Abstract
Members of the histone-like nucleoid-structuring (H-NS) family of proteins have been shown to play important roles in silencing gene expression and in nucleoid compaction. In Pseudomonas aeruginosa, the two H-NS family members MvaT and MvaU are thought to bind the same AT-rich regions of the chromosome and function coordinately to control a common set of genes. Here we present evidence that the loss of both MvaT and MvaU cannot be tolerated because it results in the production of Pf4 phage that superinfect and kill cells or inhibit their growth. Using a ClpXP-based protein depletion system in combination with transposon mutagenesis, we identify mutants of P. aeruginosa that can tolerate the depletion of MvaT in an ΔmvaU mutant background. Many of these mutants contain insertions in genes encoding components, assembly factors, or regulators of type IV pili or contain insertions in genes of the prophage Pf4. We demonstrate that cells that no longer produce type IV pili or that no longer produce the replicative form of the Pf4 genome can tolerate the loss of both MvaT and MvaU. Furthermore, we show that the loss of both MvaT and MvaU results in an increase in expression of Pf4 genes and that cells that cannot produce type IV pili are resistant to infection by Pf4 phage. Our findings suggest that type IV pili are the receptors for Pf4 phage and that the essential activities of MvaT and MvaU are to repress the expression of Pf4 genes.
INTRODUCTION
The histone-like nucleoid-structuring protein (H-NS) is a key global regulator of transcription that is found in many different Gram-negative bacteria. This small and abundant DNA-binding protein functions to repress the expression of hundreds of target genes and is also thought to play a role in DNA compaction, possibly through its well-documented ability to bridge the DNA (13, 16, 39). To silence gene expression, H-NS is thought to first bind to specific sites on the DNA with high affinity and then oligomerize across adjacent AT-rich regions (6). The resulting nucleoprotein complex represses the expression of target genes by preventing RNA polymerase from gaining access to the promoter or possibly by trapping RNA polymerase at certain promoters (16, 17).
Genome-wide location analyses performed in both Salmonella enterica and Escherichia coli reveal that H-NS exhibits a distinct preference for AT-rich regions of the DNA (22, 33, 37, 42). By binding preferentially to and silencing these regions, H-NS is thought to provide a mechanism for limiting the potentially deleterious effects of xenogeneic DNA, such as the DNA found on pathogenicity islands and acquired by horizontal transfer (1, 17, 37, 38). Indeed, in Salmonella, the ability of H-NS to repress the expression of genes housed on pathogenicity islands, whose DNA characteristically has a higher AT content than the rest of the genome, is essential for normal cell growth (33).
Pseudomonas aeruginosa is an important opportunistic pathogen of humans that is renowned for being the principal cause of morbidity and mortality in cystic fibrosis (CF) patients (21). In the CF lung, this Gram-negative bacterium persists as a biofilm and produces a plethora of virulence factors that it uses to intoxicate the host (49). A central player in the control of virulence gene expression in P. aeruginosa is MvaT, one of the two H-NS family members found in this organism. MvaT was originally identified as a global regulator of virulence gene expression in P. aeruginosa (15), and subsequent microarray studies revealed that MvaT controls the expression of at least 150 or so genes (58, 64). MvaU is the second H-NS family member in P. aeruginosa, is less abundant than MvaT, and can interact with MvaT (58, 59). Although MvaT and MvaU bear little sequence identity to their enteric counterparts, certain predicted structural similarities and known functional similarities suggest that they belong to the H-NS family (8, 9, 13, 55, 56, 58, 59). MvaU appears to play a less prominent role in the control of gene expression in P. aeruginosa than MvaT (58, 59). However, the individual contributions of MvaT and MvaU to the control of gene expression are muddied by the fact that these proteins negatively regulate one another's production (58, 59). Genome-wide location analyses reveal that MvaT and MvaU occupy the same 100 or more AT-rich regions of the genome, and the available evidence suggests that these proteins function coordinately to regulate a common set of genes (9).
We had shown previously that cells of P. aeruginosa strain PAO1 can tolerate the loss of either MvaT or MvaU but not both (9). In particular, using a ClpXP protease-based protein depletion system (35) that we adapted for use in P. aeruginosa, we found that depletion of MvaT in cells deleted for mvaU (ΔmvaU cells) or depletion of MvaU in ΔmvaT cells resulted in either loss of viability or an inability to grow (9). Furthermore, we found that the combined loss of both MvaT and MvaU had a much greater effect on the expression of MvaT and MvaU target genes than did the loss of either MvaT or MvaU alone (9). Although it was not known why the activities of MvaT and MvaU were essential in P. aeruginosa, these prior findings raised the possibility that derepression of certain MvaT/MvaU target genes might be lethal to the cell. Here we present evidence that the essential function of MvaT and MvaU is to repress the expression of prophage genes.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
P. aeruginosa strains PAO1, PAO1 cupA lacZ, PAO1 ΔmvaT cupA lacZ, PAO1 ΔmvaU cupA lacZ, PAO1 ΔmvaU, PAO1 ΔsspB ΔmvaU, PAO1 ΔsspB cupA lacZ MvaT-V-DAS4, and PAO1 ΔsspB ΔmvaU cupA lacZ MvaT-V-DAS4 have been described previously (9, 59). Escherichia coli strain SM10 (λpir) was used to mate plasmids into P. aeruginosa and as the recipient strain for the construction of plasmid pBAM1. E. coli strain DH5αF′IQ (Invitrogen) was used as the recipient strain for the construction of all other plasmids. When growing E. coli, antibiotics were used when necessary at the following concentrations: gentamicin, 15 μg/ml; carbenicillin, 100 μg/ml. When growing P. aeruginosa, antibiotics were used when necessary at the following concentrations: gentamicin, 25 μg/ml for liquid cultures, 30 μg/ml for solid LB medium, and 60 μg/ml for Pseudomonas isolation agar (PIA) medium (BD Diagnostic Systems); tetracycline, 35 μg/ml for LB medium and 250 μg/ml for PIA medium.
Construction of strains and plasmids.
P. aeruginosa strain PAO1 ΔsspB ΔmvaU MvaT-VDAS4 synthesizes a modified version of MvaT (MvaT-VDAS4) that contains an epitope tag from the vesicular stomatitis virus glycoprotein (VSV-G) together with a so-called DAS4 depletion tag at its C terminus (9). Strain PAO1 ΔsspB ΔmvaU MvaT-VDAS4 was constructed using plasmid pEX-MvaT-VDIV to modify the native copy of the mvaT gene in strain PAO1 ΔsspB ΔmvaU such that it specified MvaT-VDAS4. Plasmid pEX-MvaT-VDIV was made in two steps. First, a 447-bp HindIII/BamHI DNA fragment from pMvaT-VDIV (9) corresponding to the 3′ portion of the mvaT gene fused in frame with DNA specifying the V-DAS4 tag was cloned into pEXG2 (47) that had been cut with HindIII and BamHI, yielding plasmid pEX-MvaT-VDIV-partial. Second, 457 bp of DNA downstream of the mvaT gene was amplified by PCR, digested with BamHI and XhoI, and cloned into BamHI- and XhoI-digested pEX-MvaT-VDIV-partial, generating plasmid pEX-MvaT-VDIV. This plasmid was then used to create the PAO1 ΔsspB ΔmvaU MvaT-VDAS4 depletion strain by allelic exchange (26). Integration of the V-DAS4 tag was confirmed by PCR, and production of the MvaT-VDAS4 fusion protein was confirmed by a Western blot analysis using an anti-VSV-G antibody (Sigma-Aldrich) as described previously (10).
Plasmid pBAM1 is a suicide delivery vector that confers resistance to carbenicillin, contains an R6K origin of replication, contains a mariner C9 transposase, and was made by replacing the transposon-housed aacC1 gene (which confers resistance to gentamicin) of plasmid pBTK30 (20) with a tetracycline resistance gene.
