid protein sequencer equipped with a model 120A phenylthiohydantoin analyzer was used for automated N-terminal degradation and online identification/quantification of phenylthiohydantoin amino acids.
Activity gel.
Proteins were separated by SDS-PAGE in gels containing 0.1% (wt/vol) carboxymethyl (CM) cellulose (CMC) (Sigma) and renatured by several washes of the gels, once with a mixture (1:4 [vol/vol]) of propan-2-ol and 20 mM potassium phosphate buffer (in the pH range 1.8 to 2.5) or alternatively 20 mM phosphate citrate buffer (in the pH range 3.0 to 5.0) for 30 min. Subsequently, the same buffers without propanol were used for a 30-min wash and replaced with fresh buffers, and the gels were incubated at 75°C for 1 h. All gels were neutralized with 50 mM potassium phosphate (pH 7) before staining with 0.1% (wt/vol) Congo red (Sigma) for 30 min and destaining with 1 M NaCl.
Enzyme assays.
An assay based on soluble chromogenic substrates (azopolymers, polysaccharides coupled with Remazol brilliant blue R; Megazyme) was used to detect the enzyme activity. AZO-CM-cellulose was the substrate during purification, and AZO-derivatized pachyman and oat-spelt xylan were used to determine the relative enzyme specificity. The activity was measured by adding 170 μl of the specific 1% (wt/vol) AZO-polysaccharide solution in 50 mM phosphate-citrate buffer to 170 μl of enzyme solution and incubating the reaction mix at 80°C for 5 to 30 min at pH 5.0. The pH was measured and adjusted in the final buffer-substrate mix before incubation. The reaction was stopped by addition of 850 μl 96% ethanol to the mixture, followed by incubation at room temperature for 10 min and centrifugation at 1,000 × g for 10 min. The absorbance of the supernatants was measured at 590 nm. One unit of enzymatic activity (RBB unit) was defined as the amount of enzyme required to increase the absorbance at 590 nm of 1 optical density per min. Background control reactions were performed with buffer and bovine serum albumin (BSA) at the same enzyme concentrations.
Alternatively, enzyme activity was measured by determining the amounts of reducing sugars released from the polysaccharide substrates. This assay was also used to evaluate the enzyme specificity toward Avicel and α-celluloses, debranched arabinan, curdlan, and lichenan and to determine the pH and temperature optima. The standard reaction mixture was composed of 50 μl 1% (wt/vol) polysaccharide substrate in 50 mM phosphate citrate buffer, pH 5.0, and 50 μl of the enzyme solution (the pH was always measured and adjusted in each final buffer-substrate mix). A mild pretreatment of Avicel and α-celluloses was necessary to obtain homogeneous suspensions: suspensions in water of the polysaccharides (0.5% [wt/vol]) were homogenized by ultrasonication until they showed no clump and by subsequent autoclaving. Sulfuric acid was added up to a 0.02% (vol/vol) final concentration to the resulting suspensions under stirring. After a second ultrasonication step, the suspensions were buffered at pH 5.0 with phosphate citrate buffer for the enzyme assay and alternatively at pH 3.0 with potassium phosphate for in situ hydrolysis by Sulfolobus cells on solid medium. Acid hydrolysis of the celluloses was checked for low increase (less than 1.0%) of reducing ends by the Somogyi-Nelson method (43), as described below.
For pH dependence, the buffers potassium phosphate in the pH range 1.8 to 2.5 and phosphate citrate in the pH range 3.0 to 7.5 were used. After 10 to 60 min of incubation at 80°C, the reactions were stopped on ice and the amount of reducing sugars released was measured at 520 nm by the Somogyi-Nelson method (43). One unit of enzymatic activity (SN unit) was defined as micromoles of sugars released per minute per milliliter.
For in situ assays, plates were prepared with basal medium containing only tryptone (0.1%) and supplemented with 0.8% (wt/vol) Gelrite (Gellan gum; Sigma). A second solution of 0.35% Gelrite in the same medium containing 0.1% CM-cellulose or pretreated cellulose (avicel or α-cellulose) was overlaid after solidification. Increasing aliquots (10 to 40 μl) of the liquid cultures grown in tryptone medium up to an OD600 of 0.8 were spotted onto the Gelrite plates drop-and-dry-wise (5-μl for each drop) in order to obtain circular spots with equal areas. Dense colonization by cells was obtained by incubating the plates for 1 week at 80°C. Staining with Congo red was essentially performed as described for activity gels.
Analysis of degradation products by thin-layer chromatography (TLC).
Enzymatic reactions were performed in solutions of 1% (wt/vol) CM-cellulose at 80°C for various incubation times (2 to 16 h) under the conditions used for the Somogyi-Nelson method. Equal enzyme amounts (enzyme units) of recombinant Sulfolobus solfataricus β-glycosidase kindly provided by Marco Moracci (Institute of Protein Biochemistry [IBP], CNR) were added in parallel samples. The solutions of cellulose were prewarmed before mixing the enzyme(s) in order to reduce viscosity.
The assay was blocked by chilling, and aliquots (60 μl) were spotted onto a silica 60 TLC plate (Macherey-Nagel), which was developed in ethyl acetate, H2O, acetic acid, isopropanol, formic acid, (25:15:10:5:1, by volume) for about 2 h. Released sugars were stained with the α-naphthol reagent after a 5-min incubation at 150°C.
RESULTS
Sequence analysis.
Three putative extracellular endoglucanase sequences have been identified in the complete genome sequence of S. solfataricus P2 (48): sso1354, sso1949, and sso2534. This work has focused on the as yet uncharacterized SSO1354 gene/protein. The first approach was to perform an in-depth sequence analysis using bioinformatic tools. The protein sequence translated from the sso1354 gene was aligned with sequences available in databanks using the FASTA 3 software program on the European Bioinformatics Institute (EBI) website (http://www.ebi.ac.uk/services/index.html). The highest identity score, 85%, was produced in the comparison with SSO1949.
Interestingly, these sequences also show some similarities in the flanking genomic regions; in particular, both sequences have open reading frames (ORFs) immediately downstream encoding characterized (SSO1353) (20) and putative (SSO1948) glycoside hydrolases. The corresponding protein sequences of SSO1353 and SSO1948 show a high identity percentage value, similar to that determined for SSO1354 and SSO1949 (∼86%). Moreover, significant alignments were also produced with other ORFs flanking SSO1354 and SSO1949, namely, sequences coding for the putative transposases SSO1946, SSO1951, and SSO1367. This analysis perfectly matches a previous study described by Brouns et al. (16). Therefore, one of two sequences was presumably generated from a duplication/insertion event, which could have been mediated by the transposable elements (ISs) mentioned (45).
