Abstract
DnaA is an AAA+ ATPase and the conserved replication initiator in bacteria. Bacteria control the timing of replication initiation by regulating the activity of DnaA. DnaA binds to multiple sites in the origin of replication (oriC) and is required for recruitment of proteins needed to load the replicative helicase. DnaA also binds to other chromosomal regions and functions as a transcription factor at some of these sites. Bacillus subtilis DnaD is needed during replication initiation for assembly of the replicative helicase at oriC and during replication restart at stalled replication forks. DnaD associates with DnaA at oriC and at other chromosomal regions bound by DnaA. Using purified proteins, we found that DnaD inhibited the ability of DnaA to bind cooperatively to DNA and caused a decrease in the apparent dissociation constant. These effects of DnaD were independent of the ability of DnaA to bind or hydrolyze ATP. Other proteins known to regulate B. subtilis DnaA also affect DNA binding, whereas much of the regulation of Escherichia coli DnaA affects nucleotide hydrolysis or exchange. We found that the rate of nucleotide exchange for B. subtilis DnaA was high and not affected by DnaD. The rapid exchange is similar to that of Staphylococcus aureus DnaA and in contrast to the low exchange rate of Escherichia coli DnaA. We suggest that organisms in which DnaA has a high rate of nucleotide exchange predominantly regulate the DNA binding activity of DnaA and that those with low rates of exchange regulate hydrolysis and exchange.
INTRODUCTION
Accurate and complete replication of DNA is essential for the propagation of genomic information. DNA replication in bacteria initiates from a single origin of replication (oriC) and depends on the conserved AAA+ ATPase DnaA (reviewed in references 13, 25, 27, 29, 35, and 45). DnaA binds both ATP and ADP, and DnaA-ATP is required for replication initiation (2, 17, 33, 47, 57). The ATP-bound form of DnaA forms oligomers that are important for promoting replication initiation (14, 18, 35, 41, 58). DnaA-ATP binds to sites in oriC and promotes the unwinding of the DNA unwinding element (DUE), which serves as a platform for assembly of the replicative helicase and the rest of the replication machinery. Wherever examined, the nucleotide-bound state of DnaA controls its activity (e.g., see references 39, 42, 57, and 63), and this can be affected by factors that alter nucleotide hydrolysis and exchange (16, 26, 28, 64). The rate of nucleotide exchange for purified Escherichia coli DnaA is relatively low, with a half-life of 45 min (57). This low inherent rate of exchange enables the modulation of DnaA activity by factors that increase the rate of nucleotide hydrolysis and/or exchange (11, 16, 26, 64).
In contrast to the widespread conservation of DnaA, other proteins required for replication initiation are less conserved. For example, steps involved in loading the replicative helicase at oriC are different between E. coli and Bacillus subtilis. In B. subtilis and other low-G+C Gram-positive bacteria, helicase loading requires the essential primosomal proteins DnaD, DnaB, and DnaI, in addition to DnaA (5–8, 12, 36, 37, 54, 59, 62, 65). DnaD is associated with oriC, and this association depends on DnaA (54, 59). Association of DnaB with oriC depends on DnaD, and finally, DnaI-mediated assembly of the helicase at oriC depends on DnaB. DnaD and DnaB bind both double- and single-stranded DNA, which may help stabilize the opening up of the origin of replication (7, 38, 68).
In addition to binding to sites in oriC, B. subtilis DnaA binds to sites in chromosomal regions outside oriC (4, 20, 24). DnaA functions as a transcription factor at some of these secondary binding regions (4, 9, 20, 24). Where tested, DnaD and DnaB are also found associated with these secondary DnaA binding regions, and this association depends on DnaA (60). However, in contrast to oriC, these secondary DnaA binding regions do not function as origins of replication, and there is no indication that they are capable of loading the replicative helicase (60).
Because of the association of DnaD with DnaA at multiple regions throughout the chromosome, we hypothesized that DnaD modulates the activity of DnaA. Some factors that affect the activity of DnaA, predominantly in E. coli, are known to alter its nucleotide-bound state (16, 26, 64). In contrast, regulators of B. subtilis DnaA, (e.g., YabA, Soj, and SirA) are known to alter its DNA binding properties (40, 53, 55, 66). Using purified proteins, we tested for effects of DnaD both on the ability of DnaA to bind DNA and on nucleotide exchange.
We found that the rate of nucleotide exchange for B. subtilis DnaA was relatively high, similar to that of DnaA from Staphylococcus aureus (32) and in contrast to the low rate of exchange for DnaA from E. coli (57). DnaD had no effect on the rate of exchange of B. subtilis DnaA. In contrast, DnaD had a marked effect on the ability of DnaA to bind DNA. Binding of DnaA-ATP to DNA fragments that contain multiple binding sites is normally highly cooperative (40, 41). We found that in the presence of DnaD, binding of DnaA to DNA was no longer cooperative and the apparent dissociation constant (Kd) for DnaA and DNA was reduced. We found that the ATPase activity of DnaA was not needed for these effects by DnaD, indicating that DnaD is not regulating the ATPase activity of DnaA. These effects of DnaD on the ability of DnaA to bind DNA are similar to the effects of YabA (40) and Soj (55), two other regulators of B. subtilis DnaA and replication initiation, and further substantiate the notion that modulation of cooperative binding and oligomerization of DnaA to DNA might be a common mechanism of regulation (40, 55).
MATERIALS AND METHODS
Purification of DnaA and DnaD.
B. subtilis DnaA (no tag) was produced in and purified from an E. coli dnaA null mutant, using a clone and strain provided by A. Albuzzi and W. F. Burkholder, essentially as described previously (17, 60). Protein was stored frozen (−80°C) in buffer containing 45 mM HEPES, pH 7.6, 0.5 mM EDTA, 10 mM magnesium acetate, 1 mM dithiothreitol (DTT), 700 mM potassium glutamate, and 20% sucrose. DnaD-His6 was produced in and purified from E. coli, essentially as described previously (60). Protein was stored at −80°C in buffer containing 50 mM Tris, pH 8, 0.1 mM EDTA, 1 mM DTT, 500 mm NaCl, and 10% glycerol. Proteins were quantified using absorbance at 280 nm.
Nucleotide exchange reactions.
Nucleotide exchange was measured using [α-32P]ATP or [14C]ADP. Exchange reaction mixtures contained 40 mM HEPES, pH 7.5, 10 mM magnesium acetate, 0.5 mM EDTA, 1 mM DTT, 150 mM potassium glutamate, 100 μg/ml bovine serum albumin (BSA), 10% glycerol, and 1 μM [α-32P]ATP or 1 μM [14C]ADP in the absence or presence of DnaD (600 nM). DnaA (300 nM) was incubated with either nucleotide for 2 h on ice in exchange buffer. Fifty microliters was removed at time zero to measure binding, and unlabeled ATP was added in excess (2 mM) and incubated at 37°C. Filter binding was used to measure the amount of radiolabeled nucleotide still bound to DnaA at each time point. Fifty-microliter aliquots were removed, placed on equilibrated nitrocellulose membranes (Millipore), and washed with buffer (40 mM HEPES, pH 7.5, 150 mM KCl, 10 mM magnesium acetate, 0.5 EDTA, 10 μg/ml BSA), and the amount of [32P]ATP or [14C]ADP remaining on the filter was measured in triplicate and averaged. The half-life was calculated using an exponential decay formula and plotted using GraphPad Prism 5 software.