Deletion constructs for pilY1 (PA4554) and PA0728 were generated by amplifying flanking regions by PCR and then splicing the flanking regions together by overlap extension PCR. The resulting PCR products were then cloned on XbaI/BamHI fragments into plasmid pEXG2 (47), yielding plasmids pEX-ΔPA4554 and pEX-ΔPA0728. Plasmid pEX-ΔPA4554 was used to create strains PAO1 ΔsspB ΔmvaU ΔpilY1 and PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4, containing deletions of the pilY1 gene, by allelic exchange. Plasmid pEX-ΔPA0728 was used to create strains PAO1 ΔsspB ΔmvaU ΔPA0728, PAO1 ΔsspB ΔmvaU ΔpilY1 ΔPA0728 MvaT-VDAS4, and PAO1 ΔsspB ΔmvaU ΔPA0728 MvaT-V-DAS4, containing deletions of the PA0728 gene, by allelic exchange. Deletions were confirmed by PCR.
The PAO1 ΔsspB ΔmvaU ΔpilY1 attTn7::Ptac-pilY1 MvaT-VDAS4 strain contains a construct which directs the synthesis of the PilY1 protein under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible tac promoter stably integrated into the genome in single copy at the attTn7 locus. To make this strain, we first replaced the lac promoter of plasmid pUC18-mini-Tn7T-LAC (12) with the stronger tac promoter (14), creating plasmid pUC18-mini-Tn7T-tac. A SacI-SpeI-digested DNA fragment containing a consensus Shine-Dalgarno sequence and the P. aeruginosa pilY1 gene was then inserted downstream of the tac promoter on pUC18-mini-Tn7T-tac, generating plasmid pUC18-mini-Tn7T-tac-pilY1. This plasmid was used to make strain PAO1 ΔsspB ΔmvaU ΔpilY1 attTn7::Ptac-pilY1 MvaT-VDAS4 by site-specific recombination with strain PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4 as the recipient (12).
The PAO1 ΔsspB ΔmvaU ΔPA0728 attTn7::Ptac-PA0728 MvaT-VDAS4 strain contains a construct which directs the synthesis of the PA0728 protein under the control of the IPTG-inducible tac promoter stably integrated into the genome in single copy at the attTn7 locus. A SacI/KpnI PCR-amplified DNA fragment specifying a consensus Shine-Dalgarno sequence and the P. aeruginosa PA0728 gene was inserted downstream of the tac promoter on pUC18-mini-Tn7T-tac, generating plasmid pUC18-mini-Tn7T-tac-PA0728. This plasmid was used to make strain PAO1 ΔsspB ΔmvaU ΔPA0728 attTn7::Ptac-PA0728 MvaT-VDAS4 by site-specific recombination with strain PAO1 ΔsspB ΔmvaU ΔPA0728 MvaT-VDAS4 as the recipient (12).
Plasmid pEX2-ΔmvaT was made by subcloning the portion of plasmid pEX-ΔmvaT (59) that contains the flanking regions of the mvaT gene into plasmid pEXG2 (47). Strains containing in-frame deletions of both mvaT and mvaU (PAO1 ΔmvaT ΔmvaU) were then made by allelic exchange using plasmid pEX2-ΔmvaT and strain PAO1 ΔmvaU as the recipient. Note that colonies of the PAO1 ΔmvaT ΔmvaU double mutant strains took several days to appear on LB agar sucrose counterselection plates at 30°C and that 10 independent isolates of the same strain were taken for study. Deletion of mvaT in each of these isolates was confirmed by PCR.
Construction of the transposon mutant library and selection and identification of transposon mutants that tolerate the loss of both MvaT and MvaU.
A library of transposon insertion mutants was generated by mobilizing pBAM1 from E. coli SM10 (λ pir) into the PAO1 ΔsspB ΔmvaU MvaT-VDAS4 depletion strain by filter mating (67). Transconjugants were selected on PIA plates supplemented with 250 μg/ml tetracycline. Approximately 2 × 105 mutants were pooled for the screen. Plasmid pV-SspB, directing the IPTG-inducible synthesis of V-SspB and conferring resistance to gentamicin (9), was introduced into the library of mutants by electroporation. Mutants that survived depletion of MvaT-VDAS4 were selected on LB agar plates supplemented with 30 μg/ml gentamicin and 2 mM IPTG. The location of the transposon in each mutant was determined by arbitrary PCR followed by DNA sequencing (20).
Twitching motility.
Subsurface twitching motility was assayed essentially as described previously (3). Bacteria were stab inoculated through a thin layer of LB agar (1% agar) supplemented with 2 mM IPTG to the bottom of the petri dish. After incubation for ∼22 h at 37°C, the twitching capacity was examined by removing the agar, washing away the unattached cells with water, and staining the attached cells with Coomassie blue (PhastGel Blue R; Sigma-Aldrich).
RNA isolation and qRT-PCR.
To examine the effects of MvaT depletion on gene expression in the absence or presence of MvaU (see Fig. 3C), cells of P. aeruginosa strains PAO1 ΔsspB ΔmvaU cupA lacZ MvaT-V-DAS4 and PAO1 ΔsspB cupA lacZ MvaT-V-DAS4 carrying either plasmid pV-SspB or plasmid pPSV35 were grown as previously described in the presence of IPTG for 30 min (9). To examine the effects of the mvaT and mvaU deletions on gene expression (see Fig. 3C), cells of P. aeruginosa strains PAO1 cupA lacZ, PAO1 ΔmvaT cupA lacZ, and PAO1 ΔmvaU cupA lacZ were grown to mid-log phase as described previously (9). RNA isolation, cDNA synthesis, and quantitative real-time reverse transcriptase PCR (qRT-PCR) to measure the abundance of a particular transcript relative to that of the clpX transcript were as described previously (9).
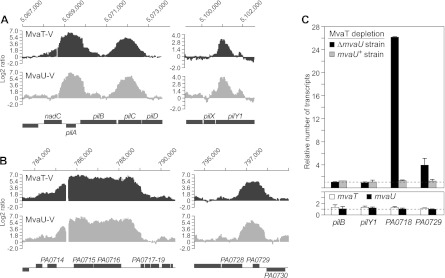
Fig 3.
The combined loss of MvaT and MvaU results in increased expression of Pf4 genes but has no effect on expression of type IV pilus genes. (A) VSV-G-tagged MvaT (MvaT-V) and VSV-G-tagged MvaU (MvaU-V) associate with type IV pilus genes, as determined by ChIP-chip. (B) VSV-G-tagged MvaT (MvaT-V) and VSV-G-tagged MvaU (MvaU-V) associate with Pf4 genes, as determined by ChIP-chip. The graphs in panels A and B depict the log2 ratio values of enrichment of DNA regions obtained with the indicated VSV-G-tagged proteins. The genomic location of the DNA is indicated at the top of each graph, and the corresponding genetic locus is indicated at the bottom. The log2 ratio values presented in panels A and B were normalized and averaged across three replicate arrays from data obtained in a previous study (9) (C) (Upper graph) Effect on gene expression of depleting MvaT in the presence (mvaU+ strain) and absence (ΔmvaU strain) of MvaU. Abundance of transcripts in cells of PAO1 ΔsspB ΔmvaU MvaT-V-DAS4 (ΔmvaU strain) carrying plasmid pV-SspB relative to that in cells carrying the empty control plasmid pPSV35 (black bars). Abundance of transcripts in cells of PAO1 ΔsspB MvaT-V-DAS4 (mvaU+ strain) carrying plasmid pV-SspB relative to that in cells carrying the empty control plasmid pPSV35 (gray bars). The dotted line at the bottom of the graph represents transcript abundance in cells containing the empty control vector pPSV35. (Lower graph) Effect of mvaT and mvaU deletions on expression of the indicated target genes. Abundance of transcripts in cells of an ΔmvaT mutant strain (white bars) and in cells of an ΔmvaU mutant strain (black bars) relative to that in cells of an isogenic wild-type strain (indicated by the dotted line). Transcripts were quantified by qRT-PCR. Error bars represent relative expression values calculated at ±1 standard deviation (SD) from the mean threshold cycle (ΔΔCT). Experiments were performed at least twice, and a representative data set is shown.