The inspection of the databanks also revealed only 26% identity between SSO1354 and the other putative endo-β-1-4-endoglucanase, CelS. Furthermore, proteins producing the most significant similarity scores (excluding SSO1949) were found to be of thermophilic bacterial origin, particularly from the genus Thermotoga (about 30% identity, on average, with both T. maritima and T. neapolitana Cel12A).
The presence of the C-terminal glyco_hydro_12 conserved domain was identified by the analysis of the SSO1354 sequence with the PFAM database program (available at the website www.sanger.ac.uk) (27). Therefore, the protein can be assigned to family 12 of glycoside hydrolase (GH) (29). Enzymes with four different catalytic activities belong to GH family 12 (clan C): cellulase (EC 3.2.1.4), xyloglucan-specific endo-β-1,4-glucanase (EC 3.2.1.151), licheninase (EC 3.2.1.73), and xyloglucan:xyloglucosyl transferase (EC 2.4.1.207); these enzymes have a retaining mechanism, in which two glutamic acid residues act as nucleophile and proton donor residues.
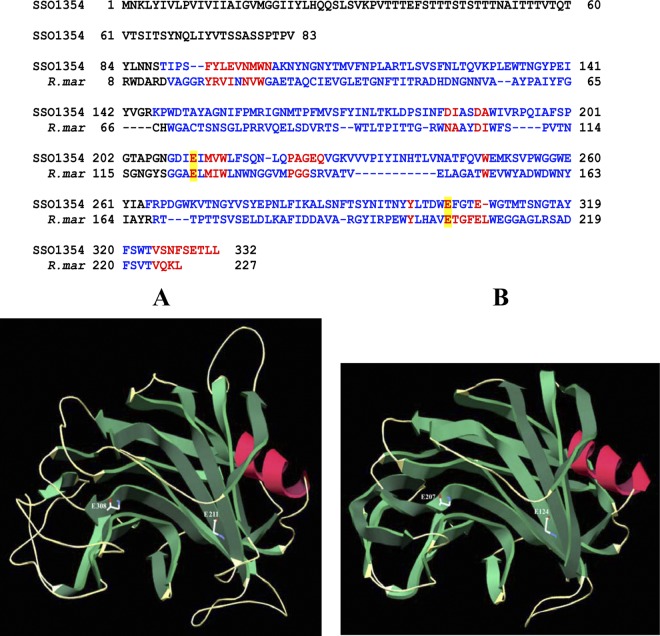
The structure of highly thermostable family 12 endoglucanase (Cel12A) from the thermophilic bacterium Rhodothermus marinus has been solved (21). This enzyme has been shown to have one domain made of two β-sheets and one α-helix with a beta-jelly roll fold, resembling the structure of xylanase family 11. The catalytic domain of SSO1354 enzyme shares significant residues with Cel12A (27.6% identity and 49.8% similarity), as previously found for SSO1949 (31). The sequence similarity allowed homology modeling of SSO1354 using the cellulase from R. marinus as a template (Fig. 1); only the modeling of the catalytic domain (residue positions 76 to 327) could be performed, since the N-terminal region does not show homology to either Cel12A or other known endoglucanases except SSO1949. The structural modeling combined with the sequence- and structure-based alignment clearly indicates the presence of a typical GH clan C fold in the SSO1354 protein (14), with an active site which closely resembles that of other cellulases of family 12. By structural modeling, two putative catalytic glutamate residues, Glu-211 and Glu-310, were also identified as being within the active site and highly conserved in other GH sequences. On the basis of known structures and well-characterized mechanisms of cellulases of GH family 12, Glu-211 should act as a nucleophile and very likely Glu-310 displays the acid-base function. As with several endoglucanases belonging to the GH family 12, SSO1354 does not possess any carbohydrate binding motif (CBM).
Fig 1.
Structural model of SSO1354 endo-β-1-4-glucanase. SSO1354 was aligned with cellulase from Rhodothermus marinus (R. mar). The complete sequence is indicated only for SSO1354. The N-terminal region (amino acids [aa] 1 to 83) has no sequence similarity to all known cellulases, including the one from R. marinus. High-consensus and low-consensus residues are indicated, respectively, in red and blue. The catalytic glutamate residues Glu-211 and Glu-308 are highly conserved and are boxed in yellow. Therefore, the structure (A) of the SSO1354 catalytic domain (aa 76 to 327) could be modeled around the template (B) of the cellulase from R. marinus (PDB no. 1H0B) using the EsyPred3D software program. The β-strands are colored in green and α-helices in red, and the glutamate residues in the catalytic domain are indicated by their positions.
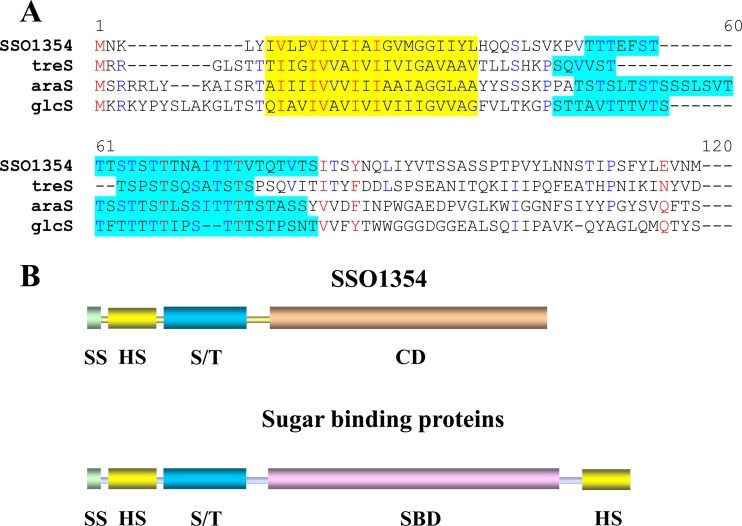
Interestingly, a more focused analysis of the N-terminal stretch of SSO1354, unique among all endoglucanases, highlighted a significant sequence and organization similarity with the corresponding amino acid regions in the sugar binding proteins (SBPs) from S. solfataricus AraS, GlcS, and TreS. These proteins, which respectively bind arabinose, glucose, and trehalose, are the subunits of ATP binding cassette (ABC) transporters responsible for sugar uptake in S. solfataricus (25).