Gel shift assays.
The DNA template from the oriC region (dnaA promoter region) used for the gel shift assays was an end-labeled 400-bp fragment, 382 bp of which corresponded to chromosomal DNA from the part of the oriC region that is upstream from dnaA. The fragment was generated by PCR using primers OCB23 (5′-CCGGAATTCTTTTTTTAGTATCCACAGAGG-3′) and OCB24 (5′-CGCGGATCCCTTTTCTTAGAAAATGGC-3′) and B. subtilis chromosomal DNA as the template. Allowing for one mismatch from the DnaA binding site consensus sequence (5′-TTATNCACA-3′), this fragment contains eight consensus DnaA binding sites. The DNA template from upstream of yydA used for gel shift assays was an end-labeled 228-bp fragment generated by PCR using primers WKS167 (5′-CCCACAGCCTGTGAATTATG-3′) and WKS168 (5′-CGTAGGCCGAAAGTCGTTTG-3′). Allowing for 1 mismatch, this fragment contains four consensus DnaA binding sites. It is important to note that the sequence requirements for binding DnaA are not well defined and that this estimate of the number of potential binding sites is likely an underestimate as DnaA is also likely to bind sequences with more than one mismatch from consensus (17).
The PCR products were purified on columns (Qiagen) and end labeled with [γ-32P]ATP using T4 polynucleotide kinase. The labeled DNA fragment was separated from free ATP with a G50 column (GE). DnaA was incubated with 2.5 mM ATP for 2 h on ice before being used in gel shift reaction mixtures containing 40 mM HEPES, pH 7.6, 10 mM KCl, 140 mM potassium glutamate, 10 mM magnesium acetate, 2.5 mM ATP, 0.5 mM EDTA, 1 mM DTT, 50 μg/ml BSA, 20% glycerol, and 50 pM DNA probe in the presence or absence of DnaD-His6 (300 nM) for 20 min at room temperature.
To determine an appropriate concentration of DnaD to use, we measured the effects of different concentrations of DnaD-His6 on the electrophoretic mobility of the DNA fragment from the oriC region in the presence of 10 nM DnaA-ATP, or with no DnaA (Fig. 1). DnaD-His6 was used at 25, 50, 100, 200, and 300 nM. At these concentrations of DnaD-His6, there was little or no change in the electrophoretic mobility of the DNA fragment in the absence of DnaA. However, in the presence of DnaA, there was a change in the gel shift beginning at 50 nM DnaD-His6. We chose to use 300 nM DnaD because there was a large change in the gel shift in the presence of DnaA but little or no change in its absence.
Fig 1.
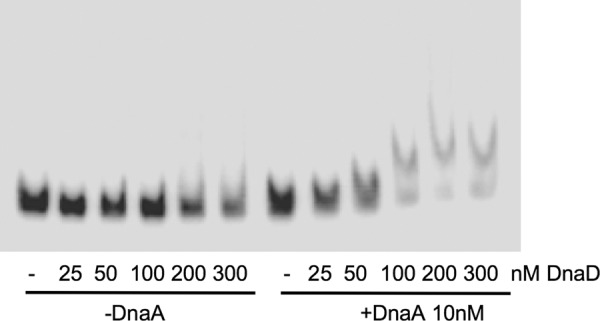
Effects of different concentrations of DnaD on DnaA binding to oriC. Representative gel of the radiolabeled DNA probe from the oriC region with different amounts of DnaD-His, in the absence of DnaA (left six lanes) or the presence of 10 nM DnaA-ATP (right six lanes). Concentrations of DnaD-His are 0 (−), 25, 50, 100, 200, or 300 nM and are indicated below each lane.
The binding reactions were run on a 5% polyacrylamide gel with 2.5% glycerol run in 0.5× Tris-buffered EDTA (TBE) at approximately 12 V/cm for 3 h. Gels were imaged on a Typhoon scanner (GE Healthcare), and GraphPad Prism 5 software was used to plot binding curves. Data were plotted and fitted to the Hill equation [y = (m1 × xn)/(Kdn + xn)], where y is the % DNA bound at any given DnaA concentration, x is the DnaA concentration, m1 is maximal binding (100%), Kd is the apparent dissociation constant (concentration at which 50% of DNA is bound, determined from data), and n is the Hill coefficient. All experiments were done in triplicate. Data presented are averages of triplicates ± standard errors.
ATPase assays.
ATPase assays were carried out using [γ-32P]ATP as the substrate, and products were separated by thin-layer chromatography (TLC). Reaction mixtures (50 μl) contained 100 nM DnaA, 50 mM Tris (pH 7), 5 mM magnesium acetate, 1 mm DTT, 100 ng/ml BSA, 10% glycerol, and 1 μM ATP (1/1,000 [γ-32P]ATP) and, where added, 1 μM PCR product from the oriC fragment containing eight DnaA binding sites. At indicated time points, reactions were stopped with 2 volumes of stop buffer (0.5% SDS, 250 mM NaCl, 25 mM EDTA) and reaction mixtures were spotted on cellulose TLC plates. Products were separated with 0.5 M LiCl, 1 M formic acid. Plates were dried and exposed to a phosphostorage screen. Free radiolabeled orthophosphate and ATP were measured, and percent hydrolysis was calculated. All experiments were done in triplicate, and data are presented as the averages ± standard errors.
Nucleotide binding assays.
Nucleotide binding was measured using [α-32P]ATP. DnaA (100 nM) and ATP (1 μM) were incubated for 30 min at room temperature in 50-μl reaction mixtures containing 40 mM HEPES, pH 7.5, 10 mM magnesium acetate, 0.5 mM EDTA, 1 mM DTT, 150 mM potassium glutamate, 100 μg/ml BSA, and 10% glycerol. The reaction mixtures were placed on equilibrated nitrocellulose membranes (Millipore) and washed with buffer (40 mM HEPES, pH 7.5, 150 mM KCl, 10 mM magnesium acetate, 0.5 mM EDTA, 10 μg/ml BSA), and radioactivity was measured by filter binding as described for the nucleotide exchange assay. All experiments were done in triplicate, and data are presented as the averages ± standard errors. The apparent Kd for ATP binding to DnaA was 29 nM (data not shown), similar to previous reports (17).
RESULTS
DnaD does not affect nucleotide exchange for DnaA.