Bacteriophage plaque assays.
Pf4 phage plaque assays were performed as described by Rice et al. with minor modifications (46). Overnight cultures were centrifuged and filter sterilized using a 0.45-μm-pore-size filter. The cell-free supernatants were spotted onto an LB top agar medium containing 0.8% (wt/vol) agar seeded with P. aeruginosa strain PAO1 ΔsspB ΔmvaU ΔPA0728 MvaT-VDAS4 or PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4 recipient cells from overnight cultures (approximately 1 × 108 cells/ml). The plates were incubated at 37°C overnight. To confirm production of the replicative form (RF) of bacteriophage Pf4, an 839-bp region of DNA formed upon circularization of the Pf4 genome was amplified by PCR with primers Pf4F and Pf4R as described previously (61).
RESULTS
Identification of transposon insertion mutants that tolerate the loss of both MvaT and MvaU.
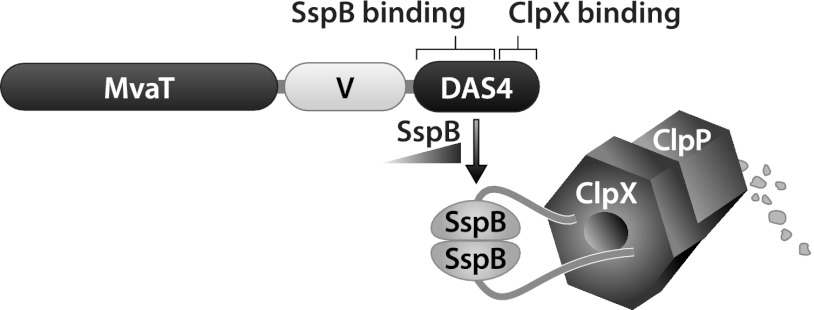
We reasoned that MvaT and MvaU are essential in the absence of the partner regulator because the uncontrolled expression of a subset of MvaT/MvaU target genes results in lethality. To test whether this might be the case, we sought to identify transposon insertion mutants that could tolerate the loss of both MvaT and MvaU. Our strategy for identifying such mutants involved mutagenizing a strain of PAO1 in which the mvaU gene was deleted and then selecting for those mutants that survived following the depletion of MvaT (i.e., that survived when both MvaU and MvaT were absent). In order to be able to deplete MvaT in a controlled fashion, we made use of the ClpXP protease-based protein depletion system we had used previously to deplete MvaT in P. aeruginosa (9) (Fig. 1). This system involves the fusion of a small peptide tag, the so-called DAS4 tag, to the C terminus of a protein of interest. The DAS4 tag contains a low-affinity binding site for ClpX and a high-affinity binding site for the adaptor protein SspB that feeds the tagged protein to the ClpXP protease complex (35). This enables a tagged protein to be degraded by ClpXP at a rate that is determined by the intracellular concentration of SspB (35). The MvaT depletion strain we used for these experiments (PAO1 ΔsspB ΔmvaU MvaT-VDAS4) carries a deletion of the native sspB gene and a deletion of the native mvaU gene and harbors a modified mvaT gene that specifies MvaT with both a vesicular stomatitis virus glycoprotein (VSV-G) epitope tag and a DAS4 tag at its C terminus (MvaT-VDAS4) (Fig. 1).
Fig 1.
Schematic representation of the ClpXP-based protein depletion system. The MvaT-VDAS4 protein is depicted. V corresponds to the VSV-G epitope tag. The DAS4 tag allows for depletion of MvaT-VDAS4 in a manner that is dependent upon the intracellular concentration of the adaptor protein SspB that feeds MvaT-VDAS4 to the ClpXP protease complex.
To identify mutants that can tolerate the loss of both MvaT and MvaU, cells of the PAO1 ΔsspB ΔmvaU MvaT-VDAS4 depletion strain were first mutagenized with a mariner transposon. Cells of the resulting transposon mutant library were transformed with plasmid pV-SspB, which directs the IPTG-controlled synthesis of SspB with an N-terminal VSV-G tag (V-SspB) (9). Transformants were then plated on LB agar plates containing IPTG to deplete MvaT-VDAS4. Table 1 indicates the location of the transposon in each of 61 unique insertion mutants of the depletion strain that grew in the presence of IPTG and in which depletion of MvaT-VDAS4 was confirmed by Western blotting (data not shown). The majority of mutants contained insertions in genes encoding components, assembly factors, or regulators of type IV pili, which have been implicated in both virulence and biofilm formation in P. aeruginosa (43, 54) and are responsible for a form of surface-associated motility, referred to as twitching motility, that is mediated by cycles of pilus extension and retraction (7, 34, 50) (Table 1). Another prominent class of mutants that tolerated the loss of both MvaT and MvaU comprised those with insertions within the Pf4 prophage (61, 62) (Table 1).
Table 1.
Locations of transposons in mutants of PAO1 that can tolerate the depletion of MvaT in an ΔmvaU mutant background
| Locus type and namea | No. of mutantsb | Description of gene product (reference[s]) |
|---|---|---|
| Type IV pilus related | ||
| pilY1 | 17 | Pilus adhesin and regulator of pilus extension/retraction (23, 41) |
| pilB | 5 | Cytoplasmic ATPase involved in pilus extension (11, 57) |
| pilC | 5 | Required for pilus biogenesis (40) |
| pilQ | 4 | Secretin (44) |
| pilA | 2 | Major pilin subunit (52, 53) |
| pilF | 2 | Lipoprotein that promotes assembly of the secretin pore (30) |
| pilM | 2 | Component of pilus assembly complex (4) |
| pilW | 2 | Minor pilin subunit (19) |
| fimV | 2 | Peptidoglycan-binding protein that promotes assembly of the secretin pore (48, 63) |
| pilZ | 1 | Required for pilus biogenesis (2) |
| pilE | 1 | Minor pilin subunit (19) |
| pilN | 1 | Component of pilus assembly complex (4) |
| pilP | 1 | Component of pilus assembly complex (4) |
| pilR | 1 | Response regulator required for pilA expression (27, 29) |
| PA1611 | 1 | Hybrid sensor kinase that influences twitching motility (32) |
| Pf4 phage related | ||
| PA0716-PA0717 ig | 2 | Intergenic region |
| PA0716 | 1 | Component of ABC transporter (36) |
| PA0717 | 1 | Hypothetical protein of phage Pf4 (36) |
| PA0718 | 1 | Hypothetical protein of phage Pf4 (36) |
| PA0726 | 1 | Hypothetical protein of phage Pf4 (36) |
| Other | ||
| PA0714 | 1 | Hypothetical protein (66) |
| PA1613 | 1 | Hypothetical protein (66) |
| gacA | 1 | Response regulator (66) |
| gacS | 1 | Sensor kinase (66) |
| PA2501-PA2502 ig | 1 | Intergenic region |
| lecA-PA2570.1 ig | 1 | Intergenic region |
| clpX-lon ig | 1 | Intergenic region |
| tig-clpP ig | 1 | Intergenic region |
ig, a transposon inserted into the intergenic region between the two indicated genes.
Refers to the number of unique insertion mutants obtained.
Cells that no longer produce type IV pili or bacteriophage Pf4 can tolerate the loss of both MvaT and MvaU.