The sequence alignment shows that SSO1354 shares overall domain architecture with the sugar binding proteins (Fig. 2), with a highly conserved hydrophobic region at the N terminus of all proteins compared. A significantly similar S-T-rich stretch is also present in all sequences aligned and connects the hydrophobic region to the catalytic site in the case of the endoglucanases and to the sugar binding modules in the case of the transporters. This lipophilic domain serves as a transmembrane anchor segment. Unlike SSO1354, the sugar binding proteins have an additional hydrophobic stretch at the C terminus, with an identical function.
Fig 2.
Structural organization of SSO1354 and sugar binding proteins (SBPs). SBPs and SSO1354 endoglucanase share a similar organization, which probably reflects a similar function of the N-terminal modules. (A) Alignment of N-terminal amino acid sequences. High-consensus and low-consensus residues are indicated, respectively, in red and blue; putative transmembrane segments are highlighted in yellow, and the long hydrophobic stretch rich in hydroxylated amino acids is indicated in light blue. (B) Schematic protein structures. SS, signal peptide; HS, putative transmembrane hydrophobic segment; S/T, linker region with high percentage of hydroxylated amino acids (mainly serine and threonine); CD, catalytic domain; SBD, sugar binding domain.
The SSO1354 sequence was also analyzed for putative N-glycosylation sites, and 11 residues were found to match the consensus Asn-X-Ser/Thr of the eukaryotic extracellular proteins.
Expression of the sso1354 gene in E. coli.
The expression of sso1354 sequence was undertaken in the conventional mesophilic host E. coli with the aim to obtain higher and reproducible production of the enzyme compared to that of the natural source, S. solfataricus. Initially, the whole coding sequence of SSO1354 fused with a sequence coding for an (His)×6 tag at the N terminus was cloned into the pET28c expression vector; this vector was used to transform E. coli BL21-CodonPlus(DE3) RIL cells. The endoglucanase activity and the presence of a specific SSO1354 polypeptide were undetectable in the transformed cells. It was hypothesized that the putative membrane-anchoring N-terminal peptide could impair the correct folding of the SSO1354 polypeptide and hence favor rapid degradation of the misfolded protein.
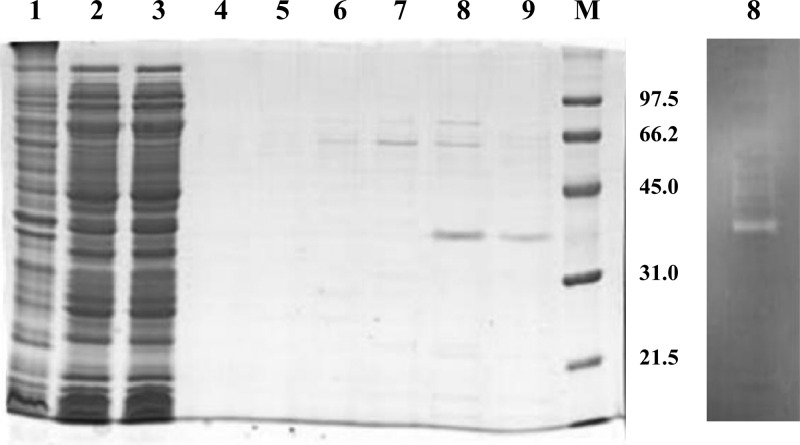
The SSO1354 DNA sequence was therefore reamplified starting from codon 28 and cloned into pET28c, in order to obtain an enzyme version lacking this peptide stretch, and fused to an N-terminal 6× His tag. This strategy resulted in successful production of recombinant enzyme in E. coli cells, with a sufficient yield for characterization. The expression of the sso1354 gene was visualized by SDS-PAGE as a main product of 35.5 kDa in mass, in good agreement with the theoretical mass of 35,154 Da. The identity of the protein was confirmed by automated Edman sequencing. The activity of the enzyme was qualitatively determined by zymographic assays that revealed a specific cellulase activity band (Fig. 3) absent in cell extracts of untransformed E. coli.
Fig 3.
Expression of the sso1354 gene in E. coli. Cell extracts of BL21 E. coli cells transformed with pET28c-1354 after growth and IPTG induction were loaded on a nickel affinity chromatography spin column for the purification of the His-tagged protein. The protein fractions from all chromatographic steps were analyzed by SDS-PAGE followed by Coomassie staining: marker (M), membrane extract (lane 1), crude cytosolic extract (2), flowthrough (3), wash (4), and elution with 10, 50, 100, 250, and 500 mM imidazole (5, 6, 7, 8, and 9). An aliquot from the 250 mM imidazole elution step (lane 8) was analyzed on a zymographic gel (containing 0.1% CMC), and the cellulase activity band was revealed with Congo red after renaturation.
SSO1354 produced in E. coli was optimally active at high temperature and low pH (80°C and pH 2.5), but unfortunately it was also unstable and showed a very low specific activity compared to the wild type enzyme. In fact, the specific activity (measured as the formation of reducing ends on CM-cellulose at the optimum pH and temperature) of the purified enzyme was only 1.2 U/mg, whereas the total activity in the cell membrane extract of S. solfataricus (grown in medium YCA, namely, under conditions not repressive for activity on CMC [38]) was about 0.3 U/mg.
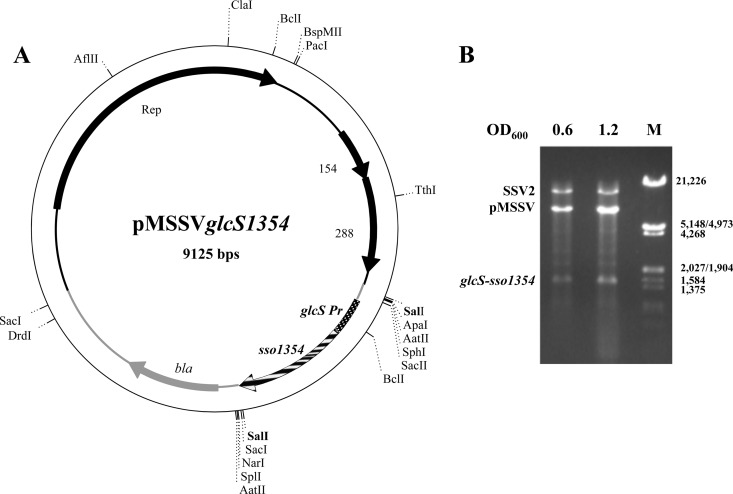
Cloning of expression cassette glcS-sso1354 in the pMSSV vector and expression in S. solfataricus.
The SSO1354 coding sequence, including a downstream sequence coding for a 6× His tag, was cloned downstream of the glcS gene promoter. This gene has been demonstrated to be actively transcribed in S. solfataricus under a variety of culture medium compositions (39), and hence its strong promoter was chosen to allow high-level gene expression without imposing stress on the transformed cells.