We found that the rate of nucleotide exchange for DnaA was relatively high and that DnaD had no detectable effect on this rate. To measure nucleotide exchange, we incubated purified DnaA with radiolabeled ADP or ATP. The amount of radioactive nucleotide that remained associated with DnaA was measured at various times after addition of excess unlabeled ATP at 37°C, and the amount of nucleotide that was released was calculated. For both DnaA-ADP (Fig. 2A) and DnaA-ATP (Fig. 2B), the radioactive nucleotide was released with a half-life of 5 min. The addition of DnaD-His6 (Materials and Methods) had no detectable effect on this half-life (Fig. 2). Based on these results, we conclude that the rate of nucleotide exchange for B. subtilis DnaA is relatively high compared to the 45-min half-life of exchange for E. coli DnaA (57) and that DnaA-ATP is regenerated from DnaA-ADP in the absence of any other cellular factors. The relatively high rate of nucleotide exchange is similar to that of DnaA from S. aureus (32).
Fig 2.
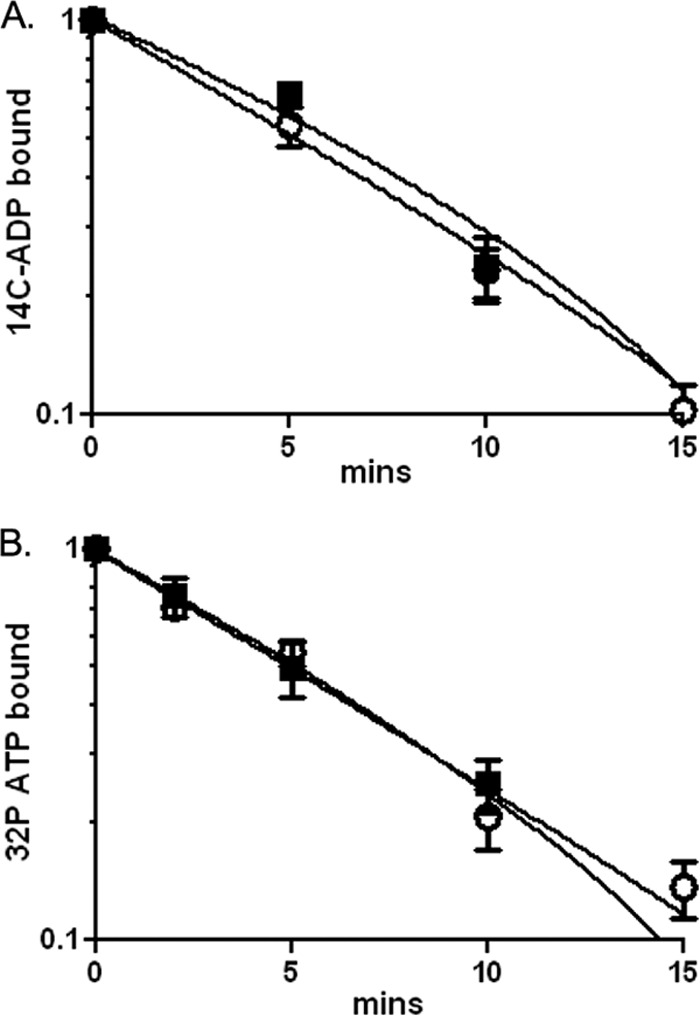
DnaD does not affect nucleotide exchange by DnaA. The amount of [14C]ADP (A) or [32P]ATP (B) bound to DnaA (300 nM) at various times after addition of unlabeled ATP (2 mM) at 37°C was measured by filter binding in the absence (open circles) and presence (filled squares) of DnaD-His6 (600 nM). Data are averages of triplicates ± standard errors and are normalized to the starting amount of radioactivity in the absence of unlabeled ATP.
DnaD increases affinity and reduces cooperativity of DnaA-ATP binding to DNA.
Given the in vivo association of DnaD with DnaA binding sites around the chromosome (60), we tested for effects of DnaD on the ability of DnaA to bind DNA using gel electrophoretic mobility shift assays. DnaA-ATP bound to a DNA fragment from the oriC region with an apparent dissociation constant (Kd) of 27 nM (Fig. 3A and C). Binding was highly cooperative and had a Hill coefficient of 8. These results are consistent with previous findings (40).
Fig 3.
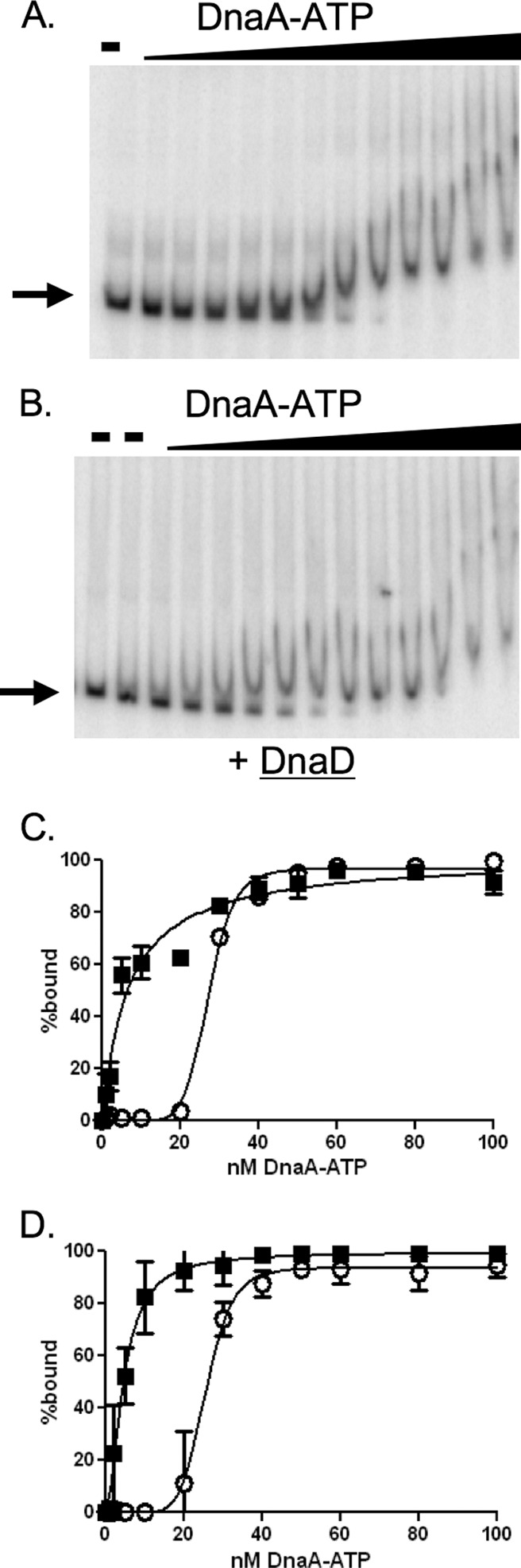
DnaD inhibits cooperative binding of DnaA to DNA. Representative gels and binding curves measuring binding of DnaA-ATP to DNA (50 pM) with and without purified DnaD-His6 (300 nM) are shown. DnaA concentrations used were 0, 1, 2, 5, 10, 20, 30, 40, 50, 60, 80, 100, and 200 nM. (A and B) Representative gels with increasing concentrations of DnaA-ATP incubated with template DNA from the oriC region in the absence (A) or presence (B) of DnaD-His6. Probe with no added protein is shown in the first lane of both panels. Probe with DnaD-His6 and no DnaA is shown in the second lane of panel B. (C and D) Data from three independent gel shift assays using template DNA from the oriC region (C) or the yydA region (D) are plotted as percent DNA bound versus the concentration of DnaA-ATP, in the absence (open circles) and presence (filled squares) of DnaD-His6. In experiments with the DNA fragment from the oriC region (C), the calculated Hill coefficient for DnaA-ATP was 8.6 in the absence of DnaD-His6 and 1 in the presence of DnaD-His6. The apparent Kd for DnaA-ATP was 27 nM in the absence and 6.6 nM in the presence of DnaD-His6. In experiments with the DNA fragment from the yydA region (D), the calculated Hill coefficient for DnaA-ATP was 6 in the absence of DnaD-His6 and 1 in the presence of DnaD-His6. The apparent Kd for DnaA-ATP was 25 nM in the absence and 5 nM in the presence of DnaD-His6.