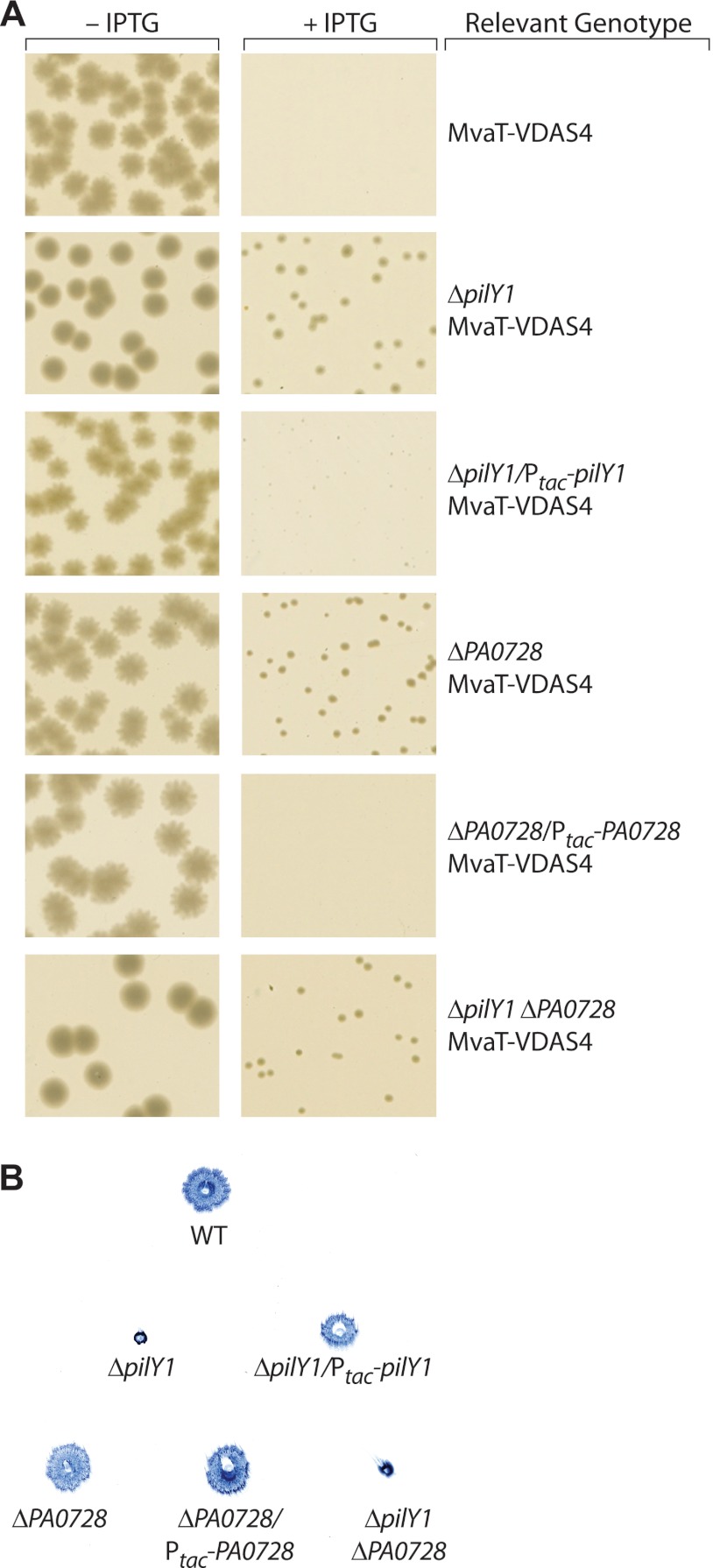
To confirm that the loss of type IV pili or the loss of the replicative form of bacteriophage Pf4 results in cells that can tolerate the loss of both MvaT and MvaU, we tested the effect of deleting the pilY1 and PA0728 genes, as pilY1 is known to be required for the production of type IV pili and PA0728 is predicted to be important for the production of Pf4; pilY1 encodes a protein that is typically required for type IV pilus biogenesis (23, 41) and was hit in our transposon mutant screen (Table 1), whereas PA0728 is a gene whose ortholog in P. aeruginosa strain PA14 has been shown to be required for production of the replicative form of phage Pf5, a phage that is highly related to Pf4 (36). We therefore deleted the pilY1 and PA0728 genes in our PAO1 ΔsspB ΔmvaU MvaT-VDAS4 depletion strain, creating strains PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4 and PAO1 ΔsspB ΔmvaU ΔPA0728 MvaT-VDAS4, respectively. We also constructed two control strains. The first of these was a derivative of the PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4 depletion strain in which the pilY1 gene is supplied in trans under the control of an IPTG-inducible promoter from the Tn7 attachment site on the PAO1 chromosome (PAO1 ΔsspB ΔmvaU ΔpilY1 attTn7::Ptac-pilY1 MvaT-VDAS4). The second control strain was a derivative of the PAO1 ΔsspB ΔmvaU ΔPA0728 MvaT-VDAS4 depletion strain in which the PA0728 gene is supplied in trans under the control of an IPTG-inducible promoter from the Tn7 attachment site (PAO1 ΔsspB ΔmvaU ΔPA0728 attTn7::Ptac-PA0728 MvaT-VDAS4). We then transformed cells of the PAO1 ΔsspB ΔmvaU MvaT-VDAS4 depletion strain and cells of each of the corresponding mutant and control strains with plasmid pV-SspB and asked whether depletion of MvaT-VDAS4 can be tolerated in these cells by plating transformants on LB agar plates that either did or did not contain IPTG. The results depicted in Fig. 2A show that cells of the PAO1 ΔsspB ΔmvaU MvaT-VDAS4 depletion strain containing plasmid pV-SspB gave rise to colonies in the absence but not the presence of IPTG. This is consistent with our previous demonstration that depletion of MvaT cannot be tolerated in cells of an ΔmvaU mutant strain (9). Figure 2A also shows that the depletion of MvaT can be tolerated in ΔmvaU mutant cells that harbor deletions of either pilY1 or PA0728 and that the effects of the pilY1 or PA0728 deletion can be complemented by supplying pilY1 or PA0728, respectively, in trans. Similar results were obtained with a pilC mutant derivative of the PAO1 ΔsspB ΔmvaU MvaT-VDAS4 depletion strain (data not shown). Taken together, these findings suggest that cells that no longer produce type IV pili or cells that no longer produce the replicative form of Pf4 can tolerate the loss of both MvaT and MvaU.
Fig 2.
Deletion of pilY1 or PA0728 results in cells that can tolerate the loss of both MvaT and MvaU. (A) Colonies from cells of MvaT depletion strains carrying the plasmid pV-SspB that were grown overnight at 37°C on LB agar plates that either contained IPTG (+IPTG) or did not (−IPTG); images are from separate agar plates. All cells contained deletions of sspB and mvaU, and additional relevant genotypes are indicated. MvaT-VDAS4, PAO1 ΔsspB ΔmvaU MvaT-VDAS4; ΔpilY1 MvaT-VDAS4, PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4; ΔpilY1/Ptac-pilY1 MvaT-VDAS4, PAO1 ΔsspB ΔmvaU ΔpilY1 attTn7::Ptac-pilY1 MvaT-VDAS4; ΔPA0728 MvaT-VDAS4, PAO1 ΔsspB ΔmvaU ΔPA0728 MvaT-VDAS4; ΔPA0728/Ptac-PA0728 MvaT-VDAS4, PAO1 ΔsspB ΔmvaU ΔPA0728 attTn7::Ptac-PA0728 MvaT-VDAS4; ΔpilY1 ΔPA0728 MvaT-VDAS4, PAO1 ΔsspB ΔmvaU ΔpilY1 ΔPA0728 MvaT-VDAS4. (B) Effects of pilY1 and PA0728 deletions on twitching motility. Cells were grown on LB agar plates containing IPTG; the image is of colonies of Coomassie blue-stained cells attached to the bottom of a single petri dish. WT, wild-type PAO1; ΔpilY1, PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4; ΔpilY1/Ptac-pilY1, PAO1 ΔsspB ΔmvaU ΔpilY1 attTn7::Ptac-pilY1 MvaT-VDAS4; ΔPA0728, PAO1 ΔsspB ΔmvaU ΔPA0728 MvaT-VDAS4; ΔPA0728/Ptac-PA0728, PAO1 ΔsspB ΔmvaU ΔPA0728 attTn7::Ptac-PA0728 MvaT-VDAS4; ΔpilY1 ΔPA0728, PAO1 ΔsspB ΔmvaU ΔpilY1 ΔPA0728 MvaT-VDAS4. Experiments were performed at least three times, and representative data sets are shown.