The glcS-sso1354 cassette was inserted into the polycloning site of the pMSSV vector (7) by directional cloning. The resultant pMSSVglcS1354 (Fig. 4A) vector could be propagated without rearrangements in E. coli. The plasmid was transferred by electroporation into S. solfataricus GθW cells infected with the helper virus SSV2, which guarantees the maintenance and propagation of the virus/plasmid hybrid pSSVx and its derivative vectors (11). After several propagation steps, successful transformation was confirmed by electrophoresis analysis of extrachromosomal DNA digests (Fig. 4B).
Fig 4.
pMSSVglcS1354 map and vector propagation in S. solfataricus cells. (A) The map of the pMSSV expression vector containing the glcS promoter fused with the SSO1354 coding sequence is shown. (B) SalI digestions of extrachromosomal DNAs prepared from equal amounts of S. solfataricus GθW/SSV2 cells transformed with the expression vector. The cultures were harvested at the two different growth stages, indicated as optical density values at 600 nm.
The cellulase activity was subsequently visualized by a zymographic assay, indicating that the pMSSVglcS1354 vector was successfully capable of driving the expression of the sso1354 gene. Distribution of the enzyme as cell bound and released was dependent on the growth phase of the cultures, with the membrane-associated fraction constituting about 90% up to the mid-exponential stage. In contrast, most of the cellulase activity was found in the supernatants of steady-state cultures, but it still can be ascribed to the membrane-bound enzyme solubilized upon cell autolysis and/or partial membrane breakdown. In fact, cesium chloride isopycnic gradients of concentrated conditioned medium confined the SSO1354 activity to a single middle band, namely, to a density compatible with protein-lipid complexes rather than single free proteins. These findings also suggest that no proteolytic cell activity had occurred at this stage of growth, since all enzyme activity was recovered and found not interspersed across the gradient.
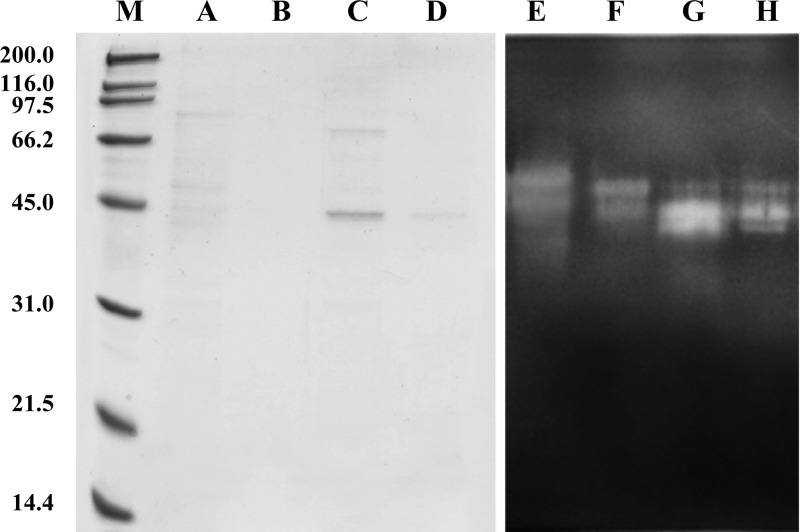
In the interest of keeping the procedure simple, SSO1354 was partially purified in the free released form by ammonium sulfate precipitation of conditioned medium and nickel affinity chromatography (Fig. 5). An apparent molecular mass of 45 kDa could be attributed to the activity band in zymograms on SDS-PAGE gels. This value was higher than expected (37.2 kDa), indicating that the protein could have some posttranslational modification, such as glycosylation.
Fig 5.
Expression of sso1354 sequence in S. solfataricus. Proteins of the supernatant from transformed S. solfataricus cells were precipitated with ammonium sulfate (90% saturation); the pellet was resuspended and loaded onto an anionic exchange column. The fractions showing cellulase activity were pooled and loaded onto nickel affinity spin columns. Samples from the nickel affinity chromatography were analyzed by SDS-PAGE followed by Coomassie blue and enzyme staining: marker (M), crude extract (A and E), wash (B and F), elution with 250 mM imidazole (C and G), and elution with 500 mM imidazole (D and H). After renaturation, cellulase activity was revealed by zymography on a gel containing 0.1% CMC stained with Congo red.
It is worth mentioning that the outer membrane extracts of recombinant S. solfataricus contained not only most of the SSO1354 activity but also sugar binding proteins as the major polypeptides present. In fact, automated Edman sequencing performed on the membrane proteins after separation on SDS-PAGE gels identified TreS and GlcS, as well as the main S-layer protein, SlaA, in the most distinct bands, similar to what was previously reported by Elferink et al. (25). Automated 6-step N-terminal sequencing of PVDF-blotted purified SSO1354 produced the amino acid sequence IVLVPI, which confirmed protein identity and defined the processing of only the first 5 residues (MNKLY). No other overlapping sequence was determined, demonstrating that a specific and not generic processing proteinase action occurred. This result provided a further indication of structural similarity with sugar ABC transporters (25), particularly with the TreS (trehalose binding) protein, which is cleaved after an even shorter stretch of four residues (5). Therefore, the hydrophobic region at the N terminus is not excised and hence is not a signal peptide for the release of the protein outside the cell. Besides the highly positively charged residue(s) at the extreme N terminus and the hydrophobic region immediately after the cleavage site, no apparent consensus matching the one defined for pilin processing could be identified. The cleavage mode of a larger number of substrates for type IV prepilin-like signal peptidase, such as pilin and other sugar binding proteins (5), in Sulfolobus should be defined to include (or not) SSO1354 as well as TreS among them.
Characterization of recombinant enzyme produced by S. solfataricus.
The SSO1354 protein was confirmed to be thermoacidophilic, with a pH optimum of 4.0 and an optimum temperature of 90°C. These values are much closer to those determined for the overall cellulosic activity expressed by wild-type S. solfataricus, than to those for the recombinant enzyme produced by E. coli. Moreover, the specific activity was about 200-fold higher (223.5 U/mg versus 1.2 U/mg), confirming that correct processing and posttranslational modification by the natural host are indispensable prerequisites for correct enzyme performance.