We measured the effects of DnaD-His6 on the ability of DnaA to bind to DNA. DnaD-His6 alone (300 nM) did not have detectable binding activity under these assay conditions (Fig. 1 and 3B), as previously reported (60). However, addition of DnaD-His6 to DNA and DnaA-ATP substantially altered the binding properties of DnaA-ATP to DNA. The apparent dissociation constant in the presence of DnaD was approximately 7 nM, compared to 27 nM in the absence of DnaD. There was a concomitant loss of cooperative binding as the Hill coefficient decreased from 8 in the absence of DnaD to 1 in its presence (Fig. 3; Table 1). This decrease in the Hill coefficient and decrease in the apparent binding constant are consistent with DnaA-ATP binding independently to multiple sites in the DNA fragment.
Table 1.
Summary of DNA binding by wild-type and mutant DnaAa
| Parameter | Mean ± SD in absence or presence of DnaDb |
|||||
|---|---|---|---|---|---|---|
| DnaA |
DnaA(K157A) |
DnaA(D215A) |
||||
| − | + | − | + | − | + | |
| Apparent Kd (nM) | 27 ± 0.4 | 6.6 ± 1 | 36 ± 0.8 | 17 ± 1 | 12 ± 0.3 | 5.7 ± 0.3 |
| Hill coefficient | 8.6 ± 0.9 | 1.0 ± 0.1 | 5.3 ± 0.5 | 2.7 ± 0.4 | 5.2 ± 0.4 | 2.2 ± 0.2 |
Results presented in Fig. 3C, 4A, and 4B are summarized. The DNA template was a fragment containing sequences upstream from oriC with eight DnaA binding sites (≤1 mismatch from consensus).
DnaD-His6 was either added at 300 nM (+) or absent (−).
The DNA fragment used in these experiments was derived from sequences upstream of dnaA in the oriC region. We found that DnaD had a similar effect on the ability of DnaA-ATP to bind to a DNA fragment from a different chromosomal region. In addition to the oriC region, DnaA is found associated with several chromosomal regions in vivo (20, 24), including the region between yydA and yydS (4, 20, 24), two genes of unknown function. This region has also been implicated in regulating DNA replication by recruiting DnaA away from oriC (51). Therefore, we isolated a DNA fragment from the region upstream of yydA that contains four DnaA binding sites with ≤1 mismatch to the consensus and tested the ability of DnaD to affect DnaA binding to this region. DnaA-ATP bound to this fragment with an apparent dissociation constant of 25 nM (Fig. 3D). Binding was cooperative with a Hill coefficient of 6, indicative of binding to 6 possible DnaA sites. Addition of DnaD-His6 (300 nM) to these reaction mixtures decreased the apparent dissociation constant to 5 nM and reduced the Hill coefficient to 1, indicating that there was essentially no cooperative binding in the presence of DnaD. These effects are similar to those on the binding of DnaA-ATP to the DNA fragment from the oriC region upstream from dnaA (Fig. 3). Together, these results indicate that DnaD affects binding of DnaA to DNA fragments from the oriC region and at least one origin-distal region. Since DnaD is found at multiple chromosomal regions bound by DnaA in vivo (60), we suspect that DnaD similarly affects DnaA binding at these regions.
We have not been able to detect any changes in the footprint of DnaA on DNA in the presence compared to that in the absence of DnaD (unpublished results). This is likely because at the high concentrations of DnaA needed to observe a footprint (17), the addition of DnaD has no detectable effect on binding (Fig. 3). At lower concentrations of DnaA, where DnaD does influence binding, we suspect that there is a population of DNA molecules with different sites occupied by DnaA, thereby not producing any obvious protection in a footprint experiment but still capable of generating a change in electrophoretic mobility.
Characterization of DnaA mutants defective in ATPase activity.
Since the rate of ADP exchange for ATP of B. subtilis DnaA is relatively high and unaffected by DnaD, and DnaD affects the ability of DnaA-ATP to bind DNA, we hypothesized that DnaD would affect DnaA mutants that are defective in nucleotide hydrolysis and/or binding. To test this, we made two different mutations in dnaA. One mutation is in the conserved Walker A motif; changes the lysine at amino acid 157 to alanine, DnaA(K157A); and is predicted to reduce nucleotide binding (21). We also made a mutation in the conserved Walker B motif that changes the glutamate at amino acid 215 to alanine, DnaA(D215A), and is predicted to alter nucleotide hydrolysis (21). We purified the mutant proteins and tested them in vitro.
Both DnaA(K157A) and DnaA(D215A) were defective in ATP hydrolysis. We measured the rate of ATP hydrolysis by using [γ-32P]ATP and measuring the release of orthophosphate (Materials and Methods). The rate of ATP hydrolysis by wild-type DnaA was 1.8 mol of ATP hydrolyzed per mole of DnaA per hour. Upon addition of DNA, the rate of hydrolysis increased approximately 6-fold to 12 mol of ATP hydrolyzed per mole of DnaA per hour. These rates of ATP hydrolysis and the effects of DNA are consistent with previously published data for DnaA from E. coli and S. aureus (32, 57). In contrast to the wild-type protein, DnaA(K157A) and DnaA(D215A) had rates of ATP hydrolysis of approximately 0.07 and 0.1 mol of ATP per mole of DnaA per hour, respectively (Table 2).
Table 2.
Summary of ATP hydrolysis and binding by wild-type and mutant DnaAa
| Protein | ATPase | ATPase + DNA | ATP binding |
|---|---|---|---|
| DnaA | 1.8 ± 0.25 | 11.9 ± 2.2 | 0.4 ± 0.05 |
| DnaA(K157A) | 0.07 ± 0.002 | 0.3 ± 0.04 | 0.02 ± 0.01 |
| DnaA(D215A) | 0.1 ± 0.2 | 0.3 ± 0.01 | 0.4 ± 0.11 |
The rate of ATP hydrolysis (ATPase) is presented as the number of moles of ATP hydrolyzed per mole of DnaA per hour. Where indicated, the 400-bp DNA fragment from the oriC region that was used for the gel shift assays was added (1 μM). The amount of ATP bound (ATP binding) is presented as moles of ATP per mole of DnaA.