The results depicted in Fig. 2A show that although ΔpilY1 and ΔPA0728 mutants can tolerate the loss of both MvaT and MvaU, deletion of pilY1 alone or deletion of PA0728 alone appears to be insufficient to allow cells to grow at wild-type rates when both MvaT and MvaU are absent (as suggested by the difference in colony size observed between plates that contain IPTG and plates that do not). We therefore next asked whether combining both the pilY1 and PA0728 deletions would allow cells to grow at rates closer to wild-type rates when both MvaU and MvaT are absent. Cells of the PAO1 ΔsspB ΔmvaU MvaT-VDAS4 depletion strain containing deletions of both the pilY1 and PA0728 genes (PAO1 ΔsspB ΔmvaU ΔpilY1 ΔPA0728 MvaT-VDAS4) and carrying the pV-SspB plasmid gave rise to colonies on IPTG-containing medium that were no larger than those formed by cells of the corresponding individual ΔpilY1 or ΔPA0728 mutant depletion strain containing the same plasmid (Fig. 2A). This finding suggests that the effects of the pilY1 and PA0728 deletions on the growth of cells that lack both MvaT and MvaU are dependent upon one another. Note, however, that this apparent dependency was not able to be explained through an effect of Pf4 on the production or function of type IV pili, as deletion of the PA0728 gene, unlike deletion of the pilY1 gene, has no effect on twitching motility (Fig. 2B).
Loss of both MvaT and MvaU results in an increase in expression of Pf4 genes.
We have previously used chromatin immunoprecipitation coupled with high-density DNA microarrays (ChIP-chip) to identify those regions of the P. aeruginosa chromosome that are associated with MvaT and MvaU (9). Analysis of these chromosomal regions revealed that both MvaT and MvaU are directly associated with a subset of the genes that encode components of the type IV pilus (Fig. 3A) and are directly associated with a subset of the Pf4 genes (the Pf4 prophage spans from PA0715 to PA0729) (Fig. 3B). In particular, we found that both MvaT and MvaU were associated with the pilA-pilD genes, the pilY1 gene, the PA0714-PA0717 genes, and the PA0728 gene through the PA0729-PA0730 intergenic region (Fig. 3AB); consistent with our previous finding that MvaT and MvaU associate with AT-rich regions of DNA (9), the pilA gene has a GC content of 48.7%, the pilC gene has a GC content of 54.8%, the pilY1 gene has a GC content of 62.7%, the PA0715 gene has a GC content of 39.1%, the PA0716 gene has a GC content of 39.3%, and the PA0729 gene has a GC content of 51.7%, whereas the average GC content for a gene in P. aeruginosa strain PAO1 is 66.6% (51). We therefore wanted to test the possibility that MvaT and MvaU are essential in the absence of the partner regulator because the uncontrolled expression of either certain type IV pilus genes or certain Pf4 genes results in lethality or inhibition of cell growth.
We first asked whether the expression of the pilB, pilY1, PA0718, and PA0729 genes (all direct targets of MvaT and MvaU) increased when both MvaT and MvaU were absent. For these experiments, we used a previously described pair of isogenic ΔmvaU and mvaU+ strains in which MvaT could be depleted upon induction of V-SspB synthesis (9). Like the MvaT depletion strain described earlier, one of these MvaT depletion strains (PAO1 ΔsspB ΔmvaU cupA lacZ MvaT-V-DAS4) contained a deletion of mvaU, whereas the other (PAO1 ΔsspB cupA lacZ MvaT-V-DAS4) did not. (Note that the MvaT depletion strains PAO1 ΔsspB ΔmvaU MvaT-VDAS4, described here, and PAO1 ΔsspB ΔmvaU cupA lacZ MvaT-V-DAS4, described previously, are similar to one another but differ with respect to the DNA sequence found immediately downstream of the modified mvaT gene.) Cells of these MvaT depletion strains were transformed with plasmid pV-SspB and an empty control plasmid and grown in LB to mid-log phase. Cultures were then grown in the presence of IPTG (to induce synthesis of V-SspB in those cells that contained the appropriate vector) for an additional 30 min, and RNA was isolated. The abundance of the pilB, pilY1, PA0718, and PA0728 transcripts was then determined following depletion of MvaT in the mvaU+ and ΔmvaU mutant cells by quantitative real-time RT-PCR (qRT-PCR). The abundance of the same transcripts was also quantified in the control cells in which MvaT was not depleted. The results presented in Fig. 3C show that depletion of MvaT results in an ∼26-fold increase in expression of PA0718 and an ∼4-fold increase in expression of PA0729 only in the absence of MvaU (i.e., only in cells containing the ΔmvaU allele). Moreover, quantification of the PA0718 and PA0728 transcripts in cells of a wild-type strain, in cells of an ΔmvaT mutant strain, and in cells of an ΔmvaU mutant strain revealed that deletion of mvaT or deletion of mvaU had little effect on expression of either PA0718 or PA0729 (Fig. 3C). However, depletion of MvaT did not result in any appreciable change in expression of the pilB or pilY1 gene, regardless of whether MvaU was present, and neither did deletion of mvaT or mvaU (Fig. 3C). Depletion of MvaT did not result in any appreciable change in expression of the pilA gene either (data not shown). Thus, although MvaT and MvaU associate directly with pilA, pilB, pilY1, PA0718, and PA0729, the loss of both MvaT and MvaU results only in an increase in the expression of the PA0718 and PA0729 genes. These findings raise the possibility that an increase in expression of Pf4 genes alone may account for the loss of viability or inhibition of cell growth that occurs in the absence of both MvaT and MvaU.
Pf4 phage cannot form plaques on cells that no longer produce type IV pili.
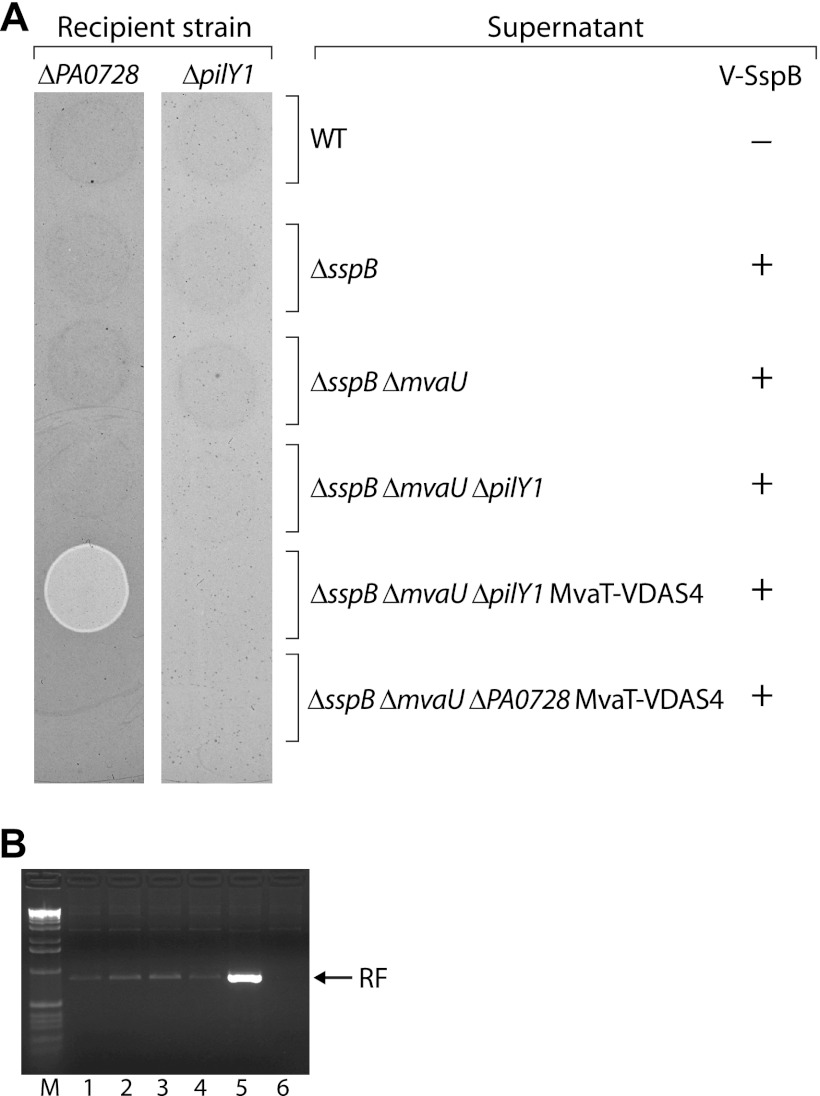
The increase in expression of Pf4 genes that occurs upon the loss of both MvaT and MvaU may, in principle, result in an increase in the single-stranded replicative form of Pf4 and a concomitant increase in the production of Pf4 phage. The resulting phage may subsequently infect cells, leading to cell death or the inhibition of cell growth, explaining why the loss of both MvaT and MvaU results in a loss of viability or inhibition of cell growth and why mutants that cannot produce the replicative form of Pf4 can tolerate the loss of both MvaT and MvaU. Indeed, the addition of purified Pf4 phage to cells of P. aeruginosa strain PAO1 has been shown to result in cell killing by a mechanism that is not understood (62). If the production of Pf4 phage were responsible for the loss of viability in cells that lack both MvaT and MvaU, then how would the loss of type IV pili allow cells to tolerate the loss of both MvaT and MvaU? Type IV pili serve as receptors for a number of bacteriophages, and type IV pili from P. aeruginosa strain PAK have been shown to bind directly to the Pf4-like bacteriophage Pf1 (28). Therefore, in order to explain why type IV pilus mutants can tolerate the loss of both MvaT and MvaU, we hypothesized that type IV pili serve as the receptors for Pf4 phage. Mutants that no longer produce type IV pili would be expected to produce Pf4 phage in the absence of both MvaT and MvaU but