The enzyme was active toward carboxymethyl (CM)-cellulose, as well as several other polymers: lichenan (a linear glucan of {1→3, 1→4}-β-glycosidic bonds in a ratio of 1:2) with a 120% activity with respect to CM-cellulose; debranched arabinan (a polymeric chain of 1,5-α-linked l-arabinofuranosyl residues) with a 25% activity; curdlan and pachyman (both polymers of 1→3-β-linked d-glucosyl residues), with, respectively, a 26% and 22% activity; oat spelt xylan (1→4-β-linked polymer contain predominantly d-xylose) with a 25% activity. Activities on intact microcrystalline Avicel and amorphous α-cellulose were 1 and 3%, increasing to approximately 10 and 15%, respectively, when the substrates were acid pretreated, as described. The low activity detected with intact celluloses can be ascribed to the lack of a cellulose binding motif in the sequence of SSO1354. However, for the most convincing proof of cellulase activity, both wild-type and recombinant Sulfolobus cells were seeded on solid medium containing the dispersed substrates. After growth, clear halos of hydrolysis were formed only around the spots colonized by the recombinant Sulfolobus cells overexpressing SSO1354 (Fig. 6). Furthermore, the SSO1354 enzyme did not hydrolyze p-nitrophenyl-β-d-cellobioside and p-nitrophenyl-β-d-cellotrioside, as observed for the other S. solfataricus endoglucanase SSO1949, namely, it has no exocellulase activity, confirming that SSO1354 is mainly an endo-1,4-β glucanase. Despite what has been previously reported (38), the third putative endoglucanase gene, sso2534 (celS), cloned and overexpressed in a similar fashion in S. solfataricus revealed that the overproduced enzyme was not endowed with any activity toward highly polymeric substrates, such as soluble and insoluble celluloses and hemicelluloses (data not shown). This discrepancy in cellulose activity attribution could be explained by the fact that the genome sequence of S. solfataricus was not yet available at the time of the work carried out by Limauro et al. (38) and hence by the lack of information about two additional endoglucanase genes besides celS in S. solfataricus.
Fig 6.
In situ hydrolysis of α-cellulose by Sulfolobus cells. Aliquots (10 to 40 μl, as indicated) of late-logarithmically grown Sulfolobus solfataricus cultures were spotted onto Gelrite/tryptone plates containing a homogeneously mixed, acid-pretreated α-cellulose suspension. After colonization of the areas, plates were stained with Congo red to visualize depolymerization halos. Upper areas of the plate are wild-type cells. Lower areas are cells transformed with the sso1354 gene.
Therefore, the specificity profile together with in situ hydrolysis of substrates by recombinant overexpressing cells clearly indicates that SSO1354 is preferentially a glucanase enzyme and is responsible for the main cellulase activity of Sulfolobus solfataricus.
pH and temperature dependence of recombinant SSO1354 activity, investigated in the range of 1.8 to 7.5 and 37 to 90°C, indicated a temperature optimum of about 90°C; at this temperature, the highest activity was found at pH 4.5. Moreover, the recombinant endoglucanase showed strong thermostability; when incubated at 90°C, the enzyme had a half-life of 180 min.
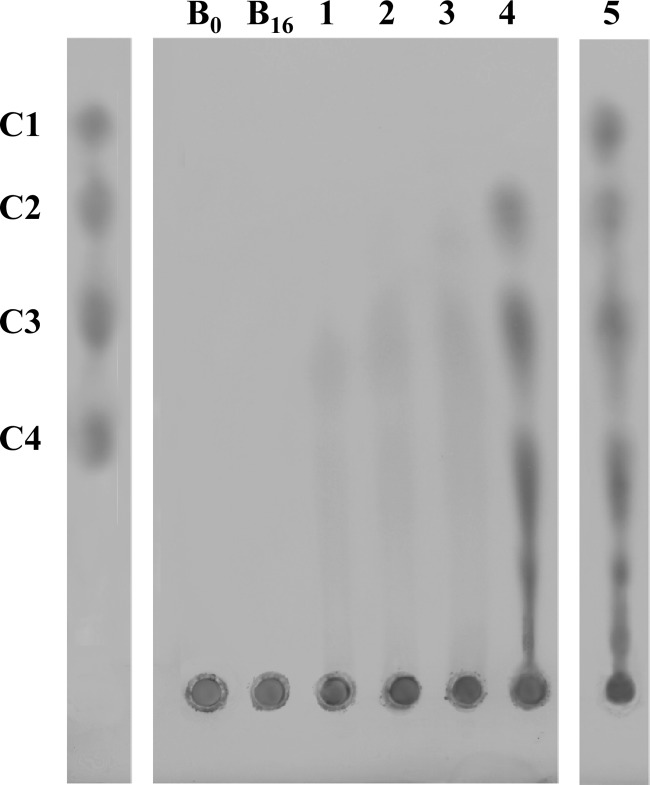
Qualitative activity detection by thin-layer chromatography analysis of the hydrolyzed cellulose products confirmed that the enzyme possesses endocellulasic activity, with cellobiose and cellotriose as the final hydrolysis products in a time-dependent accumulation over 2.0- to 16.0-h incubations. As further proof, additional hydrolysis resulting in released glucose occurred only upon addition of the β-glycosidase enzyme from S. solfataricus to the reaction mixture (Fig. 7).
Fig 7.
Thin-layer chromatography analysis of cellulose hydrolysis products. 1% CM-cellulose solutions were incubated at 80°C with SSO1354 enzyme and with a 1:1 (enzyme units) mixture of SSO1354 and S. solfataricus β-glycosidase. Aliquots of the reaction mixtures with SSO1354 (lanes 1 to 4), with SSO1354 and β-glycosidase (lane 5), and without (lanes B0 and B16) enzyme(s) were withdrawn at different time points and spotted onto silica plates. Reaction products were separated with a mixture of ethyl acetate, H2O, acetic acid, isopropanol, and formic acid (25:15:10:5:1) and visualized with α-naphthol after incubation for 5 min at 120°C. Lane M, mix of cello-oligomers (3 μl of a solution with a concentration of 5 mM [each] oligomer); B0 and B16, cellulose solutions in buffer without enzyme(s) after no and 16 h of incubation, respectively; 1, 2 h of incubation, 2, 4 h of incubation, 3,8 h of incubation, 4, 16 h of incubation, 5, 16 h of incubation with the mixture of the two enzymes.