As expected, the DnaA(K157A) mutant was defective and the DnaA(D215A) mutant had normal ATP binding. At saturating ATP concentrations, we found that wild-type protein bound 0.4 molecules of ATP per molecule of DnaA (Table 2), consistent with previous reports for DnaA from E. coli (0.48 and 0.55) (10, 57) but greater than a previous report for B. subtilis DnaA (0.17) (17). The DnaA(D215A) mutant had ATP binding (Table 2) that was indistinguishable from that of the wild-type protein. In contrast, the DnaA(K157A) mutant appeared to bind 0.02 molecules of ATP per molecule of DnaA (Table 2), consistent with little or no ATP binding.
DnaD affects DnaA mutants defective in ATPase activity and nucleotide binding.
We determined the effects of DnaD on the ability of the mutant DnaA proteins to bind DNA and compared the binding properties to those of wild-type DnaA. The mutant DnaA that binds ATP but is defective in hydrolysis [DnaA(D215A)] bound DNA with an apparent Kd of 12 nM, compared to 27 nM for the wild-type protein (Fig. 4A). Binding to DNA was cooperative and had a Hill coefficient of approximately 5 (Fig. 4A). Addition of DnaD reduced the apparent Kd to approximately 6 nM and the Hill coefficient to approximately 2 (Fig. 4A; Table 1). The mutant DnaA that is defective in binding ATP [DnaA(K157A)] has an apparent Kd of 36 nM and a Hill coefficient of approximately 5 (Fig. 4B). Addition of DnaD reduced the apparent Kd to 17 nM and the Hill coefficient to approximately 3 (Fig. 4B; Table 1). Together, these results indicate that the effects of DnaD on the ability of DnaA to bind DNA do not require ATP binding or hydrolysis.
Fig 4.
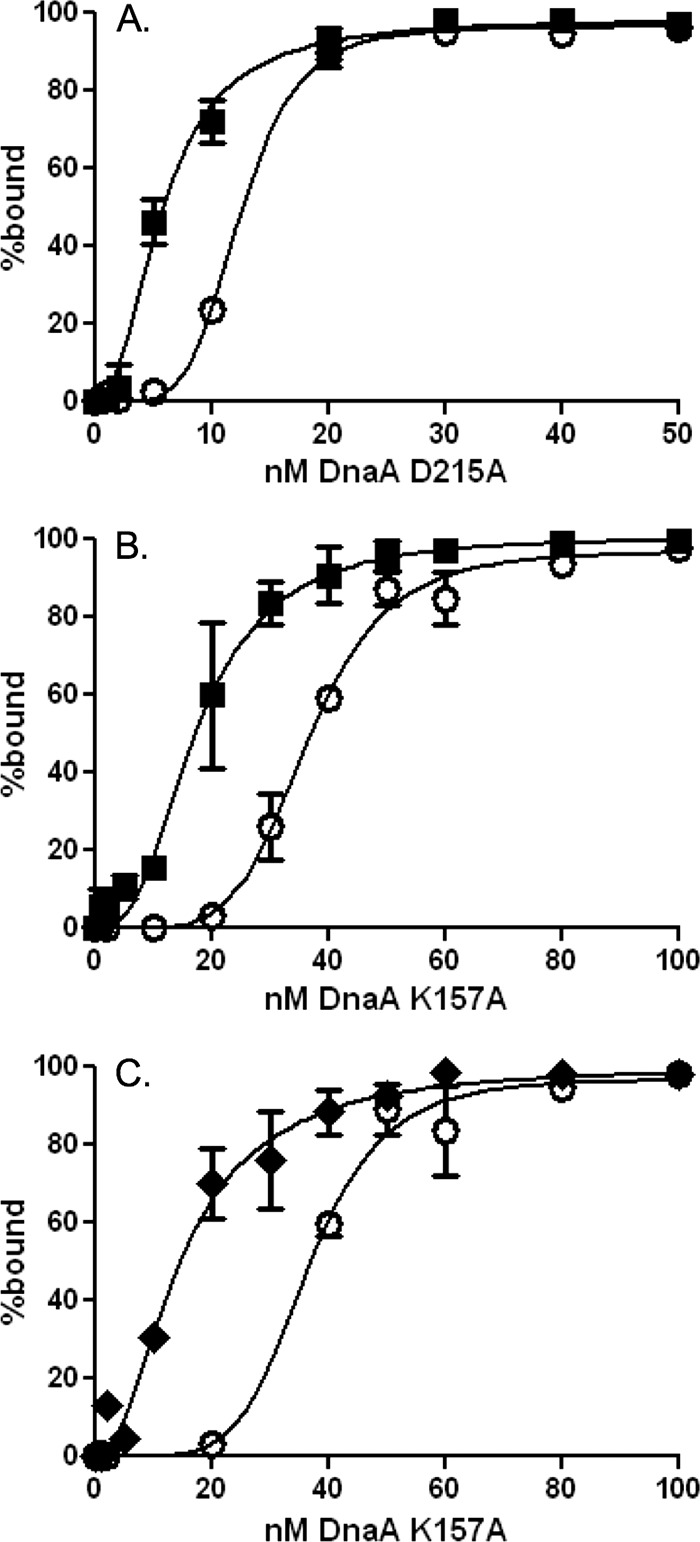
Effects of DnaD and YabA on DnaA binding to DNA are independent of ATPase activity. Binding curves of DnaA mutants defective in ATP hydrolysis, DnaA(D215A)-ATP (A), and ATP binding, DnaA(K157A) (B and C), within the DNA fragment from the oriC region. DnaA concentrations tested were 0, 1, 2, 5, 10, 20, 30, 40, 50, 60, 80, 100, and 200 nM. (A and B) Binding in the absence (open circles) and presence (filled squares) of DnaD-His6 (300 nM). (C) Binding in the absence (open circles) and presence (filled diamonds) of His6-YabA (700 nM). For the DnaA mutant defective in ATP binding, DnaA(K157A), the presence of YabA reduced the Hill coefficient from 5.6 to 2 and the apparent Kd from 36 nM to 14.7 nM.
Previously, we found that YabA, a negative regulator of replication initiation, inhibited cooperativity of DnaA while reducing the apparent Kd (40). We tested the effects of YabA on the DnaA(K157A) mutant and found that the addition of YabA (700 nM) reduced the apparent Kd from 36 nM to 14.7 nM and the Hill coefficient from 5.6 to 2 (Fig. 4C). These results are consistent with the previous reports of the effects of YabA on the ability of DnaA to bind DNA (40), are comparable to the effects of DnaD, and suggest that the two may regulate DnaA by similar mechanisms.
DISCUSSION
In many bacteria, the conserved replication initiator DnaA is a target for the control of replication initiation. DnaA is also a transcription factor, and many of the factors that modulate its activity in replication initiation are likely to affect its activity as a transcription factor. Mechanisms regulating DnaA have been most studied with E. coli and its close relatives. However, the proteins and mechanisms used by E. coli are largely limited to the proteobacteria and are not found in Gram-positive organisms like B. subtilis. Likewise, some of the proteins and mechanisms used by B. subtilis are not found in E. coli and other proteobacteria. Results presented here indicate that B. subtilis DnaD is a regulator of DnaA. Below, we discuss the possible role of DnaD in regulation of DnaA and the different properties of DnaA that might lead to differences in its regulation in different organisms.