would be resistant to infection by these phage since they would lack the requisite receptor. To test this possibility, we first attempted to isolate Pf4 phage from the supernatant of a culture of PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4 cells carrying plasmid pV-SspB following depletion of MvaT. As controls, we also attempted to isolate Pf4 phage from the supernatants of cultures of (i) PAO1 cells carrying the empty vector pPSV35, (ii) PAO1 ΔsspB cells carrying pV-SspB, (iii) PAO1 ΔsspB ΔmvaU cells carrying pV-SspB, (iv) PAO1 ΔsspB ΔmvaU ΔpilY1 cells carrying pV-SspB, and (v) PAO1 ΔsspB ΔmvaU ΔPA0728 MvaT-VDAS4 cells carrying pV-SspB. The resulting supernatants were then spotted onto LB agar plates containing top agar seeded with either a recipient PAO1 strain containing a deletion of PA0728 or a recipient PAO1 strain containing a deletion of pilY1. The results depicted in Fig. 4A show that only the supernatant isolated from cells of the PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4 strain in which MvaT was depleted gives rise to a region of clearing on cells of the ΔPA0728 recipient strain. Similar results were obtained when cells of the wild-type PAO1 strain were used as recipients (data not shown). This is consistent with the idea that infectious Pf4 phage are produced by cells of the PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4 strain upon depletion of MvaT (i.e., only in the absence of both MvaT and MvaU) but not by cells of the PAO1 ΔsspB ΔmvaU ΔPA0728 MvaT-VDAS4 strain upon depletion of MvaT. The finding that the PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4 strain produces Pf4 phage upon depletion of MvaT (Fig. 4A) but tolerates depletion of MvaT (Fig. 2A) suggests that it is infection by Pf4, and not the production of Pf4 phage itself, that is responsible for either the cell death or the inhibition of cell growth that occurs upon the loss of both MvaT and MvaU.
Fig 4.
Infectious Pf4 phage produced following depletion of MvaT in an ΔmvaU mutant background cannot form plaques on ΔpilY1 mutant cells. (A) Cell-free supernatants isolated from cultures of the indicated cells that either did (+) or did not (−) synthesize V-SspB were spotted onto top agar plates seeded with either cells of a ΔPA0728 mutant or cells of a ΔpilY1 mutant. Plates were incubated overnight at 37°C. The ΔPA0728 recipient strain was PAO1 ΔsspB ΔmvaU ΔPA0728 MvaT-VDAS4, whereas the ΔpilY1 mutant was PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4. Supernatants were isolated from cultures of PAO1 cells carrying the empty vector pPSV35 (WT), from PAO1 ΔsspB cells carrying pV-SspB, from PAO1 ΔsspB ΔmvaU cells carrying pV-SspB, from PAO1 ΔsspB ΔmvaU ΔpilY1 cells carrying pV-SspB, from PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4 cells carrying pV-SspB, and from PAO1 ΔsspB ΔmvaU ΔPA0728 MvaT-VDAS4 cells carrying pV-SspB. Only the supernatant isolated from PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4 cells carrying pV-SspB gives rise to a region of clearing on cells of the ΔPA0728 recipient strain due to the presence of Pf4 phage; the region of clearing appears lighter than the rest of the plate in this image because it contains fewer cells. (B) Agarose gel of PCR products amplified from the replicative form (RF) of phage Pf4 that was present in the following cells after growth in the presence of IPTG: lane 1, PAO1 cells carrying the empty vector pPSV35; lane 2, PAO1 ΔsspB cells carrying pV-SspB; lane 3, PAO1 ΔsspB ΔmvaU cells carrying pV-SspB; lane 4, PAO1 ΔsspB ΔmvaU ΔpilY1 cells carrying pV-SspB; lane 5, PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4 cells carrying pV-SspB; lane 6, PAO1 ΔsspB ΔmvaU ΔPA0728 MvaT-VDAS4 cells carrying pV-SspB. Lane M is a marker lane. The results indicate that depletion of MvaT in cells of an ΔmvaU mutant strain results in an increase in the replicative form of phage Pf4. Experiments in panels A and B were performed three times, and representative data sets are shown.
The results depicted in Fig. 4A also show that the same supernatant that gave rise to a region of clearing on cells of the ΔPA0728 recipient strain does not give rise to a region of clearing on cells of the ΔpilY1 recipient strain. This is consistent with the idea that ΔpilY1 mutants are resistant to infection by Pf4. To confirm that cells of the PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4 strain in which MvaT was depleted contained more of the replicative form of phage Pf4 than cells in which MvaT was not depleted, we used PCR to semiquantitatively measure the abundance of the replicative form of PF4. The results depicted in Fig. 4B show that compared to the control cells (lanes 1 to 4), the replicative form of Pf4 is more abundant in cells of the PAO1 ΔsspB ΔmvaU ΔpilY1 MvaT-VDAS4 strain in which MvaT was depleted (lane 5). Note that the replicative form of Pf4 was detectable in all the control cells that were tested (lanes 1 to 4) except for those containing a deletion of PA0728 (lane 6), consistent with the idea that PA0728 is essential for the production of the replicative form of Pf4 (36).
Taken together, our findings suggest that the loss of both MvaT and MvaU results in an increase in expression of Pf4 genes and concomitant production of infectious Pf4 phage. These phages then go on to infect those cells that are producing them in a manner that is dependent upon the presence of the type IV pilus. Superinfection by these phages subsequently results in either cell death or inhibition of cell growth.
Mutants that contain deletions of both mvaT and mvaU are defective for twitching motility or do not produce the replicative form of Pf4.
A previous report demonstrated that cells of P. aeruginosa strain PAO1 that contain deletions of both the mvaT and mvaU genes are viable (31). This finding appears to contradict our findings that MvaT and MvaU are essential in the absence of the partner regulator (9) (Fig. 2A). We therefore wondered whether cells that contain deletions of both mvaT and mvaU might be viable because they contain suppressor mutations that either render them incapable of producing type IV pili or render them incapable of producing the replicative form of Pf4. To test this idea, we deleted the mvaT gene from cells of a previously constructed PAO1 ΔmvaU mutant strain (59) and asked whether any of the resulting mutants exhibited defects in twitching motility (indicating that they no longer produced functional type IV pili) or no longer produced the replicative form of Pf4. Of the 10 independent PAO1 ΔmvaU ΔmvaT mutants that we constructed and tested, nine were completely defective for twitching motility and one no longer produced detectable amounts of the replicative form of Pf4 (see Fig. S1 in the supplemental material). These findings, taken together with our other findings, suggest that cells containing deletions of both mvaT and mvaU are viable because they contain suppressor mutations that either affect the ability to produce type IV pili or affect the ability to produce the replicative form of phage Pf4.