DISCUSSION
In this work, the gene coding for the putative extracellular endoglucanase SSO1354 from the hyperthermophilic archaeon S. solfataricus was overexpressed in homologous and heterologous fashion, and an extensive sequence analysis was performed, with the aim of determining the biotechnological potentiality of this enzyme. The recombinant enzyme produced in the archaeal host showed an activity optimum at high temperature and acidic pH combined with a marked resistance under the same conditions. These extraordinary features, together with the activity on a broad range of polymeric substrates, clearly pointed to the biotechnological relevance of the enzyme. The effort to expand the repertoire of different enzymatic activities for industrial applications is particularly channeled toward cellulolytic enzymes, which have become increasingly appealing over the last few years in the field of green/white biotechnology. The use of cellulases, like SSO1354, able to work with long operational stability under harsh conditions could significantly boost the development of a sustainable process to obtain biofuels from biomass, addressing one of the most urgent priorities of modern society, the need to find a green, economically feasible solution to replace fossil fuels (1, 13, 52, 53, 54).
The sequence analysis of SSO1354 and the experimental data collected revealed some interesting details about the structural/functional properties of the enzyme. Differently from what has previously been indicated for the closely related SSO1949, a cell-bound localization of the enzyme was also found. The catalytic domain is linked by an S-T-rich stretch to the transmembrane region, creating an architecture that could provide the enzyme with the necessary flexibility. In fact, this motif has been previously demonstrated to be fundamental for catalytic activity in the closely related SSO1949 enzyme (31), its absence causing inactivation of the endoglucanase. Likely, the enzyme possesses a secretion leader, as predicted by SignalP (and also indicated in the 1949 characterization study), but it is equipped with a leader system similar to that found in sugar binding proteins. AraS and GlcS, like other S. solfataricus solute binding proteins, have an unusual leader peptide that resembles bacterial type IV prepilin signal sequences and is cleaved by the S. solfataricus homolog of bacterial type IV prepilin peptidases, named PibD (5). SSO1354, similarly to the other sugar binding protein TreS, possesses a shorter putative signal sequence that matches, even if only in part, the relatively conserved consensus sequence defined as recognizable by PibD. However, PibD has been demonstrated to display quite broad substrate specificity and to tolerate some mutations in the four-residue core (K/R-G/A-L/I/F-S/T/A) around the cleavage site. This assumption is supported by the N-terminal sequencing of the recombinant polypeptide produced homologously by S. solfataricus: the only fragment processed and cleaved out was the stretch comprising the first five N-terminal residues, immediately upstream of the hydrophobic region.
The sequence comparison with three cell-bound sugar binding proteins (SBPs) of S. solfataricus, together with the activity data about the enzyme localization and N-terminal sequencing, demonstrates that the enzyme stays bound to the membrane via anchoring mediated by the hydrophobic N-terminal leader and that it is released into the medium only as a component of membrane fragments produced by the stress of prolonged cell culturing. SSO1354 is present mostly in the outer cell membrane subfraction, in which the sugar binding proteins are the major proteins. Sugar binding proteins of S. solfataricus are essential components together with ABC transporters for the uptake of sugars. They can be detergent extracted from membranes as heterogeneous protein complexes that also contain the main S-layer protein, and intriguingly, SSO1354 is also colocated with these larger structures. Recently, functional expression of sugar binding proteins at the cell surface has been demonstrated to require the so-called bindosome assembly system (Bas) (58, 59). In fact, Bas deletion mutants have been shown to impair, among others, the function of glucose uptake. Although little is known about the periplasmic archaeal space, it has been reasonably hypothesized that the association of the bindosome and solute binding proteins at the cell surface may allow a more efficient retrieval of sugars (and/or oligopeptides) from the environment and enrich sugars from the medium between the S-layer and the cytoplasmic membrane. The anchoring to the outer cell membrane and the vicinity with these protein complexes makes this hypothesis also appropriate for SSO1354 endoglucanase: it can increase glucose oligosaccharide concentration in this “quasiperiplasm” of S. solfataricus and hence directly provide the sugar transport complexes with substrates to take up with significantly reduced diffusion and loss. It can also be assumed that the other two endoglucanases from S. solfataricus, which also show a similar N-terminal structure, have the same cell localization and distribution of SSO1354. These findings open new and intriguing perspectives on the strategy adopted by a hyperthermophilic archaeon like S. solfataricus to coordinate and organize the activities involved in the breakdown and uptake of a specific polymeric nutrient. In fact, colocalization of the endoglucanases and the sugar binding proteins could be advantageous, particularly for cells living in habitats where complex carbohydrates are a major but scarce energy source. In this regard it is also worth noting that despite the lack of a cellulose binding domain, SSO1354 is the only endoglucanase of S. solfataricus endowed with cellulose-hydrolyzing activity. Interestingly, cellulase activity increases when the substrate is partially destructured under (sulfuric acidic and hot) conditions mimicking those found in the natural habitat of Sulfolobus cells.
The comparison of the recombinant versions of SSO1354 produced in E. coli and S. solfataricus highlighted some interesting differences. When produced in E. coli, SSO1354 displayed maximum activity at low pH and high temperature as expected but unfortunately also showed a marked instability and low specific activity. The enzyme expressed in S. solfataricus was instead highly thermoresistant (half-life of 3 h at 90°C) and also more stable than the SSO1949 enzyme (which was characterized after production in E. coli) (31). The effect on thermostability was most probably due to posttranslational modifications, such as N-glycosylation, which is present in extracellular proteins produced in Sulfolobus (17), and so it appeared for SSO1354, whereas the polypeptide produced in E. coli is surely not glycosylated. In general, the contribution of glycosylation in increasing the resistance of proteins to extreme conditions is well documented (8, 28, 30, 33, 34, 44). Archeal N-linked glycans are structurally simple, being composed of 3 to 5 sugar residues that are preferentially unbranched. Interestingly, recent studies have demonstrated that more complex structures, namely, branched glycan trees, can be found attached to N-glycosylation sites of cell surface proteins only in (hyper)thermophilic archaea, as reviewed by Albers and Meyer (4). These findings would suggest that increasing degree and complexity of glycosylation comprise a posttranslational modification selected by evolution to provide extra stability even to proteins from hyperthermophiles. This also demonstrates that conventional mesophilic bacterial hosts, unable to posttranslationally modify recombinant proteins, cannot be exhaustively successful for production of extracellular proteins from archeal (hyper)thermophiles. Therefore, effective and more sophisticated homologous expression systems are indispensable not only to better understand gene function and protein sorting in vivo but also for the production of fully active and best-performing enzymes, particularly in view of their industrial applications. Further experiments aimed at fully understanding the exact impact of posttranslational modifications on the activity/stability, in particular, and the molecular determinants of adaptation of this extracellular model enzyme to the extreme environment, in general, are under way.
ACKNOWLEDGMENTS
We thank Patrizia Contursi (at Dipartimento di Biologia Strutturale e Funzionale, University of Naples Federico II) for helpful discussion and valuable advice on Sulfolobus culturing and manipulation. We are also grateful to Vito Carratore (at IBP, CNR) for the automated N-terminal protein degradation and for the analysis of protein sequencing data.