Role of DnaD in replication initiation and regulation of DnaA.
DnaD is found in B. subtilis and other low-G+C-content Gram-positive bacteria but not in E. coli and other Gram-negative bacteria. DnaD is required for replication initiation (5, 6), interacts with DnaA (23, 60), and is needed to recruit the helicase loading protein DnaB to oriC (7, 59). DnaD is found associated with the oriC region of the chromosome (54, 59) and many other chromosomal regions that also bind DnaA (60). The association of DnaD with these regions is dependent on DnaA (59, 60).
We found that DnaD decreases the apparent Kd of DnaA-ATP for DNA and also decreases the cooperativity of DnaA binding to DNA. These effects could indicate that DnaD functions either as an activator or as a repressor of DnaA, or both. For many regulators, the phenotype caused by a null mutation typically indicates that regulation is positive or negative. Unfortunately, dnaD is essential and null mutations are not viable. Temperature-sensitive and other conditional loss-of-function dnaD mutants result in increased association of DnaA with chromosomal regions (4, 20), consistent with DnaD normally functioning to reduce binding of DnaA to DNA. However, these mutations also lead to a decrease in replication initiation. Analyzing effects of overexpressing DnaD is also problematic because overexpression of DnaD causes a severe growth defect that is independent of replication initiation from oriC (38). Mutations in other genes that cause a decrease in replication initiation also cause an increase in DnaA activity (4, 20), making it difficult to discern if the effects of DnaD are direct, are due to changes in replication initiation, or both.
It is well established that DnaD is essential and has a positive role in replication initiation. During replication initiation, DnaA-ATP binds cooperatively to many sites in oriC (e.g., see references 18, 39, and 41). By analogy to E. coli, it is likely that the temporal order of binding of DnaA-ATP to sites in oriC is important for open complex formation (39, 41). In B. subtilis, association of DnaD with the oriC region requires DnaA. DnaD is then required for association of DnaB and subsequent loading of the replicative helicase (6, 7, 54, 59). It is possible that the effects of DnaD on the binding of DnaA to oriC (increase in apparent affinity) could also stimulate replication initiation by maintaining DnaA bound to oriC. However, we think that this is unlikely if ordered and cooperative binding of DnaA is important for replication initiation.
In addition to its known positive role in replication initiation, we postulate that DnaD also serves to negatively regulate replication initiation through its effects on the ability of DnaA to bind DNA. The ability of DnaD to inhibit cooperative binding of DnaA-ATP to DNA is similar to the effect of two other negative regulators of replication initiation, YabA (40) and Soj (55).
We postulate that immediately before or after replication initiation, DnaD helps keep DnaA inactive at oriC by inhibiting cooperative binding. This activity of DnaD as a negative regulator of DnaA could be modulated by changes in the amount of available DnaD during the replication cycle. For example, the amount of DnaD available to interact with DnaA at oriC could change during a replication cycle, perhaps due to association of DnaD with other proteins or chromosomal regions (60) or due to possible changes in its synthesis or stability. We have not yet tested these possibilities.
It is also possible that inhibition of DnaA by DnaD is relieved by the replication initiation protein DnaB. That is, DnaD might be keeping DnaA inactive at oriC until proper assembly of additional parts of the replication initiation complex. For example, DnaD is needed to recruit DnaB (part of the helicase loader) to oriC (59) and other chromosomal regions bound by DnaA (60). Association of DnaB might alter interactions between DnaA and DnaD, relieving the putative inhibitory effect mediated by DnaD, enabling replication initiation. Preliminary attempts to test this in vitro have not been successful, perhaps because of a possible role of the membrane in interactions between DnaB and DnaD (54).
Emerging theme in the regulation of DnaA.
One of the emerging themes of regulation of B. subtilis DnaA and replication initiation is the role of regulators that directly alter the ability of DnaA to bind DNA. Including DnaD, there are now at least four regulators of this type. The production and activities of the regulators are differentially controlled, and each regulator is likely to be important at different times during the growth and replication cycles. The regulators are also likely to be partly redundant.
(i) SirA.
SirA is a negative regulator of DnaA that is produced during entry into stationary phase and the initiation of sporulation (52, 66). SirA likely interacts with domain I of DnaA, and it inhibits the ability of DnaA to bind sequences in oriC in vivo (53).
(ii) YabA.
YabA was identified in a yeast two-hybrid screen for interactors with replication proteins (48). YabA is produced during growth and interacts with both DnaA and DnaN, the processivity clamp of DNA polymerase (48, 49). Like DnaD, YabA reduces the apparent Kd and cooperativity of DnaA binding to DNA in vitro, and these effects are independent of the ATPase activity of DnaA (40). Also like DnaD, YabA is found associated with chromosomal regions that are bound by DnaA in vivo and this association is DnaA dependent (40). In addition, YabA is found associated with replication forks during ongoing replication (19, 49, 61), and this association is likely due to interaction between YabA and DnaN. The interaction between DnaN and YabA likely functions to reduce the ability of YabA to negatively regulate DnaA and replication initiation (40, 49, 61) and could couple relief of YabA-mediated inhibition to the release of DnaN from the replisome during replication termination (40).
(iii) Soj.
B. subtilis Soj is expressed during growth and is a member of the ParA family of chromosome partitioning proteins involved in chromosome and plasmid partitioning. Soj is a negative regulator of replication initiation (34) and DnaA (46, 55, 56). Soj inhibits the ability of DnaA to form a helix on DNA, independently of the ATPase activity of DnaA (55). The inhibitory effects of Soj on replication initiation appear to be relieved by Spo0J (56), perhaps coupling an aspect of replication control to chromosome organization or partitioning (46, 56).
Diverse mechanisms controlling DnaA in different organisms.
The mechanisms used to control DnaA are diverse. Using E. coli and B. subtilis as examples, there are some common mechanisms and some striking differences. In both of these organisms, and many others, DnaA represses its own transcription (e.g., see references 1, 3, 20, 50, and 67), establishing a homeostatic regulatory loop. In addition, there are DnaA binding sites outside oriC that function to titrate DnaA away from oriC (51, 60). In E. coli, the datA locus binds DnaA and appears to help limit the amount of DnaA available for replication initiation (31, 43, 44). Similarly, in B. subtilis, there are six chromosomal regions outside oriC that have clusters of DnaA binding sites (20, 24, 51). At least one of these clusters seems to function to help limit the amount of DnaA available for replication initiation (51).