DISCUSSION
MvaT and MvaU have shared functions that are essential in P. aeruginosa (9). We have obtained evidence that in P. aeruginosa strain PAO1, the essential activity of these H-NS family members is to repress expression of Pf4 genes. MvaT and MvaU associate directly with a portion of the Pf4 genome, and we found that the loss of both MvaT and MvaU results in an increase in the expression of Pf4 genes and in the production of infectious Pf4 phage. These phages subsequently infect cells, leading to cell death or the inhibition of cell growth. In support of this idea, we found that cells that can no longer produce Pf4 phage can tolerate the loss of both MvaT and MvaU, as can cells that no longer produce type IV pili because type IV pili likely act as the receptors for Pf4 phage.
We hypothesized that MvaT and MvaU are essential in the absence of the partner regulator because the uncontrolled expression of a subset of MvaT/MvaU target genes results in lethality. Indeed, we had shown previously that the combined loss of MvaT and MvaU had a much greater effect on the expression of MvaT and MvaU target genes than did the loss of either MvaT alone or MvaU alone (9). This is presumably because MvaT and MvaU associate with the same regions of the chromosome and because these H-NS family members control each other's production (9, 59); the increased intracellular concentration of MvaT found in ΔmvaU mutant cells and the increased intracellular concentration of MvaU found in ΔmvaT mutant cells likely obscure the individual contributions of MvaT and MvaU to the regulation of specific target genes (59). We found that depletion of MvaT in an ΔmvaU mutant background resulted in a large increase in expression of Pf4 genes that were found to be associated with MvaT and MvaU in our ChIP-chip studies. We also found that depletion of MvaT in an ΔmvaU mutant background resulted in the production of infectious Pf4 phage and that cells lacking the PA0728 gene, which is essential for making the replicative form of Pf4, grew in the absence of both MvaT and MvaU. Taken together, these findings suggest that the essential activity of MvaT and MvaU is to repress expression of the Pf4 genes.
We performed a genetic screen for mutants that can tolerate the loss of both MvaT and MvaU. The most common class of mutant identified in this screen contained a transposon inserted into a gene encoding a component, an assembly factor, or a regulator of type IV pili. We do not think that uncontrolled expression of the type IV pilus genes is what accounts for the loss of viability or inhibition of cell growth that occurs in the absence of MvaT and MvaU. Although the loss of both MvaT and MvaU resulted in derepression of Pf4 gene expression, it had no detectable effect on the expression of the pilA, pilB, and pilY1 genes despite both MvaT and MvaU being associated with all three of these genes. As H-NS family members are known to contribute to nucleoid compaction (60), it is possible that the binding of MvaT and MvaU to type IV pilus genes contributes to chromosome organization. To explain the results of the genetic screen, we propose that type IV pili serve as the receptors for Pf4 phage and that cells that no longer produce type IV pili can tolerate the loss of MvaT and MvaU because they are immune to superinfection by Pf4. Consistent with this idea, we found that the PF4 phage produced by ΔmvaU ΔpilY1 cells upon depletion of MvaT could not form plaques on a ΔpilY1 recipient strain. Indeed, several other studies support the notion that type IV pili can act as receptors for phage Pf4. Phage Pf1 (from a strain of P. aeruginosa other than PAO1), which is highly related to Pf4, has been shown to infect cells through type IV pili (25) and has been shown to interact directly with type IV pili isolated from P. aeruginosa strain PAK (28).
Our findings that the ΔpilY1 mutant produces infectious Pf4 phage in the absence of MvaT and MvaU yet is itself resistant to infection by Pf4 suggest that it is infection by Pf4, as opposed to production of Pf4, that results in cell death or the inhibition of cell growth when MvaT and MvaU are absent. This is consistent with the previous demonstration that the addition of purified Pf4 phage to cells of a biofilm results in cell death (62). Although it is not yet known why infection by Pf4 phage results in lethality, it is known that Pf4 phage are produced by cells in biofilms (45, 62, 65) and that cells that lack the Pf4 prophage fail to undergo autolysis in mature biofilms and form smaller microcolonies than wild-type cells (46). Superinfection by Pf4 therefore appears to influence biofilm formation, possibly through an effect on cell lysis, and it has been suggested that the effect of Pf4 on biofilm development may explain why Pf4 plays a role in the virulence of P. aeruginosa (46).
We found that cells containing deletions of both mvaT and mvaU likely contain suppressor mutations that render cells either incapable of producing functional type IV pili (i.e., defective for twitching motility) or incapable of producing the replicative form of Pf4. These findings resolve the apparent discrepancy between our earlier findings obtained through protein depletion, that the loss of both MvaT and MvaU cannot be tolerated (9), and the previous demonstration that cells containing deletions of both mvaT and mvaU are viable (31). We note that, consistent with the findings reported here, activation of Pf4 prophage was observed previously in cells containing deletions of both mvaT and mvaU (31).
H-NS family members appear to be essential in several different bacteria (5, 18, 24, 37). In Salmonella, in which the ability of H-NS to repress the expression of genes on pathogenicity islands is key to the fitness of the cell, H-NS does not appear to associate with prophage DNA, presumably because the AT content of these regions is similar to that of the rest of the genome and, thus, not sufficiently high (33). We have obtained evidence that in P. aeruginosa strain PAO1, the ability of the H-NS family members MvaT and MvaU to associate with prophage DNA and repress the expression of phage genes is essential. However, repression of Pf4 gene expression evidently is not the only important function of MvaT and MvaU. Cells lacking both MvaT and MvaU that contain suppressor mutations that influence either type IV pilus or Pf4 production do not appear to grow as well as wild-type cells. Our suppressor analysis therefore suggests that there are other important functions of MvaT and MvaU that remain to be determined.
Supplementary Material
ACKNOWLEDGMENTS
We thank Herbert Schweizer for plasmids, Bryan McGuffie for making the pBAM1 plasmid used for transposon mutagenesis and for help making the supplementary figure, Kirsty McFarland for making plasmid pUC18-mini-Tn7T-tac, Renate Hellmiss for artwork, Heather McManus for discussions, and Ann Hochschild for comments on the manuscript.
This work was supported by National Institutes of Health grant AI069007 (to S.L.D.).