This work was partially funded by the Agenzia Spaziale Italiana (project MoMa no. 1/014/06/0).
Footnotes
Published ahead of print 13 July 2012
REFERENCES
- 1. Aden A, et al. 2002. Lignocellulosic biomass to ethanol process design and economics utilizing co-current dilute acid prehydrolysis and enzymatic hydrolysis for corn stover. NREL publication no. TP-510-32438; National Renewable Energy Laboratory, Golden, CO [Google Scholar]
- 2. Adsul MG, Singhvi MS, Gaikaiwari SA, Gokhale DV. 2011. Development of biocatalysts for production of commodity chemicals from lignocellulosic biomass. Bioresour. Technol. 102:4304–4312 [DOI] [PubMed] [Google Scholar]
- 3. Agbor VB, Cicek N, Sparling R, Berlin A, Levin DB. 2011. Biomass pretreatment: fundamentals toward application. Biotechnol. Adv. 29:675–685 [DOI] [PubMed] [Google Scholar]
- 4. Albers SV, Meyer BH. 2011. The archaeal cell envelope. Nat. Rev. Microbiol. 9:414–426 [DOI] [PubMed] [Google Scholar]
- 5. Albers SV, Szabó Z, Driessen AJ. 2003. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J. Bacteriol. 185:3918–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anish R, Rahman MS, Rao M. 2007. Application of cellulases from an alkalothermophilic Thermomonospora sp. in biopolishing of denims. Biotechnol. Bioeng. 96:48–56 [DOI] [PubMed] [Google Scholar]
- 7. Aucelli T, Contursi P, Girfoglio M, Rossi M, Cannio R. 2006. A spreadable, non-integrative and high copy number shuttle vector for Sulfolobus solfataricus based on the genetic element pSSVx from Sulfolobus islandicus. Nucleic Acids Res. 34:e114 doi: 10.1093/nar/gkl615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bai Y, et al. 2010. Expression of an extremely acidic beta-1,4-glucanase from thermoacidophilic Alicyclobacillus sp. A4 in Pichia pastoris is improved by truncating the gene sequence. Microb. Cell Fact. 9:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balan V, Bals B, Chundawat SP, Marshall D, Dale BE. 2009. Lignocellulosic biomass pretreatment using AFEX. Methods Mol. Biol. 581:61–77 [DOI] [PubMed] [Google Scholar]
- 10. Ballesteros M, et al. 2010. Ethanol production from the organic fraction obtained after thermal pretreatment of municipal solid waste. Appl. Biochem. Biotechnol. 161:423–431 [DOI] [PubMed] [Google Scholar]
- 11. Bartolucci S, Rossi M, Cannio R. 2003. Characterization and functional complementation of a nonlethal deletion in the chromosome of a beta-glycosidase mutant of Sulfolobus solfataricus. J. Bacteriol. 185:3948–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blanch HW, Simmons BA, Klein-Marcuschamer D. 2011. Biomass deconstruction to sugars. Biotechnol. J. 6:1086–1102 [DOI] [PubMed] [Google Scholar]
- 13. Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MW, Kelly RM. 2008. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr. Opin. Biotechnol. 19:210–217 [DOI] [PubMed] [Google Scholar]
- 14. Bourne Y, Henrissat B. 2001. Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr. Opin. Struct. Biol. 11:593–600 [DOI] [PubMed] [Google Scholar]
- 15. Brodeur G, et al. 2011. Chemical and physicochemical pretreatment of lignocellulosic biomass: a review. Enzyme Res. 2011:787532 doi: 10.4061/2011/787532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brouns SJJ, et al. 2005. The hyperthermophilic archaeon Sulfolobus: from exploration and exploitation, p 261–276 In Inskeep B, McDermott TR. (ed), Geothermal biology and geochemistry in Yellowstone National Park. Montana State University, Bozeman, MT [Google Scholar]
- 17. Calo D, Kaminski L, Eichler J. 2010. Protein glycosylation in Archaea: sweet and extreme. Glycobiology 20:1065–1076 [DOI] [PubMed] [Google Scholar]
- 18. Cannio R, Di Prizito N, Rossi M, Morana A. 2004. A xylan-degrading strain of Sulfolobus solfataricus: isolation and characterization of the xylanase activity. Extremophiles 8:117–124 [DOI] [PubMed] [Google Scholar]
- 19. Chen Y, et al. 2012. Reducing acid in dilute acid pretreatment and the impact on enzymatic saccharification. J. Ind. Microbiol. Biotechnol. 39:691–700 [DOI] [PubMed] [Google Scholar]
- 20. Cobucci-Ponzano B, et al. 2010. A new archaeal beta-glycosidase from Sulfolobus solfataricus: seeding a novel retaining beta-glycan-specific glycoside hydrolase family along with the human non-lysosomal glucosylceramidase GBA2. J. Biol. Chem. 285:20691–20703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Crennell SJ, Hreggvidsson GO, Nordberg Karlsson E. 2002. The structure of Rhodothermus marinus Cel12A, a highly thermostable family 12 endoglucanase, at 1.8 A resolution. J. Mol. Biol. 320:883–897 [DOI] [PubMed] [Google Scholar]
- 22. Dashtban M, Maki M, Leung KT, Mao C, Qin W. 2010. Cellulase activities in biomass conversion: measurement methods and comparison. Crit. Rev. Biotechnol. 30:302–309 [DOI] [PubMed] [Google Scholar]
- 23. Delucchi MA. 2010. Impacts of biofuels on climate change, water use, and land use. Ann. N. Y. Acad. Sci. 1195:28–45 [DOI] [PubMed] [Google Scholar]
- 24. Du J, Shao Z, Zhao H. 2011. Engineering microbial factories for synthesis of value-added products. J. Ind. Microbiol. Biotechnol. 38:873–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Elferink MG, Albers SV, Konings WN, Driessen AJ. 2001. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol. Microbiol. 39:1494–1503 [DOI] [PubMed] [Google Scholar]
- 26. Elkins JG, Raman B, Keller M. 2010. Engineered microbial systems for enhanced conversion of lignocellulosic biomass. Curr. Opin. Biotechnol. 21:657–662 [DOI] [PubMed] [Google Scholar]
- 27. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guerrero-Olazarán M, Rodríguez-Blanco L, Carreon-Treviño JG, Gallegos-López JA, Viader-Salvadó JM. 2010. Expression of a Bacillus phytase C gene in Pichia pastoris and properties of the recombinant enzyme. Appl. Environ. Microbiol. 76:5601–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henrissat B, Davies GJ. 1997. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7:637–644 [DOI] [PubMed] [Google Scholar]