One of the most notable differences between regulation of E. coli DnaA and that of B. subtilis DnaA is the stimulation of nucleotide binding and hydrolysis in E. coli. One of the primary mechanisms used by E. coli to inhibit the activity of DnaA is called RIDA (regulatory inactivation of DnaA) and uses a protein called Hda (26, 28). Hda interacts with E. coli DnaN (β-clamp) and stimulates nucleotide hydrolysis by DnaA, thereby stimulating conversion of the replication-competent DnaA-ATP to the inactive DnaA-ADP (64). E. coli also has specific DnaA-reactivating sequences that directly promote nucleotide exchange to generate DnaA-ATP from DnaA-ADP (16). The stimulated rate of nucleotide exchange for E. coli DnaA (15) is about the same as the basal rate for B. subtilis DnaA. E. coli also has a protein called DiaA that stimulates replication initiation by stimulating binding by DnaA-ATP (22, 30).
In contrast to the mechanisms used by E. coli to regulate DnaA and replication initiation, B. subtilis is not known to regulate nucleotide hydrolysis or exchange. Rather, the primary mechanisms for controlling B. subtilis DnaA affect its binding to DNA (40, 53, 55), probably by inhibiting formation of multimeric DnaA structures (helix formation) on the DNA and preventing cooperative binding to sites in oriC (40, 55). No known regulator of DnaA in B. subtilis affects nucleotide hydrolysis or exchange.
Clearly, different organisms use different mechanisms to control the activity of DnaA and replication initiation. We suggest that the multiple mechanisms may have evolved in different organisms, in part, due to the different rates of nucleotide exchange. For organisms like E. coli where DnaA has a relatively low rate of nucleotide exchange, stimulation of nucleotide hydrolysis and exchange is likely to be a predominant mode of regulation. In contrast, for organisms like B. subtilis and S. aureus where DnaA has a relatively high rate of nucleotide exchange, the predominant modes of regulation of DnaA affect DNA binding and cooperativity.
ACKNOWLEDGMENTS
We thank T. Baker, A. Olivares, and members of the Grossman lab for useful discussions and C. Lee, J. L. Smith, C. Seid, M. Laub, and L. Simmons for comments on the manuscript.
This work was supported, in part, by Public Health Service grant GM41934 to A.D.G.
Footnotes
Published ahead of print 20 July 2012
REFERENCES
- 1. Atlung T, Clausen ES, Hansen FG. 1985. Autoregulation of the dnaA gene of Escherichia coli K12. Mol. Gen. Genet. 200:442–450 [DOI] [PubMed] [Google Scholar]
- 2. Bramhill D, Kornberg A. 1988. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 52:743–755 [DOI] [PubMed] [Google Scholar]
- 3. Braun RE, O'Day K, Wright A. 1985. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell 40:159–169 [DOI] [PubMed] [Google Scholar]
- 4. Breier AM, Grossman AD. 2009. Dynamic association of the replication initiator and transcription factor DnaA with the Bacillus subtilis chromosome during replication stress. J. Bacteriol. 191:486–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bruand C, Ehrlich SD, Janniere L. 1995. Primosome assembly site in Bacillus subtilis. EMBO J. 14:2642–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bruand C, Farache M, McGovern S, Ehrlich SD, Polard P. 2001. DnaB, DnaD and DnaI proteins are components of the Bacillus subtilis replication restart primosome. Mol. Microbiol. 42:245–255 [DOI] [PubMed] [Google Scholar]
- 7. Bruand C, et al. 2005. Functional interplay between the Bacillus subtilis DnaD and DnaB proteins essential for initiation and re-initiation of DNA replication. Mol. Microbiol. 55:1138–1150 [DOI] [PubMed] [Google Scholar]
- 8. Bruck I, O'Donnell M. 2000. The DNA replication machine of a gram-positive organism. J. Biol. Chem. 275:28971–28983 [DOI] [PubMed] [Google Scholar]
- 9. Burkholder WF, Kurtser I, Grossman AD. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104:269–279 [DOI] [PubMed] [Google Scholar]
- 10. Carr KM, Kaguni JM. 1996. The A184V missense mutation of the dnaA5 and dnaA46 alleles confers a defect in ATP binding and thermolability in initiation of Escherichia coli DNA replication. Mol. Microbiol. 20:1307–1318 [DOI] [PubMed] [Google Scholar]
- 11. Crooke E, Castuma CE, Kornberg A. 1992. The chromosome origin of Escherichia coli stabilizes DnaA protein during rejuvenation by phospholipids. J. Biol. Chem. 267:16779–16782 [PubMed] [Google Scholar]
- 12. Davey MJ, O'Donnell M. 2003. Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 13:R594–R596 [DOI] [PubMed] [Google Scholar]
- 13. Duderstadt KE, Berger JM. 2008. AAA+ ATPases in the initiation of DNA replication. Crit. Rev. Biochem. Mol. Biol. 43:163–187 [DOI] [PubMed] [Google Scholar]
- 14. Felczak MM, Kaguni JM. 2004. The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J. Biol. Chem. 279:51156–51162 [DOI] [PubMed] [Google Scholar]
- 15. Fujimitsu K, Katayama T. 2004. Reactivation of DnaA by DNA sequence-specific nucleotide exchange in vitro. Biochem. Biophys. Res. Commun. 322:411–419 [DOI] [PubMed] [Google Scholar]
- 16. Fujimitsu K, Senriuchi T, Katayama T. 2009. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 23:1221–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fukuoka T, Moriya S, Yoshikawa H, Ogasawara N. 1990. Purification and characterization of an initiation protein for chromosomal replication, DnaA, in Bacillus subtilis. J. Biochem. 107:732–739 [DOI] [PubMed] [Google Scholar]
- 18. Fuller RS, Funnell BE, Kornberg A. 1984. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell 38:889–900 [DOI] [PubMed] [Google Scholar]
- 19. Goranov AI, Breier AM, Merrikh H, Grossman AD. 2009. YabA of Bacillus subtilis controls DnaA-mediated replication initiation but not the transcriptional response to replication stress. Mol. Microbiol. 74:454–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goranov AI, Katz L, Breier AM, Burge CB, Grossman AD. 2005. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc. Natl. Acad. Sci. U. S. A. 102:12932–12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hanson PI, Whiteheart SW. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519–529 [DOI] [PubMed] [Google Scholar]
- 22. Ishida T, et al. 2004. DiaA, a novel DnaA-binding protein, ensures the timely initiation of Escherichia coli chromosome replication. J. Biol. Chem. 279:45546–45555 [DOI] [PubMed] [Google Scholar]
- 23. Ishigo-Oka D, Ogasawara N, Moriya S. 2001. DnaD protein of Bacillus subtilis interacts with DnaA, the initiator protein of replication. J. Bacteriol. 183:2148–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ishikawa S, et al. 2007. Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method. DNA Res. 14:155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaguni JM. 2006. DnaA: controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 60:351–375 [DOI] [PubMed] [Google Scholar]
- 26. Katayama T, Kubota T, Kurokawa K, Crooke E, Sekimizu K. 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94:61–71 [DOI] [PubMed] [Google Scholar]
- 27. Katayama T, Ozaki S, Keyamura K, Fujimitsu K. 2010. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8:163–170 [DOI] [PubMed] [Google Scholar]
- 28. Kato J, Katayama T. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kawakami H, Katayama T. 2010. DnaA, ORC, and Cdc6: similarity beyond the domains of life and diversity. Biochem. Cell Biol. 88:49–62 [DOI] [PubMed] [Google Scholar]