Footnotes
Published ahead of print 20 July 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Ali SS, Xia B, Liu J, Navarre WW. 2012. Silencing of foreign DNA in bacteria. Curr. Opin. Microbiol. 15:175–181 [DOI] [PubMed] [Google Scholar]
- 2. Alm RA, Bodero AJ, Free PD, Mattick JS. 1996. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 178:46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alm R, Mattick JS. 1995. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol. Microbiol. 16:485–496 [DOI] [PubMed] [Google Scholar]
- 4. Ayers M, et al. 2009. PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J. Mol. Biol. 394:128–142 [DOI] [PubMed] [Google Scholar]
- 5. Banos RC, Pons JI, Madrid C, Juarez A. 2008. A global modulatory role for the Yersinia enterocolitica H-NS protein. Microbiology 154:1281–1289 [DOI] [PubMed] [Google Scholar]
- 6. Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. 2007. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 14:441–448 [DOI] [PubMed] [Google Scholar]
- 7. Bradley DE. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 26:146–154 [DOI] [PubMed] [Google Scholar]
- 8. Castang S, Dove SL. 2010. High-order oligomerization is required for the function of the H-NS family member MvaT in Pseudomonas aeruginosa. Mol. Microbiol. 78:916–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castang S, McManus HR, Turner KH, Dove SL. 2008. H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl. Acad. Sci. U. S. A. 105:18947–18952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Charity JC, Blalock LT, Costante-Hamm MM, Kasper DL, Dove SL. 2009. Small molecule control of virulence gene expression in Francisella tularensis. PLoS Pathog. 5:e1000641 doi:10.1371/journal.ppat.1000641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chiang P, et al. 2008. Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT, and PilU. Microbiology 154:114–126 [DOI] [PubMed] [Google Scholar]
- 12. Choi KH, et al. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443–448 [DOI] [PubMed] [Google Scholar]
- 13. Dame RT, et al. 2005. DNA bridging: a property shared among H-NS-like proteins. J. Bacteriol. 187:1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Boer HA, Comstock LJ, Vasser M. 1983. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc. Natl. Acad. Sci. U. S. A. 80:21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diggle SP, Winzer K, Lazdunski A, Williams P, Camara P. 2002. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 184:2576–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dorman CJ. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391–400 [DOI] [PubMed] [Google Scholar]
- 17. Dorman CJ. 2007. H-NS, the genome sentinel. Nat. Rev. Microbiol. 5:157–161 [DOI] [PubMed] [Google Scholar]
- 18. Ellison DW, Miller VL. 2006. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J. Bacteriol. 188:5101–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giltner CL, Habash M, Burrows LL. 2010. Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J. Mol. Biol. 398:444–461 [DOI] [PubMed] [Google Scholar]
- 20. Goodman AL, et al. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745–754 [DOI] [PubMed] [Google Scholar]
- 21. Govan JRW, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grainger DC, Hurd D, Goldberg MD, Busby SJW. 2006. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 34:4642–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heiniger RW, Winther-Larsen HC, Pickles RJ, Koomey M, Wolfgang MC. 2010. Infection of human mucosal tissue by Pseudomonas aeruginosa requires sequential and mutually dependent virulence factors and a novel pilus-associated adhesin. Cell. Microbiol. 12:1158–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heroven AK, Nagel G, Tran HJ, Parr S, Dersch P. 2004. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol. Microbiol. 53:871–888 [DOI] [PubMed] [Google Scholar]
- 25. Hill DF, Short NJ, Perham RN, Petersen GB. 1991. DNA sequence of the filamentous bacteriophage Pf1. J. Mol. Biol. 218:349–364 [DOI] [PubMed] [Google Scholar]
- 26. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 27. Hobbs M, Collie ES, Free PD, Livingston SP, Mattick JS. 1993. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 7:669–682 [DOI] [PubMed] [Google Scholar]
- 28. Holland SJ, Sanz C, Perham RN. 2006. Identification and specificity of pilus adsorption proteins of filamentous bacteriophages infecting Pseudomonas aeruginosa. Virology 345:540–548 [DOI] [PubMed] [Google Scholar]
- 29. Ishimoto KS, Lory S. 1992. Identification of pilR, which encodes a transcriptional activator of the Pseudomonas aeruginosa pilin gene. J. Bacteriol. 174:3514–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koo J, et al. 2008. PilF is an outer membrane lipoprotein required for multimerization and localization of the Pseudomonas aeruginosa type IV pilus secretin. J. Bacteriol. 190:6961–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li C, Wally H, Miller SJ, Lu C-D. 2009. The multifaceted proteins MvaT and MvaU, members of the H-NS family, control arginine metabolism, pyocyanin synthesis, and prophage activation in Pseudomonas aeruginosa PAO1. J. Bacteriol. 191:6211–6218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin CT, et al. 2006. Identification of an HptB-mediated multi-step phosphorelay in Pseudomonas aeruginosa PAO1. Res. Microbiol. 157:169–175 [DOI] [PubMed] [Google Scholar]
- 33. Lucchini S, et al. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:e81 doi:10.1371/journal.ppat.0020081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mattick JS. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289–314 [DOI] [PubMed] [Google Scholar]
- 35. McGinness K, Baker T, Sauer R. 2006. Engineering controllable protein degradation. Mol. Cell 22:701–707 [DOI] [PubMed] [Google Scholar]
- 36. Mooij MJ, et al. 2007. Characterization of the integrated filamentous phage Pf5 and its involvement in small-colony formation. Microbiology 153:1790–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Navarre WW, et al. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238 [DOI] [PubMed] [Google Scholar]
- 38. Navarre WW, McClelland M, Libby SJ, Fang FC. 2007. Silencing of xenogeneic DNA by H-NS—facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 21:1456–1471 [DOI] [PubMed] [Google Scholar]
- 39. Noom MC, Navarre WW, Oshima T, Wuite GJ, Dame RT. 2007. H-NS promotes looped domain formation in the bacterial chromosome. Curr. Biol. 17:R913–R914 [DOI] [PubMed] [Google Scholar]
- 40. Nunn D, Bergman S, Lory S. 1990. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J. Bacteriol. 172:2911–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Orans J, et al. 2010. Crystal structure analysis reveals Pseudomonas PilY1 as an essential calcium-dependent regulator of bacterial surface motility. Proc. Natl. Acad. Sci. U. S. A. 107:1065–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N. 2006. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 13:141–153 [DOI] [PubMed] [Google Scholar]
- 43. O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304 [DOI] [PubMed] [Google Scholar]
- 44. Pelicic V. 2008. Type IV pili: e pluribus unum? Mol. Microbiol. 68:827–837 [DOI] [PubMed] [Google Scholar]
- 45. Petrova OE, Schurr JR, Schurr MJ, Sauer K. 2011. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol. Microbiol. 81:767–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rice SA, et al. 2009. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 3:271–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 102:8006–8011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Semmler ABT, Whitchurch CB, Leech AJ, Mattick JS. 2000. Identification of a novel gene, fimV, involved in twitching motility in Pseudomonas aeruginosa. Microbiology 146:1321–1332 [DOI] [PubMed] [Google Scholar]
- 49. Singh PK, et al. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764 [DOI] [PubMed] [Google Scholar]
- 50. Skerker JM, Berg HC. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U. S. A. 98:6901–6904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stover CK, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 52. Strom MS, Lory S. 1986. Cloning and expression of the pilin gene of Pseudomonas aeruginosa PAK in Escherichia coli. J. Bacteriol. 165:367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Strom MS, Lory S. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565–596 [DOI] [PubMed] [Google Scholar]
- 54. Tang H, Kays M, Prince A. 1995. Role of Pseudomonas aeruginosa pili in acute pulmonary infection. Infect. Immun. 63:1278–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tendeng C, Bertin PN. 2003. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol. 11:511–518 [DOI] [PubMed] [Google Scholar]
- 56. Tendeng C, Soutourina OA, Danchin A, Bertin PN. 2003. MvaT proteins in Pseudomonas spp.: a novel class of H-NS-like proteins. Microbiology 149:3047–3050 [DOI] [PubMed] [Google Scholar]
- 57. Turner LR, Lara JC, Nunn DN, Lory S. 1993. Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 175:4962–4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vallet I, et al. 2004. Biofilm formation in Pseudomonas aeruginosa: fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J. Bacteriol. 186:2880–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vallet-Gely I, Donovan KE, Fang R, Joung JK, Dove SL. 2005. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 102:11082–11087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang W, Li G-W, Chen C, Xie XS, Zhuang X. 2011. Chromosome organization by a nucleoid-associated protein in live bacteria. Science 333:1445–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Webb JS, Lau M, Kjelleberg S. 2004. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:8066–8073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Webb JS, et al. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wehbi H, et al. 2011. The peptidoglycan-binding protein FimV promotes assembly of the Pseudomonas aeruginosa type IV pilus secretin. J. Bacteriol. 193:540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Westfall LW, et al. 2006. mvaT mutation modifies the expression of the Pseudomonas aeruginosa multidrug efflux operon mexEF-oprN. FEMS Microbiol. Lett. 255:247–254 [DOI] [PubMed] [Google Scholar]
- 65. Whiteley M, et al. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864 [DOI] [PubMed] [Google Scholar]
- 66. Winsor GL, et al. 2011. Pseudomonas genome database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39:D596–D600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wong SM, Mekalanos JJ. 2000. Genetic footprinting with mariner-based transposition in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 97:10191–10196 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.