- 30. Huang P, Chen C, Mague SD, Blendy JA, Liu-Chen LY. 2011. A common single nucleotide polymorphism A118G of the mu opioid receptor alters its N-glycosylation and protein stability. Biochem. J. doi: 10.1042/BJ20111050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang Y, Krauss G, Cottaz S, Driguez H, Lipps G. 2005. A highly acid-stable and thermostable endo-beta-glucanase from the thermoacidophilic archaeon Sulfolobus solfataricus. Biochem. J. 385:581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keegstra K. 2010. Plant cell walls. Plant Physiol. 154:483–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim YO, Kim HW, Lee JH, Kim KK, Lee SJ. 2006. Molecular cloning of the phytase gene from Citrobacter braakii and its expression in Saccharomyces cerevisiae. Biotechnol. Lett. 28:33–38 [DOI] [PubMed] [Google Scholar]
- 34. Koseki T, et al. 2006. N-linked oligosaccharides of Aspergillus awamori feruloyl esterase are important for thermostability and catalysis. Biosci. Biotechnol. Biochem. 70:2476–2480 [DOI] [PubMed] [Google Scholar]
- 35. Kuhad RC, Gupta R, Singh A. 2011. Microbial cellulases and their industrial applications. Enzyme Res. 2011:280696 doi: 10.4061/2011/280696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kumar R, Singh S, Singh OV. 2008. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J. Ind. Microbiol. Biotechnol. 35:377–391 [DOI] [PubMed] [Google Scholar]
- 37. Lee KJ, Marcus SE, Knox JP. 2011. Cell wall biology: perspectives from cell wall imaging. Mol. Plant. 4:212–219 [DOI] [PubMed] [Google Scholar]
- 38. Limauro D, Cannio R, Fiorentino G, Rossi M, Bartolucci S. 2001. Identification and molecular characterization of an endoglucanase gene, celS, from the extremely thermophilic archaeon Sulfolobus solfataricus. Extremophiles 5:213–219 [DOI] [PubMed] [Google Scholar]
- 39. Lubelska JM, Jonuscheit M, Schleper C, Albers SV, Driessen AJ. 2006. Regulation of expression of the arabinose and glucose transporter genes in the thermophilic archaeon Sulfolobus solfataricus. Extremophiles 10:383–391 [DOI] [PubMed] [Google Scholar]
- 40. Lynd LR, et al. 2008. How biotech can transform biofuels. Nat. Biotechnol. 26:169–172 [DOI] [PubMed] [Google Scholar]
- 41. Malherbe S, Cloete TE. 2002. Lignocellulose biodegradation: fundamentals and applications. Rev. Environ. Sci. Biotechnol. 1:105–114 [Google Scholar]
- 42. McCann MC, Carpita NC. 2008. Designing the deconstruction of plant cell walls. Curr. Opin. Plant Biol. 11:314–320 [DOI] [PubMed] [Google Scholar]
- 43. Nelson N. 1944. A photometric adaptation of the Somogyi method for the determination of glucose. J. Biol. Chem. 153:375–380 [Google Scholar]
- 44. Öberg F, et al. 2011. Glycosylation increases the thermostability of human aquaporin 10 protein. J. Biol. Chem. 286:31915–31923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Redder P, Garrett RA. 2006. Mutations and rearrangements in the genome of Sulfolobus solfataricus P2. J. Bacteriol. 188:4198–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rollin JA, Zhu Z, Sathitsuksanoh N, Zhang YH. 2011. Increasing cellulose accessibility is more important than removing lignin: a comparison of cellulose solvent-based lignocellulose fractionation and soaking in aqueous ammonia. Biotechnol. Bioeng. 108:22–30 [DOI] [PubMed] [Google Scholar]
- 47. Schubert C. 2011. Renewable energy: making fuels for the future. Nature 474:531–533 [DOI] [PubMed] [Google Scholar]
- 48. She Q, et al. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U. S. A. 98:7835–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Solomon BD. 2010. Biofuels and sustainability. Ann. N. Y. Acad. Sci. 1185:119–134 [DOI] [PubMed] [Google Scholar]
- 50. Somerville C, Youngs H, Taylor C, Davis SC, Long SP. 2010. Feedstocks for lignocellulosic biofuels. Science 13:790–792 [DOI] [PubMed] [Google Scholar]
- 51. Templeton DW. 2010. Compositional analysis of lignocellulosic feedstocks. 2. Method uncertainties. J. Agric. Food Chem. 58:9054–9062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Turner P, Mamo G, Karlsson EN. 2007. Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb. Cell. Fact. 6:9 doi: 10.1186/1475-2859-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Unsworth LD, van der Oost J, Koutsopoulos S. 2007. Hyperthermophilic enzymes-stability, activity and implementation strategies for high temperature applications. FEBS J. 274:4044–4056 [DOI] [PubMed] [Google Scholar]
- 54. U.S. Energy Information Administration. 2010. Annual energy outlook. 2010 DOE/EIA-0383. U.S. Energy Information Administration, Washington, DC [Google Scholar]
- 55. Wilson DB. 2009. Cellulases and biofuels. Curr. Opin. Biotechnol. 20:295–299 [DOI] [PubMed] [Google Scholar]
- 56. Wilson DB. 2011. Microbial diversity of cellulose hydrolysis. Curr. Opin. Microbiol. 14:259–263 [DOI] [PubMed] [Google Scholar]
- 57. Zhao J, Li X, Qu Y. 2006. Application of enzymes in producing bleached pulp from wheat straw. Bioresour. Technol. 97:1470–1476 [DOI] [PubMed] [Google Scholar]
- 58. Zolghadr B, Klingl A, Rachel R, Driessen AJ, Albers SV. 2011. The bindosome is a structural component of the Sulfolobus solfataricus cell envelope. Extremophiles 15:235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zolghadr B, Weber S, Szabó Z, Driessen AJ, Albers SV. 2007. Identification of a system required for the functional surface localization of sugar binding proteins with class III signal peptides in Sulfolobus solfataricus. Mol. Microbiol. 64:795–806 [DOI] [PubMed] [Google Scholar]
- 60. Zhu JY, Pan X, Zalesny RS., Jr 2010. Pretreatment of woody biomass for biofuel production: energy efficiency, technologies, and recalcitrance. Appl. Microbiol. Biotechnol. 87:847–857 [DOI] [PubMed] [Google Scholar]