- 30. Keyamura K, et al. 2007. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 21:2083–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kitagawa R, Ozaki T, Moriya S, Ogawa T. 1998. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12:3032–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kurokawa K, et al. 2009. Rapid exchange of bound ADP on the Staphylococcus aureus replication initiation protein DnaA. J. Biol. Chem. 284:34201–34210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T. 1999. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 18:6642–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee PS, Grossman AD. 2006. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol. Microbiol. 60:853–869 [DOI] [PubMed] [Google Scholar]
- 35. Leonard AC, Grimwade JE. 2010. Regulating DnaA complex assembly: it is time to fill the gaps. Curr. Opin. Microbiol. 13:766–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Y, et al. 2004. Identification of temperature-sensitive dnaD mutants of Staphylococcus aureus that are defective in chromosomal DNA replication. Mol. Genet. Genomics 271:447–457 [DOI] [PubMed] [Google Scholar]
- 37. Marsin S, McGovern S, Ehrlich SD, Bruand C, Polard P. 2001. Early steps of Bacillus subtilis primosome assembly. J. Biol. Chem. 276:45818–45825 [DOI] [PubMed] [Google Scholar]
- 38. Marston FY, et al. 2010. When simple sequence comparison fails: the cryptic case of the shared domains of the bacterial replication initiation proteins DnaB and DnaD. Nucleic Acids Res. 38:6930–6942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McGarry KC, Ryan VT, Grimwade JE, Leonard AC. 2004. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. U. S. A. 101:2811–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Merrikh H, Grossman AD. 2011. Control of the replication initiator DnaA by an anti-cooperativity factor. Mol. Microbiol. 82:434–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miller DT, et al. 2009. Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proc. Natl. Acad. Sci. U. S. A. 106:18479–18484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mizushima T, et al. 1998. Site-directed mutational analysis for the ATP binding of DnaA protein. Functions of two conserved amino acids (Lys-178 and Asp-235) located in the ATP-binding domain of DnaA protein in vitro and in vivo. J. Biol. Chem. 273:20847–20851 [DOI] [PubMed] [Google Scholar]
- 43. Morigen, Lobner-Olesen A, Skarstad K. 2003. Titration of the Escherichia coli DnaA protein to excess datA sites causes destabilization of replication forks, delayed replication initiation and delayed cell division. Mol. Microbiol. 50:349–362 [DOI] [PubMed] [Google Scholar]
- 44. Morigen, Molina F, Skarstad K. 2005. Deletion of the datA site does not affect once-per-cell-cycle timing but induces rifampin-resistant replication. J. Bacteriol. 187:3913–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mott ML, Berger JM. 2007. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 5:343–354 [DOI] [PubMed] [Google Scholar]
- 46. Murray H, Errington J. 2008. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135:74–84 [DOI] [PubMed] [Google Scholar]
- 47. Nishida S, et al. 2002. A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidence from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J. Biol. Chem. 277:14986–14995 [DOI] [PubMed] [Google Scholar]
- 48. Noirot-Gros MF, et al. 2002. An expanded view of bacterial DNA replication. Proc. Natl. Acad. Sci. U. S. A. 99:8342–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Noirot-Gros MF, et al. 2006. Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 103:2368–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ogura Y, Imai Y, Ogasawara N, Moriya S. 2001. Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J. Bacteriol. 183:3833–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Okumura H, et al. 2012. Regulation of chromosomal replication initiation by oriC-proximal DnaA-box clusters in Bacillus subtilis. Nucleic Acids Res. 40:220–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rahn-Lee L, Gorbatyuk B, Skovgaard O, Losick R. 2009. The conserved sporulation protein YneE inhibits DNA replication in Bacillus subtilis. J. Bacteriol. 191:3736–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rahn-Lee L, Merrikh H, Grossman AD, Losick R. 2011. The sporulation protein SirA inhibits the binding of DnaA to the origin of replication by contacting a patch of clustered amino acids. J. Bacteriol. 193:1302–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rokop ME, Auchtung JM, Grossman AD. 2004. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol. Microbiol. 52:1757–1767 [DOI] [PubMed] [Google Scholar]
- 55. Scholefield G, Errington J, Murray H. 2012. Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 31:1542–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Scholefield G, Whiting R, Errington J, Murray H. 2011. Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol. Microbiol. 79:1089–1100 [DOI] [PubMed] [Google Scholar]
- 57. Sekimizu K, Bramhill D, Kornberg A. 1987. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell 50:259–265 [DOI] [PubMed] [Google Scholar]
- 58. Simmons LA, Felczak M, Kaguni JM. 2003. DnaA protein of Escherichia coli: oligomerization at the E. coli chromosomal origin is required for initiation and involves specific N-terminal amino acids. Mol. Microbiol. 49:849–858 [DOI] [PubMed] [Google Scholar]
- 59. Smits WK, Goranov AI, Grossman AD. 2010. Ordered association of helicase loader proteins with the Bacillus subtilis origin of replication in vivo. Mol. Microbiol. 75:452–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smits WK, Merrikh H, Bonilla CY, Grossman AD. 2011. Primosomal proteins DnaD and DnaB are recruited to chromosomal regions bound by DnaA in Bacillus subtilis. J. Bacteriol. 193:640–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Soufo CD, et al. 2008. Cell-cycle-dependent spatial sequestration of the DnaA replication initiator protein in Bacillus subtilis. Dev. Cell 15:935–941 [DOI] [PubMed] [Google Scholar]
- 62. Soultanas P. 2002. A functional interaction between the putative primosomal protein DnaI and the main replicative DNA helicase DnaB in Bacillus. Nucleic Acids Res. 30:966–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Speck C, Weigel C, Messer W. 1999. ATP- and ADP-DnaA protein, a molecular switch in gene regulation. EMBO J. 18:6169–6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Su'etsugu M, Shimuta TR, Ishida T, Kawakami H, Katayama T. 2005. Protein associations in DnaA-ATP hydrolysis mediated by the Hda-replicase clamp complex. J. Biol. Chem. 280:6528–6536 [DOI] [PubMed] [Google Scholar]
- 65. Velten M, et al. 2003. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol. Cell 11:1009–1020 [DOI] [PubMed] [Google Scholar]
- 66. Wagner JK, Marquis KA, Rudner DZ. 2009. SirA enforces diploidy by inhibiting the replication initiator DnaA during spore formation in Bacillus subtilis. Mol. Microbiol. 73:963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang QP, Kaguni JM. 1987. Transcriptional repression of the dnaA gene of Escherichia coli by dnaA protein. Mol. Gen. Genet. 209:518–525 [DOI] [PubMed] [Google Scholar]
- 68. Zhang W, Allen S, Roberts CJ, Soultanas P. 2006. The Bacillus subtilis primosomal protein DnaD untwists supercoiled DNA. J. Bacteriol. 188:5487–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]


